Abstract
Growing evidence indicates a relationship between telomere length (TL) and the stage, prognosis, and treatment responsiveness of hematopoietic malignancies. However, the relationship between TL and the risk of hematologic malignancies remains unclear, considering the vulnerability of observational studies to potential confounding and reverse causation. A two-sample bidirectional Mendelian randomization (MR) analysis was conducted utilizing publicly available genome-wide association study data to assess whether TL was causally associated with the risk of hematologic malignancies. The inverse variance weighted approach was used as the primary assessment approach to evaluate the effects of the causes, augmented by the weighted median and MR-Egger methods. Cochran’s Q test, MR-Egger intercept test, MR-Pleiotropy Residual Sum and Outlier test, and leave-one-out analysis were performed to evaluate sensitivity, heterogeneity, and pleiotropy. According to forward MR estimations, longer TL was related to an increased risk of acute lymphocytic leukemia (OR = 2.690; P = 0.041), chronic lymphocytic leukemia (OR = 2.155; P = 0.005), multiple myeloma (OR = 1.845; P = 0.024), Hodgkin lymphoma (OR = 1.697; P = 0.014), and non–Hodgkin lymphoma (OR = 1.737; P = 0.009). Specific types of non–Hodgkin lymphoma were also associated with TL. The reverse MR results revealed that hematologic malignancies had no effect on TL. This MR analysis revealed an association between longer TL and an increased risk of specific hematologic malignancies, indicating a potential role of TL in risk evaluation and management in hematologic malignancies.
Significance:
In contrast to observational studies, this study uncovered the reliable causal relationships between TL and hematologic malignancies, emphasizing the potential role of telomeres in tumor development. TL maintenance may offer a promising strategy to reduce the risk of hematologic malignancies.
Introduction
Hematologic malignancies are among the most prevalent and rapidly developing malignant tumors, posing an enormous threat to human health (1). However, the development of effective prediction, treatment, and prevention strategies for hematologic malignancies encounter considerable challenges, primarily due to their unclear nature of pathogenesis. Although research has indicated that radiation (1), smoking (2), and chemical carcinogens (3) may increase the risk of developing hematologic malignancies, the exact cause of hematologic malignancies remains unclear, and there is no reliable approach for predicting hematologic malignancies.
Telomeres are unique DNA–protein complexes located at the ends of chromosomes, consisting of a G-rich DNA sequence with associated proteins. Human telomeres are typically composed of TTAGGG repeats, and their primary function is to prevent chromosomal DNA degradation, terminal fusion loss, and aberrant recombination, thus maintaining chromosome stability and integrity (4). Telomeres progressively shorten with cellular division in normal human cells, limiting their lifespan and serving as a critical barrier against excessive proliferation and malignant transformation. In contrast, telomere maintenance is a hallmark of human malignant cells and is necessary for their limitless proliferation (5). Multiple investigations suggest that telomere length (TL) may be important in the development and progression of cancer, particularly hematologic malignancies (6). According to research, TL in acute myeloid leukemia (AML; ref. 7), chronic myeloid leukemia (CML; refs. 8, 9), non–Hodgkin lymphoma (NHL; ref. 10), and other hematologic malignancies is longer than TL in normal blood cells. However, the causal associations between TL and hematologic malignancies remain controversial, and various observational studies have yielded conflicting data about the impact of TL on malignancy. Shorter TL has been shown in several studies to be a probable predictor of AML (11, 12), CML (13), acute lymphoblastic leukemia (ALL; refs. 14, 15), chronic lymphoblastic leukemia (CLL; refs. 16, 17), and NHL (18). The inadequacy of the research methodologies utilized is one plausible explanation for the emergence of these conflicting results. The question of whether TL alteration is a cause or a consequence of the development of hematologic malignancies remains unanswered.
Mendelian randomization (MR) is an instrumental variable (IV) method that uses single-nucleotide polymorphisms (SNP) as IVs to infer causal links between two traits, with the advantages of minimizing confounders and bias for reverse causation (19). The MR study provides an evaluation of causality that can aid in the prevention of divergence caused by confounding factors. MR studies have been widely used to investigate the causal relationships between potential risk factors and disease risk, such as TL and cancer (20), body mass index and diabetes (21), smoking and stroke (22), and so forth. However, the bidirectional causal association between TL and hematologic malignancies has yet to be thoroughly elucidated. In this study, we used a bidirectional two-sample MR method, leveraging its unique advantages, to investigate the potential bidirectional association between TL and hematologic malignancies, aiming to provide evidence about the role of telomeres in potential targets for prediction, prevention, and treatment in hematopoietic malignancies.
Materials and Methods
Study design
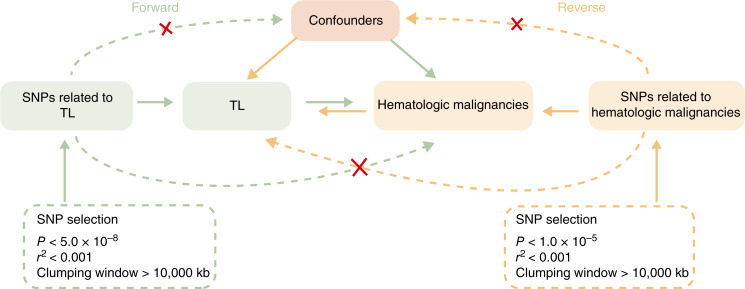
The bidirectional MR analysis between TL and hematologic malignancies followed the “STROBE-MR” protocol, using the approach depicted in Fig. 1. Forward and reverse MR analyses were performed to investigate the bidirectional causal relationships between TL and hematologic malignancies. Based on summary statistics from the genome-wide association study (GWAS), SNPs associated with exposure were identified and utilized as IVs to assess causality with the outcome. In the forward MR analysis, TL was chosen as the exposure, with hematologic malignancies serving as the outcome. Conversely, in the reverse MR analysis, hematologic malignancies were chosen as the exposure, with TL serving as the outcome. This MR analysis was conducted based on the fundamental hypotheses that underlie MR: (i) IVs of interest exposures are highly correlated with exposures; (ii) IVs of interest exposures are irrelevant to confounders of interest exposure–outcome associations; and (iii) IVs only affect outcomes by influencing the interest exposure.
Figure 1.
Principle diagram of MR analysis of the bidirectional causal relationship between TL and hematologic malignancies. In the forward MR analysis, TL was chosen as the exposure, with hematologic malignancies serving as the outcome. In the reverse MR analysis, hematologic malignancies were chosen as the exposure, with TL serving as the outcome.
Datasets and selection of IVs
We obtained summary statistics of TL from GWAS data on European ancestry from the Integrative Epidemiology Unit (IEU) OpenGWAS Project (ID: ieu-b-4879; Supplementary Table S1) consisting of 472,174 individuals. The GWAS information for primary lymphoid and hematopoietic malignant neoplasms, eight hematologic malignancies (AML, ALL, CML, CLL, multiple myeloma, Hodgkin lymphoma, NHL, and leukemia of unspecified cell type), and eight classifications of NHL (follicular lymphoma, nonfollicular lymphoma, diffuse large B-cell lymphoma, Waldenström macroglobulinemia/lymphoplasmacytic lymphoma, mantle cell lymphoma, marginal zone B-cell lymphoma, mature T/NK lymphomas, and other and unspecified types of NHL) were obtained from the FinnGen GWAS database (23).The GWAS cohorts exclusively consist of individuals of European ancestry (Supplementary Table S1), and the criterion of the participants can be found on FinnGen (www.finngen.fi/en). To further validate the effectiveness of the results across different populations, we additionally obtained GWAS data from the UK Biobank on IEU OpenGWAS Project (Supplementary Table S1), encompassing GWAS data of leukemia, lymphoid leukemia, and myeloid leukemia on European ancestry.
For IVs representing exposure, SNPs that passed the genome-wide significance threshold were subsequently clumped with other related SNPs within 10,000 kb based on the 1000 Genomes Project’s evaluation of linkage disequilibrium using r2 < 0.001. In the forward MR analysis, the TL significance threshold was selected at 5 × 10−8, and the IVs that were significantly associated with the outcome (P < 5 × 10−8) were excluded. In the reverse MR analysis, we adjusted the significance threshold to 1 × 10−5 in order to prevent unreliable outcomes caused by an absence of SNPs because there were limited IVs for hematopoietic malignancies (Fig. 1). Subsequently, we harmonized the exposure and outcome data to eliminate strand mismatches and guarantee the alignment of SNP effect sizes. F-statistics were used to estimate the strength of IVs. F > 10 indicated strong instrument strength, and IVs with F < 10 were excluded (24). The PhenoScanner v2 (http://www.phenoscanner.medschl.cam.ac.uk/) was used to exclude variations related to phenotypes that might be associated with hematologic malignancies in the forward MR analysis, as well as those associated with TL in the reverse MR analysis (25, 26).
Statistical analysis
To evaluate the potential bidirectional causal connections between TL and hematologic malignancies, we conducted a two-sample bidirectional MR analysis using the inverse variance weighted (IVW) method, which is most commonly used in MR studies and offers robust causal estimates in the absence of directional pleiotropy. Additionally, the MR-Egger and weighted median (WM) methods were used for verifying the results. The MR-Egger regression was used for estimating the level of bias induced by directional pleiotropy. To detect and exclude outliers, the Mendelian Randomization Pleiotropy Residual Sum and Outlier (MR-PRESSO) and leave-one-out analyses were used. The heterogeneity was estimated by IVW and MR-Egger methods via Cochran’s Q statistics and funnel plots. The OR and 95% confidence interval (CI) were utilized to estimate the causal relationship between TL and hematologic malignancies. R version 4.3.1 and its package “TwoSampleMR (version 0.6.4)” were used for statistical analysis.
Data availability
The datasets analyzed during the current study are available in the FinnGen (www.finngen.fi/en) and IEU OpenGWAS (gwas.mrcieu.ac.uk/) databases.
Results
The effect of TL on hematologic malignancies
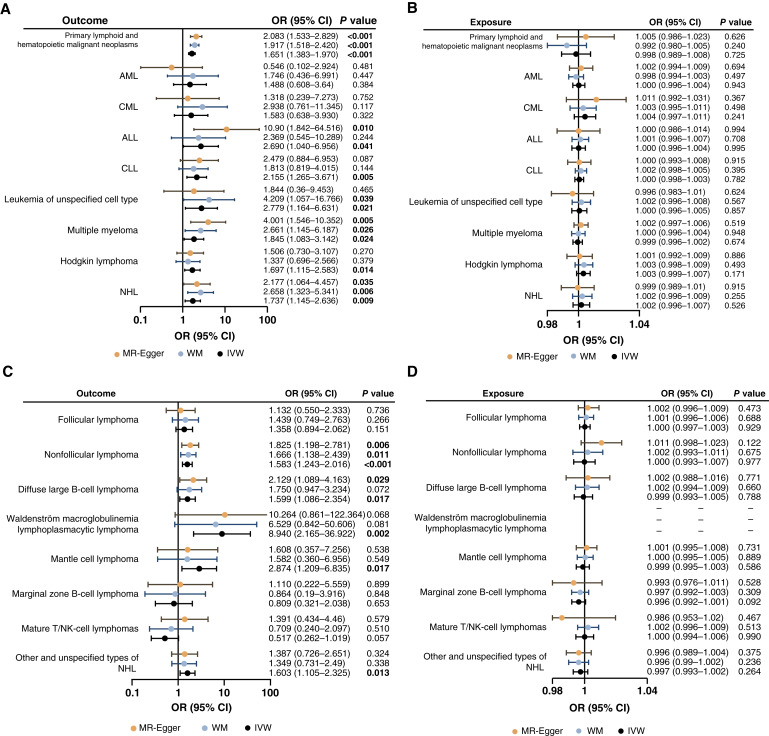
As shown in Fig. 2A and Table 1, the result of the IVW analysis demonstrated that TL was significantly associated with primary lymphoid and hematopoietic malignant neoplasms (OR = 1.650; 95% CI, 1.383–1.970; P = 2.69 × 10−8). In detail, IVW results indicated that TL was associated with leukemia of unspecified cell type (OR = 2.779; 95% CI, 1.164–6.631; P = 0.021) and lymphoid hematologic malignancies, including ALL (OR = 2.690; 95% CI, 1.040–6.956; P = 0.041), CLL (OR = 2.155; 95% CI, 1.265–3.671; P = 0.005), multiple myeloma (OR = 1.845; 95% CI, 1.083–3.142; P = 0.024), Hodgkin lymphoma (OR = 1.697; 95% CI, 1.115–2.583; P = 0.014), and NHL (OR = 1.737; 95% CI, 1.145–2.636; P = 0.009; Fig. 2A; Table 1; Supplementary Fig. S1). The results of the MR-Egger and WM methods displayed consistency (OR > 1), subsequently reinforcing the reliability of the analysis. On contrast, TL was not significantly associated with myeloid leukemia, including AML (OR = 1.488; 95% CI, 0.608–3.640; P = 0.384) and CML (OR = 1.583; 95% CI, 0.638–3.930; P = 0.322), as confirmed by MR-Egger and WM methods. As a result, longer TL was predicted to be related to an increased risk of primary lymphoid and hematopoietic malignant neoplasms, including lymphoid hematologic malignancies and leukemia of unspecified cell type.
Figure 2.
Forest plots showing the forward and reverse causal associations between TL and hematopoietic malignancies. The dots in this forest plot show the value of OR, and the horizontal lines demonstrate the range of 95% CI. A, Causal association between TL and different types of hematopoietic malignancies. B, Reverse causal association between TL and different types of hematopoietic malignancies. C, Causal association between TL and different types of NHL. D, Reverse causal association between TL and different types of NHL.
Table 1.
The forward MR analysis on TL and hematologic malignancies
| Outcome | nSNP | Methods | P value∗ | OR | CI (95%) | P (heterogeneity) | P (pleiotropy) | MR-PRESSO | |
|---|---|---|---|---|---|---|---|---|---|
| MR-Egger | IVW | ||||||||
| Primary lymphoid and hematopoietic malignant neoplasms | 116 | MR-Egger | 7.53E−6 | 2.083 | 1.533–2.829 | 0.000 | 0.000 | 0.073 | 0.008 |
| WM | 4.61E−8 | 1.917 | 1.518–2.420 | ||||||
| IVW | 2.69E−8 | 1.651 | 1.383–1.970 | ||||||
| AML | 112 | MR-Egger | 0.481 | 0.546 | 0.102–2.924 | 0.330 | 0.307 | 0.170 | 0.297 |
| WM | 0.447 | 1.746 | 0.436–6.991 | ||||||
| IVW | 0.384 | 1.488 | 0.608–3.640 | ||||||
| CML | 115 | MR-Egger | 0.752 | 1.318 | 0.239–7.273 | 0.096 | 0.106 | 0.804 | 0.112 |
| WM | 0.117 | 2.938 | 0.761–11.345 | ||||||
| IVW | 0.322 | 1.583 | 0.638–3.930 | ||||||
| ALL | 115 | MR-Egger | 0.010 | 10.901 | 1.842–64.516 | 0.748 | 0.692 | 0.071 | 0.713 |
| WM | 0.244 | 2.369 | 0.545–10.289 | ||||||
| IVW | 0.041 | 2.690 | 1.040–6.956 | ||||||
| CLL | 115 | MR-Egger | 0.087 | 2.479 | 0.884–6.953 | 0.943 | 0.950 | 0.756 | 0.956 |
| WM | 0.144 | 1.813 | 0.819–4.015 | ||||||
| IVW | 0.005 | 2.155 | 1.265–3.671 | ||||||
| Leukemia of unspecified cell type | 114 | MR-Egger | 0.465 | 1.844 | 0.360–9.453 | 0.879 | 0.887 | 0.562 | 0.882 |
| WM | 0.039 | 4.209 | 1.057–16.766 | ||||||
| IVW | 0.021 | 2.779 | 1.164–6.631 | ||||||
| Multiple myeloma | 117 | MR-Egger | 0.005 | 4.001 | 1.546–10.352 | 0.252 | 0.198 | 0.058 | 0.191 |
| WM | 0.026 | 2.661 | 1.145–6.187 | ||||||
| IVW | 0.024 | 1.845 | 1.083–3.142 | ||||||
| Hodgkin lymphoma | 118 | MR-Egger | 0.270 | 1.506 | 0.730–3.107 | 0.316 | 0.336 | 0.691 | 0.344 |
| WM | 0.379 | 1.337 | 0.696–2.566 | ||||||
| IVW | 0.014 | 1.697 | 1.115–2.583 | ||||||
| NHL | 117 | MR-Egger | 0.035 | 2.177 | 1.064–4.457 | 0.058 | 0.061 | 0.449 | 0.058 |
| WM | 0.006 | 2.658 | 1.323–5.341 | ||||||
| IVW | 0.009 | 1.737 | 1.145–2.636 | ||||||
∗Bold values indicate significant results.
The Cochran’s Q test, applied in conjunction with the IVW and MR-Egger methods, indicated that there was no heterogeneity in the IVs in the eight hematologic malignancies (P > 0.05), whereas there was heterogeneity in the primary lymphoid and hematopoietic malignant neoplasms (P < 0.05). Furthermore, the leave-one-out analysis demonstrated that the results remained stable regardless of the removal of any single SNP (Supplementary Fig. S2). As depicted in the funnel plot (Supplementary Fig. S3), the symmetrical distribution of effect size variations around the point estimate suggested that there was no discernible evidence of horizontal pleiotropy. Moreover, the MR-PRESSO analysis did not detect any potential instrumental outliers at the nominal significance level of 0.05 in the eight hematologic malignancies. Using the MR-Egger regression intercept method, we found no evidence of horizontal pleiotropy for TL in hematologic malignancies with P > 0.05.
The effect of hematologic malignancies on TL
The reverse causality bias may exist due to the potential impact on TL in hematologic malignancies and the possibility that an IV causes the outcome which in turn causes the exposure (27). Consequently, a reverse MR analysis was performed to exclude the possibility of reverse causality bias. The IVW analysis indicated no significant causal association between hematologic malignancies and TL (P > 0.05). The MR-Egger and WM methods were used as well, and the results concurred with the IVW analysis (Fig. 2B; Supplementary Fig. S4; Supplementary Table S2).
The MR-Egger regression, MR-PRESSO, and funnel plot analyses revealed no possible pleiotropy (P > 0.05). The Cochran’s Q test of IVW and MR-Egger methods confirmed no heterogeneity (P > 0.05) in the IVs (Supplementary Table S2). In conclusion, these results demonstrated that the causal relationship between TL and hematologic malignancies was stable and reliable, with no significant confounders and bias.
The associations between NHL and TL
In the preceding analysis, we discoursed a casual effect of TL on NHL, and we further investigated the relevance of TL for the classification of NHL. NHL accounts for 85% of all lymphoma cases and has several subtypes, including follicular lymphoma, nonfollicular lymphoma, diffuse large B-cell lymphoma, lymphoplasmacytic lymphoma (Waldenström macroglobulinemia), mantle cell lymphoma, marginal zone B-cell lymphoma, mature T/NK lymphomas, and other unspecified types. The effect of TL on the risk of developing each kind of lymphoma was further investigated. The results of the forward MR analysis suggested an association between TL and the risk of nonfollicular lymphoma (P = 1.98 × 10−4), diffuse large B-cell lymphoma (P = 0.017), lymphoplasmacytic lymphoma (Waldenström macroglobulinemia; P = 0.002), mantle cell lymphoma (P = 0.017), and other and unspecified types of NHL (P = 0.013; Fig. 2C; Table 2; Supplementary Fig. S5). The results indicated that longer TL was predicted to be associated with an elevated risk of developing these types of NHL. However, no significant association was observed between TL and the risk of follicular lymphoma (P = 0.151), marginal zone B-cell lymphoma (P = 0.653), or mature T/NK-cell lymphoma (P = 0.057).
Table 2.
The forward MR analysis on TL and the classification NHL
| Outcome | nSNP | Methods | P value∗ | OR | CI (95%) | P (heterogeneity) | P (pleiotropy) | MR-PRESSO | |
|---|---|---|---|---|---|---|---|---|---|
| MR-Egger | IVW | ||||||||
| Follicular lymphoma | 108 | MR-Egger | 0.736 | 1.132 | 0.550–2.333 | 0.068 | 0.073 | 0.547 | 0.083 |
| WM | 0.266 | 1.439 | 0.749–2.763 | ||||||
| IVW | 0.151 | 1.358 | 0.894–2.062 | ||||||
| Nonfollicular lymphoma | 110 | MR-Egger | 0.006 | 1.825 | 1.198–2.781 | 0.730 | 0.737 | 0.419 | 0.722 |
| WM | 0.011 | 1.666 | 1.138–2.439 | ||||||
| IVW | 1.98E−4 | 1.583 | 1.243–2.016 | ||||||
| Diffuse large B-cell lymphoma | 109 | MR-Egger | 0.029 | 2.129 | 1.089–4.163 | 0.791 | 0.789 | 0.309 | 0.807 |
| WM | 0.072 | 1.750 | 0.947–3.234 | ||||||
| IVW | 0.017 | 1.599 | 1.086–2.354 | ||||||
| Waldenström macroglobulinemia, lymphoplasmacytic lymphoma | 110 | MR-Egger | 0.068 | 10.264 | 0.861–122.364 | 0.068 | 0.077 | 0.894 | 0.075 |
| WM | 0.081 | 6.529 | 0.842–50.606 | ||||||
| IVW | 0.002 | 8.940 | 2.165–36.922 | ||||||
| Mantle cell lymphoma | 110 | MR-Egger | 0.538 | 1.608 | 0.357–7.256 | 0.743 | |||
| WM | 0.549 | 1.582 | 0.360–6.956 | 0.740 | 0.743 | 0.358 | |||
| IVW | 0.017 | 2.874 | 1.209–6.835 | ||||||
| Marginal zone B-cell lymphoma | 109 | MR-Egger | 0.899 | 1.110 | 0.222–5.559 | 0.227 | 0.243 | 0.639 | 0.250 |
| WM | 0.848 | 0.864 | 0.190–3.916 | ||||||
| IVW | 0.653 | 0.809 | 0.321–2.038 | ||||||
| Mature T/NK-cell lymphomas | 110 | MR-Egger | 0.579 | 1.391 | 0.434–4.460 | 0.379 | |||
| WM | 0.510 | 0.709 | 0.240–2.097 | 0.465 | 0.382 | 0.044 | |||
| IVW | 0.057 | 0.517 | 0.262–1.019 | ||||||
| Other and unspecified types of NHL | 110 | MR-Egger | 0.324 | 1.387 | 0.726–2.651 | 0.907 | 0.915 | 0.594 | 0.915 |
| WM | 0.338 | 1.349 | 0.731–2.490 | ||||||
| IVW | 0.013 | 1.603 | 1.105–2.325 | ||||||
∗Bold values indicate significant results.
We performed a MR reverse analysis to exclude the possibility of reverse causation. The findings indicated that the onset of follicular lymphoma, nonfollicular lymphoma, diffuse large B-cell lymphoma, mantle cell lymphoma, marginal zone B-cell lymphoma, mature T/NK-cell lymphomas, and other and unspecified types of NHL did not lead to changes in TL (Fig. 2D; Supplementary Fig. S6; Supplementary Table S3). However, an exception was observed in Waldenström macroglobulinemia (lymphoplasmacytic lymphoma), which yielded no results due to the absence of SNPs (Supplementary Table S3).
The results of the leave-one-out analysis exhibited stability (Supplementary Fig. S7), and upon investigation of heterogeneity and pleiotropy, the results conclusively demonstrated that there was no evidence of either heterogeneity or horizontal pleiotropy in both the forward and reverse MR analyses (Supplementary Fig. S8; Supplementary Table S3; P > 0.05).
Replicated validation analysis
To further validate the effectiveness of the results in this study across different populations and datasets, we conducted MR analysis using GWAS data from the UK Biobank. The results showed that TL was significantly associated with leukemia (OR = 1.003; P = 2.35 × 10−6) and lymphoid leukemia (OR = 1.002; P = 1.45 × 10−8) but not associated with myeloid leukemia (Supplementary Table S4; P = 0.316), which aligns with the results above.
Discussion
The relationship between TL and cancer risk has been the focus of various arguments in epidemiologic research, with inconsistent findings across cancer types (28). In this study, we conducted a MR analysis to investigate the association between TL and hematologic malignancies, ultimately uncovering a correlation between longer TL and an increased risk of certain hematologic malignancies.
TL plays a crucial role in maintaining genomic integrity and preventing the onset of cellular senescence or apoptosis. Numerous studies have suggested that TL may serve as a potential predictor of risk for hematologic malignancies. Studies have discovered that genetic variants associated with longer TL increase the risk of developing AML, CML, CLL, and NHL (7–10). However, some studies have contrasting results, indicating that individuals with shorter TL may have a higher risk of AML, ALL, CML, CLL, NHL, and other hematologic malignancies (11–13, 16–18).The reason for the contrasting results may be that traditional observational studies could be influenced by residual confounding, as they may be prone to bias from reverse causality and influenced by TL-related demographic and lifestyle factors. Additionally, studies have indicated that TL may undergo modifications, such as lengthening or shortening, following the onset of hematologic malignancies (29, 30). Consequently, retrospective studies face further limitations due to bias, as TL measurements are often conducted after diagnosis and/or treatment.
To address these concerns, we used the MR analysis approach, which differs significantly from observational studies. Our MR analysis enabled us to study the causal relationship between TL and hematologic malignancies, and the results were less prone to confounding factors and reverse causation than traditional observational methods. Similarly, two MR analyses independently corroborated a significant association between longer TL and an increased risk of hematopoietic malignancies (31, 32). Our research expands on this by investigating a broader spectrum of hematopoietic malignancies, using bidirectional MR analysis to eliminate potential causal reverse relationships, and subsequently validating these findings in an additional population. In this study, we found that TL was associated exclusively with lymphoid and hematopoietic malignant neoplasms, solely in the context of lymphatic system cancers, including ALL, CLL, Hodgkin lymphoma, and NHL (nonfollicular lymphoma, diffuse large B-cell lymphoma, lymphoplasmacytic lymphoma, and mantle cell lymphoma). The utilization of diverse MR methods, along with the detection of heterogeneity and pleiotropy, and replicated validations ensures the reliability of the results. However, there was heterogeneity in primary lymphoid and hematopoietic malignant neoplasms, potentially attributed to their encompassing of a wide spectrum of hematologic malignancies.
TL is a complex trait that is influenced by an intricate interplay of individual genetics, age, gender, epigenetics, environmental exposures, and a myriad of uncertain interacting factors (33). As cells divide, telomeres progressively shorten, and once a critical threshold is reached, they often trigger cellular senescence or apoptosis. During carcinogenesis, cells adopt pathways that enhance telomerase expression through telomerase reverse transcriptase (TERT) and/or utilize the alternative lengthening of telomeres (ALT) pathway to extend telomeres and continue dividing (34). A larger clonal expansion due to long TL would increase the risk of acquiring mutations and ultimately cause malignant transformation (28). Multiple genetic and epigenetic mechanisms, including amplifications, structural variants, promoter mutations, and promoter methylation (epigenetic modification), have been shown to induce TERT expression (33). ALT functions independently of telomerase and relies somewhat on homologous recombination (35). Disruptions in the regulation of TERT and ALT can lead to variations in TL among individuals, thereby increasing the risk of mutations and malignant transformation in hematopoietic malignancies.
We observed a significant correlation between TL and hematologic malignancies, which helped us grasp the malignancy–TL association. Our findings indicate that TL is linked to the risk of specific hematologic malignancies, suggesting its potential as a risk prediction tool or as a target for disease prevention interventions. Moreover, some studies indicate that lifestyle characteristics such as cigarette smoking (36, 37), diet (38, 39), physical activity (40, 41), and body mass index (42) are correlated with TL, which encourages people to adopt healthier lifestyles. TL maintenance may serve as a promising strategy to reduce the risk of hematologic malignancies through lifestyle modifications or targeted TL management. Furthermore, targeting telomere maintenance represents a compelling opportunity for malignancy treatment, though further investigation is required (43).
However, there were some limitations in this study. First, the genetic relationship between TL and the risk of developing hematologic malignancies was investigated in individuals of European descent, thereby potentially limiting its generalizability to other ethnicities. Second, the sample size of certain hematologic malignancies in the FinnGen database was relatively small, and the subtypes of leukemia in the UK Biobank have not been further differentiated. Consequently, the findings should be further validated in larger, ethnically diverse cohorts to ensure their generalizability and reliability. Last, the approach we used relies on the three fundamental assumptions underlying MR analysis, and failures of these hypotheses may distort the degree to which the association between genetic longitude and sickness is established.
In conclusion, we found that longer TL was associated with an increased risk of specific hematologic malignancies, emphasizing the crucial role of telomeres in tumor development. Consequently, individuals with elongated TL should maintain heightened awareness of their susceptibility to lymphatic system cancers. The precise role and underlying mechanisms of telomere in the risk of hematologic malignancies, as well as the efficacy of telomere maintenance in preventing and treating these malignancies, are crucial areas that require further investigation.
Supplementary Material
Supplemental Figure 1. Scatter plot of single nucleotide polymorphism potential effects on TL and hematopoietic malignancies.
Supplemental Figure 2. Forest plot of the results of the leave-one-out analysis for forward MR analysis between TL and hematologic malignancies.
Supplemental Figure 3. Funnel plot for TL displays the estimation obtained through the utilization of the inverse of the standard error of the causal estimate, with each individual SNP serving as a tool.
Supplemental Figure 4. Scatter plot of single nucleotide polymorphism potential effects on hematopoietic malignancies and TL.
Supplemental Figure 5. Scatter plot of single nucleotide polymorphism potential effects on TL and non-Hodgkin lymphoma.
Supplemental Figure 6. Scatter plot of single nucleotide polymorphism potential effects on non-Hodgkin lymphoma and TL.
Supplemental Figure 7. Forest plot of the results of the leave-one-out analysis for forward MR analysis between TL and non-Hodgkin lymphoma.
Supplemental Figure 8. Funnel plot for TL displays the estimation obtained through the utilization of the inverse of the standard error of the causal estimate, with each individual SNP serving as a tool.
Supplemental Table 1. GWAS data information of TL and hematologic malignancies
Supplemental Table 2. The reverse MR analysis on telomere length and hematologic malignancies
Supplemental Table 3. The reverse MR analysis on telomere length and the classification non-Hodgkin lymphoma
Supplemental Table 4. The Replicated MR analysis on telomere length and hematologic malignancies
Supplemental Figure Legend
Acknowledgments
This study was supported by Shandong Provincial Natural Science Foundation, Grant/Award Number: ZR2023QH062.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
No disclosures were reported.
Authors’ Contributions
G. Jiang: Data curation, formal analysis, funding acquisition, writing–original draft. L. Cao: Software, investigation, methodology, writing–review and editing. Y. Wang: Writing–review and editing. L. Li: Writing–review and editing. Z. Wang: Writing–review and editing. H. Zhao: Writing–review and editing. Y. Qiu: Conceptualization, writing–review and editing. B. Feng: Conceptualization, supervision, writing–review and editing.
Ethics Approval and Consent to Participate
This study was based on publicly available summary statistics, and all original GWAS studies were approved by the ethics committee.
References
- 1. Bosch de Basea Gomez M, Thierry-Chef I, Harbron R, Hauptmann M, Byrnes G, Bernier MO, et al. Risk of hematological malignancies from CT radiation exposure in children, adolescents and young adults. Nat Med 2023;29:3111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Larsson SC, Burgess S. Appraising the causal role of smoking in multiple diseases: a systematic review and meta-analysis of Mendelian randomization studies. EBioMedicine 2022;82:104154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pogribny IP, Beland FA. DNA methylome alterations in chemical carcinogenesis. Cancer Lett 2013;334:39–45. [DOI] [PubMed] [Google Scholar]
- 4. Revy P, Kannengiesser C, Bertuch AA. Genetics of human telomere biology disorders. Nat Rev Genet 2023;24:86–108. [DOI] [PubMed] [Google Scholar]
- 5. Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 2017;18:175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lansdorp PM. Telomeres, aging, and cancer: the big picture. Blood 2022;139:813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sullivan SM, Cole B, Lane J, Meredith JJ, Langer E, Hooten AJ, et al. Predicted leukocyte telomere length and risk of myeloid neoplasms. Hum Mol Genet 2023;32:2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ojha J, Codd V, Nelson CP, Samani NJ, Smirnov IV, Madsen NR, et al. Genetic variation associated with longer telomere length increases risk of chronic lymphocytic leukemia. Cancer Epidemiol Biomarkers Prev 2016;25:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wysoczanska B, Dratwa M, Gebura K, Mizgala J, Mazur G, Wrobel T, et al. Variability within the human TERT gene, telomere length and predisposition to chronic lymphocytic leukemia. Onco Targets Ther 2019;12:4309–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Machiela MJ, Lan Q, Slager SL, Vermeulen RC, Teras LR, Camp NJ, et al. Genetically predicted longer telomere length is associated with increased risk of B-cell lymphoma subtypes. Hum Mol Genet 2016;25:1663–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghaffari SH, Shayan-Asl N, Jamialahmadi AH, Alimoghaddam K, Ghavamzadeh A. Telomerase activity and telomere length in patients with acute promyelocytic leukemia: indicative of proliferative activity, disease progression, and overall survival. Ann Oncol 2008;19:1927–34. [DOI] [PubMed] [Google Scholar]
- 12. Schratz KE, Haley L, Danoff SK, Blackford AL, DeZern AE, Gocke CD, et al. Cancer spectrum and outcomes in the Mendelian short telomere syndromes. Blood 2020;135:1946–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohyashiki K, Iwama H, Tauchi T, Shimamoto T, Hayashi S, Ando K, et al. Telomere dynamics and genetic instability in disease progression of chronic myeloid leukemia. Leuk Lymphoma 2000;40:49–56. [DOI] [PubMed] [Google Scholar]
- 14. Karow A, Haubitz M, Oppliger Leibundgut E, Helsen I, Preising N, Steiner D, et al. Targeting telomere biology in acute lymphoblastic leukemia. Int J Mol Sci 2021;22:6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. da Mota THA, Camargo R, Biojone ER, Guimarães AFR, Pittella-Silva F, de Oliveira DM. The relevance of telomerase and telomere-associated proteins in B-acute lymphoblastic leukemia. Genes 2023;14:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steinbrecher D, Jebaraj BMC, Schneider C, Edelmann J, Cymbalista F, Leblond V, et al. Telomere length in poor-risk chronic lymphocytic leukemia: associations with disease characteristics and outcome. Leuk Lymphoma 2018;59:1614–23. [DOI] [PubMed] [Google Scholar]
- 17. Jebaraj BM, Kienle D, Lechel A, Mertens D, Heuberger M, Ott G, et al. Telomere length in mantle cell lymphoma. Blood 2013;121:1184–7. [DOI] [PubMed] [Google Scholar]
- 18. Widmann TA, Herrmann M, Taha N, König J, Pfreundschuh M. Short telomeres in aggressive non-Hodgkin's lymphoma as a risk factor in lymphomagenesis. Exp Hematol 2007;35:939–46. [DOI] [PubMed] [Google Scholar]
- 19. Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, et al. Mendelian randomization. Nat Rev Methods Primers 2022;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haycock PC, Burgess S, Nounu A, Zheng J, Okoli GN, Bowden J, et al. Association between telomere length and risk of cancer and non-neoplastic diseases: a Mendelian randomization study. JAMA Oncol 2017;3:636–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedron S, Kurz CF, Schwettmann L, Laxy M. The effect of BMI and type 2 diabetes on socioeconomic status: a two-sample multivariable Mendelian randomization study. Diabetes Care 2021;44:850–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Larsson SC, Burgess S, Michaëlsson K. Smoking and stroke: a Mendelian randomization study. Ann Neurol 2019;86:468–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023;613:508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burgess S, Thompson SG; CRP CHD Genetics Collaboration . Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 2011;40:755–64. [DOI] [PubMed] [Google Scholar]
- 25. Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 2016;32:3207–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019;35:4851–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aviv A, Anderson JJ, Shay JW. Mutations, cancer and the telomere length paradox. Trends Cancer 2017;3:253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roka K, Solomou EE, Kattamis A. Telomere biology: from disorders to hematological diseases. Front Oncol 2023;13:1167848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nogueira BMD, Machado CB, Montenegro RC, DE Moraes MEA, Moreira-Nunes CA. Telomere length and hematological disorders: a review. In Vivo 2020;34:3093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fang T, Zhang Z, Ren K, Zou L. Genetically determined telomere length as a risk factor for hematological malignancies: evidence from Mendelian randomization analysis. Aging (Albany NY) 2024;16:4684–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Liu Q, Liang S, Yao M, Zheng H, Hu D, et al. Genetically predicted telomere length and the risk of 11 hematological diseases: a Mendelian randomization study. Aging (Albany NY) 2024;16:4270–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsatsakis A, Oikonomopoulou T, Nikolouzakis TK, Vakonaki E, Tzatzarakis M, Flamourakis M, et al. Role of telomere length in human carcinogenesis (Review). Int J Oncol 2023;63:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Savage SA. Telomere length and cancer risk: finding Goldilocks. Biogerontology 2024;25:265–78. [DOI] [PubMed] [Google Scholar]
- 35. Sobinoff AP, Pickett HA. Alternative lengthening of telomeres: DNA repair pathways converge. Trends Genet 2017;33:921–32. [DOI] [PubMed] [Google Scholar]
- 36. Astuti Y, Wardhana A, Watkins J, Wulaningsih W; PILAR Research Network . Cigarette smoking and telomere length: a systematic review of 84 studies and meta-analysis. Environ Res 2017;158:480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park S, Kim SG, Lee S, Kim Y, Cho S, Kim K, et al. Causal linkage of tobacco smoking with ageing: Mendelian randomization analysis towards telomere attrition and sarcopenia. J Cachexia Sarcopenia Muscle 2023;14:955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crous-Bou M, Molinuevo JL, Sala-Vila A. Plant-rich dietary patterns, plant foods and nutrients, and telomere length. Adv Nutr 2019;10(Suppl_4):S296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Canudas S, Becerra-Tomás N, Hernández-Alonso P, Galié S, Leung C, Crous-Bou M, et al. Mediterranean diet and telomere length: a systematic review and meta-analysis. Adv Nutr 2020;11:1544–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mundstock E, Zatti H, Louzada FM, Oliveira SG, Guma FT, Paris MM, et al. Effects of physical activity in telomere length: systematic review and meta-analysis. Ageing Res Rev 2015;22:72–80. [DOI] [PubMed] [Google Scholar]
- 41. Güneşliol BE, Karaca E, Ağagündüz D, Acar ZA. Association of physical activity and nutrition with telomere length, a marker of cellular aging: a comprehensive review. Crit Rev Food Sci Nutr 2023;63:674–92. [DOI] [PubMed] [Google Scholar]
- 42. Müezzinler A, Zaineddin AK, Brenner H. Body mass index and leukocyte telomere length in adults: a systematic review and meta-analysis. Obes Rev 2014;15:192–201. [DOI] [PubMed] [Google Scholar]
- 43. Gao J, Pickett HA. Targeting telomeres: advances in telomere maintenance mechanism-specific cancer therapies. Nat Rev Cancer 2022;22:515–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Scatter plot of single nucleotide polymorphism potential effects on TL and hematopoietic malignancies.
Supplemental Figure 2. Forest plot of the results of the leave-one-out analysis for forward MR analysis between TL and hematologic malignancies.
Supplemental Figure 3. Funnel plot for TL displays the estimation obtained through the utilization of the inverse of the standard error of the causal estimate, with each individual SNP serving as a tool.
Supplemental Figure 4. Scatter plot of single nucleotide polymorphism potential effects on hematopoietic malignancies and TL.
Supplemental Figure 5. Scatter plot of single nucleotide polymorphism potential effects on TL and non-Hodgkin lymphoma.
Supplemental Figure 6. Scatter plot of single nucleotide polymorphism potential effects on non-Hodgkin lymphoma and TL.
Supplemental Figure 7. Forest plot of the results of the leave-one-out analysis for forward MR analysis between TL and non-Hodgkin lymphoma.
Supplemental Figure 8. Funnel plot for TL displays the estimation obtained through the utilization of the inverse of the standard error of the causal estimate, with each individual SNP serving as a tool.
Supplemental Table 1. GWAS data information of TL and hematologic malignancies
Supplemental Table 2. The reverse MR analysis on telomere length and hematologic malignancies
Supplemental Table 3. The reverse MR analysis on telomere length and the classification non-Hodgkin lymphoma
Supplemental Table 4. The Replicated MR analysis on telomere length and hematologic malignancies
Supplemental Figure Legend
Data Availability Statement
The datasets analyzed during the current study are available in the FinnGen (www.finngen.fi/en) and IEU OpenGWAS (gwas.mrcieu.ac.uk/) databases.