Abstract
The recombinant bifunctional protein HELP-UnaG (HUG) is a fusion product of the Human Elastin-like Polypeptide (HELP) with the bilirubin-binding fluorescent protein UnaG. HUG is used for the fluorometric detection of bilirubin in serum and a variety of biological fluids and extracts. Here we describe a detailed method for the standardized production and purification of HUG from E. coli extracts on a laboratory scale. This method takes advantage of the HELP-specific thermoreactive behavior that enables the separation of HUG from complex E. coli extracts by repeated precipitation/re-dissolution steps at near physiological temperature.
-
•
The method is based on the inverse thermal transition process.
-
•
The “green” method is affordable for basic laboratories and can be easily transferred to new users.
Keywords: HUG, Human elastin-like fusion proteins, Inverse thermal transition, Protein purification method
Method name: Lab-scale production of the recombinant fusion protein HUG
Graphical abstract
Specifications table
| Subject area: | Biochemistry, Genetics and Molecular Biology |
| More specific subject area: | Macromolecular Chemistry/Biopolymer |
| Name of your method: | Lab-scale production of the recombinant fusion protein HUG |
| Name and reference of original method: | N/A |
| Resource availability: | Reagents and Equipment are listed in the Materials and Reagents section |
Background
The genetically engineered protein HUG is the product of the fusion of the Human Elastin-like Polypeptide (HELP) [1], with the eel protein UnaG, which was identified and isolated from the muscle of the Japanese eel [2]. UnaG is a free fatty acid binding protein that binds unconjugated bilirubin (BR) highly specifically and selectively and emits strong fluorescence [[2], [3], [4]]. The HELP sequence originates from the most repetitive region of human tropoelastin. HELP gene consists of a part of exon 23, which encodes the cross-linking domain, and exon 24, which encodes the hydrophobic domain [5]. The recombinant HELP protein was fused to the UnaG eel protein to obtain a chimeric, bifunctional polypeptide, which was named HUG. The synthetic gene was obtained by in-frame insertion of the coding UnaG sequence at the C-terminus of the HELP domain. The fusion sequence was then expressed in Escherichia coli (E. coli). The chimeric product binds bilirubin with high affinity, resulting in fluorescence emission [6,7]. HUG is used in a fluorometric assay to determine bilirubin concentration [8] in various experimental models of translational medicine, such as in isolated, perfused rat liver [9], in cancer cell line cultures [10], and in human plasma [8]. HUG is not commercially available, so its production depends on the standardized laboratory-scale protocol described here. The purification method takes advantage of an outstanding property of elastin-like proteins, namely their inverse thermal transition behavior. These proteins dissolved in an aqueous environment aggregate into particles at temperatures close to 40 °C and can be collected after centrifugation. Only these components can be dissolved from the pellet in cold water, while most of the other components remain precipitated. This process is called the inverse transition cycling (ITC) [11]. The resulting product is then structurally and functionally characterized following an established quality control procedure [8].
Method details
Materials and reagents
All reagents were purchased from Merck, unless otherwise specified.
-
-
Expression vector pHUG, containing the fusion HUG gene [6]
-
-
C3037 E. coli strain (New England Biolabs)
-
-
Agar
-
-
Ampicillin
-
-
Chloramphenicol
-
-
Isopropyl-β-D-1-thiogalactopyranoside (IPTG)
-
-
Tryptone
-
-
Yeast extract
-
-
Sodium chloride (NaCl)
-
-
Potassium dihydrogen phosphate (KH2PO4)
-
-
Dipotassium phosphate (K2HPO4)
-
-
Glycerol
-
-
Trizma base
-
-
Ethylenediaminetetraacetic acid (EDTA)
-
-
Triton X-100
-
-
Hydrochloric acid (HCl)
-
-
β-mercaptoethanol (β-ME)
-
-
Ethanol
-
-
Phenylmethylsulphonyl fluoride (PMSF)
-
-
Sodium deoxy-taurocholate
-
-
Tween-20
-
-
Bilirubin (BR)
-
-
Bovine serum albumin fraction V (BSA)
-
-
HELP (lab production according to Bandiera 2020)
Equipment
-
-
Multi-purpose water purification system (Crystal EX, Androna®)
-
-
Magnetic stirrer (Icamag® Rec-G)
-
-
Shaker 709, Asal
-
-
Thermostat Cabinet
-
-
Homogenizer OV5, VELP Scientifica
-
-
APV model 1000 homogenizer, SPX Brand
-
-
Centrifuge Avanti J-26 XP Beckman coulter
-
-
Centrifuge Eppendorf 5804 R
-
-
RadWAG WTB-2000 Balance
-
-
Analytical Balance ABT 120–4NM (KERN 770)
-
-
Centrifuge Rotor JA-14
-
-
Centrifuge Rotor 25.5 Eppendorf
-
-
IKA Vortex Shaker
-
-
Cary 4E, Varian
-
-
SynergyH1, Biotek platereader
-
-
Falcon tubes (15 mL)
-
-
Glass flask for bacterial culture (250 mL)
-
-
Glass flask for bacterial culture (1 L)
-
-
Nalgene Centrifuge Bottles with Caps‚ PPCO (250 mL)
-
-
Quartz couvette
-
-
Black 96-well plates (Nunc®)
Recipes
Luria Bertani (LB) broth and agar
-
•
10 g tryptone
-
•
5 g yeast extract
-
•
10 g NaCl
-
•
(only for LB agar) 20 g of agar
Bring volume to 1.0 L with MilliQ water.
Sterilized at 121 °C.
Store at room temperature until use.
Terrific broth (TB)
-
•
24 g tryptone
-
•
48 g yeast extract
Bring the volume up to 2.0 L with MilliQ water.
Adjust the pH value to 7.2 and then bring the final volume to 2.1 L with MilliQ water.
Sterilize at 121 °C.
Store at room temperature until use.
If only half of the volume is used, store the bottle at 4 °C for a maximum of 4 days after opening.
Tryptone and yeast extract are both sticky and take too much time long to dissolve,; start dissolving with smaller less quantities amounts and add them gradually.
Phosphate-glycerol buffer (PGB)
-
•
6.93 g KH2PO4
-
•
37.6 g K2HPO4
-
•
12.5 mL glycerol
Bring volume to 300 mL with MilliQ water.
Sterilized at 121 °C.
Terrific broth medium (TBm)
Mix 2.1 L of TB with 0.3 L of PGB immediately before use.
Extraction buffer (50 mM Tris/HCl pH 8, 250 mM NaCl, 0.1 mM EDTA, 0.1 %v/v Triton X-100)
-
•
50 mL 1 M Tris/HCl pH 8
-
•
50 mL 5 M NaCl
-
•
200 µL 0.5 M EDTA (to achieve getting maximum solubility, the pH value must be 8 and EDTA must be in salt form and not as an acid)
-
•
1 mL Triton X-100
Bring volume to 1 L with MilliQ water.
Store at 4 °C until use.
Solubilization buffer (0.1 mM sodium deoxy-taurocholate and 0.05 % v/v tween-20)
-
•
50 mg sodium deoxy-taurocholate
-
•
50 µL Tween-20
Bring volume to 100 mL with MilliQ water.
Store at 4 °C until use.
50 mg/ml Ampicillin
-
•
50 mg ampicillin powder
-
•
1 mL MilliQ water
Store at −20 °C until use.
35 mg/ml Chloramphenicol
-
•
35 mg chloramphenicol powder
-
•
1 mL absolute ethanol
Store at −20 °C until use.
0.1 M IPTG
-
•
23.8 mg IPTG powder
-
•
1 mL MilliQ water
Store at −20 °C until use.
Phenylmethylsulfonyl fluoride (PMSF)
-
•
2 mL ethanol
-
•
PMSF powder
Add PMSF until a the solute appears as a solid precipitate.
Software
GraphPad Prism 10.1.0 (264) (GraphPad Softwares, Boston, MA, USA).
Graphical abstract and Schemes were created with BioRender.com.
STEP 1
Selection of the expressing clone
1. The E. coli strain C3037 was transformed with the HUG plasmid (pHUG) according to the manufacturer's instructions.
Note: This step should be performed every 3 months, as the clone is not stable for long in glycerol.
2. The transformed bacteria were plated and allowed to grow on an LB agar plate containing 50 µg/mL ampicillin and 70 µg/mL chloramphenicol overnight at 37 °C.
3. Several clones (individually labeled) were expanded in 3 mL LB broth containing 50 µg/mL ampicillin and 70 µg/mL chloramphenicol (LB complete) and incubated overnight at 37 °C.
4. 1 mL of the overnight culture was then diluted in 10 mL LB complete and allowed to grow with shaking at 37 °C until O.D.600 = 0.8 - 1.0 was reached.
5. A sample (0.4 mL) of bacterial culture (pre-induction) was taken and centrifuged at 10,000 rpm (or 15,344 g) for 5 min. The pellet was resuspended in Laemmli loading buffer (1X) for further electrophoretic analysis.
6. Induction of the bacterial culture was made by adding 0.01 mL of 0.1 M IPTG (0.09 mM) allowing it to grow under shaking at 37 °C for 5 h.
7. A sample (0.2 mL) of the induced bacterial culture (post-induction) was taken and centrifuged at 10,000 rpm (or 15,344 g) for 5 min. The pellet was resuspended in Laemmli loading buffer (1X) for further electrophoretic analysis.
Note: The volumes of the samples before and after induction are different because they take into account bacterial growth during the induction period.
Only cultures expressing the HUG protein (see Fig. 1) were utilized for the preparative scale production.
Fig. 1.
Protein expression profile of the induced bacterial culture. Aliquots of the bacterial cultures obtained from 4 clones were taken before (0.4 mL) and after (0.2 mL) induction with IPTG. Lane 1: molecular weight markers; lane 2: the HELP elastin-like protein purified by ITC; lanes 3–5–7–9: bacterial extract before induction; lanes 4–6–8–10: bacterial extract after IPTG induction.
STEP 2
Preparation and storage of the bacterial biomass containing HUG
1. Preparation of a starter culture from the selected clone (Scheme 1). A single clone culture (0.4 mL) was added to 300 mL complete LB broth. The starter culture was incubated at 37 °C overnight, before use.
Note: Use fresh transformed bacteria or glycerol-preserved bacteria that have been stored at - 20 °C for no longer than 3 months.
2. Preparation of the preparative expression culture. Several aliquots of the starter culture (40 mL) were added to 6 flasks each containing 400 mL TBm (see recipe). The bacterial cultures were incubated at 37 °C with shaking (Shaker 709, Asal), and bacterial growth was monitored by measuring the optical density (O.D.) at λ = 600 nm (Cary 4E, Varian). The optimal O.D. range to induce HUG expression in the cultured clone is 0.8 – 1.0, which was reached after about two hours of incubation.
3. Induction of the expanded culture. When the expanded culture reached O.D.600 = 0.8 - 1.0, it was induced by adding 0.4 mL of 0.1 M IPTG (0.09 mM) to each flask to obtain the E. coli culture expressing the HUG polypeptide.
4. Collection of the E. coli biomass. Five hours after induction, the 2.4 L of bacterial culture were divided into twelve 250 mL centrifuge bottles and then centrifuged at 8000 rpm (9820 g) for 20 min at 8 °C (Beckman rotor JA-14).
5. Storage of the collected biomass. The pellets obtained from previous point 4 (Fig. 2) were resuspended in the extraction buffer, pooled to a final volume of 120 mL and stored at −20 °C.
Scheme 1.
Sequence for the recovery of pelleted E. coli cells expressing HUG.
Fig. 2.
Bacterial pellets as appeared after the centrifugation.
The procedure described in STEP 2 requires 2 days, i.e. preparation of the starter culture on day 1 and overnight growth followed by preparative expression on day 2 that requires about 9–10 h.
STEP 3
Preparation of crude cell extracts containing HUG
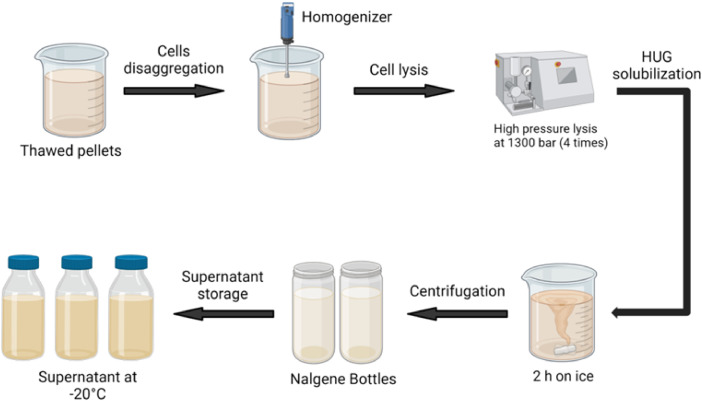
When preparing the crude cell extract containing HUG, it is particularly important to observe the temperature specifications in the individual steps (Scheme 2).
1. Suspension of the biomass. Twelve frozen (- 20 °C) pellets from 2.4 L of expression culture were thawed and added with 400 mL extraction buffer containing 0.5 mL PMSF.
2. Pellet disaggregation. The resuspended pellets were treated by the OV5 homogenizer (Velp Scientifica), operated for 3 cycles, each for 1 min with 1 min interval (Fig. 3A and B).
Note: The suspension should be kept on ice during the entire procedure.
3. Bacterial cell lysis. The suspension was subjected to high-pressure bacterial cell disruption (APV model 1000 homogenizer, SPX Brand). Using additional extraction buffer (about 300 mL), the solution was passed through this device four times at a pressure of approximately 1300 bar. The final volume of the sample was 700 mL.
Note: The homogenizer should be operated in a cold room (4 °C) to avoid protein precipitation during high-pressure mediated bacterial lysis.
4. HUG solubilization in the bacterial lysate. The final bacterial lysate was supplemented with 0.5 mL of β-ME and stirred on ice for 2 h.
Note: This is a critical step as incomplete solubilization of the HUG protein will affect affects the final yield.
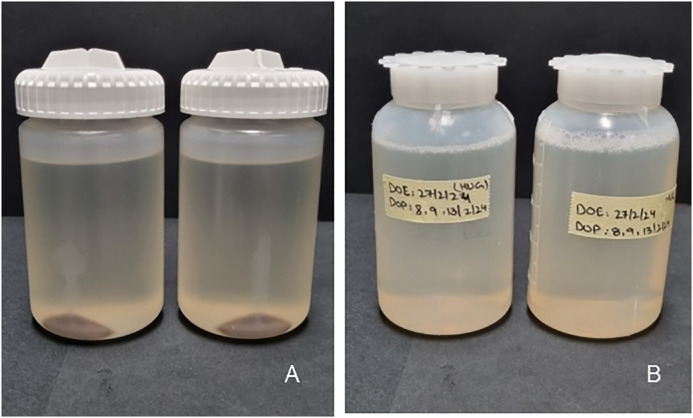
5. Clarification of the bacterial lysate. The bacterial lysate (700 mL) was divided into four Nalgene bottles and centrifuged (Beckman rotor JA-14) at 10,000 rpm (or 15,344 g) at 8 °C for 30 min (Fig. 4A).
6. Storage of the supernatant. The collected supernatants were pooled and divided into 3 bottles (approx. 240 mL) and stored at −20 °C (Fig. 4B).
Note: The supernatants can be stored at −20 °C for at least 1 month until the purification procedure.
The entire preparation of crude extracts takes about 5 h.
Scheme 2.
Overview of crude extracts preparation.
Fig. 3.
(A). Homogenization of bacterial pellets in extraction buffer by the OV5 homogenizer. (B) Appearance of mixture after disruption of bacterial cells.
Fig. 4.
Example of bacterial lysate clarification. (A) Separation of HUG from bacterial debris after centrifugation. (B) Collected supernatant ready to be stored at −20 °C.
STEP 4
Purification of HUG
The purification procedure exploits the thermal inversion properties of the HUG, therefore it is important to strictly follow the indications relating to the temperature specifications in the various steps.
All the following purification steps refer to one bottle containing 240 mL of extract (Scheme 3).
1. Temperature- and NaCl-dependent precipitation of HUG. The clarified cell extract was transferred to a beaker and about 160 mL of 5 M NaCl were added (Fig. 5A) under gentle stirring until turbidity remained constant (Fig. 5B).
The suspension was divided into 2 Nalgene centrifuge tubes and then the precipitation step was completed at 42 °C in a water bath for 15 min.
Note: Excessive addition of NaCl solution should be avoided as the aggregation process also depends on the protein concentration.
2. HUG collection. The heated Nalgene tubes were centrifuged at 10,000 rpm (or 15,344 g) at 30 °C for 30 min (Beckman rotor JA-14). After centrifugation, the supernatant was discarded and the tubes were placed upside down on paper towel for no >5 min (Fig. 6).
Note: It is important to turn the Nalgene upside down to remove all the supernatant, which contains a high concentration of salt.
The pellet must not desiccate at this stage in order to avoid difficulties with subsequent dissolution.
3. HUG resuspension. Each pellet was dissolved stepwise in about 25 mL of ice-cold solubilizing buffer; then all suspended solutions were pooled in a covered beaker and stirred on ice for 2 h.
Note: This step is critical, as incomplete dissolution of the HUG will result in protein loss.
Optimal dissolution of the pellet was achieved with sodium deoxy-taurocolate and Tween 20 in aqueous solution. This solubilizing buffer removes the lipid that may be associated with UnaG, a known fatty acid binding protein.
If the pellet is difficult to dissolve, additional 60 mL of solubilization buffer can be added to the pooled suspension.
4. Clarification of the HUG suspension. The cold suspension was placed in centrifuge tubes and centrifuged at 10,000 rpm (or 15,344 g) at 8 °C for 10 min (Rotor 25.5 Eppendorf) (Fig. 7). The supernatant was recovered and the pellets were discarded.
5. HUG coacervation and precipitation. The supernatant was heated to 42 °C until a slight turbidity appeared (Fig. 8A), and then about 40 mL of 5 M NaCl were added under gentle stirring until turbidity remained constant (Fig. 8B). Then the solution was placed in a Nalgene bottle and incubated at 42 °C for 15 min to obtain the complete aggregation of HUG (Fig. 8C).
Note: If dissolution of the HUG pellet (see point 3 above) required additional 60 mL of solubilization buffer, coacervation can be achieved by increasing the volume of 5 M NaCl accordingly (up to 90 mL).
6. HUG collection. The HUG pellet was obtained by centrifugation at 8500 rpm (or 11,086 g) at 30 °C for 20 min (Beckman rotor JA-14). The supernatant was discarded and the bottles were placed upside down on a paper towel for a maximum of >5 min to remove the excess liquid (Fig. 9). Each HUG pellet was then resuspended with about 60 mL of ice-cold milliQ water by adding low volumes stepwise.
Note: The resulting HUG pellet may appear weak as it is dispersed on the bottle wall and not concentrated on the bottom. Therefore, make sure that you carefully recover all the material.
7. Freeze-drying of HUG. The pellet resuspended with milliQ water was further stirred on ice for half an hour and then frozen at - 80 °C overnight and lyophilized. The dry protein (Fig. 10) was stored at −20 °C.
Scheme 3.
Sequential steps for purification of HUG.
Fig. 5.
HUG precipitation starting from a 2 × 240 mL cell extract. (A) Supernatant from extraction before precipitation. (B) Supernatant after adding 2 × 160 mL 5 M NaCl and precipitate appearance.
Fig. 6.
Pellets obtained after the first coacervation step.
Fig. 7.

Insoluble materials removed by centrifugation.
Fig. 8.
Supernatant at 42 °C. (A) Before the addition of NaCl; (B) After the addition of NaCl; (C) After 15 min at 42 °C in the presence of NaCl, leading to complete coacervation.
Fig. 9.

Pellet after final centrifugation.
Fig. 10.

Freeze-dried HUG protein.
The process of purification of HUG takes almost 6 h and the subsequent freeze-drying takes at least 3 days. The HUG yield under laboratory conditions was between 125 and 270 mg protein per liter of bacterial culture.
Method validation
In order to evaluate the degree of purification, integrity, and functionality of the purified HUG, its characterization was performed using three methods, as described below (Scheme 4).
(1) Determination of HUG amount in the freeze-dried material
Scheme 4.
Outline of quality controls of purified HUG.
The content of HUG in the freeze-dried material was determined by direct UV–VIS spectroscopy. The presence of the aromatic amino acids tryptophan, tyrosine and phenylalanine in the UnaG domain can be used to evaluate the absorbance of HUG water solution at λ = 280 nm. The exact concentration of HUG was calculated using the experimental extinction coefficient of 18,747 M-1 cm-1, obtained as shown in Fig. 11. Typical absorbance of 2 mg/mL HUG working solution was 0.529 (± 0.040) at λ = 280 nm.
Fig. 11.
UV–vis spectra of serially diluted HUG solutions in water. The absorbance values at λ =280 nm were plotted against the HUG concentration. The angular coefficient of the curve represents the experimental extinction coefficient of HUG in water (ε = 18,747 ± 1262; R2 = 0.9728).
Note: The actual HUG concentration might be lower than the gravimetrically determined value, due to impurities. The actual HUG concentration used to prepare the working solutions for the nanoscale bilirubin assay (Sist et al., 2023) is determined by UV spectroscopy and using ε = 18,747 M-1 cm-1.
(2) Determination of purified HUG electrophoretic mobility
The HUG sample was analyzed on a 10 % SDS-PAGE, in order to identify its major band and the possible presence of degradation products, according to the following protocol:
-
-
Prepare 10 % polyacrylamide-SDS running gel and a 3.75 % stacking gel.
-
-
Prepare a 2 mg/mL HUG solution in water.
-
-
Dilute 8 µL of this HUG solution with 8 µL of 2X Loading Buffer.
-
-
Load 10 µL HUG sample.
-
-
Load 4 µL molecular weight marker.
-
-
Apply the following electrophoretic conditions: 70 V along the stacking gel, 140 V along the running gel.
-
-
Stain the gel with Coomassie blue dye.
-
-
Destain the gel using an ethanol/acetic acid solution.
Molecular weight markers (lane 1) were loaded to determine the molecular weight of the samples. We used HELP (lane 2) as a appropriate reference for HUG mobility and the band observed in lanes 3–4–5 loaded with purified HUG was approximately 66 kDa (Fig. 12). Since the electrophoretic mobility of HELP analyzed by SDS-PAGE (calculated M.W. 44,886 Da) corresponds to an apparent molecular weight of approximately 50 kDa, the estimated value of 66 kDa is consistent with the full-length HUG molecule (calculated M.W. 60,406 Da).
(3) Determination of HUG bilirubin-binding activity
Fig. 12.

SDS-PAGE analysis of three different purified HUG preparations.
The bilirubin-binding activity of the purified HUG was tested by recording its fluorescence emission in the presence of an excess bilirubin (160 nM) in PBS containing 0.4 g/L BSA. The assay specifically evaluates the ability of each batch of HUG to bind bilirubin and emit a fluorescent signal that is directly proportional to the amount of HUG. The specific fluorescence (A.U./µg HUG) enables to compare the purity of different production batches.
The test was performed in black 96-well plates (Nunc®) as follows:
-
-
Prepare a solution of 160 nM BR in PBS-BSA 0.4 g/L, pH 7.4
-
-
Add 10 µL of HUG solutions with increasing concentrations (0.01 – 0.8 mg/mL) directly into the wells in 4 replicates.
-
-
Add fixed volumes (0.2 mL) of 160 nM BR solution to each well.
-
-
Incubate the 96-well plate at 25 °C for 2 h in the dark.
-
-
Measure the fluorescence intensity (λex = 485 nm, λem = 528 nm; gain 100, reading height 2.50 mm; T = 25 °C) in a benchtop multiplate reader (Synergy H1; BioTek, Winooski, VT, USA).
-
-
Record the mean fluorescence value of the blank (160 nM BR solution without HUG) and subtract this value from the emitted HUG·BR fluorescence.
-
-
Plot the fluorescence data as a function of HUG (µg) and fit the data by linear regression analysis and obtain the angular coefficient, representing is the specific fluorescence emission of HUG (Fig. 13).
Fig. 13.
Fluorescence values as a function of HUG (µg) in the presence of bilirubin excess.
Note: Prepare the bilirubin standard solutions in dim light and cover them with aluminium foil until analysis. Use freshly prepared solutions in order to avoid the slow degradation of bilirubin.
The intrinsic fluorescence of the solvent (160 nM BR in PBS-BSA 0.4 g/L, pH 7.4) was approximately 1000 AU and was always subtracted from the HUG·BR signal that ranged from 2600 to 94,000 AU. The fluorescence intensity per µg of HUG is obtained by the regression coefficient and the value is 11,663 ± 196 (A.U./µg).
Final considerations
The proposed protocol is simple, relatively fast and selective. However, there are two important issues that need to be emphasised. First, ageing of the clone of the transformed bacteria leads to a reduction in the final yield. This can be easily circumvented by using freshly transformed bacteria that have not been stored for more than three months. Secondly, the appropriate amount of NaCl to be added to start the HUG precipitation step may vary depending on the concentration of HUG in the solution. Since this cannot be predicted with certainty, the operator must monitor the increase in turbidity of the solution and stop adding salt when the turbidity is stable. At this point, HUG aggregation is completed by raising the temperature to 42 °C. This skill is achieved after repeating this protocol 2–4 times.
The average yield of 500 mg HUG can be obtained from 2.4 L of bacterial culture. The procedure requires the use of one person for 5 days of experimental work when the bacterial clone is already available. Then another 3 days are required for freeze-drying. Considering that the standard protocol for nanoscale fluorometric analysis of bilirubin requires 1 mg of HUG for a 96-well plate [8], it can be calculated that up to 500 96-well plates can be prepared with the same HUG production batch. Assuming that each sample (standard or biological sample) is analysed in quadruplicate, an average of 12,000 samples can be analysed with 500 mg of pure HUG polymer.
Limitations
None.
CRediT authorship contribution statement
Paola Sist: Resources, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. Suman Saeed: Resources, Investigation, Writing – original draft, Visualization. Federica Tramer: Investigation, Methodology, Validation, Writing – review & editing. Antonella Bandiera: Conceptualization, Methodology, Writing – review & editing. Sabina Passamonti: Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was funded by the European Union via complementary programs, such as: 1) NextGenerationEU - Italian National Recovery and Resilience Plan (PNRR), which funded project iNEST - Interconnected Nord-Est Innovation Ecosystem (iNEST ECS0000004-CUP J43C22000320006) and a PhD fellowship (PNRR, D.M. 352/2022, code 38-033-33-DOT1333098-2758; CUP J92B22001560007), the latter co-funded by Centro Ricerche Scientifiche Dott. Dino Paladin, Trieste, Italy (recipient Ms Suman Saeed). 2) European Regional Development Fund, Interreg VI-A Italy-Slovenia 2021–2027, which funded project Agrotur+, code ITA-SI0100048.
Footnotes
Related research article: N/A.
For a published article: None.
Data availability
Data will be made available on request.
References
- 1.Bandiera A., Sist P., Urbani R. Comparison of thermal behavior of two recombinantly expressed human elastin-like polypeptides for cell culture applications. Biomacromolecules. 2010;11(12):3256–3265. doi: 10.1021/bm100644m. [DOI] [PubMed] [Google Scholar]
- 2.Kumagai A., Ando R., Miyatake H., Greimel P., Kobayashi T., Hirabayashi Y., Shimogori T., Miyawaki A. A bilirubin-inducible fluorescent protein from eel muscle. Cell. 2013;153(7):1602–1611. doi: 10.1016/j.cell.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 3.Shitashima Y., Shimozawa T., Asahi T., Miyawaki A. A dual-ligand-modulable fluorescent protein based on UnaG and calmodulin. Biochem. Biophys. Res. Commun. 2018;496:872e879. doi: 10.1016/j.bbrc.2018.01.134. [DOI] [PubMed] [Google Scholar]
- 4.Eczacioglu N., Ulusu Y., Gokce İ., Lakey J.H. Investigation of mutations (L41F, F17M, N57E, Y99F_Y134W) effects on the TolAIII-UnaG fluorescence protein’s unconjugated bilirubin (UC-BR) binding ability and thermal stability properties. Prep. Biochem. Biotechnol. 2021;52(4):365–374. doi: 10.1080/10826068.2021.1952597. [DOI] [PubMed] [Google Scholar]
- 5.Bandiera A., Taglienti A., Micali F., Pani B., Tamaro M., Crescenzi V., Manzini G. Expression and characterization of human-elastin-repeat-based temperature-responsive protein polymers for biotechnological purposes. Biotechnol. Appl. Biochem. 2005;42(3):247–256. doi: 10.1042/BA20050114. [DOI] [PubMed] [Google Scholar]
- 6.Bandiera A., Corich L., Tommasi S., De Bortoli M., Pelizzo P., Stebel M., Paladin D., Passamonti S. Human elastin-like polypeptides as a versatile platform for exploitation of ultrasensitive bilirubin detection by UnaG. Biotechnol. Bioeng. 2020;117(2):354–361. doi: 10.1002/bit.27217. [DOI] [PubMed] [Google Scholar]
- 7.Sist P., Bandiera A., Urbani R., Passamonti S. Macromolecular and solution properties of the recombinant fusion protein HUG. Biomacromolecules. 2022;23(8):3336–3348. doi: 10.1021/acs.biomac.2c00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sist P., Tramer F., Bandiera A., Urbani R., Redenšek Trampuž S., Dolžan V., Passamonti S. Nanoscale bilirubin analysis in translational research and precision medicine by the recombinant protein HUG. Int J Mol Sci. 2023;24(22):16289. doi: 10.3390/ijms242216289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelizzo P., Stebel M., Medic N., Sist P., Vanzo A., Anesi A., Vrhovsek U., Tramer F., Passamonti S. Cyanidin 3-glucoside targets a hepatic bilirubin transporter in rats. Biomed. Pharmacother. 2023;157 doi: 10.1016/j.biopha.2022.114044. [DOI] [PubMed] [Google Scholar]
- 10.Tonelotto V., Costa-Garcia M., O’Reilly E., Smith K.F., Slater K., Dillon E.T., Pendino M., Higgins C., Sist P., Bosch R., Passamonti S., Piulats J.M., Villanueva A., Tramer F., Vanella L., Carey M., Kennedy B.N. 1,4-dihydroxy quininib activates ferroptosis pathways in metastatic uveal melanoma and reveals a novel prognostic biomarker signature. Cell Death Discov. 2024;10(1):70. doi: 10.1038/s41420-023-01773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colomina-Alfaro L., Marchesan S., Stamboulis A., Bandiera A. Smart tools for antimicrobial peptides expression and application: the elastic perspective. Biotechnol. Bioeng. 2023;120(2):323–332. doi: 10.1002/bit.28283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.