Abstract
Earlier reports from this laboratory have shown that the promiscuous transactivator infected-cell protein 0 (ICP0) binds and stabilizes cyclin D3, that the binding site maps to aspartic acid 199 (D199), and that replacement of D199 with alanine abolishes binding and reduces the capacity of the mutant virus to replicate in quiescent cells or to cause mortality in mice infected by a peripheral site. The objective of this report was to investigate the role of cyclin D3 in the biology of ICP0. We report the following results. (i) Wild-type ICP0 activates cyclin D-dependent kinase 4 (cdk4) and stabilizes cyclin D1 although ICP0 does not interact with this cyclin. (ii) The D199A mutant virus (R7914) does not activate cdk4 or stabilize cyclin D1, and neither the wild-type nor the mutant virus activates cdk2. (iii) Early in infection of human embryonic lung (HEL) fibroblasts both wild-type and D199A mutant ICP0s colocalize with PML, and in these cells the ND10 nuclear structures are dispersed. Whereas wild-type ICP0 is transported to the cytoplasm between 3 and 9 h. after infection, ICPO containing the D199A substitution remains quantitatively in the nucleus. (iv) To examine the interaction of ICP0 with cyclin D3, we used a previously described mutant carrying a wild-type ICP0 but expressing cyclin D3 (R7801) and in addition constructed a virus (R7916) that was identical except that it carried the D199A-substituted ICP0. Early in infection with R7801, ICP0 colocalized with cyclin D3 in structures similar to those containing PML. At 3 h after infection, ICP0 was translocated to the cytoplasm whereas cyclin D3 remained in the nucleus. The translocation of ICP0 to the cytoplasm was accelerated in cells expressing cyclin D3 compared with that of ICP0 expressed by wild-type virus. In contrast, ICP0 carrying the D199A substitution remained in the nucleus and did not colocalize with cyclin D3. These studies suggest the following conclusions. (i) ICP0 brings to the vicinity of ND10 cyclin D3 and, in consequence, an activated cdk4. The metabolic events occurring at or near that structure and involving cyclin D3 cause the translocation of ICP0 to the cytoplasm. (ii) In the absence of the cyclin D3 binding site in ICP0, cyclin D3 is not brought to ND10, cyclin D is not stabilized, and the function responsible for the translocation of ICP0 is not expressed, and in quiescent HEL fibroblasts the yields of virus are reduced.
Infected-cell protein 0 (ICP0) of herpes simplex virus 1 (HSV-1) acts as a promiscuous transactivator (reviewed in references 31 and 32). Alone, or more effectively in combination with ICP4, another regulatory protein, ICP0 activates genes introduced by infection or transfection (13, 27, 30). ICP0 is particularly important for viral replication in cells infected at low multiplicity but appears to be nonessential for viral replication (33, 37). The mechanism by which ICP0 accomplishes this task is unknown. A rich, extensive literature has documented several important features of ICP0. They are summarized in the following paragraphs.
The 775-amino-acid protein is translated from a spliced mRNA. The three exons encoding ICP0 contain 19, 222, and 534 codons. The protein is extensively posttranslationally processed by both cellular and viral protein kinases and is nucleotidylylated by casein kinase II (3, 26). ICP0 contains a zinc RING finger located in exon II (8).
Early in infection, ICP0 colocalizes with PML, a component of a structure known as ND10 (23). The function of ND10 is not known, but it has been suggested that this structure acts as some kind of a cellular repressor. ICP0 has been shown to degrade the PML isoforms conjugated to SUMO-1 or PIC1 and cause the disruption of ND10 (9, 22, 11). Also, ICP0 has been shown to bind a ubiquitin-specific protease and divert it to ND10, possibly to cleave SUMO-1 from PML (10).
ICP0 also appears to play a role in the destabilization of the regulatory subunit of the cellular DNA-dependent protein kinase (20) and in the degradation of centromeric protein C (CENP-C) (12). This protein plays a key role in the assembly of the kinetochore; in uninfected cells degradation of this protein results in delayed transition of metaphase to anaphase. The zinc RING finger is required for the association of ICP0 with the kinetochore, but the actual structure to which ICP0 binds is unknown. The mutation which abolishes the degradation is in a region that overlaps the site required for binding the ubiquitin-specific protease near the carboxyl terminus of ICP0.
This laboratory reported that ICP0 binds two additional proteins. The first, elongation factor 1δ (EF-1δ), is responsible for ADP-ATP exchange. Consistent with this finding, ICP0 was shown to be translocated into cytoplasm sometime between 3 and 9 h after infection depending on the cell type (16). The significance that HSV places on EF-1δ is underscored by two observations. First, a truncated ICP0 polypeptide containing the binding site interfered with in vitro synthesis of a reporter protein. Second, the UL13 protein kinase phosphorylates EF-1δ and this protein is also phosphorylated in cells infected with representative beta- and gammaherpesviruses (18, 19). The second protein bound by ICP0 is cyclin D3. In this instance the binding site was mapped to aspartic acid 199 (D199), and furthermore it was shown that cyclin D3 colocalizes with ICP0 early in infection and that the binding of ICP0 does not interfere with the ability of cyclin D3 to phosphorylate its main substrate, the retinoblastoma protein (17). In a subsequent study (38) it was shown that the replacement of D199 with alanine (D199A) abolished the interaction with cyclin D3 and that a recombinant virus (R7914) carrying the D199A substitution exhibits reduced growth in quiescent human embryonic lung (HEL) fibroblasts and reduced virulence when administered to mice by peripheral routes. The need for cyclin D3 appears to be shared by other herpesviruses. Thus, the simian herpesvirus saimiri and its distant cousin human herpesvirus 8 both encode functional homologs of D-type cyclins (4, 15, 21, 25). A central question is the role of cyclin D3 in the biology of HSV.
The focus of the experiments described in this report was twofold. First, we compared the D199A mutant with wild-type virus with respect to the status of cyclin D and its partner, cyclin-dependent kinase 4 (cdk4). We also compared the localization of wild-type and mutant ICP0 in infected cells. Last, we compared wild-type virus with a recombinant carrying wild-type ICP0 and the cyclin D3 gene. The results indicate that the interaction of ICP0 with cyclin D3 correlates with two events: the stabilization and activation of G1-phase cyclins, even though cdk2 is not activated, and the translocation of ICP0 from the nucleus to the cytoplasm.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% newborn calf serum. Human 143 thymidine kinase-negative (143TK−) cells were originally obtained from Carlo Croce. HEL fibroblasts were obtained from Aviron (Mountain View, Calif.) and grown in DMEM supplemented with 10% fetal bovine serum. To arrest HEL fibroblasts in G1, 50% confluent cultures were rinsed with phosphate-buffered saline (PBS) and stored at room temperature for 7 days in DMEM supplemented with 0.25% fetal bovine serum. The cultures were incubated in the same medium at 37°C for 1 day prior to their use. HSV-1 strain F [HSV-1(F)], a limited-passage isolate, is the prototype strain used in this laboratory (7). The construction and phenotypic properties of recombinant viruses R7914, R7915, and R7801 have been described earlier (17, 38).
Plasmids.
Plasmids pRB4986 and pRB5266 were described elsewhere and have been used in a yeast two-hybrid analysis (17, 38). Plasmid pRB5162, containing the Egr1-driven human cyclin D3 gene, was described earlier (17) and was used to construct cyclin D3-expressing recombinant viruses R7801 and R7916. Plasmids pBH1035A and pBH1035B were independent clones of a PCR-amplified human cyclin D1 gene obtained from an Epstein-Barr virus-transformed human peripheral blood lymphocyte cDNA library (gift from Aviron). The human cyclin D1 gene was amplified by standard PCR with oligonucleotide primers designed to amplify the 0.9-kb gene with EcoRI (5′) and BamHI (3′) sites. The amplified DNA fragment was purified and ligated to yeast vector pGAD424 in frame with its activation domain.
Construction of recombinant viruses.
The construction of recombinant virus R7801, possessing the human cyclin D3 gene inserted within the thymidine kinase (TK) locus has been described previously (17). Recombinant R7916 was constructed by cotransfection of rabbit skin cells with intact R7914 viral DNA and pRB5162. TK− recombinant viruses were selected on 143TK− cells overlaid with DMEM containing 5% newborn calf serum and 40 μg of bromodeoxyuridine per ml of medium as described previously (29). Recombinants were plaque purified after two rounds of selection and were verified by both hybridization of electrophoretically separated restriction digests of progeny viral DNA and immunoblotting of R7916-infected cell lysates for cyclin D3 with anti-cyclin D3 antibody (Pharmingen catalog no. 14781C).
Yeast two-hybrid β-galactosidase analysis.
A yeast two-hybrid system described in a earlier publication (17) was employed to determine whether cyclin D1 interacts with ICP0 in the same fashion as does cyclin D3. Yeast vector pGBT9 encoding wild-type ICP0 exon 2 (pRB4986) or the D199A mutant (pRB5266) fused to the GAL4 binding domain was cotransformed with the pGAD424 vector encoding either human cyclin D1 or cyclin D3 into yeast strain Y190 and plated on SD synthetic medium lacking tryptophan and leucine. Individual cotransformants, after the freeze-fracturing of the cells, were tested for β-galactosidase activity.
Cell infection.
Cells grown in 150-cm2 flasks, unless otherwise stated, were exposed to 5 × 108 PFU of the appropriate virus (multiplicity of infection of 10) in 5 ml of 199V on a rotary shaker at 37°C. After 2 h, the inoculum was replaced with fresh medium and incubated at 37°C until cells were harvested.
Immunoblotting of electrophoretically separated cell lysates.
Cell lysates were harvested at the times indicated in Results in the following manner. The medium was replaced with 5 ml of ice-cold PBS, and the cells were stored on ice for 10 min and then harvested by scraping. After centrifugation, the cell pellet was rinsed, pelleted again by centrifugation in 1 ml of PBS, and then solubilized in disruption buffer (2% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 6.8], 3% sucrose, 5% β-mercaptoethanol, and bromophenol blue). The extract was sonicated, boiled for 5 min, and subjected to electrophoresis on 12% bisacrylamide gels before being transferred to nitrocellulose membranes. The membranes were then blocked for 1 h with 5% nonfat dry milk and reacted with the appropriate primary antibody overnight at 4°C. Monoclonal antibodies to human cyclin D1 and D3 (Pharmingen catalog no. 66271A and 14781C, respectively) and human p21 (Santa Cruz Biotechnology, Santa Cruz, Calif.; catalog no. sc-817) were diluted 1:1,000 in PBS containing 1% bovine serum albumin (BSA) and 0.05% Tween 20. Secondary antibodies (peroxidase-conjugated goat anti-mouse [1:1,000] and goat anti-rabbit [1:3,000] antibodies; Sigma) were applied for 2 h. All rinses were done in PBS containing 0.05% Tween 20. Immunoblots were developed by enhanced chemiluminescence according to instructions supplied by the manufacturer (Pierce).
RNA isolation and RPA.
Total RNA was isolated from HEL cells in 150-cm2 flasks with a Trizol reagent (Gibco BRL) according to instructions supplied by the manufacturer. Precipitated total RNA was resuspended in RNase-free water, quantified, and stored at −80°C. The RNase protection assay (RPA) (RPA II; Ambion) consists of annealing a labeled probe with purified sample RNA followed by RNase digestion of unprotected RNA. A multiprobe set (Pharmingen; Riboquant hCYC-1, catalog no. 45352) was [α-32P]CTP labeled with the aid of a T7 transcription system (Ambion; T7 MAXIscript, catalog no. 1310). Specifically, 1 μl of RiboQuant multiprobe template was transcribed with T7 polymerase and 50 μCi of [α-32P]CTP for 1 h at 37°C followed by digestion with 2 U of DNase I for 30 min at 37°C. After digestion, the probe was extracted with phenol-chloroform-isoamyl alcohol and precipitated. Labeled probe was resuspended in 50 μl of RPA II hybridization solution (Ambion). Isolated total RNA (10 μg) was hybridized overnight at 56°C with a labeled RNA probe (2.5 × 105 cpm) set complementary to the target cyclin RNA to be detected. After hybridization, the mixture was treated with the appropriate amount of RNase A-RNase T1 mixture (1:500) to degrade unhybridized probe. Labeled probe protected by hybridization with cRNA was separated on 5% polyacrylamide gels and detected by autoradiography.
pRB kinase assay.
HeLa cells were seeded on 25-cm2 flasks and allowed to adhere for 1 h and then were rinsed to remove unattached cells and were mock infected or infected with 2 × 107 PFU of R7914 or HSV-1(F) in 1 ml of 199V on a rotary shaker at 37°C. After 2 h, the inoculum was aspirated and replaced with 5 ml of DMEM with 10% newborn calf serum. Flasks were incubated at 37°C until harvested at the times indicated in Results. Harvested cells were resuspended in lysis buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40, 400 mM NaCl, 0.1% Sodium orthovanadate, 10 mM NaF, 2 mM dithiothreitol [DTT], TPCK [tolylsulfonyl phenylalanyl chloromethyl ketone], TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone], and phenylmethylsulfonyl fluoride for 1 h on ice. Insoluble material was then removed by centrifugation, and protein concentrations were determined by Bradford assay (Bio-Rad). Equivalent amounts of protein from cell lysates were subjected to immunoprecipitation. Cell lysates were reacted with rabbit preimmune sera for 2 h and then mixed with 50 μl of 50% protein A slurry for 1 h. The samples were centrifuged, and supernatant fluid was collected and reacted with the anti-cdk4 antibody (Santa Cruz Biotechnology) overnight at 4°C. Immunoprecipitated cdk4 was recovered by the addition of 20 μl of 50% protein A slurry for 1 h and rinsed twice with lysis buffer, twice with low-salt buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40, 1 mM NaCl, and 2 mM DTT), and twice with incomplete kinase buffer (50 mM Tris [pH 7.4], 10 mM MgCl2, and 5 mM DTT). Forty microliters-of complete kinase buffer was then added to each sample (2 μg of glutathione S-transferase [GST]-pRb [Santa Cruz Biotechnology], 10 μM ATP, and 20 μCi of [γ-32P]ATP in incomplete kinase buffer), and samples were incubated at 30°C for 20 min. The reaction was stopped by the addition of 13 μl of 4 × disruption buffer, and the reaction mixture was heated for 5 min at 95°C. The reaction mixtures were subjected to electrophoresis in 10% bisacrylamide gels, transferred to a nitrocellulose membrane, and subjected to autoradiography. The amount of radiolabeled product was quantified with the aid of a Molecular Dynamics Storm 860 PhosphorImager.
Immunofluorescence.
HEL fibroblasts seeded on glass slides (Cell-Line, Newfield, N.J.) were exposed to 10 PFU of the HSV-1(F) parent strain or the R7914 recombinant per cell and incubated at 37°C for the time intervals specified in Results. Cells were fixed in ice-cold methanol for 2 h and blocked in PBS containing 1% BSA and 20% normal human serum, followed by an overnight reaction with the primary antibody at 4°C. The primary antibody consisted of rabbit anti-ICP0 exon 2 (diluted 1:1,000) and mouse anti-PML catalog no. sc-966; Santa Cruz Biotechnology) diluted 1:200 in PBS containing 1% BSA and 10% normal human serum. After overnight incubation, the slides were rinsed three times in PBS and reacted for 1 h with a 1:400 dilution of goat anti-mouse immunoglobulin G (IgG) conjugated to Texas red (Molecular Probes, Eugene, Oreg.) and a 1:160 dilution of goat anti-rabbit IgG conjugated to fluorescein isothiocyanate (Sigma). Slides were washed as described above and mounted in PBS containing 90% glycerol and 1 mg of p-phenylenediamine per ml. The slides were examined in a Zeiss confocal microscope. Digitized images of the fluorescent-antibody-stained cells were acquired with software provided by Ziess.
RESULTS
ICP0 mediates the stabilization of both cyclin D1 and cyclin D3.
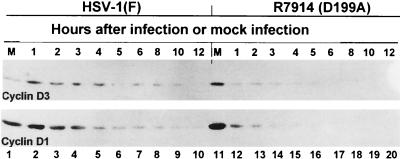
An earlier report (17) showed that ICP0 mediates the stabilization of cyclin D3 in HSV-1-infected cells. The objective of the experiments described in this section was to determine whether HSV-1 specifically targets cyclin D3 or whether other D-type cyclins are stabilized. In this series of experiments replicate cultures of HEL fibroblasts were mock infected or exposed to 10 PFU of HSV-1(F) or the D199A mutant (R7914) per cell. The cultures were harvested at 1, 2, 3, 4, 5, 6, 8, 10, or 12 h after infection, solubilized, subjected to electrophoresis in denaturing gels, and reacted with anti-cyclin D1 or a cyclin D3-specific antibody as described in Materials and Methods. The results were as follows. (i) As previously reported, cyclin D3 was stabilized in wild-type virus-infected cells as late as 10 h after initiation of infection (Fig. 1, lane 9), whereas in R7914-infected, quiescent HEL fibroblasts, cyclin D3 could not be detected after 6 h of infection (Fig. 1, lane 17). (ii) Cyclin D1 was detected as late as 12 h in cells infected with HSV-1(F) but was not detectable in cells infected with the D199A virus after 4 h after infection. Cyclin D2 was not tested in these series of experiments because it could not be detected or was not present in appreciable amounts in HEL fibroblasts.
FIG. 1.
Photographs of immunoblots of electrophoretically separated lysates of quiescent HEL fibroblast cells mock infected (lanes 1 and 11) or infected with HSV-1(F) (lanes 2 to 10) or R7914 (lanes 12 to 20). The lysates were harvested at the times indicated, subjected to electrophoresis on SDS-12% polyacrylamide gels, transferred to nitrocellulose, and reacted with mouse monoclonal antibodies against cyclins D1 and D3.
Exon 2 domain of the ICP0 of HSV-1(F) reacts with cyclin D3 but not with cyclin D1 in the yeast two-hybrid system.
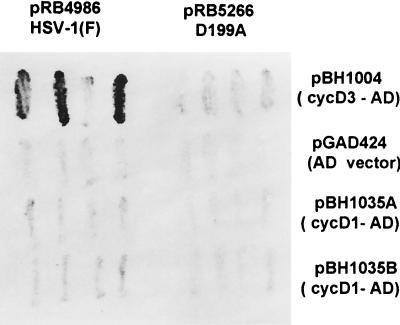
The interaction between cyclin D3 and ICP0 was first detected in the yeast two-hybrid system and was then verified in pulldown experiments (17). In view of the results presented above showing that ICP0 stabilizes both cyclins D3 and D1, it was of interest to determine whether cyclin D1 interacts with ICP0 in the yeast two-hybrid system. In essence we repeated the experiments described by Van Sant et al. (38) except that we included two independently derived plasmids containing the cyclin D1 fused to the activation domain. Specifically, yeast cells were cotransformed with a vector encoding wild-type or mutant ICP0 exon 2 fused to a GAL4 binding domain and a vector encoding either human cyclin D1 or cyclin D3 fused to a GAL4 activation domain. Individual cotransformants able to grow on synthetic media lacking tryptophan and leucine were analyzed for β-galactosidase activity. The results of one of several experiments are shown in Fig. 2. Consistent with results published earlier, cyclin D3 interacted with ICP0. Cyclin D1 did not interact with ICP0 in any of the multiple assays carried out to date.
FIG. 2.
Photographs of independent Y190 yeast cotransformants analyzed for β-galactosidase activity. Yeast vector pGBT9 encoding wild-type ICP0 exon 2 (pRB4986) or the D199A mutant (pRB5266) fused to the GAL4 binding domain was cotransformed with the pGAD424 vector encoding either human cyclin D1 or cyclin D3 into yeast, plated on SD synthetic medium lacking tryptophan and leucine, freeze-fractured, and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
HSV-1(F) does not induce D-type cyclin RNA synthesis.
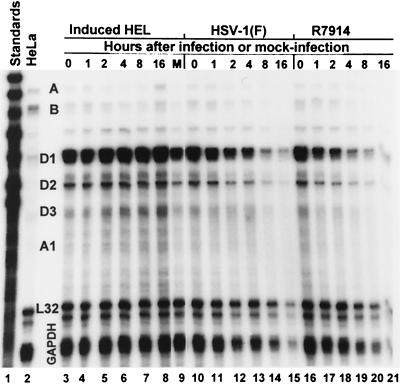
Inasmuch as we could not demonstrate a direct interaction between ICP0 and cyclin D1, the question of whether HSV induces the transcription of cyclin D family members arose. The experiments were also motivated by other reasons. First, ICP0 by itself or in the presence of ICP4 has been reported to induce the expression of both HSV and non-HSV genes in transient assay systems (27, 30). Second, a recent article suggested that this activation of gene expression by HSV-1 ICP0 occurs at the levels of mRNA synthesis (14). Conceivably, the D199A substitution within the coding sequences of ICP0 in R7914 could affect this function of ICP0. To test this hypothesis, we carried out an RPA as described in Materials and Methods. A probe set (Pharmingen catalog no. 45352), representing many human cyclins, was [α32P]CTP labeled by a T7-driven in vitro transcription system. Total RNA (10 μg) isolated from HEL cells mock infected or exposed to 10 PFU of HSV-1(F) or the recombinant R7914 at the times indicated in Fig. 3 was hybridized overnight at 56°C with 2.5 × 105 cpm of the radiolabeled RNA probe set complementary to the target cyclin RNA to be detected. After hybridization, the samples were incubated with an RNase mixture to degrade unhybridized probe. Labeled probe, protected by complementary RNA within the sample, was separated by nondenaturing gel electrophoresis and visualized by autoradiography. The assay measured the levels of RNA of cyclin D1, D2, and D3 as well as those of cyclin A, B, and C. The RPA was done with RNA extracted from mock-infected quiescent HEL fibroblasts induced into cell cycle progression by the addition of serum as a control (Fig. 3, lanes 3 to 8) or virally infected quiescent HEL fibroblasts (Fig. 3, lanes 10 to 21). The results (Fig. 3) were quantified with the aid of the ImageQuant program of the Molecular Dynamics PhosphorImager and are shown in Table 1. Lane 1 represents RNA probes that have not been digested with RNase. Lane 2 represents the probe hybridized to control mRNA from HeLa cells and then digested with RNase. It should be noted that HeLa cells do not contain detectable amounts of cyclin D2 RNA (lane 2). Protected cyclin D2 RNA would be expected to be 181 nucleotides long.
FIG. 3.
Autoradiographic image of RPA of levels of total human cyclin RNA isolated from quiescent HEL fibroblast cells induced with 10% fetal bovine serum and mock infected (lanes 3 to 9) or quiescent HEL fibroblasts infected with HSV-1(F) (lanes 10 to 15) or R7914 (lanes 16 to 21). Total RNAs from HEL fibroblasts harvested at the times indicated were annealed to the radiolabeled cyclin probe set, and then unprotected RNA was digested with RNase. The labeled probe protected by hybridization with complementary RNA was electrophoretically separated on 5% nondenaturing polyacrylamide gels and detected by autoradiography. The cyclin species are indicated between lanes 2 and 3.
TABLE 1.
Quantification of protected cyclin RNA speciesa
| RNA species | Virus | Radioactivity at indicated time (h) after initiation of infection
|
|||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 4 | 8 | 16 | ||
| Cyclin A | Mock | 1.00 | 1.25 | 0.63 | 0.38 | 0.75 | 2.88 |
| HSV-1(F) | 1.00 | 1.00 | 0.88 | 0.75 | 0.80 | 0.00 | |
| R7914 | 1.00 | 0.88 | 1.13 | 1.13 | 1.00 | 0.00 | |
| Cyclin B | Mock | 1.00 | 1.32 | 0.96 | 0.56 | 0.68 | 1.24 |
| HSV-1(F) | 1.00 | 1.16 | 0.84 | 0.72 | 0.52 | 0.00 | |
| R7914 | 1.00 | 1.36 | 1.04 | 0.76 | 0.52 | 0.00 | |
| Cyclin D1 | Mock | 1.00 | 0.97 | 1.30 | 1.16 | 1.58 | 1.29 |
| HSV-1(F) | 1.00 | 0.83 | 0.79 | 0.60 | 0.33 | 0.42 | |
| R7914 | 1.00 | 0.78 | 0.68 | 0.45 | 0.37 | 0.36 | |
| Cyclin D2 | Mock | 1.00 | 0.97 | 1.22 | 0.97 | 1.21 | 1.09 |
| HSV-1(F) | 1.00 | 0.90 | 0.88 | 0.76 | 0.49 | 0.39 | |
| R7914 | 1.00 | 0.90 | 0.71 | 0.67 | 0.56 | 0.49 | |
| Cyclin D3 | Mock | 1.00 | 0.92 | 1.37 | 1.22 | 1.58 | 1.54 |
| HSV-1(F) | 1.00 | 0.74 | 1.10 | 0.62 | 0.57 | 0.15 | |
| R7914 | 1.00 | 0.73 | 0.81 | 0.59 | 0.31 | 0.12 | |
| Cyclin A1 | Mock | 1.00 | 1.22 | 1.14 | 1.25 | 1.41 | 1.30 |
| HSV-1(F) | 1.00 | 0.56 | 0.65 | 0.47 | 0.45 | 0.00 | |
| R7914 | 1.00 | 0.54 | 0.37 | 0.35 | 0.30 | 0.00 | |
Quantification was performed on protected species in total RNA isolated from quiescent HEL cells induced with 10% fetal bovine serum or mock infected or infected as described in Results. Radioactivity was quantified with the aid of a Storm 860 phosphorimager and ImageQuant software and normalized to amounts detected at time zero (beginning of the experiment).
The results of the RPAs indicate that uninfected HEL cells induced to enter cell cycle progression by the addition of serum reestablished appropriate cyclin RNA levels by 2 h coincident with a reduction in inactive cyclin A and cyclin B (Fig. 3, lane 5) and an induction of D-type cyclin RNA (Fig. 3, lanes 5 to 8, and Table 1). The D-type cyclin RNAs peaked at 8 h after serum addition (Table 1). Cyclin A RNA levels reached maximal levels at 16 h after induction (lane 8), indicating cell progression through S phase, followed by a rise in cyclin B levels (compare Fig. 3, lanes 7 and 8). In contrast to what was found for uninfected cells, analyses of RNA extracted from both HSV-1(F)- and R7914-infected cells show a steady decline in all cyclin RNAs tested (Fig. 3, lanes 11 to 15 and 17 to 21, respectively) as well as a decrease in the levels of RNA of L32 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) housekeeping genes. Table 1 summarizes cyclin RNA levels. These results indicate the following: (i) existing RNA is rapidly degraded, most likely as a consequence of the action of vhs protein, (ii) cyclin D RNAs were not induced by viral infection, and (iii) the levels of RNA detected in cells infected with the D199A mutant do not differ significantly from those detected in cells infected with the wild-type parent virus.
The proteasomal inhibitor MG132 blocks the degradation of D-type cyclin proteins in R7914-infected cells.
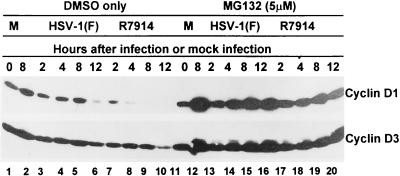
Inasmuch as we could not demonstrate that wild-type virus induces de novo synthesis of D-type cyclin RNA after infection, the question of whether the disappearance of D-type cyclins in D199A mutant-infected cells is the consequence of protein degradation arose (24, 28). In this series of experiments, replicate cultures of HEL cells were mock infected or exposed to 10 PFU of HSV-1(F) or R7914 per cell and were either treated with MG132 (5 μM) or exposed to dimethyl sulfoxide (DMSO) only. The cells were harvested at 2, 4, 8, or 12 h after infection, solubilized, subjected to electrophoresis in denaturing gels, and reacted with antibodies specific for cyclin D1 and D3 as described in Materials and Methods. The results (Fig. 4) indicate that, in contrast to what was found for DMSO-treated cells, cyclin D1 and D3 levels in cells treated with MG132 and infected with either R7914 or HSV-1(F) continued to accumulate for at least 12 h after infection.
FIG. 4.
Photographs of immunoblots of electrophoretically separated quiescent HEL cell lysates treated with proteasomal inhibitor MG132 (lanes 11 to 20) or only with DMSO (lanes 1 to 10). Quiescent HEL cells were either mock infected (lanes 1 and 2 and 11 and 12) or infected with HSV-1(F) (lanes 3 to 6 and 13 to 16) or R7914 (lanes 7 to 10 and 17 to 20). The lysates were harvested at the times indicated, subjected to electrophoresis on SDS-12% polyacrylamide gels, transferred to nitrocellulose, and reacted with mouse monoclonal antibodies against cyclins D1 and D3.
The results presented in this section indicate that the accelerated disappearance of cyclins D1 and D3 in cells infected with the R7914 mutant was due to proteasomal degradation.
Stabilization of D-type cyclins by ICP0 in wild-type HSV-1(F) correlates with enhanced cdk4 kinase activity in vitro.
The results shown in Fig. 1 indicate that a single amino acid substitution that abrogated the ability of ICP0 to bind and stabilize cyclin D3 also resulted in the loss of the capacity of ICP0 to stabilize cyclin D1 even though it did not interact with this cyclin. D-type cyclins are known to associate with cyclin-dependent kinases to form an active kinase complex that advances cell cycle progression (34, 35). An earlier publication from this laboratory indicated that ICP0 did not affect the activity of cyclin D3-dependent kinase (cdk4) in vitro (17). A central question of considerable interest was whether the activity of cdk4 extracted from infected cells is concordant with the relative amounts of cyclin D1 or D3 accumulating in these cells. In this series of experiments replicate HeLa cell cultures synchronized as described in Materials and Methods were mock infected or exposed to 10 PFU of HSV-1(F) or the D199A mutant (R7914) per cell. At times indicated in Fig. 5, cultures were harvested and cdk4-specific kinase complexes were immunoprecipitated by the sequential addition of anti-cdk4 antibody to the lysates followed by capture with protein A-Sepharose. After exhaustive rinsing, the immune complexes were incubated with the GST-retinoblastoma protein substrate (39) in a kinase buffer supplemented with [γ-32P]ATP. The reactions were stopped by addition of disruption buffer, and the reaction mixtures were subjected to electrophoresis in a denaturing gel, transferred to a nitrocellulose membrane, and subsequently analyzed by radiography. The results were as discussed in the following paragraphs.
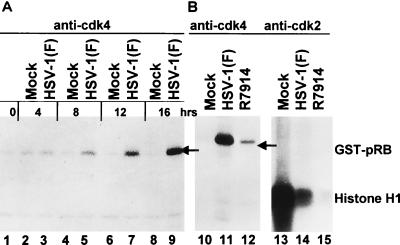
FIG. 5.
(A) Autoradiographic image of electrophoretically separated [γ-32P]ATP-labeled GST-pRB fusion protein. Synchronized HeLa cells were mock infected (lanes 1, 2, 4, 6, and 8) or infected with HSV-1(F) (lanes 3, 5, 7, and 9). The cells were harvested at the times indicated, solubilized, electrophoretically separated in denaturing gel, transferred to a nitrocellulose sheet, and reacted with polyclonal anti-cdk4. Immunoprecipitates were incubated with the substrate GST-pRB in a kinase buffer supplemented with [γ-32P]ATP. The reaction mixtures were subjected to electrophoresis in a denaturing 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and subjected to autoradiography. The radiolabeled product was collected on a phosphorimager. (B) Lanes 10 to 12, autoradiographic image of electrophoretically separated GST-pRB labeled with [γ-32P]ATP by cdk4 immune precipitated from synchronized HeLa cells harvested 12 h after mock infection (lane 10) or infection with HSV-1(F) (lane 11) or R7914 (lane 12). Experimental details were the same as for panel A. Lanes 13 to 15, autoradiographic image of electrophoretically separated histone H1 labeled with [γ-32P]ATP by cdk2 immune precipitated from synchronized HeLa cells 12 h after mock infection (lane 13) or infection with HSV-1(F) (lane 14) or with R7914 (lane 15). Experimental details were the same as for panel A.
cdk4 activity was detected in mock-infected cells at 4 h after synchronization, but it decreased at 8 h and at later time intervals (Fig. 5A, lanes 2, 4, 6, and 8). In cells infected with wild-type virus, the cdk4 activity was similar to that in mock-infected cells at 4 h but continued to increase and exceeded by manyfold the peak activity in mock-infected cells by 16 h after synchronization and infection (Fig. 5A, lanes 3, 5, 7, and 9).
Comparison of cdk4 activities immune precipitated from cells harvested 12 h after synchronization and mock infection or infection with HSV-1(F) or R7914 as described above indicated that the activity of the cyclin D-dependent enzyme was greatly reduced in R7914-infected cells compared to that in wild-type virus-infected cells but was higher than that in mock-infected cells (Fig. 5B, lanes 10 to 12).
Ehmann et al. have reported that cdk2 activity is not enhanced in viral infection (6). To test whether the D199A mutant affects cdk2 activity, replicate mock-infected or infected cells were harvested at 12 h after synchronization. cdk2-specific kinase complexes were immunoprecipitated with anti-cdk2 antibody and collected with protein A-Sepharose. The kinase reactions were carried out with histone H1 as a substrate. The results (Fig. 5B, lanes 13 to 15) indicate that cdk2 activity was reduced in infected cells relative to that in mock-infected cells. The decrease was even more striking in cells infected with the R7914 mutant (Fig. 5, lanes 13 to 15).
The enhanced cdk4 activity in wild-type virus-infected cells correlates with reduced p21 levels.
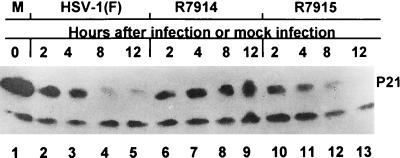
One function of activated cdk4 is to phosphorylate p21. In turn, phosphorylated p21 is recognized by the SCF (Skp1/Cullin 1/F-box) E3 ligase, ubiquitinated, and targeted for proteasomal degradation, allowing cycle progression (5, 28, 41). The status of p21 is therefore a good indicator of the activity of cdk4 kinase. To test the effect of HSV-1 on p21, we selected quiescent HEL fibroblasts since in these cells the level of p21 is expected to be high (5). In these experiments, replicate cultures of quiescent HEL cells were mock infected or exposed to 10 PFU of wild-type HSV-1(F) or recombinant virus R7914 or R7915 per cell. The cultures were harvested at 2, 4, 8, or 12 h after infection, solubilized, subjected to electrophoresis in denaturing gels, and reacted with the anti-p21 monoclonal antibody. The results shown in Fig. 6 were as follows: (i) p21 levels decreased drastically between 4 and 8 h after infection with wild-type virus or with recombinant R7915, in which the D199A mutation was restored, and (ii) in HEL fibroblasts infected with the D199A mutant, R7914, p21 levels continued to increase throughout the 12-h interval of the experiment.
FIG. 6.
Photographic image of immunoblots of electrophoretically separated lysates of quiescent HEL cells mock infected (lane 1) or infected with HSV-1(F) (lanes 2 to 5), R7914 (lanes 6 to 9), or R7915 in which ICP0 was restored to the wild-type form (lanes 10 to 13). The lysates were harvested at the times indicated, subjected to electrophoresis on an SDS-12% polyacrylamide gel, transferred to nitrocellulose, and reacted with a mouse monoclonal antibody against cyclin-dependant kinase inhibitor p21.
We draw several conclusions from these series of experiments. (i) In the experimental systems in which it was tested, wild-type HSV-1 stabilized cyclins D1 and D3 and activated cdk4 kinase. Consistent with the presence of an active cdk4, the levels of p21 decreased. (ii) In cells infected with the D199A mutant, R7914, cyclins D1 and D3 were not stabilized, cdk4 was not active, and p21 levels were maintained. (iii) In cells infected with either virus, the activity of cdk2 was reduced relative to that in mock-infected cells.
The localization of wild-type and mutant ICP0 in the infected cells.
Studies on parent strain HSV-1(F) showed that (i) wild-type ICP0 was translocated from the nucleus of HEL fibroblasts into the cytoplasm beginning approximately 3 h after infection and (ii) early in infection, ICP0 colocalized with cyclin D3 in the nucleus (17). An earlier report from this laboratory also described the construction of a recombinant virus (R7801) carrying the cyclin D3 gene under the control of the Egr1 promoter. To define the role of the D199 locus in ICP0, it was of interest to determine the localization of ICP0 at various times after infection and to examine the interaction of ICP0 carrying the D199A mutation with PML and cyclin D3. To meet this objective, we constructed, as described in Materials and Methods, a mutant (R7916) by inserting the cyclin D3 gene under the control of the Egr1 promoter into R7914 in a manner exactly identical to that for R7801 described earlier (17). All of the experiments described in this section were done on HEL fibroblasts grown on four-well slides and mock infected or exposed to 10 PFU of wild-type or mutant viruses per cell. The cells were examined and photographed at a magnification of ×100. Enumerations were done by counting infected cells in adjacent fields at a magnification of ×64. The results of these studies are summarized in the following paragraphs.
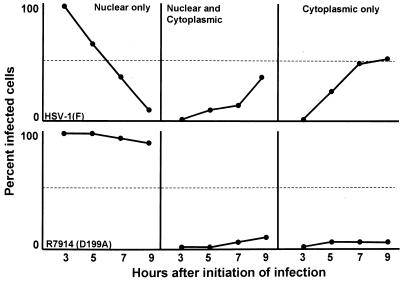
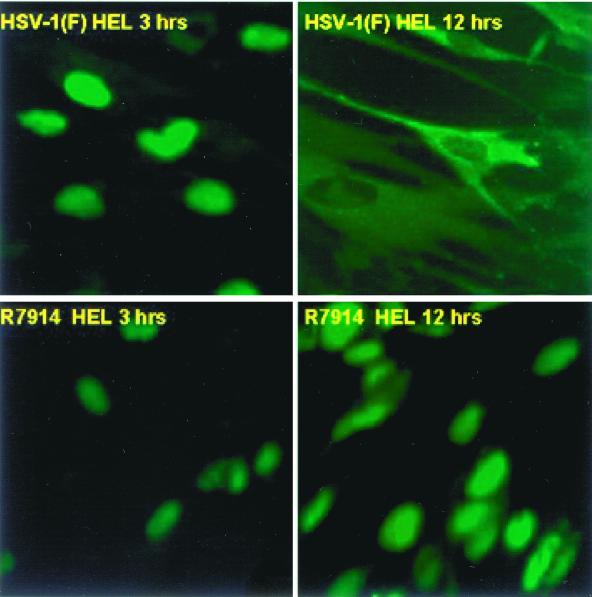
Examination of HEL fibroblasts at intervals between 3 and 9 h after infection revealed that wild-type ICP0 was translocated into the cytoplasm after 3 h of infection. As shown in Fig. 7, ICP0 gradually shifted from a totally nuclear localization to either an exclusively cytoplasmic localization or both a cytoplasmic and a nuclear localization. In contrast, in HEL fibroblasts infected with the D199A mutant, ICP0 remained localized in the nucleus and only a small fraction of cells exhibited ICP0 in trace amounts in the cytoplasm. ICP0 carrying the D199A mutation was retained in the nucleus even as late as 15 h after infection (Fig. 8).
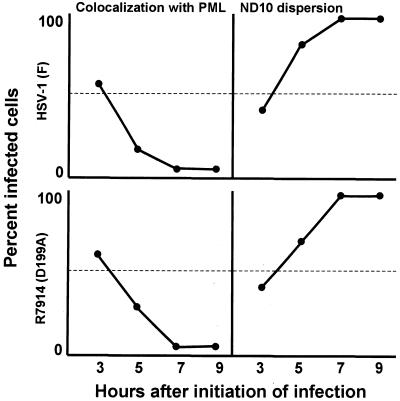
FIG. 7.
Quantification of wild-type and mutant ICP0 translocation to the cytoplasm as a function of time in infected HEL fibroblast cells. A minimum of 200 infected cells were counted per data point.
FIG. 8.
Digital images of HSV-1(F) and recombinant R7914-infected HEL cells reacted with antibody to ICP0 at early and late times of infection. The infected cells were fixed 3 or 12 h after infection and reacted with polyclonal rabbit antibody to ICP0 and then reacted with anti-rabbit IgG conjugated to fluorescein isothiocyanate. The single-color images were captured with a Zeiss confocal microscope and software provided by Ziess. The digitized images were not modified subsequent to capture.
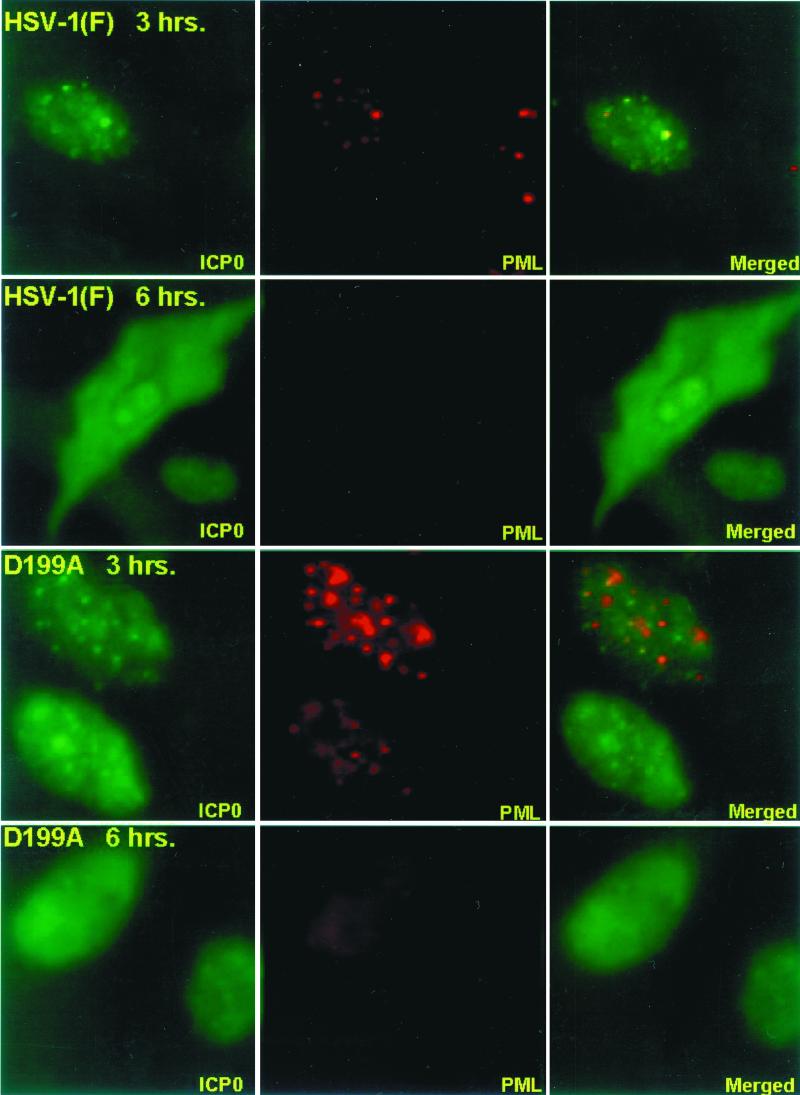
HEL fibroblasts were fixed and reacted with anti-PML and anti-ICP0 antibodies as described in Materials and Methods at intervals after infection with wild-type virus or the D199A mutant, R7914. The results of the examination of the stained cells, summarized in Fig. 9, were that both wild-type and mutant ICP0 colocalized with structures containing PML. In both sets of cultures, colocalization occurred in approximately 60% of cells at 3 h after infection, but this value decreased to less than 5% by 7 h after infection. In both sets of cultures the ND10 structures declined with time and were virtually undetectable by 7 h after infection. In terms of gross appearance, the cells infected with the D199A mutant could not be differentiated from wild-type virus-infected cells with respect to colocalization of ICP0 with PML and disappearance of ND10 structures (Fig. 10).
FIG. 9.
Quantification of wild-type and mutant D199A ICP0 colocalization with PML and subsequent ND10 dispersion in infected HEL fibroblasts as a function of time. A minimum of 200 infected cells were counted per data point.
FIG. 10.
Digital images of HEL fibroblasts infected with HSV-1(F) or R7914 (D199A mutant) and reacted with antibodies to ICP0 and PML. The infected cells were fixed at 3 and 6 h after infection and reacted with a mouse monoclonal antibody to PML and a rabbit polyclonal antibody to ICP0. The secondary antibodies were anti-mouse IgG conjugated to Texas red and rabbit IgG conjugated to fluorescein isothiocyanate. Left and middle columns, single-color images captured separately; right column, merged images. The yellow color visualized in the overlaid image represents colocalization of ICP0 and PML. The images were captured with a Zeiss confocal microscope and software provided by Ziess. The digitized images were not modified subsequent to capture.
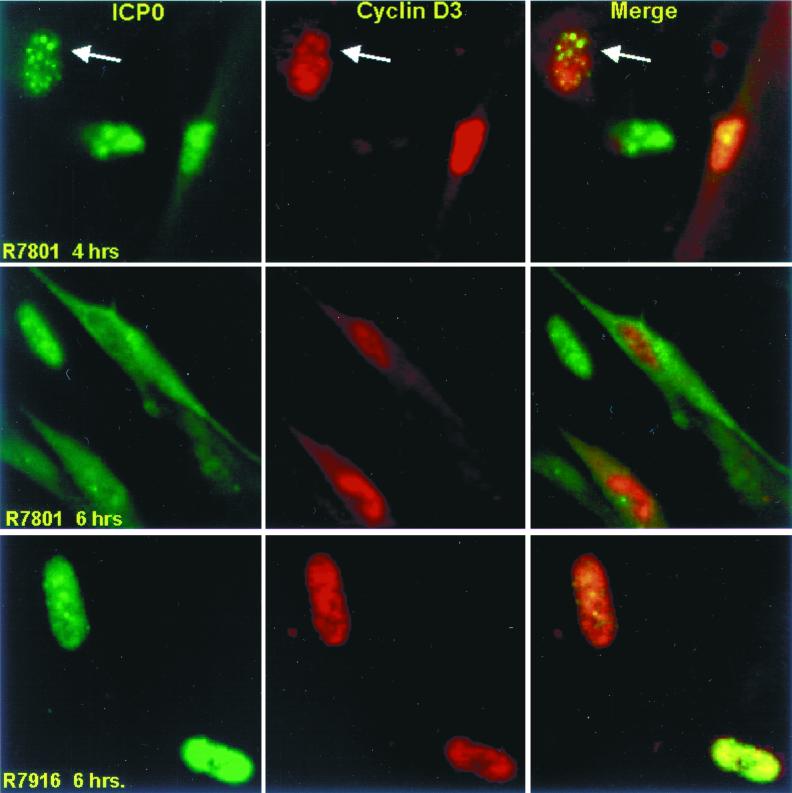
The levels of cyclin D3 in either mock-infected or wild-type virus-infected cells were too low to be detected by immunofluorescence, and we resorted to the use of viruses carrying the cyclin D3 gene to determine its localization. As shown in Fig. 11, wild-type ICP0 expressed by R7801 localized in nuclei at 4 h after infection but was present in both nuclei and cytoplasm at 6 h after infection. Cyclin D3 localized exclusively to the nucleus throughout this interval. In many cells, especially early in infection, cyclin D3 formed small dense structures that colocalized with wild-type ICP0 (Fig. 11, top row). In contrast to what was found for R7801 expressing wild-type ICP0, cyclin D3 expressed by mutant R7916 did not colocalize with the ICP0 carrying the D199A substitution. In a number of mutant virus-infected cells, both ICP0 and cyclin D3 formed dense structures but these generally did not coincide. Moreover the mutant ICP0 was not exported from the nucleus.
FIG. 11.
Digital images of HEL fibroblasts infected with wild-type (R7801) and D199A mutant (R7916) viruses expressing cyclin D3 and reacted with antibodies to ICP0 and cyclin D3. The infected cells were fixed 4 or 6 h after infection and reacted with antibodies and processed as described in the legend to Fig. 10. The digital images were not modified subsequent to capture. The arrows point to ICP0 and cyclin D3 colocalized in the nucleus of the cell shown in the upper left corner of each panel.
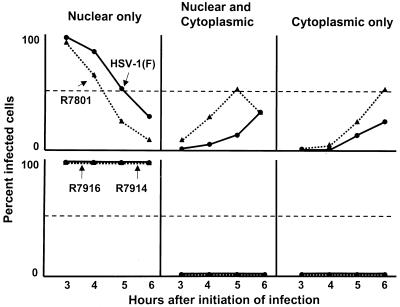
A striking observation that impacts on the potential function of cyclin D3 in HSV-1 infection emerged from comparisons of the distributions of ICP0 at various times after infection. In this series of experiments we determined the distribution of ICP0 in cells expressing cyclin D3 and in cells infected with wild-type virus. The results (Fig. 12) show that, in cells expressing cyclin D3, ICP0 was transported into the cytoplasm more rapidly than in cells infected with wild-type virus.
FIG. 12.
Cellular localization of ICP0 in HEL fibroblasts. (Top) Cells infected with wild-type HSV-1(F) or with R7801 expressing cyclin D3 and a wild-type α0 gene. (Bottom) Cells infected with R7914 (D199A ICP0 mutant) and R7916 carrying the cyclin D3 gene. A minimum of 150 infected cells were counted per data point.
We conclude from this series of experiments the following. ICP0s encoded by the wild-type and R7914 mutant viruses cannot be differentiated with respect to early localization in nuclei, colocalization with PML, and degradation of ND10 structures. They differ with respect to localization late in infection in that ICP0 carrying the D199A mutation is not translocated into the cytoplasm. In cells infected with recombinant virus R7801, which encodes and expresses cyclin D3, ICP0 was translocated more rapidly into the cytoplasm than in cells infected with wild-type virus.
DISCUSSION
The salient features of this report are three sets of observations. The first concerned D cyclins and their partners. The results of the studies that we present is that in quiescent HEL fibroblasts infected with wild-type virus both cyclins D3 and D1 were stabilized and that cdk4, their partner, actively phosphorylated the retinoblastoma protein, the natural substrate of the cyclin D-cdk4 complex. In contrast, in replicate HEL fibroblast cultures infected with R7914 carrying the D199A substitution in ICP0, the D cyclins were not stabilized and cdk4 was inactive. To further substantiate the activation of cdk4, we showed that, in wild-type virus-infected cells, p21, the regulator of cdk4 activity, rapidly disappeared whereas, in mock-infected cells and in cells infected with the D199A mutant, the levels of p21 remained high. Finally, the results show that, in R7914-infected cells, cyclins D1 and D3 were targeted for degradation. Thus, in cells infected with the D199A mutant and maintained in the presence of proteasomal inhibitor MG132, cyclins D3 and D1 were not degraded. Furthermore, the presence of cyclins D1 and D3 after infection was not due to de novo transcription of the corresponding cellular genes. There are two issues regarding these findings.
The first stems from the evidence that, although ICP0 stabilized cyclin D1, it did not interact with it in any of the assays. One hypothesis to explain the data is that the binding of cyclin D3 by ICP0 stabilizes it and causes the formation of an active complex with cdk4. This in turn results in the maintenance of cyclin D1, possibly through cycling of D-type cyclins in their complex with cdk4. This may also explain the small decrease in cyclin D3 observed during the first few hours after infection (17, 36). The data exclude the possibility that cdk4 is activated by HSV-1 independently of stabilization of cyclin D3 since cdk4 is not activated by the D199A mutant.
The second issue concerns the reason why D cyclins and their partner, cdk4, are activated. The sum total of available data indicate that the objective is not the transactivation of cellular S-phase genes since cdk2 is not activated and since the E2F family of proteins appear to be posttranslationally modified or transported to compartments in which they cannot function to induce S-phase protein synthesis (2, 6). The available data indicate that, early in infection, during the nuclear phase of ICP0, this protein colocalizes with cyclin D3 and by extension, with cdk4, in structures similar to those in which ICP0 colocalizes with PML. For reasons not yet understood, HSV-1 brings to these structures a cyclin D-cdk4 complex that targets a novel substrate. That HSV-1 may use cyclin-dependent kinases to benefit viral replication emerged recently from analyses of viral gene expression in cells transfected with and expressing a dominant-negative cdc2 homolog. Cells transfected with this construct failed to express a subset of γ2 genes, exemplified by US11, which depend on ICP22 and UL13 protein kinase for optimal expression. Activation of cdc2 cyclin-dependent kinase is also dependent on ICP22 and UL13 protein kinase (1).
The second key observation to emerge from these studies is that ICP0 carrying the D199A substitution failed to be transported to the cytoplasm. The cytoplasmic phase of ICP0 localization was first reported by Kawaguchi et al. (16) but received scant attention in light of the many and varied functions of ICP0 in nuclei. Recent studies in this laboratory (P. Lopez, C. Van Sant, and B. Roizman, submitted for publication) indicate that ICP0 is transported into the cytoplasm in cells expressing α genes only, that ICP0 is actively retained in the nucleus by post-α-gene expression, and that translocation of ICP0 requires the onset of viral DNA synthesis and is an active, reversible function. In the earlier study (38), it was shown that D199A mutant virus replicated as well as wild-type virus in dividing cells but yielded 10-fold-less virus in quiescent HEL fibroblasts than wild-type virus. As noted above, in cells infected with the D199A mutant, cyclin D3 did not colocalize with ICP0. We conclude from these observations that failure to bind cyclin D3, and by extension to activate cdk4, correlates with reduced replication in quiescent HEL fibroblasts and with failure to export ICP0 to the cytoplasm.
The third key observation to emerge from these studies is that, in HEL fibroblasts infected with a recombinant virus containing a wild-type ICP0 and the cyclin D3 gene, the translocation of ICP0 from the nucleus into the cytoplasm was accelerated compared to that in wild-type virus-infected cells. In HEL fibroblasts infected with R7916 carrying ICP0 with the D199A substitution and the identical cyclin D3 gene, ICP0 was retained in the nucleus notwithstanding the accumulation of cyclin D3. The unambiguous conclusion is that the mere presence of cyclin D3 did not lead to the translocation of mutant ICP0 to the cytoplasm. Cyclin D3 had to be bound and brought to the nuclear structures containing PML in order to ensure that the translocation of ICP0 took place.
In essence, ICP0 at the D199 locus encodes a function whose consequences are reflected in a series of events best described as seemingly disparate and wide ranging. The ultimate consequence of these events is optimal viral replication in quiescent cells. Not all of the intermediate steps designed to achieve this goal are known, but they initiate with the mobilization of cyclin D3 by ICP0 to specific sites in the nucleus and terminate with accumulation of ICP0 in the cytoplasm. The apparently key role of cyclin D3 in this process is strengthened by the observation that other herpesviruses either stabilize a D cyclin or encode their own functional homolog (4, 25). It is interesting and reflective of the density of information encoded into ICP0 that a single amino acid substitution abolished the entire chain of events. It is also of interest to note that the functions encoded by the D199 locus dovetail with, but do not overlap, the functions mapped by Everett and colleagues in the carboxyl-terminal domain of ICP0 (10). The ICP0 carrying the D199A mutation colocalizes with PML, and ND10 is effectively dispersed in cells infected with the mutant virus.
The sum total of the studies reported to date on ICP0 creates an image of a protein that binds and brings into a single structure early in infection several proteins whose function is to enable effective viral replication. The consequences of this act are a series of events that are crucial to this pathway and some that may well reflect unintended consequences. Many of the net effects on the stabilization of cyclin D3 remain to be sorted out.
ACKNOWLEDGMENTS
We than Alice P. W. Poon for a careful reading of the manuscript.
These studies were aided by grants from the National Cancer Institute (CA47451, CA71933, and CA78766) from the United States Public Health Service.
REFERENCES
- 1.Advani S J, Brandimarti R, Weichselbaum R R, Roizman B. The disappearance of cyclins A and B and the increase in activity of the G2/M-phase cellular kinase cdc2 in herpes simplex virus 1-infected cells require expression of the α22/US1.5 and UL13 viral genes. J Virol. 2000;74:8–15. [PMC free article] [PubMed] [Google Scholar]
- 2.Advani S J, Weichselbaum R R, Roizman B. The role of cdc2 in the expression of herpes simplex virus genes. Proc Natl Acad Sci USA. 2000;97:10996–11001. doi: 10.1073/pnas.200375297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 4.Chang Y, Moore P S, Talbot I, Boshoff C H, Zarlowska T, Godden-Kent D, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 5.Deshaies R J. SCF and Cullin/RING H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 6.Ehmann G L, McLean T I, Bachenheimer S L. Herpes simplex virus type 1 infection imposes a G(1)/S block in asynchronously growing cells and prevents G(1) entry in quiescent cells. Virology. 2000;267:335–349. doi: 10.1006/viro.1999.0147. [DOI] [PubMed] [Google Scholar]
- 7.Ejercito P, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 8.Everett R D, Barlow P, Milne A, Luisi B, Orr A, Hope G, Lyon D. A novel arrangement of zinc-binding residues and secondary structure in the C3CH4 motif of an alpha herpes virus protein family. J Mol Biol. 1993;234:1038–1047. doi: 10.1006/jmbi.1993.1657. [DOI] [PubMed] [Google Scholar]
- 9.Everett R D, Maul G G. HSV-1 IE protein Vmw 100 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett R D, Earnshaw W C, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelman I H, Silverstein S. Herpes simplex virus immediate-early promoters are responsive to virus and cell trans-acting factors. J Virol. 1987;61:2286–2296. doi: 10.1128/jvi.61.7.2286-2296.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung J U, Stager M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Van Sant C, Roizman B. Eukaryotic elongation factor 1δ is hyperphosphorylated by the protein kinase encoded by the UL13 gene of herpes simplex virus 1. J Virol. 1998;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaguchi Y, Matsumura T, Roizman B, Hirai K. Cellular elongation factor 1δ is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J Virol. 1999;73:4456–4460. doi: 10.1128/jvi.73.5.4456-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lees-Miller S P, long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Lee H, Yoon D W, Albrecht J C, Fleckstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maul G G, Everett R D. The nuclear localization of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 23.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 24.Murray A. Cyclin ubiquitination: the destructive end of mitosis. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- 25.Nicholas J, Cameron K R, Honess R W. Herpesvirus saimiri encodes homologues of G protein-coupled receptor and cyclins. Nature. 1992;355:362–365. doi: 10.1038/355362a0. [DOI] [PubMed] [Google Scholar]
- 26.Ogle W O, Ng T I, Carter K L, Roizman B. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology. 1997;235:406–413. doi: 10.1006/viro.1997.8710. [DOI] [PubMed] [Google Scholar]
- 27.O'Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early gene proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters J-M. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 29.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: α gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 30.Quinlan M P, Knipe D M. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roizman B. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc Natl Acad Sci USA. 1996;93:11307–11312. doi: 10.1073/pnas.93.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roizman B, Sears A E. The replication of herpes simplex viruses. In: Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 2231–2295. [Google Scholar]
- 33.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherr C J. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 35.Sherr C J. Cancer cell cycle. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 36.Song B, Liu J J, Yeh K C, Knipe D M. Herpes simplex virus infection blocks events in the G1 phase of the cell cycle. Virology. 2000;267:326–334. doi: 10.1006/viro.1999.0146. [DOI] [PubMed] [Google Scholar]
- 37.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 38.Van Sant C, Kawaguchi Y, Roizman B. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 40.Zhong-Kang Y, Gervais J L M, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]