Abstract
Previous biochemical data identified a host cell fraction, designated RAF-2, which stimulated influenza virus RNA synthesis. A 48-kDa polypeptide (RAF-2p48), a cellular splicing factor belonging to the DEAD-box family of RNA-dependent ATPases previously designated BAT1 (also UAP56), has now been identified as essential for RAF-2 stimulatory activity. Additionally, RAF-2p48 was independently identified as an influenza virus nucleoprotein (NP)-interacting protein, NPI-5, in a yeast two-hybrid screen of a mammalian cDNA library. In vitro, RAF-2p48 interacted with free NP but not with NP bound to RNA, and the RAF-2p48–NP complex was dissociated following addition of free RNA. Furthermore, RAF-2p48 facilitated formation of the NP-RNA complexes that likely serve as templates for the viral RNA polymerase. RAF-2p48 was shown, in both in vitro binding assays and the yeast two-hybrid system, to bind to the amino-terminal region of NP, a domain essential for RNA binding. Together, these observations suggest that RAF-2p48 facilitates NP-RNA interaction, thus leading to enhanced influenza virus RNA synthesis.
The genome of influenza A virus consists of eight single-stranded RNA segments of negative polarity. These viral RNA (vRNA) segments exist as ribonucleoprotein (vRNP) complexes with nucleocapsid proteins (NP) and viral RNA polymerases as components. Each RNA segment contains highly conserved 3′- and 5′-terminal untranslated regions which function as regulatory signals for transcription and replication of the genome. The partially hybridized terminal regions have been referred to as panhandle (9), fork, hook, and corkscrew (14, 15, 25, 44) forms. In vRNP complexes prepared from purified virions, viral RNA polymerase is found bound to the panhandle region (33), and NP is bound to vRNA such that each NP monomer occupies approximately 20 nucleotides (6, 56).
Studies using the vRNP isolated from virions have revealed that viral RNA polymerase and NP are essential for transcription (3, 21, 24). The viral RNA polymerase consists of PB2, PB1, and PA subunits and is capable of initiating primer-dependent RNA synthesis (20, 27). However, for synthesis of full-length RNA, NP is required (21, 22). Transcription is initiated by recognition by PB2 of the cap structure of nuclear pre-mRNA. PB2 truncates the capped RNA at 10 to 13 bases downstream from the 5′ end (3, 42). After the capped oligonucleotide is cleaved, it serves as a primer for viral mRNA synthesis catalyzed by PB1 (17). Elongation of the RNA chain proceeds until the polymerase reaches a polyadenylation signal, consisting of five to seven U residues located near the 5′-terminal region of the vRNA (29). The viral RNA polymerase polyadenylates the nascent RNA chain, possibly by a slippage mechanism at the U stretch (43). Replication of vRNA is a primer-independent two-step reaction: first, cRNAs are synthesized from vRNA templates; and second, the progeny vRNAs are amplified from cRNA templates. Genetic analyses suggest that PA participates in the replication process. However, vRNP complexes isolated from virions are incapable of catalyzing replication reactions.
It has been reported that in vitro RNA synthesis systems utilizing nuclei or nuclear extracts prepared from infected cells are capable of supporting transcription for viral mRNA synthesis and viral genome replication wherein both cRNA and progeny vRNA synthesis occurs (2, 8, 47, 52). However, vRNP complexes prepared from nuclear extracts of infected cells through centrifugation, or those prepared from solubilized virions, cannot catalyze replication and catalyze transcription with efficiency lower than that obtained using crude nuclear extracts. Addition of either the supernatant fraction, separated from the vRNP complexes, or free NP restores the replication activity of vRNP. Also, addition of supernatant fractions depleted of free NP by treatment with anti-NP antibody does not restore the activity. Therefore, free NP and/or a factor(s) associated with NP is presumed to be required for cRNA and progeny vRNA synthesis.
Since the systems described above are dependent on endogenous vRNA templates, precise replication and transcription mechanisms including those for initiation reactions are difficult to assess. To address this issue, we constructed a novel in vitro vRNA synthesis system using nuclear extracts prepared from infected cells and an exogenous model influenza virus genome RNA (31, 48). The artificial viral genome, consisting of the 5′- and 3′-terminal regions of the eighth segment, contains the cis-acting signals essential for transcription and replication. The system can also be reconstituted using two complementing fractions: vRNP complexes from purified virions, and nuclear extracts from uninfected cells. This suggests that viral RNA polymerase and NP are essential and other factors present in host cells are required for efficient RNA synthesis. In fact, we have previously identified host factors which influence influenza virus RNA synthesis using a biochemical complementation assay in which fractions from uninfected HeLa cell nuclear extracts are added to vRNPs (31, 48). Among the recovered fractions, we found significant stimulatory activity for the RNA polymerase in a fraction that is not adsorbed to a phosphocellulose column. This fraction stimulates RNA synthesis from both vRNA and cRNA templates. The fraction containing the stimulatory activity was further separated into two distinct fractions, designated RNA polymerase-activating factor 1 (RAF-1) and RAF-2.
Here we further purified RAF-2 and showed that highly purified RAF-2 contains 48- and 36-kDa polypeptides, designated RAF-2p48 and RAF-2p36, respectively. RAF-2p48 was found to be identical to BAT1/UAP56, a cellular splicing factor belonging to the DEAD-box family of RNA-dependent ATPases. The 48-kDa RAF-2 component, BAT1/UAP56, was separately identified as an NP-interacting protein, designated NPI-5, in a yeast two-hybrid screen of a mammalian cDNA library (38). Biochemical and yeast two-hybrid analyses reveal that RAF-2p48 interacts with the amino-terminal region of NP, and biochemical analyses indicate that RAF-2p48 facilitates formation of NP-RNA complexes. These data suggest that RAF-2p48 is a host cell factor that regulates influenza virus RNA synthesis.
MATERIALS AND METHODS
In vitro vRNA synthesis system.
vRNP containing 50 ng of NP/μl was prepared from purified influenza A/PR/8/34 virus as described previously (56). Micrococcal nuclease-treated vRNP (mnRNP) was prepared by incubation of vRNP at 30°C for 2 h with 1 U of micrococcal nuclease (Roche Molecular Biochemicals)/μl in the presence of 1 mM CaCl2 (45). The nuclease was inactivated by adding a final concentration of 3 mM EGTA. The 53-nucleotide-long model vRNA (53-mer Vwt; 5′-AGUAGAAACAAGGGUGUUUUUUCAUAUCAUUUAAACUUCACCCUGCUUUUGCU-3′) (39) was synthesized by transcription with MEGAscript T7 kits (Ambion) and synthetic DNA template as described elsewhere (48). In vitro RNA synthesis was carried out at 30°C for 60 min in a final volume of 25 μl in the presence of 50 mM HEPES-NaOH (pH 7.9), 3 mM MgCl2, 50 mM KCl, 1.5 mM dithiothreitol, 500 μM each ATP, GTP, and CTP, 25 μM UTP, 5 μCi of [α-32P]UTP (400 Ci/mmol), 10 U of RNasin, 25 μg of actinomycin D/ml, 250 μM ApG, 5 ng of a 53-nucleotide-long model RNA template of negative polarity, and RNP cores (10 ng of NP equivalents) in the presence or absence of host factor fractions. RNA products were purified, subjected to 10% polyacrylamide gel electrophoresis (PAGE) in the presence of 50% urea, and visualized by autoradiography. For determination of the level of RNA synthesis, an autoradiogram was scanned with NIH Image analyzing system.
Purification of RAF-2.
The buffer (buffer H) used for purification of RAF-2 contained 50 mM HEPES-NaOH (pH 7.9), 20% (vol/vol) glycerol, and 1 mM dithiothreitol plus the appropriate concentration of KCl or (NH4)2SO4. The uninfected HeLa cell nuclear extracts were prepared by a method described previously (10). The purification scheme started with nuclear extracts containing 34 mg of protein. Nuclear extracts were loaded onto a phosphocellulose column (P11; 10-ml bed volume; Whatman) equilibrated with buffer H containing 0.05 M KCl. The material that was not adsorbed to the column was loaded onto a Mono Q HR 5/5 column (Amersham Pharmacia Biotech) equilibrated with buffer H containing 0.05 M KCl. The column was washed with 0.15 M KCl, and proteins adsorbed to the anion exchanger were eluted with buffer H containing 0.4 M KCl. The 0.4 M KCl eluate was adjusted with buffer H containing 1 M (NH4)2SO4 so as to contain 0.5 M (NH4)2SO4 equivalents of salt. The RAF-2 activity was not adsorbed to a hydrophobic column (phenyl-Superose HR 5/5; Amersham Pharmacia Biotech) equilibrated with buffer H containing 0.5 M (NH4)2SO4. The RAF-2 fraction was diluted with buffer H so as to contain 0.15 M KCl equivalents of salt and applied to a Mono Q column equilibrated with buffer H containing 0.15 M KCl. RAF-2 activity was eluted with a linear gradient of 0.15 to 0.4 M KCl, concentrated by a small Mono Q column (Mono Q PC 1.6/5; Amersham Pharmacia Biotech), and subsequently separated through a gel filtration column (Superose 12 PC 3.2/30; Amersham Pharmacia Biotech).
Yeast two-hybrid screen and mapping.
Saccharomyces cerevisiae strain EGY48 (MATa trp1 ura3 his3 LEU::pLexAop6-LEU2), plasmids pEG202, pSH18-34, and pRFHM1, and the HeLa cell cDNA library were generously provided by R. Brent (Harvard Medical School). pLexA-NP was generated as described elsewhere (37).
The yeast two-hybrid screen has been described previously (37). Briefly, the screen was performed by transforming the HeLa cDNA library, in which the cDNAs were cloned as fusions to the B42 transcriptional activation domain, into an EGY48-derived yeast strain (R100) harboring plasmid pLexA-NP, in which the pLexA DNA-binding domain is fused to the influenza virus NP protein. Potential NP-interacting proteins encoded by cDNAs were identified based on the ability to confer upon the R100 yeast the ability to grow in the absence of leucine. An interaction between a cDNA-encoded protein and NP would mediate formation of a complex between the B42 transcriptional activation domain and the LexA DNA-binding domain such that activation of the LEU2 gene would occur, allowing rescue of the yeast from leucine auxotrophy. Library plasmids encoding potential interactors were isolated from the yeast, recovered by electroporation into Escherichia coli MH3 cells, and selected on 1× A+amp+glucose (37) plates.
Specificity of the NP interaction with the cDNA encoding RAF-2p48 (BAT1/UAP56/NPI-5) was confirmed by transformation of the cDNA-encoding plasmid into a derivative of the R100 strain. Introduction of the plasmid encoding RAF-2p48, plasmid pLexA-NP, and plasmid pSH18-34, which encodes β-galactosidase under the control of a LexA-regulated GAL1 promoter, resulted in induction of β-galactosidase activity. Introduction of the RAF-2p48 cDNA plasmid into a yeast strain which was transformed with both pSH18-34 plus pRFHM1, a plasmid encoding the LexA DNA-binding domain fused to a transcriptionally inert fragment of the Drosophila melanogaster bicoid protein, did not result in induction of β-galactosidase expression. By similar methods, the RAF-2p48 cDNA was found not to interact in the yeast two-hybrid system with the influenza A/PR/8/34 (H1N1) virus NS1 protein (data not shown).
The ability of RAF-2p48 to interact with NP deletion mutants was determined by assessing the ability of the mutants, when fused to the LexA DNA-binding domain, to activate β-galactosidase expression using the methods described above. The NP mutants were generated by PCR and cloned as LexA fusions in plasmid pEG202 using standard methods. The mutants were confirmed by DNA sequencing.
Preparation of recombinant proteins.
The plasmid constructs used in this study were confirmed by DNA sequencing. The full-length RAF-2p48 cDNA was cloned from HeLa cell total RNA as follows. Total RNA was prepared from 106 HeLa cells by the guanidine thiocyanate method (5); then single-stranded complementary DNA was synthesized with Moloney murine leukemia virus reverse transcriptase (TOYOBO) and oligo(dT) primers. The double-stranded RAF-2p48 cDNA was PCR amplified using LA-Taq polymerase (TaKaRa), with a portion of the cDNA as template and two specific primers, 5′-CCGGATCCATGGCAGGAACGATGTGGACAATGAG-3′ and 5′-CCGGATCCTGCAGCTACCGTGTCTGTTCAATGTAGGAGG-3′, corresponding to RAF-2p48 amino-terminal and RAF-2p48 carboxyl-terminal regions, respectively. For preparation of the hexahistidine-tagged RAF-2p48 (His-p48) expression vector, the amplified cDNA fragments were digested with PstI and BamHI and then cloned into pQE-9 vector (Qiagen) that was predigested with the same enzymes. A cDNA for recombinant RAF-2p48 mutant Arg380→Gln380 was constructed by PCR using a primer containing a CAG codon for Gln instead of a CGG codon for Arg. The resulting plasmids were used for transformation of E. coli M15, which harbors pREP4 for expression of the lac repressor protein (12).
For preparation of the glutathione S-transferase (GST)-tagged RAF-2p48 (GST-ρ48) expression vector (pGEX-p48), the RAF-2p48 cDNA fragment was prepared by digestion of the PCR product with BamHI, followed by cloning into pGEX-2T vector (Amersham Pharmacia Biotech) and amplification of the resulting plasmid in E. coli DH5α. After confirmation of the DNA sequence, pGEX-p48 was introduced into E. coli BL21. For preparation of GST-p48 deletion mutants, pGEX-p48 was digested with BamHI and BglII, resulting in three DNA fragments that correspond to the vector, the amino terminus of RAF-2p48, and the carboxy terminus of RAF-2p48. For pGEX-p48-N, the DNA fragment corresponding to the amino-terminal region of RAF-2p48 was cloned into pGEX-2T at the BamHI site. For pGEX-p48-C, cohesive ends of the pGEX-2T vector fragment and the 547-bp-long DNA fragment corresponding to the carboxyl-terminal region of RAF-2p48 were blunted with the Klenow fragment (TOYOBO) before subsequent ligation.
For preparation of recombinant NP, NP-N, and NP-C, the NP cDNA fragments were PCR amplified from plasmid pSP-NP, containing a cDNA of influenza A/PR/8/34 virus, using KOD DNA polymerase (TOYOBO) and a combination of specific primers 5′-GGAATTCATATGGCGTCTCAAGGCACCAAACG-3′, 5′-GGAATTCTTAATTATCGTATTCCTCTGCATTGTCTCCG-3′, 5′-GGAATTCTTATGTTCCAACTCCTTTGACTGCAGCAC-3′, and 5′-GGAATTCATATGGTGATGGAATTGGTCAGAATGATCAAAC-3′. The primers contain EcoRI sites, allowing the cDNA fragments to be cloned into pGEX-2T at EcoRI sites. For the preparation of histidine-tagged NP, we used plasmid pET-14b (Novagen).
The histidine- and GST-tagged proteins were purified using Ni-nitrilotriacetic acid and glutathione-conjugated resin, respectively, treated with RNase A (200 ng/μl), and further purified with a Mono Q column by KCl gradient elution.
Preparation of rabbit anti-RAF-2p48 antibody.
Affinity-purified recombinant His-p48 was further purified by sodium dodecyl sulfate (SDS)-PAGE and used for immunization. Polyclonal rabbit antiserum against RAF-2p48 was generated by immunization of a female rabbit (New Zealand White; Japan SLC, Inc.) with 250 μg of His-p48 in complete Freund's adjuvant (Sigma), followed by two boosts of 150 μg of His-p48 in incomplete Freund's adjuvant (Sigma) at 2-week intervals. For immunoblotting analysis, antiserum was used at a dilution of 1:1,000. The anti-RAF-2p48 antibody was purified from antiserum against RAF-2p48 by blot affinity purification (36). RAF-2p48 antigen (50 μg) was blotted onto a polyvinylidene difluoride (PVDF) membrane (Millipore) and blocked with 10% bovine serum albumin (BSA) in PBS. The PVDF membrane strip was incubated in 1 ml of the antiserum against RAF-2p48 at 4°C for 12 h, followed by washing with 1% Tween 20 in PBS. The anti-RAF-2p48 antibody was eluted from the strip with 300 μl of 0.2 M glycine (pH 2.8) at 4°C for 2 min, immediately followed by addition of BSA to 10% and dialysis against 500 ml of PBS at 4°C for 6 h. For indirect immunofluorescence assay, purified rabbit anti-RAF-2p48 antibody was used without dilution.
Indirect immunofluorescence assay.
HeLa cells on coverslips were infected with influenza A/PR/8/34 (H1N1) virus at a multiplicity of infection of 10 and fixed with 3% paraformaldehyde in PBS at 9 h postinfection. Permeabilization of the cells was carried out with 0.5% Triton X-100 in PBS, and the coverslips were then soaked in 1% nonfat dry milk in PBS. Samples were incubated at 4°C for 1 h with primary antibody, either affinity-purified rabbit anti-RAF-2p48 antibody or mouse anti-SC35 monoclonal antibody (PharMingen) as a spliceosome marker. After being washed twice with PBS, samples were incubated at 4°C for 30 min with secondary antibody, either rhodamine-conjugated goat anti-rabbit or fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin. After washing, samples were incubated at room temperature for 5 min with 3 μM 4′, 6′-diamidino-2-phenylindole (DAPI). Coverslips were finally mounted on glass plates, and cells were observed in a confocal laser scanning microscope (LSM410; Carl Zeiss).
GST pull-down assay.
A 30-pmol aliquot of each RNase A-treated GST-tagged recombinant protein in solution was fixed on a 10-μl bed volume of glutathione-Sepharose beads (Amersham Pharmacia Biotech). The binding reaction was carried out at 37°C for 60 min in a final volume of 100 μl containing 50 mM HEPES-NaOH (pH 7.9), 3 mM MgCl2, 50 mM KCl, 1.5 mM dithiothreitol, and the affinity beads in the presence or absence of test proteins. After the adsorption, the beads were washed three times with a wash buffer containing 20 mM Tris-HCl (pH 7.9), 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40. Proteins bound to affinity beads were eluted by boiling in SDS-PAGE loading buffer and then subjected to SDS-PAGE on a 10% gel. For the identification of RAF-2p48 and NP, rabbit anti-RAF-2p48 and anti-vRNP antisera, respectively, were used in immunoblotting analyses.
Glycerol density gradient centrifugation.
Fifty nanograms of 32P-labeled 53-mer RNA probe was mixed with 300 ng of the recombinant His-NP and incubated at 30°C for 30 min in a final volume of 50 μl. Binding was carried out in buffer containing 50 mM HEPES-NaOH (pH 7.9), 3 mM MgCl2, 50 mM KCl, 2.5 mM dithiothreitol, and 10 ng of BSA/μl and in the presence or absence of 800 ng of recombinant His-p48. Samples were layered onto a 1.25-ml 15 to 35% linear glycerol gradient in a buffer containing 50 mM HEPES-NaOH (pH 7.9), 3 mM MgCl2, 50 mM KCl, and 2.5 mM dithiothreitol. Centrifugation was carried out at 4°C for 15 h at 54,000 rpm with a TLS-55 rotor (Beckman). Fractions (100 μl each) were collected from the top of centrifuge tubes. An aliquot of each fraction (20 μl) was loaded onto a 5% polyacrylamide gel, and the RNA probe was visualized by autoradiography.
RESULTS
Identification and purification of RAF-2.
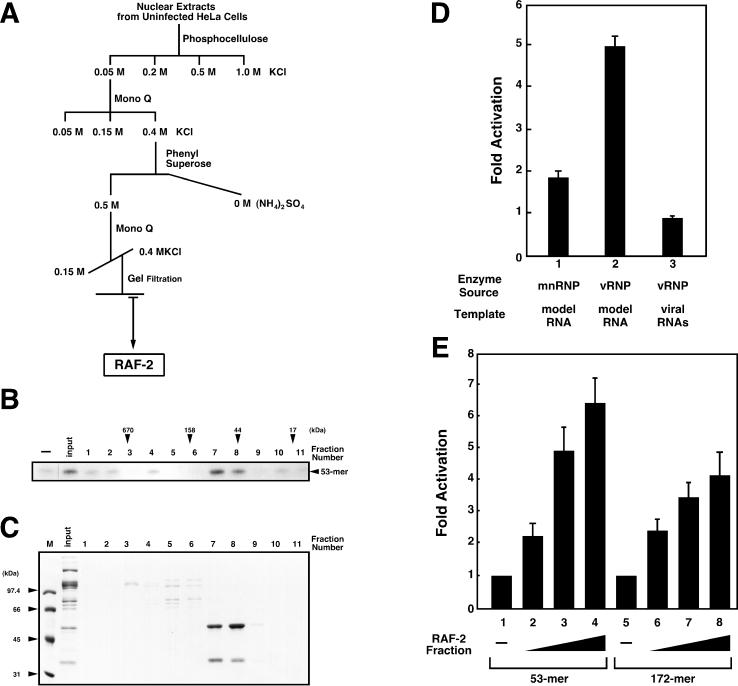
Details of the purification of RAF-2 were described previously (31) and are summarized in Fig. 1A. The chromatographic behavior of RAF-2 suggests that it is highly acidic and hydrophilic. In the final purification step, chromatography on a gel filtration column, RAF-2 was recovered in the 60- to 80-kDa fraction (Fig. 1B). SDS-PAGE revealed that the RAF-2 fraction contained approximately equal amounts of a 48-kDa and a 36-kDa polypeptide, designated RAF-2p48 and RAF-2p36, respectively (Fig. 1C). The RAF-2 fraction could stimulate about fivefold vRNA synthesis from the 53-mer model vRNA but had no effect on RNA synthesis from endogenous vRNAs derived from vRNP which was added as the enzyme source (Fig. 1D). The RAF-2 fraction could stimulate about twofold RNA synthesis from the 53-mer model vRNA when mnRNP was used as the enzyme source. These observations indicate that RAF-2 interacts with the viral RNA polymerase and/or NP and facilitates their recruitment to naked RNA templates. Since endogenous vRNAs are in advance complexed with the RNA polymerase and NP and the mnRNP supplies proteins free of RNA, RAF-2 may have less effect on RNA synthesis from endogenous vRNAs and that from the 53-mer model vRNA when mnRNP is used. Effects of RAF-2 on 53-mer and 172-mer model vRNA templates were compared (Fig. 1E). The levels of stimulation by RAF-2 from both templates are not significantly different, although RNA synthesis from the 53-mer model vRNA is slightly more than that from the 172-mer model vRNA.
FIG. 1.
Purification of RAF-2. (A) General scheme for purification of RAF-2. The details of column chromatography and the biochemical complementation assay system are described in Materials and Methods. (B) Gel filtration profile of RAF-2 activity. RNA synthesis was carried out in the absence (−) or presence of 0.34 M KCl eluate from the second Mono Q column (input, 1.5 μl) and aliquots of fractions separated through a gel filtration column (lanes 1 to 11, 1.5 μl of each). Arrowheads indicate the elution positions of molecular weight marker proteins (Bio-Rad). RAF-2 activity is observed in fractions corresponding to a molecular mass of 60 to 80 kDa (lanes 7 and 8). (C) Proteins present in the RAF-2 fraction. Five microliters of each fraction was loaded onto a 10% polyacrylamide gel containing 1% SDS. Electrophoresis was carried out, and the polypeptides were visualized by Coomassie brilliant blue staining. The RAF-2 fraction (lanes 7 and 8) contains 48-kDa and 36-kDa polypeptides as the major components (arrowheads). M, markers. (D) Stimulatory activity of RAF-2 in in vitro RNA synthesis systems containing different enzyme sources. In vitro RNA synthesis was carried out with mnRNP (lane 1) and vRNP (lanes 2 and 3). The level of RNA synthesis was determined by scanning the autoradiogram with the NIH Image analysis program. Ratios of the amounts of RNA products synthesized from the 53-mer model vRNA (lanes 1 and 2) or endogenous vRNAs (lane 3) in the presence of purified RAF-2 to those in the absence of purified RAF-2 are indicated as the average of three independent experiments. To determine the level of RNA synthesis from endogenous vRNAs, RNA products corresponding to all eight segments were included. (E) Stimulatory activity of RAF-2 in in vitro RNA synthesis systems with different RNA templates. In vitro RNA synthesis was carried out in the absence (lanes 1 and 5) or presence of 0.1 (lanes 2 and 6), 0.2 (lanes 3 and 7), and 0.4 (lanes 4 and 8) μl of purified RAF-2, using equimolar amounts of 53-mer (lanes 1 to 4) and 172-mer (lanes 5 to 8) model vRNA templates. The 172-mer RNA containing the same 5′- and 3′-terminal sequences as the 53-mer RNA was prepared as described elsewhere (54). RNA synthesis activity is shown as the ratio (the average of two independent experiments) of the amount of RNA products synthesized in the presence of RAF-2 to the amount synthesized in the absence of RAF-2.
To determine which protein, RAF-2p48 or RAF-2p36, is responsible for the activity of RAF-2, a portion of the purified RAF-2 fraction was electrophoresed on an SDS-polyacrylamide gel, and RAF-2p48 and RAF-2p36 were individually eluted from the gel and renatured (19). In the in vitro RNA synthesis system, stimulatory activity was detected for RAF-2p48 but not RAF-2p36 (data not shown). Since RAF-2p36 was not soluble and was recovered in aggregations after renaturation, we cannot rule out the possibility that RAF-2p36 itself has stimulatory activity. However, the finding that renatured RAF-2p48 and a recombinant RAF-2p48 (see below) stimulate RNA synthesis suggests that RAF-2p48 is, at least, an active component of RAF-2.
RAF-2p48 is identical to the splicing factor BAT1/UAP56 and interacts with the influenza virus NP in the yeast two-hybrid system.
To determine the identity of RAF-2p48, oligopeptides prepared from RAF-2p48 were analyzed with matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS). A portion of RAF-2 was electrophoresed on an SDS-polyacrylamide gel and electrophoretically blotted to a PVDF membrane. After staining with Ponceau S, blotted RAF-2p48 was excised and digested into oligopeptides with a lysyl endopeptidase, Achromobacter protease I (23). Comparison of the molecular masses for the oligopeptides determined by MALDI-TOF MS to those in the database indicated that all determined molecular masses are identical to those derived from a known human protein, a 428-amino-acid 48-kDa putative ATP-dependent RNA helicase designated UAP56/BAT1 (Table 1; GenBank accession number Z37166).
TABLE 1.
Molecular masses of lysyl endopeptidase-digested peptides from RAF-2p48 and amino acid sequences corresponding to the lysyl endopeptidase-digested peptide molecular mass database
| No. | Predicted amino acid sequence from database | Mass (Da)
|
||
|---|---|---|---|---|
| Observed | Theoretical | Difference | ||
| 1 | EYERFSK | 958.48 | 958.46 | +0.02 |
| 2 | EIRPVCRK | 1,058.61 | 1,058.58 | +0.03 |
| 3 | VAVFFGGLSIK | 1,137.69 | 1,137.67 | +0.02 |
| 4 | HFILDECDK | 1,177.55 | 1,177.52 | +0.03 |
| 5 | LTLHGLQQYYVK | 1,462.85 | 1,462.81 | +0.04 |
| 6 | GLAITFVSDENDAK | 1,479.77 | 1,479.73 | +0.04 |
| 7 | FMQDPMEIFVDDETK | 1,844.67 | 1,844.81 | −0.14 |
| 8 | FMQDPMEIFVDDETK…1Met-Ox | 1,860.66 | 1,860.81 | −0.15 |
| 9 | FMQDPMEIFVDDETK…2Met-Ox | 1,876.70 | 1,876.81 | −0.11 |
| 10 | NCPHIVVGTPGRILALARNK | 2,187.25 | 2,187.22 | +0.03 |
| 11 | KNCPHIVVGTPGRILALARNK | 2,315.35 | 2,315.31 | +0.04 |
| 12 | MLEQLDMRRDVQEIFRMTPHEK…1Met-Ox | 2,818.41 | 2,818.39 | +0.02 |
| 13 | MLEQLDMRRDVQEIFRMTPHEK…2Met-Ox | 2,834.38 | 2,834.39 | −0.01 |
| 14 | Ac-AENDVDNELLDYEDDEVETAAGGDGAEAPAKa | 3,264.45 | 3,264.37 | +0.08 |
The amino terminus of p48 was acetylated alanine (position 14).
Independent of the purification of RAF-2, a yeast two-hybrid screen of a HeLa cell cDNA library was performed using as bait the influenza A/PR/8/34 virus NP (37). A cDNA corresponding to amino acids 60 to 428 of RAF-2p48 was found to encode the NP-interacting protein NPI-5. The RAF-2p48/NPI-5/BAT1/UAP56 cDNA obtained in the two-hybrid screen possessed all of the conserved motifs characteristic of the DEAD-box protein family. The specificity of the NP–RAF-2p48/NPI-5/BAT1/UAP56 interaction in the yeast two-hybrid system was confirmed as described for NPI-1 and NPI-3 (37) (data not shown). Additionally, RAF-2p48/NPI-5/BAT1/UAP56 did not interact with the influenza A virus NS1 protein in the yeast two-hybrid system (data not shown).
RAF-2p48/NPI-5 was first described as BAT1 (HLA-B-associated transcript 1) protein. The BAT1 gene is located in the major histocompatibility complex gene class III region (41, 49, 50). In the recently completed nucleotide sequence of the human major histocompatibility complex gene, some disease-related genes are found in the vicinity of the BAT1 gene locus (30). The RAF-2p48 yeast homolog, Sub2 (product of the ydl084w gene), is essential for vegetative growth of the FY1679 strain (28). RAF-2p48 also appears to be an essential splicing factor required for the interaction of U2 snRNP with the pre-mRNA branch point (13). RAF-2p48, designated UAP56 (56-kDa U2AF65-associated protein) in this case, was found to interact in vitro with the linker region of splicing factor U2AF65 and to be recruited to pre-mRNA. Because RAF-2p48 contains ATP-dependent DEAD-box RNA helicase motifs in its amino acid sequence, it was hypothesized that RAF-2p48 may mediate conversion of RNA secondary structures within the U2 snRNA or the branch point (51), although direct evidence for RNA-unwinding activity of RAF-2p48/NPI-5/BAT1/UAP56 has not been reported.
RAF-2p48 is concentrated in spliceosomes in uninfected cells but not in influenza virus-infected cells.
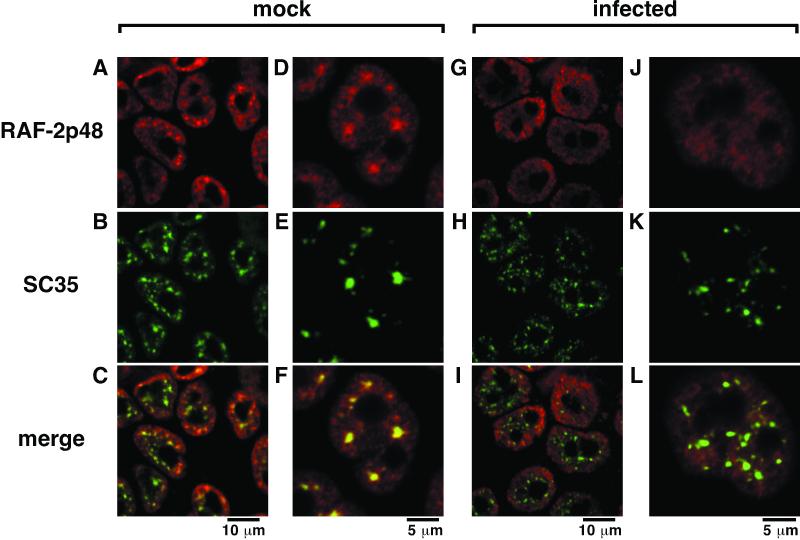
In uninfected cells, RAF-2p48 is localized in nuclei, excluded from nucleoli, and concentrated at spliceosomes where SC35, a splicing factor, is present (Fig. 2A to F). This observation is in good agreement with the notion that RAF-2p48 could be involved in RNA splicing. RAF-2p48 is observed both diffusely and as concentrated speckles in the nucleoplasm. It is possible that RAF-2p48 colocalized with SC35 is present in the splicing machinery and that diffusive RAF-2p48 is reserved for splicing reactions and/or participates in the other reactions. It has been reported that in infected cells, spliceosomes are partially destroyed (16, 54). In fact, speckles of SC35 become small and separated in infected cells (Fig. 2H and K). Interestingly, in infected cells, concentrated speckles of RAF-2p48 disappear (Fig. 2G and J), and substantially less colocalization between RAF-2p48 and SC35 is seen (Fig. 2I and L).
FIG. 2.
Localization of RAF-2p48 in influenza virus-infected cells. HeLa cells were infected with influenza A/PR/8/34 virus at a multiplicity of 10 (G to L) or mock infected (A to F). At 9 h postinfection, cells were fixed with PBS containing 3% paraformaldehyde and an indirect immunofluorescence assay was carried out as described in Materials and Methods. RAF-2p48 (A, D, G, and J) and SC35 (B, E, H, and K) were detected with rhodamine- and FITC-conjugated secondary antibodies, respectively. Images stained with rhodamine and FITC are merged (C, F, I, and L). Magnification is indicated by scale bars.
Stimulation of vRNA synthesis by RAF-2p48.
To further analyze the function of RAF-2p48, we prepared His-p48 in E. coli using a system in which the expression level of the recombinant protein is stringently regulated because of the toxicity of the protein to the host microbe (see Materials and Methods). Figure 3 shows that recombinant RAF-2p48 stimulates in vitro vRNA synthesis (lanes 7 to 11), but the stimulatory properties are different from those of the purified native RAF-2 fraction (lanes 12 to 16). The specific activity of recombinant RAF-2p48 is lower than that of the RAF-2 fraction, and adding excess amounts of the recombinant protein attenuates RNA synthesis. These results suggest that the RAF-2p48 protein is required for stimulating RNA synthesis and that an additional factor(s) such as RAF-2p36 could be required for the optimal activity of RAF-2. In addition, the recombinant RAF-2p48 mutant Arg380→Gln380 showed the same level of activity as the wild-type recombinant RAF-2p48 (lanes 2 to 6). This mutation, located in DEAD-box RNA helicase consensus motif VI, is analogous to the eIF-4A DEAD-box RNA helicase mutant Arg365→Gln365, which was defective in ATPase and helicase activities (40). This result suggests that the putative RNA-unwinding activity of RAF-2p48 may not be needed for stimulation of RNA synthesis by recombinant RAF-2p48.
FIG. 3.
Stimulatory activity of recombinant RAF-2p48. In vitro RNA synthesis was carried out in the absence (lane 1) or presence of 2, 8, 32, 128, and 512 ng of the RAF-2p48 equivalent of recombinant His-p48 mutant Arg380→Gln380 (lanes 2 to 6), recombinant His-p48 (lanes 7 to 11), or the purified native RAF-2 fraction (lanes 12 to 16). The synthesized 53-mer RNA is indicated by an arrowhead.
RAF-2p48 interacts with free NP but not with NP-RNA complexes.
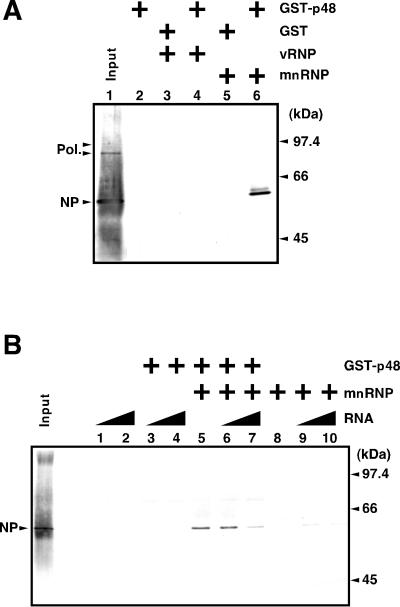
To confirm the interaction between RAF-2p48 and NP, we carried out a GST pull-down assay using GST-p48 (Fig. 4A). GST-p48 immobilized on glutathione beads was incubated with vRNP or mnRNP. In mnRNP, the RNA polymerase complex and NP are depleted of vRNA (46). GST-p48 specifically interacted with NP but not with the RNA polymerase when mnRNP was used (lane 6), while GST-p48 did not interact with either NP or the RNA polymerase when vRNP was used (lane 4). This result indicates that RAF-2p48 binds to free NP but not to the NP-RNA complex. This finding suggests that the NP–RAF-2p48 complex is dissociated by the addition of RNA. This is indeed the case (Fig. 4B). The addition of large amounts of RNA dissociated the RAF–2p48-NP complex. Simultaneously, dissociated NP was found to be associated with RNA (data not shown).
FIG. 4.
Association/dissociation of RAF-2p48 and NP. A GST pull-down assay was carried out as described in Materials and Methods, and proteins were detected by Western blotting analysis with rabbit anti-vRNP antiserum. As a positive control, 20% of input vRNP was loaded (Input). (A) Association of NP and RAF-2p48. GST (lanes 3 and 5) or GST-p48 (lanes 2, 4, and 6) was fixed on glutathione-Sepharose beads and incubated for 1 h at 37°C in the presence of a 100-ng equivalent of vRNP (lanes 3 and 4) or mnRNP (lanes 5 and 6). (B) Dissociation of RAF-2p48–NP complexes. GST (lanes 1, 2, 8, 9, and 10) or GST-p48 (lanes 3 to 7) was fixed on glutathione-Sepharose beads and incubated with mnRNP (lanes 5 to 10) at 37°C for 1 h. Unbound proteins were washed out, and beads were further incubated in the presence of 200 ng (lanes 1, 3, 6, and 9) or 1 μg (lanes 2, 4, 7, and 10) of free RNA (53-mer Vwt).
RAF-2p48 interacts with the amino terminus of NP.
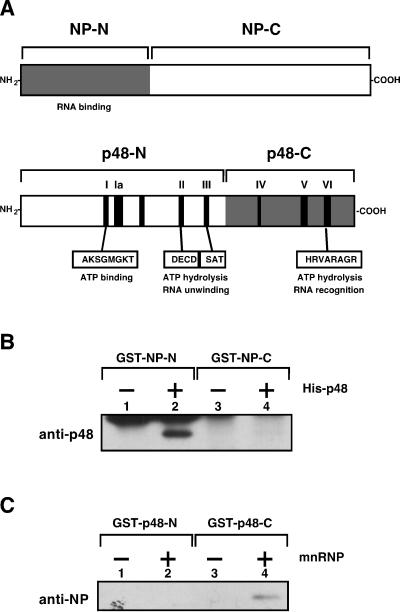
The formation of complexes between RAF-2p48 and NP and the formation of complexes between RNA and NP seem to be mutually exclusive. This suggested the possibility that the region of NP required for interaction with RAF-2p48 is located near its RNA-binding and recognition domain. To test this hypothesis, NP was divided into two regions (Fig. 5A): one consisted of the amino-terminal 188 amino acid residues (NP-N), and the other consisted of the carboxyl-terminal 320 amino acid residues (NP-C). NP-N contains a region essential for RNA binding, although NP interacts with RNA through several stretches that are widely distributed in its sequence (1, 11, 26). RAF-2p48 was also divided into two regions (Fig. 5A): the amino-terminal 248 amino acid residues (p48-N), and the remaining carboxyl-terminal 180 amino acid residues (p48-C). p48-N contains (i) the DEAD-box RNA helicase consensus motifs that participate in ATP binding and (ii) the catalytic center of RNA unwinding; p48-C contains motif VI, which is involved in RNA recognition, and other motifs (7). Recombinant proteins were purified and extensively treated with RNase A to remove RNA fragments, if any, in purified fractions (Materials and Methods). When beads fixed with deletion mutants of NP were used, His-p48 interacted with GST–NP-N (Fig. 5B, lane 2) but not with GST–NP-C (lane 4). The deletion mutants of RAF-2p48 that were fused with GST at their amino termini were fixed on beads and incubated at 37°C in the presence of mnRNP (Fig. 5C). NP interacted with GST–p48-C (lane 4) but not with GST–p48-N (lane 2).
FIG. 5.
RAF-2p48 and NP interaction domains. (A) Schematic structures of proposed RAF-2p48 and NP functional domains. Locations of the DEAD-box RNA helicase consensus motifs in RAF-2p48 are indicated by black boxes and roman numerals. NP-N, NP-C, p48-N, and p48-C indicate the mutant proteins used in panels B and C. GST pull-down assays (B and C) were carried out as described in Materials and Methods. RAF-2p48 and NP were visualized by Western blotting with rabbit anti-RAF-2p48 and anti-vRNP antisera. (B) GST–NP-N and GST–NP-C were fixed on glutathione-Sepharose beads and incubated at 37°C for 1 h in the absence (−) or presence (+) of recombinant RAF-2p48. (C) GST–p48-N and GST–p48-C were fixed on glutathione-Sepharose beads and incubated at 37°C for 1 h in the absence (−) or presence (+) of mnRNP.
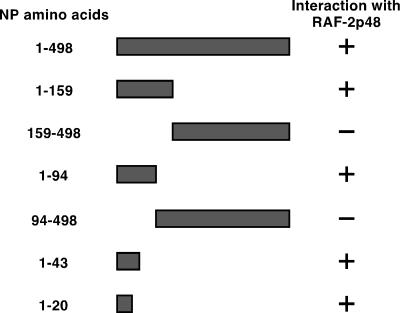
To further examine the region of NP required for interaction with RAF-2p48, a series of NP deletion mutants was generated and tested for the ability to interact with RAF-2p48 in the yeast two-hybrid system (Fig. 6). In agreement with the in vitro binding data, the amino terminus of NP was required for interaction with RAF-2p48. Further mapping defined the amino-terminal 20 amino acids of NP as sufficient for binding to RAF-2p48 (Fig. 6). These results, together with the in vitro binding data (Fig. 5), suggest that formation of the RAF-2p48–NP complex involves interaction between domains of NP and RAF-2p48 which may also be involved in RNA binding by the individual proteins.
FIG. 6.
The amino-terminal 20 amino acids of NP are required for interaction with RAF-2p48. The influenza A virus NP deletion mutants indicated were expressed in yeast as fusions with the LexA DNA-binding domain and then screened for the ability to interact with amino acids 60 to 428 of RAF-2p48 when expressed as fusions with the B42 transcriptional activation domain. Interaction was indicated by activation of the LexA-regulated β-galactosidase gene encoded on plasmid pSH18-34. β-Galactosidase gene expression was assessed by plating the transformed yeast on media containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
RAF-2p48 facilitates formation of the NP-RNA complex.
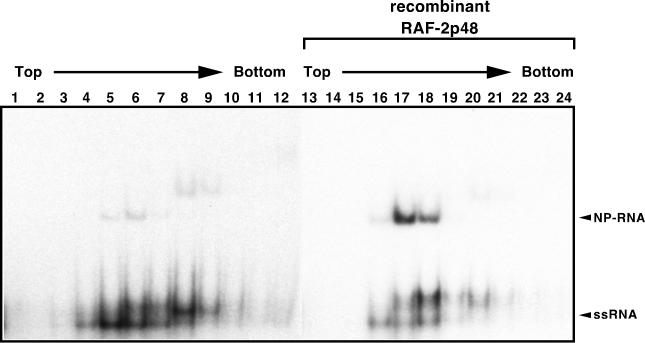
As described above, RAF-2p48 binds to the free NP and is released upon interaction of NP with RNA. Therefore, RAF-2p48 may act as a chaperon-like factor, facilitating loading of free NP onto RNA. We therefore examined the effect of RAF-2p48 on the efficiency of NP-RNA complex formation (Fig. 7). A labeled RNA probe was incubated with recombinant NP in the presence or absence of the recombinant RAF-2p48 and subjected to separation through a 15 to 35% linear glycerol density gradient. After centrifugation, aliquots were fractionated and the NP-RNA complex was separated by PAGE on a 5% polyacrylamide gel. Mobility of the RNA probe complexed with NP is slower than that of RNA. Clearly, the formation of NP-RNA complexes is increased in the presence of RAF-2p48 (compare lanes 5 and 6 and lanes 17 and 18). This result strongly suggests that RAF-2p48 functions as a chaperon for NP, facilitating formation of NP-RNA complexes.
FIG. 7.
RAF-2p48-mediated NP-RNA complex formation. 32P-labeled RNA probe mixed with recombinant NP was incubated in the absence (lanes 1 to 12) or presence (lanes 13 to 24) of recombinant RAF-2p48. After 15 to 35% glycerol density gradient centrifugation, fractions were collected from the tops of the tubes. An aliquot of each fraction was analyzed by PAGE on a 5% polyacrylamide gel, and the RNA was visualized by autoradiography. The unbound RNA probe and NP-RNA complexes (lanes 5, 6, 17, and 18) are indicated by arrowheads. The bands shown in lanes 8 and 9 with less mobility than those in lanes 5, 6, 17, and 18 could be NP-RNA complexes containing more than one NP molecule per RNA molecule.
DISCUSSION
Possible functions of RAF-2p48 in the formation of NP-RNA complexes and in the stimulation of RNA synthesis.
In this report, we have identified the 48-kDa host cell splicing factor RAF-2p48 (BAT1/UAP56/NPI-5), which belongs to the DEAD-box family of putative RNA helicases, as a factor that interacts with the influenza virus NP and stimulates the influenza virus RNA polymerase activity. RAF-2p48 was identified based on its presence in RAF-2, a fraction from uninfected host cells that stimulates influenza virus RNA synthesis. The 48-kDa RAF-2p48 protein was found to be required for RAF-2 activity. RAF-2p48 was also found to interact with free NP in vitro and with NP in the yeast two-hybrid system. Further, RAF-2p48 was found to facilitate formation of NP-RNA complexes.
In general, nucleic acid-binding basic proteins such as histones and viral basic proteins tend to aggregate and be inactive under physiological conditions in the absence of appropriate substrates such as DNA, RNA, or possibly chaperons (34). This could also be the case for free NP. Since free NP is produced in infected cells, RAF-2p48 may participate in suppression of nonspecific NP aggregation and may assist in delivery of NP to vRNA. Newly synthesized NP is transported into the nucleus with the help of nuclear import factors such as NPI-1 and -3 (also karyopherin-α1 and -α2) (32, 53). It is possible that RAF-2p48 binds NP in the nucleoplasm and facilitates its binding to vRNAs. With regard to this hypothesis, it is intriguing that the regions of NP required for binding to RAF-2p48 and for binding to the nuclear import factors NPI-1 and NPI-3 overlap. It has not been clarified whether the association of the NP with viral RNAs is autogenous or mediated by a factor(s). RAF-2p48 is a possible cellular candidate for such a cellular factor. Stimulatory effects of RAF-2 on 53-mer and 172-mer model vRNA templates do not differ greatly (Fig. 1E). This may be interpreted as indicating that RAF-2 facilitates the initial binding of NP to RNA rather than the cooperative binding of NP to RNA, the latter of which could be more apparent for the longer RNA template. For dissociation of a RAF-2p48–NP complex, the addition of relatively large amounts of RNA is required (Fig. 4B, lane 7). This result suggests that even if RAF-2p48 is involved in the arrangement of NP on RNA, there exists a cofactor(s) that assists in RNA recognition and/or translocation of the RAF-2p48–NP complex to the substrate RNA. In this regard, the translocation of RAF-2p48/UAP56 to the branch point of pre-mRNA is mediated by the splicing factor U2AF65 (13).
RAF-2p48 may be a multifunctional stimulatory factor.
RAF-2p48 contains the canonical ATP-dependent DEAD-box RNA helicase motifs (41). However, no RNA helicase activity of RAF-2p48 has been reported. Similarly, we also did not detect, under a variety of conditions, RNA-unwinding activity from the purified RAF-2 or from the recombinant RAF-2p48 protein. Furthermore, both the interaction of RAF-2p48 with NP and the RAF-2p48-mediated NP-RNA complex formation were ATP independent (data not shown), suggesting that RAF-2p48 does not require ATP for these reactions. In contrast, in vitro vRNA synthesis stimulated by the purified RAF-2 fraction required high concentrations of ATP. The Km values for ribonucleoside triphosphates in the absence of the RAF-2 fraction have been previously reported to be about 14 μM (52), suggesting that the affinities of the RNA polymerase to four kinds of ribonucleoside triphosphates as substrates of polymerization are similar. In contrast, Km of around 95 μM for ATP was detected in the presence of the RAF-2 fraction. These observations suggest that there is an ATP consumer in the purified RAF-2. In fact, the RAF-2 fraction was found to contain an ATPase activity that was stimulated in the presence of purified NP and RNA. Furthermore, RAF-2p48 and RAF-2p36 in the purified RAF-2 were phosphorylated, and the phosphorylation was stimulated in the presence of NP (data not shown). However, it is not known whether RAF-2p48, RAF-2p36, or another protein present in the RAF-2 fraction is responsible for utilization of ATP.
If RAF-2p48 participated in the vRNA-unwinding processes as an RNA helicase, there are two possible substrates for RAF-2p48. The first is the RNA duplex consisting of the template vRNA and the newly synthesized RNA; the second is the partially complementary intramolecular terminal RNA duplex. Secondary structure models for the terminal region of the influenza virus genome have been proposed (see the introduction). We are currently designing an RNA synthesis-coupled RNA-unwinding system to examine these as potential substrates for RAF-2p48 helicase activity.
Role of RAF-2p48 in vRNA synthesis and splicing.
It has been observed in influenza virus-infected cells that the localization of splicing-related host proteins such as SC35 and NS1-binding protein is altered (16, 55). Upon infection, large speckled forms of SC35 change to small punctate forms, and the localization of NS1-binding protein in spliceosomes disappears. This alteration is shown to be dependent on the presence of the viral nonstructural protein NS1. The localization pattern of RAF-2p48 (a putative splicing factor previously identified as UAP56) was also affected by influenza virus infection (Fig. 2). In infected cells, drastic inhibition of the host cellular protein production, termed the shutoff phenomenon, has been observed (18), possibly due to the inhibition of splicing and 3′-terminal processing of pre-mRNA (4, 35, 55). The alteration of RAF-2p48 localization indicates that it may be partially responsible for the splicing inhibition or concomitantly be caused by disruption of spliceosomes. Furthermore, relocalization of RAF-2p48 would contribute to the release of RAF-2p48 from spliceosomes and facilitate the involvement of RAF-2p48 in vRNA synthesis.
ACKNOWLEDGMENTS
We thank Kim Pepin for proofreading the manuscript. We thank Louis Nguyenvu for excellent technical assistance with the yeast two-hybrid experiments.
This research was supported in part by a grant-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (K.N.), a grant from the Bioarchitect Research Project of RIKEN (K.N.), a grant from Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (F.M.), and grants from the National Institutes of Health (C.F.B. and P.P.).
REFERENCES
- 1.Albo C, Valencia A, Portela A. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J Virol. 1995;69:3799–3806. doi: 10.1128/jvi.69.6.3799-3806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaton A R, Krug R M. Synthesis of the templates for influenza virion RNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:4682–4686. doi: 10.1073/pnas.81.15.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braam J, Ulmanen I, Krug R M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell. 1983;34:609–618. doi: 10.1016/0092-8674(83)90393-8. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Li Y, Krug R M. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Compans R W, Content J, Duesberg P H. Structure of the ribonucleoprotein of influenza virus. J Virol. 1972;10:795–800. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 8.del Rio L, Martinez C, Domingo E, Ortin J. In vitro synthesis of full-length influenza virus complementary RNA. EMBO J. 1985;4:243–247. doi: 10.1002/j.1460-2075.1985.tb02342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desselberger U, Racaniello V R, Zazra J J, Palese P. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 10.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elton D, Medcalf L, Bishop K, Harrison D, Digard P. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J Virol. 1999;73:7357–7367. doi: 10.1128/jvi.73.9.7357-7367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farabaugh P J. Sequence of the lacI gene. Nature. 1978;274:765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- 13.Fleckner J, Zhang M, Valcarcel J, Green M R. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 14.Flick R, Hobom G. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J Gen Virol. 1999;80:2565–2572. doi: 10.1099/0022-1317-80-10-2565. [DOI] [PubMed] [Google Scholar]
- 15.Fodor E, Pritlove D C, Brownlee G G. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J Virol. 1995;69:4012–4019. doi: 10.1128/jvi.69.7.4012-4019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortes P, Lamond A I, Ortin J. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J Gen Virol. 1995;76:1001–1007. doi: 10.1099/0022-1317-76-4-1001. [DOI] [PubMed] [Google Scholar]
- 17.Galarza J M, Peng Q, Shi L, Summers D F. Influenza A virus RNA-dependent RNA polymerase: analysis of RNA synthesis in vitro. J Virol. 1996;70:2360–2368. doi: 10.1128/jvi.70.4.2360-2368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garfinkel M S, Katze M G. How does influenza virus regulate gene expression at the level of mRNA translation? Let us count the ways. Gene Expr. 1993;3:109–118. [PMC free article] [PubMed] [Google Scholar]
- 19.Hager D A, Burgess R R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- 20.Honda A, Ishihama A. The molecular anatomy of influenza virus RNA polymerase. Biol Chem. 1997;378:483–488. [PubMed] [Google Scholar]
- 21.Honda A, Ueda K, Nagata K, Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J Biochem. 1988;104:1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- 22.Huang T S, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamatsu A, Yoshida-Kubomura N. Systematic peptide fragmentation of polyvinylidene difluoride (PVDF)-immobilized proteins prior to microsequencing. J Biochem. 1996;120:29–34. doi: 10.1093/oxfordjournals.jbchem.a021389. [DOI] [PubMed] [Google Scholar]
- 24.Kato A, Mizumoto K, Ishihama A. Purification and enzymatic properties of an RNA polymerase-RNA complex from influenza virus. Virus Res. 1985;3:115–127. doi: 10.1016/0168-1702(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 25.Kim H J, Fodor E, Brownlee G G, Seong B L. Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J Gen Virol. 1997;78:353–357. doi: 10.1099/0022-1317-78-2-353. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi M, Toyoda T, Adyshev D M, Azuma Y, Ishihama A. Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J Virol. 1994;68:8433–8436. doi: 10.1128/jvi.68.12.8433-8436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M L, Ramirez B C, Krug R M. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 1998;17:5844–5852. doi: 10.1093/emboj/17.19.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez M C, Sanchez M, Ferminan E, Dominguez A. Disruption of six Saccharomyces cerevisiae genes from chromosome IV and basic phenotypic analysis of deletion mutants. Yeast. 1998;14:1199–1208. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1199::AID-YEA309>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 29.Luo G X, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The MHC Sequencing Consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 31.Momose F, Handa H, Nagata K. Identification of host factors that regulate the influenza virus RNA polymerase activity. Biochimie. 1996;78:1103–1108. doi: 10.1016/s0300-9084(97)86736-3. [DOI] [PubMed] [Google Scholar]
- 32.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murti K G, Webster R G, Jones I M. Localization of RNA polymerases on influenza viral ribonucleoproteins by immunogold labeling. Virology. 1988;164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 34.Nagata K, Momose F, Okuwaki M. Acidic molecular chaperones: their involvement in viral genome replication and transcription. Recent Res Dev Virol. 1999;1:559–597. [Google Scholar]
- 35.Nemeroff M E, Barabino S M, Li Y, Keller W, Krug R M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 36.Olmsted J B. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981;256:11955–11957. [PubMed] [Google Scholar]
- 37.O'Neill R E, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 38.Palese P, Wang P, Wolff T, O'Neill R E. Host-viral protein-protein interactions in influenza virus replication. In: McCrae M A, et al., editors. Molecular aspects of host-pathogen interaction. Cambridge, United Kingdom: Cambridge University Press; 1997. pp. 327–340. [Google Scholar]
- 39.Parvin J D, Palese P, Honda A, Ishihama A, Krystal M. Promoter analysis of influenza virus RNA polymerase. J Virol. 1989;63:5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peelman L J, Chardon P, Nunes M, Renard C, Geffrotin C, Vaiman M, Van Zeveren A, Coppieters W, van de Weghe A, Bouquet Y, Choy W W, Strominger J L, Spies T. The BAT1 gene in the MHC encodes an evolutionarily conserved putative nuclear RNA helicase of the DEAD family. Genomics. 1995;26:210–218. doi: 10.1016/0888-7543(95)80203-x. [DOI] [PubMed] [Google Scholar]
- 42.Plotch S J, Bouloy M, Ulmanen I, Krug R M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 43.Poon L L, Pritlove D C, Fodor E, Brownlee G G. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pritlove D C, Poon L L, Devenish L J, Leahy M B, Brownlee G G. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J Virol. 1999;73:2109–2114. doi: 10.1128/jvi.73.3.2109-2114.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seong B L, Brownlee G G. A new method for reconstituting influenza polymerase and RNA in vitro: a study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology. 1992;186:247–260. doi: 10.1016/0042-6822(92)90079-5. [DOI] [PubMed] [Google Scholar]
- 46.Seong B L, Kobayashi M, Nagata K, Brownlee G G, Ishihama A. Comparison of two reconstituted systems for in vitro transcription and replication of influenza virus. J Biochem. 1992;111:496–499. doi: 10.1093/oxfordjournals.jbchem.a123786. [DOI] [PubMed] [Google Scholar]
- 47.Shapiro G I, Krug R M. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol. 1988;62:2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu K, Handa H, Nakada S, Nagata K. Regulation of influenza virus RNA polymerase activity by cellular and viral factors. Nucleic Acids Res. 1994;22:5047–5053. doi: 10.1093/nar/22.23.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spies T, Blanck G, Bresnahan M, Sands J, Strominger J L. A new cluster of genes within the human major histocompatibility complex. Science. 1989;243:214–217. doi: 10.1126/science.2911734. [DOI] [PubMed] [Google Scholar]
- 50.Spies T, Bresnahan M, Strominger J L. Human major histocompatibility complex contains a minimum of 19 genes between the complement cluster and HLA-B. Proc Natl Acad Sci USA. 1989;86:8955–8958. doi: 10.1073/pnas.86.22.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 52.Takeuchi K, Nagata K, Ishihama A. In vitro synthesis of influenza viral RNA: characterization of an isolated nuclear system that supports transcription of influenza viral RNA. J Biochem. 1987;101:837–845. doi: 10.1093/oxfordjournals.jbchem.a121950. [DOI] [PubMed] [Google Scholar]
- 53.Wang P, Palese P, O'Neill R E. The NPI-1/NPI-3 (karyopherin α) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe K, Momose F, Handa H, Nagata K. Interaction between influenza virus proteins and pine cone antitumor substance that inhibits the virus multiplication. Biochem Biophys Res Commun. 1995;214:318–323. doi: 10.1006/bbrc.1995.2290. [DOI] [PubMed] [Google Scholar]
- 55.Wolff T, O'Neill R E, Palese P. NS1-binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J Virol. 1998;72:7170–7180. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamanaka K, Ishihama A, Nagata K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J Biol Chem. 1990;265:11151–11155. [PubMed] [Google Scholar]