Abstract
Purpose
To explore the characteristics of the clinical phenotype of ARDS based on Machine Learning.
Methods
This is a study on Machine Learning. Screened cases of acute respiratory distress syndrome (ARDS) in the eICU database collected basic information in the cases and clinical data on the Day 1, Day 3, and Day 7 after the diagnosis of ARDS, respectively. Using the Calinski-Harabasz criterion, Gap Statistic, and Silhouette Coefficient, we determine the optimal clustering number k value. By the K-means cluster analysis to derive clinical phenotype, we analyzed the data collected within the first 24 h. We compared it with the survival of cases under the Berlin standard classification, and also examined the phenotypic conversion within the first 24 h, on day 3, and on day 7 after the diagnosis of ARDS.
Results
We collected 5054 cases and derived three clinical phenotypes using K-means cluster analysis. Phenotype-I is characterized by fewer abnormal laboratory indicators, higher oxygen partial pressure, oxygenation index, APACHE IV score, systolic and diastolic blood pressure, and lower respiratory rate and heart rate. Phenotype-II is characterized by elevated white blood cell count, blood glucose, creatinine, temperature, heart rate, and respiratory rate. Phenotype-III is characterized by elevated age, partial pressure of carbon dioxide, bicarbonate, GCS score, albumin. The differences in ICU length of stay and in-hospital mortality were significantly different between the three phenotypes (P < 0.05), with phenotype I having the lowest in-hospital mortality (10 %) and phenotype II having the highest (31.8 %). To compare the survival analysis of ARDS patients classified by phenotype and those classified according to Berlin criteria. The results showed that the differences in survival between phenotypes were statistically significant (P < 0.05) under phenotypic classification.
Conclusions
The clinical classification of ARDS based on K-means clustering analysis is beneficial for further identifying ARDS patients with different characteristics. Compared to the Berlin standard, the new clinical classification of ARDS provides a clearer display of the survival status of different types of patients, which helps to predict patient prognosis.
Keywords: Acute respiratory distress syndrome, Machine learning, K-means clustering analysis, Phenotype
1. Introduction
Acute respiratory distress syndrome (ARDS) is an acute respiratory system disease characterized by refractory hypoxemia and non-cardiogenic pulmonary edema [1]. A study in 2016 revealed that ARDS accounted for 10.4 % of the visits to intensive care units in 50 countries worldwide, and 40 % of patients went undiagnosed. Among the diagnosed ARDS patients, 70 % had moderate-to-severe symptoms, and severe ARDS patients had a mortality rate of 46 % [2]. In China, the mortality rate of severe ARDS patients is as high as 60 % [3]. Therefore, acute respiratory distress syndrome remains a major issue in the intensive care unit (ICU) [4].
The Berlin criteria proposed in 2012 remain the main clinical diagnostic criteria and inclusion criteria for experimental studies [5]. In terms of scientific research, many studies on ARDS have shown negative results, including large multicenter randomized controlled trials (RCTs) [6]. This shows that identifying ARDS patients based on existing Berlin standards may have certain limitations. Therefore, researchers are attempting to re-examine ARDS from more perspectives and suggest the study of ARDS phenotype accordingly. Phenotype refers to the detectable or observable similar characteristics exhibited by organisms under the interaction between genes and the environment [7]. And, experts can further categorize it into corresponding categories based on shared expressions, risk factors, etc. [8]。For example, pulmonary imaging identifies two categories of ARDS: localized and diffuse [9], which are considered the imaging phenotype of ARDS. Previously, phenotype research has played a specific role in identifying and treating many diseases, such as asthma and cancer metabolism [10,11], which also brings hope for the study of ARDS phenotype. The heterogeneous expression of ARDS in etiology, pathology, clinical manifestations, imaging manifestations, transcription, and gene characteristics [12,13] is a favorable basis for conducting phenotype research on ARDS. The existing research on the phenotype of ARDS focuses on biological subtypes, metabolomes, transcriptomics, and genetic subtypes [[14], [15], [16], [17]]. At present, many secondary analyses based on biomarkers have shown effective new classifications for ARDS, which have led to favorable results in fluid management [18], PEEP titration [19], and statin therapy [20] for ARDS patients. However, because of the limitations of the clinical use of biomarkers, genes, proteins, etc., timely and convenient operations cannot be provided for clinical workers. Therefore, there is an urgent need for phenotype research related to clinical indicators of ARDS, namely the clinical classification of ARDS.
With the development of scientific research, machine learning is increasingly being applied in healthcare, including predictive models, diagnostic models, and phenotype analysis [[21], [22], [23], [24]]. We can further divide it into supervised learning, unsupervised learning, and semi-supervised learning based on the presence or absence of data labels [25]. Compared to supervised learning, unsupervised learning does not use artificially set data labels, which it mainly identifies potential correlations and structures between high-dimensional data, divides the dataset into relevant clusters, and simplifies the dataset [26]. At present, unsupervised learning has been applied to the research of many diseases, such as osteoarthritis [27], chronic obstructive pulmonary disease [28], diabetes [29], hypertension, etc. K-means clustering analysis [30] is a type of unsupervised learning that is widely used because of its advantages of ease of use and fast training speed.
Therefore, this study used K-means clustering analysis to perform phenotype typing on clinical data of ARDS patients in large databases, exploring the reproducibility and stability of typing results, providing ideas for further research on ARDS clinical typing and data support for clinical applications.
2. Methods
2.1. The database of eICU
The eICU Collaborative Research Database is a multicenter public database developed by Philips Healthcare, covering health data of patients admitted to ICUs in 335 hospitals in the United States from 2014 to 2015. Researchers recorded over 200,000 medical records from 139,367 treated patients [31].
2.2. Simplified application and installation of an eICU database
The database application must first complete the human research course and sign the data usage agreement. After obtaining authorization, download the complete data from the official website of the eICU database and use PostgreSQL for database installation.
2.3. Data extraction
Use Navicat software to input the corresponding extraction code for relevant data extraction. GitHub official website provides some preprocessing codes, including ICU hospitalization status, laboratory indicators, and ventilator usage, for the convenience of researchers. Patient-unit-stay ID in the table is the primary identifier of the patient, distinguishing them from each hospitalization. Patient-unit-stay ID jointly identified all tables.
2.4. Inclusion criteria
-
1)
Diagnosed as "ARDS" or "acute respiratory distress syndrome" or ICD, with a 9 code of "518.82". 2) After being diagnosed with ARDS, the ICU hospitalization time is ≥ 48 h.
2.5. Exclusion criteria
-
1)
Age ≤18 years old; 2) Gender deficiency; 3) Oxygenation index (OI=PO2/FiO2) > 300; 4) Patients with missing variable values of ≥10 were tested.
2.6. Data collection
-
1)
General information: age, gender, admission time, discharge time. 2) Maximum values: creatinine, blood potassium, white blood cell count, blood glucose, pH, heart rate, respiratory rate, body temperature. Minimum values: albumin, partial pressure of oxygen, partial pressure of carbon dioxide, bicarbonate ions, blood sodium, systolic blood pressure, diastolic blood pressure, Glasgow Coma Scale (GCS), platelet count, oxygenation index, Acute Physiology and Chronic Health Status Scoring System IV (APACHE IV) score. 3) Comorbidities: hypertension, diabetes, chronic obstructive pulmonary disease. 4) Prognosis: in-hospital mortality rate, length of ICU stays.
2.7. Data processing and analysis
2.7.1. Extreme value processing
Limit all measured values to 0.02 and 0.98 percentiles respectively to eliminate the influence of extreme values on clustering analysis.
2.7.2. Missing value handling
Perform proportional statistics on missing variables and remove variables with missing ratios greater than 60 %. The missing variables belong to completely random missing cases. Therefore, we employ the Chain Equation Multiple Interpolation (MICE) method, setting 5 imputation times. After imputation, we calculate and compare the Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) for each dataset, and then select the dataset with the smallest value for both criteria for subsequent analysis.
2.7.3. Normality test and data transformation
If the sample size is greater than 5000, use Kolmogorov-Smirno test method to perform normality test on the interpolated dataset, and perform a logarithmic transformation on data values with obvious skewness distribution.
2.7.4. Correlation analysis
Using SPSS for Kendall's tau-b correlation analysis, the correlation coefficient range is [−1, +1], with values less than 0 showing negative correlation, values greater than 0 showing positive correlation, and values equal to 0 showing no correlation. We define a correlation coefficient ≥0.5 as indicating a strong correlation, and we consider P < 0.05 to be significant. If there are two sets of strongly correlated data, delete one set of data and visualize the results using Origin.
2.7.5. K-means clustering analysis
The main idea behind the K-means clustering algorithm is to randomly select sample points, assume k as the number of sample points, and use these sample points as the initial center points for clustering (cluster). For each remaining sample, divide it into clusters corresponding to the cluster center that is most similar to it based on its similarity (distance) with each cluster center; Then recalculate the average value of all samples at each cluster center as the new cluster center. The K-means clustering algorithm has the advantages of ease of use and fast training speed [32]. We comprehensively evaluate the determination of the number of clusters using the Kalinski-Halabas criterion, contour coefficients, and interval statistics. Kalinsky-Halabas criterion is defined as a good cluster with a larger inter cluster variance and a smaller intra cluster variance. Larger the result value, the tighter the intra cluster, and the more dispersed the inter cluster. Contour coefficient defines the average distance (cohesion) between an object and other samples in the same cluster, as well as the average distance (separability) between an object and all samples in the other clusters. The range of values is [−1,1], and the larger the result value, the better the cohesion and separability of the object. Taking the average of the contour coefficients of all objects yields the contour coefficient of the clustering result. We define the interval statistic as the difference in loss between the actual sample and the random sample. As the number of random clusters approaches the optimal number, the interval statistic continuously increases and changes significantly. When the number of random clusters exceeds the optimal number, the interval statistic continues to increase, but the magnitude of change slows down significantly. Use R Studio for the software and enter the corresponding code.
2.7.6. Stability analysis
Collect clinical data on the 3rd and 7th day after diagnosis of ARDS, clean the data, delete cases with ≥10 missing variables, calculate the Euclidean distance between the data and the 3 phenotype cluster centers, and assign it to the closest phenotype category based on the size of the distance. Compare the phenotypic changes within the first 24 h, 3 days, and 7 days after diagnosis.
2.7.7. Statistic analyze
Perform data statistical analysis using SPSS 26.0 and R studio software. The normality test of data is based on the sample size. If the sample size is greater than 5000, the Kolmogorov-Smirno test method is used to describe data that conforms to a normal distribution using the mean ± standard deviation (X ± S), non- normal distribution data using the median (interquartile range), and categorical variables using numbers and percentages. Using Kendall's tau-b test, we performed the correlation analysis between variables. Draw relevant charts using R language and Origin.
3. Results
3.1. Data collection
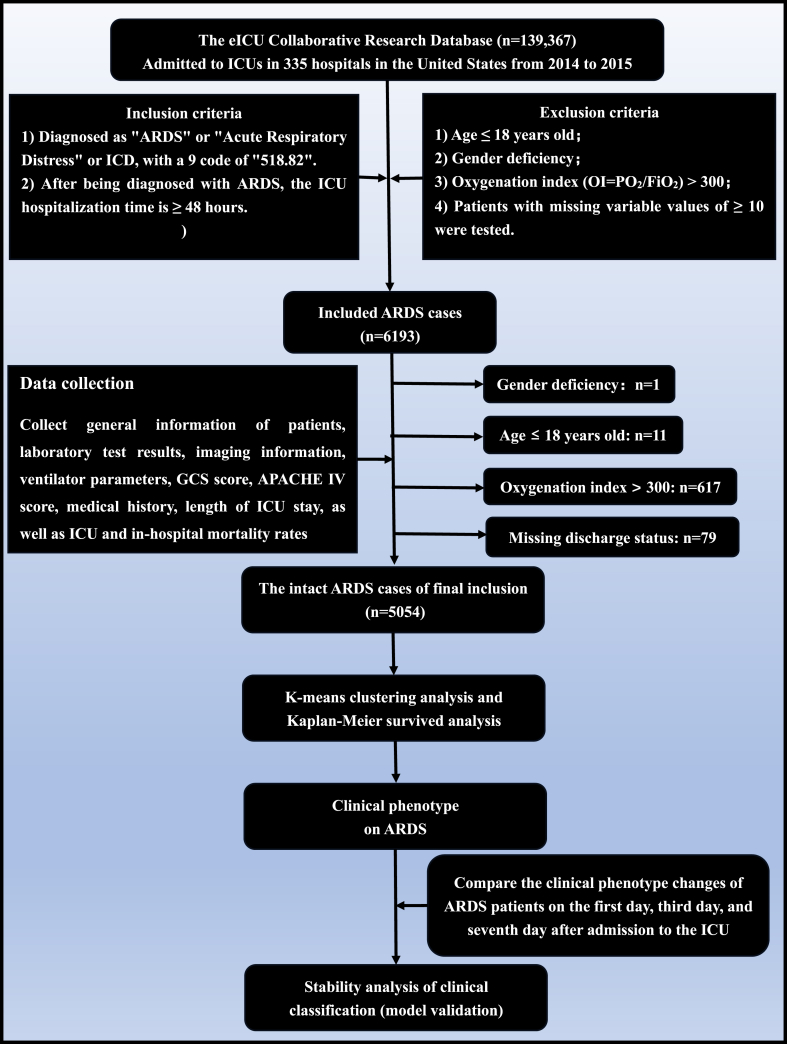
Data collection refers to Fig. 1. Based on the diagnosis name "ARDS" and the ICD-9 value, we included 6193 cases from the eICU database. We only kept the first-time ICU entry records among the 139,367 medical records. Among them, we identified 1 case with missing gender, 11 cases with age mismatch, 617 cases with oxygenation index >300, 79 cases with missing discharge information, 431 cases with ≥10 missing variables, and ultimately included 5054 cases in the study.
Fig. 1.
Flow diagram of this study.
3.2. Basic characteristics of the patients being included in this study (Table 1)
Table 1.
Basic characteristics of the patients being included in this study.
| Variables | Included ARDS cases n = 5054 |
Variables | Included ARDS cases n = 5054 |
|---|---|---|---|
| Race | Laboratory test data | ||
| Asians, cases (%) | 72 (1.4) | pH, M (Q1, Q3) | 7.41 (7.4, 7.5) |
| Caucasians, cases (%) | 3806 (75.3) | PO2, mm Hg, M (Q1, Q3) | 73 (61, 90.4) |
| Native Americans, cases (%) | 32 (0.6) | PCO2, mm Hg, M (Q1, Q3) | 39.5 (34.1, 48) |
| African Americans, cases (%) | 600 (11.9) | OI (PO2/FiO2), M (Q1, Q3) | 127 (92.9, 186.3) |
| Hispanics | 280 (5.5) | HCO3−, mmol/L, M (Q1, Q3) | 24.9 (21, 30) |
| Others, cases (%) | 264 (5.2) | Blood sugar, mg/dL, M (Q1, Q3) | 137 (112, 172) |
| Age (years); M (Q1, Q3) | 66 (54, 77) | Serum Na+, mmol/L, M (Q1, Q3) | 139 (136, 142) |
| Gender | Serum K+, mmol/L, M (Q1, Q3) | 4.1 (3.7, 4.5) | |
| Male, cases (%) | 2678 (53.0) | White blood cell, × 109/L, M (Q1, Q3) | 11.5 (8.2, 15.7) |
| Female, cases (%) | 2376 (47.0) | Platelet, × 109/L, M (Q1, Q3) | 188 (135, 257) |
| Past medical history | Serum albumin, (g/dL), M (Q1, Q3) | 2.5 (2.1, 2.9) | |
| COPD, cases (%) | 1210 (23.9) | Creatinine, (mg/dL), M (Q1, Q3) | 1.1 (0.8, 1.9) |
| Diabetes, cases (%) | 1586 (31.4) | Vital sign data | |
| Hypertension, cases (%) | 2757 (54.6) | Body Temperature, (°C), M (Q1, Q3) | 37.2 (36.9, 37.8) |
| Stratification of Berlin definition | Heart Rate (counts per minute), M (Q1, Q3) | 105 (92, 119) | |
| Mild, cases (%) | 587 (11.6) | Respiratory rate (counts per minute), M (Q1, Q3) | 29 (24, 34) |
| Moderate, cases (%) | 1135 (22.5) | Systolic blood pressure, (mm Hg), M (Q1, Q3) | 96 (85, 110) |
| Severe, cases (%) | 875 (5.4) | Diastolic blood pressure, (mm Hg), M (Q1, Q3) | 50 (42, 57) |
| No stratification, cases (%) | 2457 (48.6) | ||
| Length of ICU stay, (hours) | |||
| M (Q1,) | 120 (73, 220) | ||
| ICU mortality, cases (%) | 984 (19.5) | ||
| APACHE IV, (M, Q1∼Q3) | 65 (51,84) | ||
COPD, Chronic obstructive pulmonary disease; APACHE IV, Acute Physiology and Chronic Health Evaluation IV; pH, Hydrogen ion concentration index; GCS, Glasgow Coma Scale; ICU, Intensive Care Unit; Detect that all continuous variables do not follow a normal distribution, therefore, median (interquartile range) is used to represent, and categorical variables are expressed in terms of number of cases and percentage.
This study included 5054 cases. The race includes Asians, Caucasians, Native Americans, African Americans, and Hispanics, with Caucasians accounting for 75.3 % of the total study population. There is not much difference in the proportion of males and females in terms of gender ratio. The median age of the total study population is 66 years old, the median length of ICU stay is 120 h, and the in-hospital mortality is 19.5 %.
3.3. Derivation and analysis of clinical phenotypes in ARDS
3.3.1. Normality test and data transformation
All data show skewness, and we perform a logarithmic transformation on data values with obvious skewed distribution, including creatinine, white blood cells, glucose, urea nitrogen, and body temperature.
3.3.2. Correlation analysis
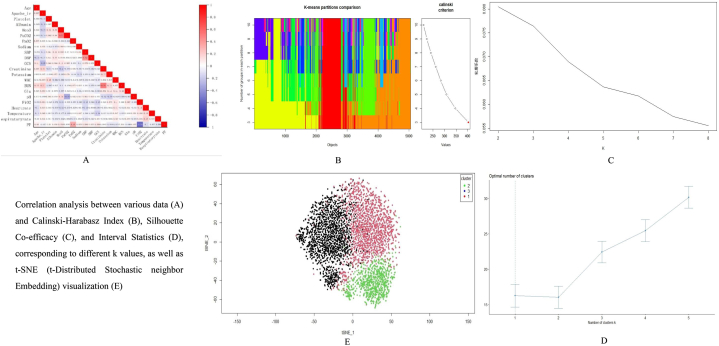
The results showed a strong correlation between oxygenation index and oxygen uptake concentration (correlation coefficient = 0.58, P < 0.05), creatinine and urea nitrogen (correlation coefficient = 0.509, P < 0.05). Therefore, we removed oxygen uptake concentration and urea nitrogen, and visualized the results using Origin (Fig. 2A).
Fig. 2.
Derivation and analysis of clinical phenotypes in ARDS.
Correlation analysis between various data (A) and Calinski-Harabasz Index (B), Silhouette Co-efficacy (C), and Interval Statistics (D), corresponding to different k values, as well as t-SNE (t-Distributed Stochastic neighbor Embedding) visualization (E).
3.3.3. K-means clustering analysis
When comparing the Calinski-Harabasz Index, contour coefficients, and interval statistics under different k values, we can see the results in Fig. 2B, C, and 2D. When k = 3, the corresponding Calinski-Harabasz Index is maximum, and when k = 2, the contour coefficient is maximum. Except for a decrease in the time interval statistics at k = 2, the time interval statistics continue to increase as k increases. After a comprehensive evaluation, we determined that the clustering effect is best when k = 3, so we set 3 as the final number of clusters. Through t-SNE dimensionality reduction (Fig. 2E), we visualized the clustering results and found no significant mixing of boundaries between the three clusters.
3.4. Data analysis
3.4.1. Characteristics of different clinical phenotypes
In the end, we got three clinical phenotypes and described their characteristics based on the mean of each phenotype center point on each indicator. The Phenotype-I is characterized by fewer abnormal laboratory indicators, higher oxygen partial pressure, oxygenation index, APACHE IV score, systolic and diastolic blood pressure, and lower respiratory rate and heart rate. Phenotype-II is characterized by higher white blood cell count, blood glucose, creatinine, body temperature, heart rate, and respiratory rate, as well as lower albumin, GCS score, bicarbonate ion, systolic blood pressure, diastolic blood pressure, and oxygenation index. Phenotype-III is characterized by higher age, carbon dioxide partial pressure, bicarbonate ion, GCS score, albumin, and lower white blood cell count, creatinine, and body temperature (Table 2).
Table 2.
The basic characteristic of three clinical phenotypes in this study.
| Phenotype-I (n = 2037) | Phenotype-II (n = 2029) | Phenotype-III (n = 988) | P值 | |
| Age, years | 64.7 | 62.4 | 67.5 | <0.05 |
| Sex | ||||
| Female, cases (%) | 980 (48.1) | 931 (45.9) | 465 (47.1) | <0.05 |
| Comorbidities | ||||
| COPD, cases (%) | 249 (20.8) | 188 (24.4) | 146 (23.2) | |
| Diabetes, cases (%) | 329 (27.5) | 265 (34.4) | 206 (32.7) | |
| Hypertension, cases (%) | 619 (51.7) | 417 (54.2) | 355 (56.3) | |
| Berlin Definition | ||||
| Mild, cases (%) | 639 (31.4) | 264 (13.0) | 159 (16.1) | |
| Moderate, cases (%) | 989 (48.6) | 806 (39.7) | 595 (60.2) | |
| Severe, cases (%) | 409 (20) | 959 (47.3) | 234 (23.7) | |
| APACHE IV | 93.9 | 54.9 | 48.0 | <0.05 |
| pH | 7.5 | 7.4 | 7.3 | <0.05 |
| Platelet, × 109/L | 157.37 | 222.2 | 248.2 | <0.05 |
| Serum albumin, (g/dL) | 2.8 | 2.1 | 2.9 | <0.05 |
| HCO3-, mmol/L | 25.81 | 18.76 | 41.4 | <0.05 |
| PCO2, mm Hg | 35.42 | 35.34 | 77.77 | <0.05 |
| PO2, mm Hg | 94.29 | 71.95 | 68.5 | <0.05 |
| Oxygenation Index | 186.1 | 83.2 | 142.8 | <0.05 |
| Serum Na+, mmol/L | 139.9 | 138.2 | 139.2 | <0.05 |
| Systolic blood pressure mm Hg | 114.36 | 81.5 | 98.9 | <0.05 |
| Diastolic blood pressure, (mm Hg) | 59.5 | 40.7 | 49.9 | <0.05 |
| Serum K+, mmol/L | 3.8 | 4.5 | 4.3 | <0.05 |
| Heart rates, counts per minute | 96.6 | 119.7 | 97.4 | <0.05 |
| Respiratory rate, times per minute | 26.4 | 33.4 | 29.9 | <0.05 |
| White blood cell, × 109/L, | 10.5 | 16.4 | 10.2 | <0.05 |
| Creatine, mg/dL | 1.1 | 2.5 | 1.0 | <0.05 |
| Blood sugar, mg/dL | 130.9 | 175.6 | 136.3 | <0.05 |
| Body temperature, °C | 37.3 | 37.8 | 36.9 | <0.05 |
| GCS Score | 5.3 | 2.3 | 6.4 | <0.05 |
COPD, Chronic obstructive pulmonary disease; APACHE IV, Acute Physiology and Chronic Health Evaluation IV; pH, Hydrogen ion concentration index; GCS, Glasgow Coma Scale; ICU, Intensive Care Unit; Detect that all continuous variables do not follow a normal distribution, therefore, median (interquartile range) is used to represent, and categorical variables are expressed in terms of number of cases and percentage.
3.4.2. Comparison of the survival analysis between clinical phenotype and Berlin definition
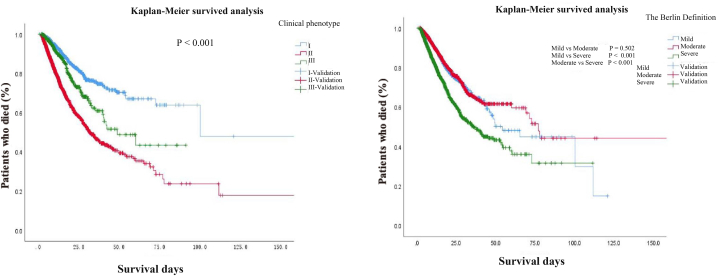
In the model we studied, there were significant differences (P < 0.05) in ICU hospitalization time and mortality among the three clinical phenotypes, with a 28-day mortality rate of 9.0 % for phenotype I, 29.0 % for phenotype II, and 12.3 % for phenotype III. The survival analysis of ARDS patients classified by clinical phenotype and Berlin criteria showed significant differences in survival between different phenotypes (P < 0.05). Patients with phenotypes I and III had significantly better survival than those with phenotypes II. According to the Berlin standard classification, there was no statistically significant difference in survival time between mild ARDS patients and moderate patients. Compared with the Berlin standard classification, the survival situation of patients with different clinical phenotypes was clearer after classification according to clinical phenotypes (Fig. 3).
Fig. 3.
Comparison of survival curves of ARDS patients stratified by clinical phenotype and Berlin criteria.
3.4.3. Clinical phenotype stability analysis
Collect the indicator results of 5054 cases on the 3rd day after diagnosis, including 146 cases with survival days less than 3 days, 2038 cases with variable deletions ≥10, and 2870 cases ultimately participating in phenotype stability analysis. Perform missing value imputation and standardization on the data, and calculate the Euclidean distance between each case of data and the phenotype center point. As shown in Table 3, among the 2870 cases, there were 1061 (37 %) patients with phenotype I, 1277 (44 %) patients with phenotype II, and 532 (19 %) patients with phenotype III.
Table 3.
Clinical phenotype changes on the third day after diagnosis of ARDS.
| Clinical | The first 24 h | The 3rd day | Transformed clinical phenotype | ||
|---|---|---|---|---|---|
| Phenotype | (n = 5054) | (n = 2870) | I | II | III |
| I | 2037 | 1061 | 266 (9.3 %) | 190 (6.6 %) | |
| II | 2029 | 1277 | 470 (16.4 %) | 151 (5.3 %) | |
| III | 988 | 532 | 211 (7.4 %) | 139 (4.8 %) | |
Among 1061 cases of phenotype I, 456 cases experienced phenotypic changes, of which 266 cases (9.3 %) transitioned to phenotype II with a worse prognosis, and 190 cases (6.6 %) transitioned to phenotype III. Among 1277 cases of phenotype II, 621 cases showed phenotypic changes, with 470 cases (16.4 %) transitioning to phenotype I and 151 cases (5.3 %) transitioning to phenotype III (Table 3). Over 70 % of the data on the 7th day of the statistics were missing. To minimize the impact on stability analysis, we did not compare the phenotype changes on the 7th day.
4. Discussion
Today, ARDS is still one disease with a high morbidity and mortality in the world [33]. Based on the eICU database, three clinical phenotypes of ARDS were derived, and there were differences conditions, laboratory indicators, and prognosis among different phenotypes. In survival analysis, patients with clinical phenotype I and phenotype III had significantly better survival than those with clinical phenotype II.
4.1. The impact of different machine learning methods on the clinical phenotype of ARDS
The current methods used for clinical phenotype analysis mainly include K-means clustering analysis and latent category analysis. This study used K-means clustering analysis to derive clinical phenotypes. Similarly, the study by Kaijiang Yu et al. [34] identified three phenotypes, among which the characteristics of phenotype II in white blood cell count, body temperature, heart rate, and respiratory rate were consistent with our study. In addition, Duggal et al. [35] also used K-means clustering analysis to perform secondary analysis on six datasets with different time and regional factors, and identified two subtypes of ARDS based on nine clinical and laboratory variables (heart rate, mean arterial pressure, respiratory rate, bilirubin, bicarbonate, creatinine, partial oxygen pressure, arterial pH, and oxygen concentration). Among them, phenotype II had higher heart rate, respiratory rate, creatinine, and bilirubin, and lower platelet and oxygen partial pressure. The changes in these indicators are also consistent with our research. However, Calfee et al. [36] used latent class analysis methods for secondary analysis, and found that the changes in creatinine, bilirubin, age oxygen partial pressure, and platelets in Class 2 models were consistent with the aforementioned studies. This shows that regardless of the statistical method used, changes in phenotype based on clinical variables on certain indicators are universal. However, different statistical methods yielded inconsistent total numbers of clusters, showing the need for further comparative analysis of different phenotype derivation methods while using the same dataset.
4.2. The impact of different datasets on clinical phenotype inference
Under the premise of using the same phenotype derivation method (using K-means clustering analysis as an example), this study mainly used the eICU database to record ICU admitted cases from over 200 hospitals in the United States from 2014 to 2015. Duggal et al.'s study used six datasets: ARMA, ALVEOLI, FACTT, EDEN, SAILS, and ART. ARMA [37] dataset records data from 10 medical centers in the United States from March 1996 to March 1999, mainly focusing on respiratory parameter settings research. ALVEOLI [38] dataset records data from 23 hospitals in the ARDS clinical trial network of the National Institute of Cardiopulmonary and Hematology from October 1999 to February 2002. FACTT [39] dataset records case data that met inclusion criteria in 20 medical centers in North America from June 2000 to October 2005. EDEN [40] dataset records patient data recruited from 44 hospitals in the acute respiratory distress syndrome clinical trial network of the National Institute of Cardiopulmonary and Hematology between January 2008 and March 2011. SAILS [41] dataset records data from 44 medical centers in the United States that met the inclusion criteria for research, starting from March 18, 2010 and officially released in 2014. ART [42] dataset records multicenter data on adults with moderate-to-severe ARDS from 120 intensive care units (ICUs) in 9 countries from November 2011 to April 2017. The collection time and studied regions of the above dataset are different, so the reasons for the different models got under the same clustering method may be attributed to regional differences, differences in medical conditions, and environmental differences.
4.3. Follow-up questions on clinical phenotype derivation
At present, phenotype research mostly uses unsupervised learning methods (including K-means clustering analysis, latent class analysis, etc.). In addition, the characteristics of unsupervised learning determine that it cannot directly provide labels for describing important features, which limits clinical implementation. Therefore, we still need to optimize it through further model training. Sinha et al. [43] conducted such a study, using GBM to develop a clinical classifier model based on the ARDS phenotype got from previous research by Calfee et al. [17] There is limited research on the stability of phenotypes. In China, Qiu Haibo et al. [44] conducted a longitudinal study using the China Intensive Care Database and foreign databases, and found that over 94 % of patients did not change their phenotype categories within 3 days of phenotype classification. Ultimately, 56.9 % of patients changed their phenotype at least once during the study period. Delluchi et al. [45] also concluded through analysis that phenotype classification has a certain stability within 3 days after determination. This differs from the results, and the reason for this may be because the eICU database does not come from professional RCT research data, and there are certain omissions in its data entry.
The existing research seems to only show that the ARDS phenotype is a short-term evaluation method, and the significance of phenotype classification for clinical application still needs to be explored.
5. Conclusions
Compared to the Berlin standard, the new clinical classification of ARDS based on Machine Learning of K-means clustering analysis provides a clearer display of the survival status of different types of patients, which helps to predict patient prognosis.
CRediT authorship contribution statement
Wei Zhang: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Linlin Wu: Resources, Project administration, Methodology, Investigation, Data curation, Conceptualization. Shucheng Zhang: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation, Formal analysis, Data curation.
Ethics approval and consent to take part
The researchers did not get the consent of the ethics committee before sourcing all data for this study from a clinical database named eICU. Since this study did not involve any additional clinical interventions in the diagnosis and treatment, the researchers did not get the informed consent of patients or their relatives.
Consent to publish
All authors have read and approved the manuscript version, and agree to submit for consideration for publication in the journal. There are no any ethical/legal conflicts involved in the article.
Availability of data and materials
We stated that all the data and materials were true and available in the study.
Take-home message
Identifying ARDS patients based on existing Berlin standards may have certain limitations. Phenotype refers to the detectable or observable similar characteristics exhibited by organisms under the interaction between genes and the environment. Compared to the Berlin standard, the new clinical classification of ARDS based on Machine Learning of K-means clustering analysis provides a clearer display of the survival status of different types of patients, which helps to predict patient prognosis.
Funding
This study was funded by Science and Technology Plan of Guizhou Province in 2020 (Foundation of Guizhou Science and Technology Cooperation [2020] 1Z061) and Zunyi Science and Technology Plan Project (Zun Shi Ke He HZ Zi (2022) 286).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank all the experts of the developed database of eICU. The authors also thank all the patients who took part in the study.
Contributor Information
Wei Zhang, Email: zhangwei_hxicu@163.com.
Linlin Wu, Email: wulinlin_icu@163.com.
Shucheng Zhang, Email: zhangchunlei761@163.com.
List of Abbreviations
| AIC | Akaike Information Criterion |
| APACHE IV | Acute Physiology and Chronic Health Evaluation IV |
| ARDS | Acute Respiratory Distress Syndrome |
| BIC | Bayesian Information Criterion |
| CHI | Calinski-Harabaz Index |
| COPD | Chronic Obstructive Pulmonary Disease |
| eICU | Telehealth Intensive Care Unit |
| GCS | Glasgow Coma Scale |
| ICU | Intensive Care Unit |
| IL-6 | Interleukin-6 |
| RCT | Randomized Controlled Trial |
References
- 1.Matthay M.A., Arabi Y.M., Siegel E.R., Ware L.B., Bos L.D.J., Sinha P., Beitler J.R., Wick K.D., Curley M.A.Q., Constantin J.M., et al. Phenotypes and personalized medicine in the acute respiratory distress syndrome. Intensive Care Med. 2020;46(12):2136–2152. doi: 10.1007/s00134-020-06296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellani G., Laffey J.G., Pham T., Fan E., Brochard L., Esteban A., Gattinoni L., van Haren F., Larsson A., McAuley D.F., et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315(8):788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 3.Liu L., Yang Y., Gao Z., Li M., Mu X., Ma X., Li G., Sun W., Wang X., Gu Q., et al. Practice of diagnosis and management of acute respiratory distress syndrome in mainland China: a cross-sectional study. J. Thorac. Dis. 2018;10(9):5394–5404. doi: 10.21037/jtd.2018.08.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Zhang L., Xi X., Zhou J.X. China critical care sepsis trial W: the association between etiologies and mortality in acute respiratory distress syndrome: a multicenter observational cohort study. Front. Med. 2021;8 doi: 10.3389/fmed.2021.739596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acute Respiratory Distress Syndrome JAMA. 2012;307(23) doi: 10.1001/jama.2012.6761. [DOI] [PubMed] [Google Scholar]
- 6.Tonelli A.R., Zein J., Adams J., Ioannidis J.P. Effects of interventions on survival in acute respiratory distress syndrome: an umbrella review of 159 published randomized trials and 29 meta-analyses. Intensive Care Med. 2014;40(6):769–787. doi: 10.1007/s00134-014-3272-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reilly J.P., Calfee C.S., Christie J.D. Acute respiratory distress syndrome phenotypes. Semin. Respir. Crit. Care Med. 2019;40(1):19–30. doi: 10.1055/s-0039-1684049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wildi K., Livingstone S., Palmieri C., LiBassi G., Suen J., Fraser J. The discovery of biological subphenotypes in ARDS: a novel approach to targeted medicine? J Intensive Care. 2021;9(1):14. doi: 10.1186/s40560-021-00528-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierrakos C., Smit M.R., Pisani L., Paulus F., Schultz M.J., Constantin J.M., Chiumello D., Mojoli F., Mongodi S., Bos L.D.J. Lung ultrasound assessment of focal and non-focal lung morphology in patients with acute respiratory distress syndrome. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.730857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gans M.D., Gavrilova T. Understanding the immunology of asthma: pathophysiology, biomarkers, and treatments for asthma endotypes. Paediatr. Respir. Rev. 2020;36:118–127. doi: 10.1016/j.prrv.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Park J.H., Pyun W.Y., Park H.W. Cancer metabolism: phenotype, signaling and therapeutic targets. Cells. 2020;9(10) doi: 10.3390/cells9102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pab Sinha, Csabc Calfee. Phenotypes in acute respiratory distress syndrome: moving towards precision medicine. [Miscellaneous Article] Curr. Opin. Crit. Care. February 2019;25(1):12–20. doi: 10.1097/MCC.0000000000000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson J.G., Calfee C.S. ARDS subphenotypes: understanding a heterogeneous syndrome. Crit. Care. 2020;24(1):102. doi: 10.1186/s13054-020-2778-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du M., Garcia J.G.N., Christie J.D., Xin J., Cai G., Meyer N.J., Zhu Z., Yuan Q., Zhang Z., Su L., et al. Integrative omics provide biological and clinical insights into acute respiratory distress syndrome. Intensive Care Med. 2021;47(7):761–771. doi: 10.1007/s00134-021-06410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bos L.D.J., Scicluna B.P., Ong D.S.Y., Cremer O., van der Poll T., Schultz M.J. Understanding heterogeneity in biologic phenotypes of acute respiratory distress syndrome by leukocyte expression profiles. Am. J. Respir. Crit. Care Med. 2019;200(1):42–50. doi: 10.1164/rccm.201809-1808OC. [DOI] [PubMed] [Google Scholar]
- 16.Viswan A., Ghosh P., Gupta D., Azim A., Sinha N. Distinct metabolic endotype mirroring acute respiratory distress syndrome (ARDS) subphenotype and its heterogeneous biology. Sci. Rep. 2019;9(1):2108. doi: 10.1038/s41598-019-39017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calfee C.S., Delucchi K., Parsons P.E., Thompson B.T., Ware L.B., Matthay M.A. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir. Med. 2014;2(8):611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Famous K.R., Delucchi K., Ware L.B., Kangelaris K.N., Liu K.D., Thompson B.T., Calfee C.S. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am. J. Respir. Crit. Care Med. 2017;195(3):331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maddali M.V., Churpek M., Pham T., Rezoagli E., Zhuo H., Zhao W., He J., Delucchi K.L., Wang C., Wickersham N., et al. Validation and utility of ARDS subphenotypes identified by machine-learning models using clinical data: an observational, multicohort, retrospective analysis. Lancet Respir. Med. 2022;10(4):367–377. doi: 10.1016/S2213-2600(21)00461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S., Lu Z., Wu Z., Xie J., Yang Y., Qiu H. Determination of a "specific population who could benefit from rosuvastatin": a secondary analysis of a randomized controlled trial to uncover the novel value of rosuvastatin for the precise treatment of ARDS. Front. Med. 2020;7 doi: 10.3389/fmed.2020.598621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu W., Wang Y., Tang J., Yu M., Yuan J., Zhang G. Developing and evaluating a machine-learning-based algorithm to predict the incidence and severity of ARDS with continuous non-invasive parameters from ordinary monitors and ventilators. Comput. Methods Progr. Biomed. 2023;230 doi: 10.1016/j.cmpb.2022.107328. [DOI] [PubMed] [Google Scholar]
- 22.Silva G.F.S., Fagundes T.P., Teixeira B.C., Chiavegatto Filho A.D.P. Machine learning for hypertension prediction: a systematic review. Curr. Hypertens. Rep. 2022;24(11):523–533. doi: 10.1007/s11906-022-01212-6. [DOI] [PubMed] [Google Scholar]
- 23.Sealfon R.S.G., Mariani L.H., Kretzler M., Troyanskaya O.G. Machine learning, the kidney, and genotype-phenotype analysis. Kidney Int. 2020;97(6):1141–1149. doi: 10.1016/j.kint.2020.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai Y., Huang X., Xia J., Zhan Q. A narrative review of progress in the application of artificial intelligence in acute respiratory distress syndrome: subtypes and predictive models. Ann. Transl. Med. 2023;11(2):128. doi: 10.21037/atm-22-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacEachern S.J., Forkert N.D. Machine learning for precision medicine. Genome. 2021;64(4):416–425. doi: 10.1139/gen-2020-0131. [DOI] [PubMed] [Google Scholar]
- 26.Eckhardt C.M., Madjarova S.J., Williams R.J., Ollivier M., Karlsson J., Pareek A., Nwachukwu B.U. Unsupervised machine learning methods and emerging applications in healthcare. Knee Surg. Sports Traumatol. Arthrosc. 2023;31(2):376–381. doi: 10.1007/s00167-022-07233-7. [DOI] [PubMed] [Google Scholar]
- 27.Angelini F., Widera P., Mobasheri A., Blair J., Struglics A., Uebelhoer M., Henrotin Y., Marijnissen A.C., Kloppenburg M., Blanco F.J., et al. Osteoarthritis endotype discovery via clustering of biochemical marker data. Ann. Rheum. Dis. 2022;81(5):666–675. doi: 10.1136/annrheumdis-2021-221763. [DOI] [PubMed] [Google Scholar]
- 28.Eckhardt C.M., Gambazza S., Bloomquist T.R., Hoff P.D., Vuppala A. Extracellular vesicle-encapsulated microRNAs as novel biomarkers of lung health. Am. J. Respir. Crit. Care Med. 2023;207(1):50–59. doi: 10.1164/rccm.202109-2208OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Cui L., Tu L., Hu X., Wang S., Shi Y., Liu J., Zhou C., Li Y., Huang J., et al. Research of the distribution of tongue features of diabetic population based on unsupervised learning Technology. Evid. base Compl. Alternative Med.: eCAM. 2022 doi: 10.1155/2022/7684714. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castela Forte J., Perner A., van der Horst I.C.C. The use of clustering algorithms in critical care research to unravel patient heterogeneity. Intensive Care Med. 2019;45(7):1025–1028. doi: 10.1007/s00134-019-05631-z. [DOI] [PubMed] [Google Scholar]
- 31.Pollard T.J., Johnson A.E.W., Raffa J.D., Celi L.A., Mark R.G., Badawi O. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An Q., Rahman S., Zhou J., Kang J.J. A comprehensive review on machine learning in healthcare industry: classification, restrictions, opportunities and challenges. Sensors (Basel, Switzerland) 2023;23(9) doi: 10.3390/s23094178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiumello D., Marino A., Cammaroto A. Practical Trends in Anesthesia and Intensive Care 2018. 2019. The acute respiratory distress syndrome: diagnosis and management; pp. 189–204. [Google Scholar]
- 34.Liu X., Jiang Y., Jia X., Ma X., Han C., Guo N., Peng Y., Liu H., Ju Y., Luo X., et al. Identification of distinct clinical phenotypes of acute respiratory distress syndrome with differential responses to treatment. Crit. Care. 2021;25(1):320. doi: 10.1186/s13054-021-03734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duggal A., Kast R., Van Ark E., Bulgarelli L., Siuba M.T., Osborn J., Rey D.A., Zampieri F.G., Cavalcanti A.B., Maia I., et al. Identification of acute respiratory distress syndrome subphenotypes de novo using routine clinical data: a retrospective analysis of ARDS clinical trials. BMJ Open. 2022;12(1) doi: 10.1136/bmjopen-2021-053297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calfee C.S., Delucchi K.L., Sinha P., Matthay M.A., Hackett J., Shankar-Hari M., McDowell C., Laffey J.G., O'Kane C.M., McAuley D.F. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir. Med. 2018;6(9):691–698. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brower R.G., Matthay M.A., Morris A., Schoenfeld D., Thompson B.T., Wheeler A. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N. Engl. J. Med. 2000;342(18):1301–1308. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 38.Brower R.G., Lanken P.N., MacIntyre N., Matthay M.A., Morris A., Ancukiewicz M., Schoenfeld D., Thompson B.T., National Heart L. Blood Institute ACTN: higher versus lower positive end-expiratory pressures in patients with the acute respiratory distress syndrome. N. Engl. J. Med. 2004;351(4):327–336. doi: 10.1056/NEJMoa032193. [DOI] [PubMed] [Google Scholar]
- 39.National Heart L., Wheeler A.P., Bernard G.R., Thompson B.T., Schoenfeld D., Wiedemann H.P., deBoisblanc B., Connors A.F., Jr., Hite R.D., et al. Pulmonary-artery versus central venous catheter to guide treatment of acute lung injury. N. Engl. J. Med. 2006;354(21):2213–2224. doi: 10.1056/NEJMoa061895. [DOI] [PubMed] [Google Scholar]
- 40.National Heart L., Rice T.W., Wheeler A.P., Thompson B.T., Steingrub J., Hite R.D., Moss M., Morris A., Dong N., et al. Initial trophic vs full enteral feeding in patients with acute lung injury: the EDEN randomized trial. JAMA. 2012;307(8):795–803. doi: 10.1001/jama.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Heart L., Blood Institute ACTN., Truwit J.D., Bernard G.R., Steingrub J., Matthay M.A., Liu K.D., Albertson T.E., Brower R.G., Shanholtz C., et al. Rosuvastatin for sepsis-associated acute respiratory distress syndrome. N. Engl. J. Med. 2014;370(23):2191–2200. doi: 10.1056/NEJMoa1401520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavalcanti A.B., Suzumura É.A., Laranjeira L.N., Paisani D.M., Damiani L.P., Guimarães H.P., Romano E.R., Regenga M.M., Taniguchi L.N.T., Teixeira C., et al. Effect of lung recruitment and titrated positive end-expiratory pressure (PEEP) vs low PEEP on mortality in patients with acute respiratory distress syndrome: a randomized clinical trial. JAMA. 2017;318(14):1335–1345. doi: 10.1001/jama.2017.14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinha P., Churpek M.M., Calfee C.S. Machine learning classifier models can identify acute respiratory distress syndrome phenotypes using readily available clinical data. Am. J. Respir. Crit. Care Med. 2020;202(7):996–1004. doi: 10.1164/rccm.202002-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H., Yu Q., Xie J., Liu S., Pan C., Liu L., Huang Y., Guo F., Qiu H., Yang Y. Longitudinal phenotypes in patients with acute respiratory distress syndrome: a multi-database study. Crit. Care. 2022;26(1):340. doi: 10.1186/s13054-022-04211-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delucchi K., Famous K.R., Ware L.B., Parsons P.E., Thompson B.T., Calfee C.S. Stability of ARDS subphenotypes over time in two randomised controlled trials. Thorax. 2018;73(5):439–445. doi: 10.1136/thoraxjnl-2017-211090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We stated that all the data and materials were true and available in the study.