Abstract
The global spread of multi-drug-resistant (MDR) bacteria is rapidly increasing due to antibiotic overuse, posing a major public health threat and causing millions of deaths annually. The present study explored the potential of nanocarriers for delivering novel and alternative antibacterial agents using nanotechnology-based approaches to address the challenge of MDR bacteria. The purpose was to enhance the solubility, stability, and targeted delivery of berberine (BER) and its synthetic derivative NR16 using Styrene-co-Maleic Acid (SMA) nanoparticles. Characterization of the nanoparticles, including dynamic light scattering (DLS) analysis, TEM, and UV/Vis absorption spectroscopy, confirmed their suitability and high stability for passive drug delivery. Antibacterial and antifungal activities were evaluated against a panel of pathogens, revealing significant inhibitory effects on Gram-positive strains; particularly BER, SMA-BER, and NR16 were active against MRSA, MSSA, VR, and VS E. faecalis, and S. epidermidis. Additionally, SMA-BER and SMA-NR16 showed promising activity against biofilm formation of S. epidermidis; while the two free drugs contributed to S. epidermidis biofilm disruption activity. Hemolysis tests and in vitro studies on human embryonic kidney cells (HEK-293) confirmed the safety profiles of the nanoparticles and free drugs. Overall, this research highlighted the potential of nanotechnology in developing effective antibacterial agents with reduced toxicity, addressing the growing threat of MDR bacterial infections.
The produced SMA-BER and SMA-NR16 nanomicelles proved to be a valid and safe tool to contrast bacterial infections and biofilm formation.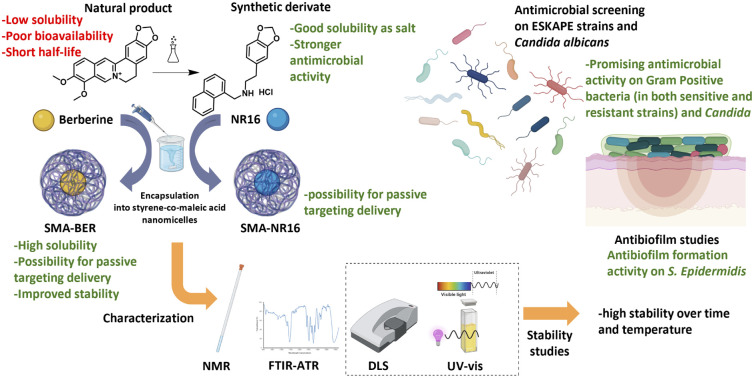
Introduction
Multidrug-resistant (MDR) bacteria are so called for their ability to resist the action of commonly used antimicrobials. Worldwide, the spread of MDR bacteria is worryingly rising due to uncritical use of antibiotics and is quickly becoming one of the major public health threats that is responsible for millions of deaths globally every year.1 Different mechanisms are involved in the development of MDR, such as drug inactivation, drug uptake reduction, genetic modification of drug targets, and drug efflux. Another mechanism of antibacterial resistance is biofilm formation.2 Biofilm formation is regulated by quorum sensing (QS), a specific type of communication system related to cell density dependent on the production of compounds defined as autoinducers. After exceeding a threshold value, the autoinducer can bind to its specific receptor and modulate the transcription or repression of virulence factors. Biofilm life cycle occurs in four stages that include adhesion, microcolony formation, growth and maturation, and finally detachment of bacteria to start a new cycle.3 The consequent formation of a matrix composed of extracellular polymeric substances (EPS) maintains nutrients and solutes diffusion needed for survival, but at the same time behaves as impenetrable barrier to larger compounds, such as most antibiotics, causing severe and difficult to treat infections.4 Furthermore, proliferation in non-living static surfaces such as catheters and prosthesis can also lead to alarming nosocomial infections.5 Establishment of biofilm may require up to 1000-fold higher concentrations of antibacterial than the normal used dosage against planktonic form. Consequently, it makes it highly difficult to cure infection by the systemic antibiotic administration.
As part of their continuous efforts in prioritizing and coordinating worldwide research towards the discovery of novel antibacterial agents, the World Health Organization (WHO) recently categorized MDR bacteria into different priority levels: critical, high, and medium, and published an overview of antibacterial clinical development pipeline.6 In this evolving panorama, bacterial strains of greatest concern are the Gram-negative Pseudomonas aeruginosa carbapenem-resistant, the Acinetobacter baumannii carbapenem-resistant and the Gram-positive methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pneumoniae, and vancomycin-resistant Enterococcus faecium (VRE), which all can lead to severe and difficult-to-treat infections. Moreover, most of the recently approved agents showed a total or partial lack of the four WHO innovation criteria including new chemical class, new target, new mechanism of action (MoA), and absence of known cross-resistance.7
Natural substances deriving from three different sources—plants, animals, and microorganisms—may have significant interest for antibacterial applications.8 Natural compounds exhibiting antibacterial activity and biocompatibility have gained attention due to increasing concerns about the safety of synthetic compounds and emerging drug resistance phenomena. In particular, medicinal plants represent one of the earliest medications by virtue of their secondary metabolites; limitless relevant bioactive substances, including many antibiotics, have been sourced from natural extracts.9 Nowadays phytochemicals still represent a wide source of derivatives endowed with strong biological activities and can be considered as novel substances and/or novel chemotypes in the fight against MDR bacteria, by targeting microbial virulence.10,11 Regrettably, phytochemicals also display several drawbacks, as their isolation and characterization is often challenging, the chemical synthesis is hardly adapted to large-scale production owing to their structural complexity, and they often exhibit an unfavorable pharmacokinetic profile.
Berberis vulgaris has long been known in folk medicine for its pleiotropic activities such as lipid-lowering and insulin-resistance improving actions, cardiovascular, antitumoral, and antibacterial activities.12 Most of these beneficial actions can be attributed to berberine (BER), one of the main secondary metabolites of B. vulgaris (Fig. 1). BER, found in roots, rhizomes, stems, and bark, possesses an isoquinoline structure belonging to the structural class of protoberberines. BER antibacterial actions have been related to interactions with bacterial membrane and DNA, and to inhibition of the filamentous temperature-sensitive Z protein (FtsZ), a well-known bacterial protein involved in cell division. In addition, BER has been shown to prevent formation of S. epidermidis and K. pneumoniae biofilm.10,13 Unfortunately, the clinical use of BER has been hampered by its very low water solubility, bioavailability (less than 1%), and short half-life.14,15 Several structural analogues of BER have been reported over the year with the aim of improving the pharmacokinetic and pharmacodynamic profile of BER; among them, an interesting series of flexible and structurally simplified BER analogues has been recently reported. Particularly, NR16 showed an interesting antibacterial activity against Gram-positive S. aureus and E. faecalis, and Gram-negative E. coli and K. pneumoniae (Fig. 1).13 However, limitations in the pharmacokinetic or pharmacodynamic profiles were shown by most of BER natural and synthetic analogues.
Fig. 1. Chemical structure of berberine (BER) and NR16.
Nanoparticle formulations have also been identified as a high-value option for the treatment of MDR bacteria.16 The application of nanostructured and/or supramolecular systems is particularly prominent in both pharmacological and analytical fields.17,18 The use of nanotechnology-based approaches to encapsulate and deliver BER and NR16 to the site of action can ameliorate their water solubility, stability, and targeting ability as well as improve their antibacterial activity. In this study, the Styrene-co-Maleic Acid (SMA) amphiphilic polymer was chosen for nanoparticle system formation, given its demonstrated history of safety and passive targeting efficacy for both tumors and inflamed tissues. Its unique amphiphilic nature makes it ideal for constructing nanosized polymeric carriers designed to enhance the parameters of bioactive molecules like BER and NR16. By encapsulating these agents within SMA nanoparticles, we aim to improve their solubility, stability, safety, and targeted delivery, paving the way for novel antibacterial therapeutic options. This approach leverages SMA's established track record in nanomedicine while offering the potential for tailored drug delivery with minimized side effects.
Experimental
Styrene-co-maleic anhydride with a number average molecular mass (Mn) ∼1600 was from Sigma-Aldrich, berberine hydrochloride was from TCI Europe (Switzerland), N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDAC) was obtained from Sigma-Aldrich Corp (St Louis, MO, USA). NR16 was synthesized as previously reported.13 HPLC system consists of a Vanquish (Thermo Fisher) equipped with two pumps, an autosampler, an online degasser, a PDA and a column compartment with temperature control. Data acquisition, analysis, and reporting were performed using the Xcalibur software (Thermo Fisher). The analysis was conducted using a reverse phase C18 column (XDB Agilent) with a 1.8 μm particle size, 4.6 mm internal diameter, and 50 mm length. An Agilent 8453 UV-Vis spectrophotometer (Agilent, Waldbronn, Germany) was used to acquire UV-Vis absorption spectra. The Fourier transform infrared (FTIR) spectral analysis was carried out on a Frontier FT-IR spectrometer (PerkinElmer) with PerkinElmer Spectrum software (version 10.4.2.279). 1H-NMR spectra were recorded on a Varian Unity Inova 500 MHz spectrometer. The chemical shifts are reported as δ (ppm) referenced to the resonance of residual HOD. Coupling constants (J) are given in Hz. Signal multiplicities are characterized as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), br (broad). For antimicrobial activity studies, the ATCC strains were obtained by the American Type Culture Collection (Manassas, VA). MacConkey NO. 3 agar (cat. no. CM0115, OXOID), MSA (cat. no. CM0085, OXOID), Sabouraud dextrose agar (cat. no. CM0041, OXOID), and TSB (cat. no. CM0129, OXOID) were purchased from ThermoFisher Scientific (Segrate, MI – Italy). Cation adjusted Mueller Hinton broth (cat. no. 212322) was purchased from Becton Dickinson (New Jersey – USA), whilst RPMI (cat. no. R6504), MOPS (cat. no. M3183), Glucose (cat. no. G5767), and DMSO (cat. no. D8418) were obtained from Sigma-Aldrich/Merck (Darmstadt – Germany). Human HEK-293 embryonic kidney cell line ATCCC number CRL-1573. Advanced RPMI 1640 (1×) growth medium was purchased from Gibco® by Life Technologies™, USA; fetal bovine serum (FBS) was purchased from SIGMA life sciences, USA, penicillin–streptomycin antibiotics were purchased from SIGMA life sciences, USA, and l-glutamine 200 mM (100×) was purchased from Gibco® by life technologies™, USA. All solvents were reagent grade and were purchased from commercial vendors (Sigma-Aldrich, Merck).
Nanoparticles formulation and characterization
SMA-hydrolysis
For SMA hydrolysis a well-known protocol was followed.19 Briefly, 2 g of SMA suspended in 200 mL of 1 M NaOH in water were heated at 70 °C under continuous stirring until the suspension turned into a clear solution.
SMA-BER formulation
SMA-BER preparation was accomplished by using a slightly adjusted protocol previously reported.19 Briefly, 100 mL of hydrolysed SMA (10 mg mL−1) were diluted to a final volume of 200 mL using deionized water and left under stirring into a beaker. The pH of the solution was taken at pH 5 using 1 M HCl and 0.1 M HCl for fine-tuning until stabilization. Meanwhile, 200 mg of BER were dissolved into 5 mL of DMSO until forming a clear solution, and 1 g of EDAC·HCl were dissolved into 5 mL of deionized water. The BER and EDAC·HCl solutions were added dropwise to the SMA solution, maintaining the pH stable at 5 for 30 min after the addition. Subsequently, the pH of the solution was raised to pH 11 using 1 M NaOH and 0.1 M NaOH for fine-tuning, under stirring for 1 h. After that time, the pH of the solution was lowered to pH 2 using 1 M HCl to allow the precipitation of the formed SMA-BER. The precipitated nanomicelles were centrifuged and washed 3 times with a solution of 0.001 M HCl. Afterwards, the nanomicelles were resuspended in deionized water and the pH was adjusted at 7.4. The solution was then purified using a dialysis bag to remove any residues of NaCl or urea products. The complete removal of the NaCl was evaluated by means of the Pharmacopeia chloride assay using HNO3 and AgNO3. The nanomicelles were freeze dried to obtain 0.947 mg of SMA-BER as a yellow powder (79% recovery).
SMA-NR16 formulation
A 100 mL solution of SMA (10 mg mL−1) hydrolysed as reported above was diluted to 100 mL using deionized water and let stir into a beaker. The pH of the solution was taken at pH 5 using 1 M HCl and 0.1 M HCl until stabilization. Meanwhile, 100 mg of NR16 were dissolved into 4 mL of DMSO until forming a clear solution, and 1 g of EDAC·HCl were dissolved into 4 mL of deionized water. The NR16 and EDAC·HCl solutions were added dropwise to the SMA solution, maintaining the pH stable at 5 for 30 min after the addition. Subsequently, the pH of the solution was adjusted to pH 11 using 1 M NaOH and 0.1 M NaOH for fine-tuning, letting it stir for 1 h. After that time, the solution was taken to pH 2 by using 1 M HCl, to precipitate the formed nanomicelles. The precipitated SMA-NR16 were centrifuged and washed 3 times with a solution of 0.001 M HCl. Subsequently, the nanomicelles were resuspended in deionized water and the pH was set at 7.4. The solution was then purified using a dialysis bag to remove any residues of NaCl or urea products. The complete removal of the NaCl was evaluated by means of the Pharmacopeia chloride assay using HNO3 and AgNO3 SMA-NR16 nanomicelles were further filtered with a 0.45 μm filter in order to remove the few small solid impurities and then freeze dried, obtaining 0.891 mg of a white powder of SMA-NR16 (74%).
Size, PDI, and zeta potential analysis
Zeta potential (ζ potential), average hydrodynamic diameter (DH, or size), and width of distribution (polydispersity index, PDI) measurements were carried out by dynamic light scattering (DLS) analysis using a Zetasizer Nano ZS (Malvern Instrument, Malvern, UK) at 25 °C in double-distilled water (DDW). Specifically, for SMA-BER and SMA-NR16, a sample of 1 mg of lyophilized micelles was dissolved in 10 mL of DDW to obtain appropriate dilution for analysis. Results are reported as the mean of three separate measurements ± the standard deviation (SD).
TEM analysis
To prepare SMA-BER and SMA-NR16 for the analysis, 10 μL of each nanoparticle solution (300 μg mL−1 in DDW) were placed on a copper grid covered by a carbon membrane. Then, samples were stained with phosphotungstic acidic solution (2% w/v) for 2 min, and analyzed at 120 kV by means of a JOEL microscope (JEM 2010, Japan). Images were acquired using a Gatan Orius camera (Gatan Inc, Pleasanton, CA, USA). ImageJ software (National Institutes of Health, MD, USA) was used to compute the mean diameter of the nanomicelles.
Loading calculation of SMA-BER and SMA-NR16 by HPLC
Chromatographic conditions
Chromatographic analyses were performed in the gradient mode.20 The eluting solutions consisting of: A (1 mM of CH3COONH4 in ultra-pure water), B (CH3CH), and C (CH3OH with 0.1% of CH3COOH) were pumped at a flow rate of 0.6 mL min−1. The gradient parameters were:
| Time (min) | % A | % B | % C |
|---|---|---|---|
| 0 | 80 | 10 | 10 |
| 10 | 10 | 80 | 10 |
| 11 | 10 | 80 | 10 |
| 13 | 80 | 10 | 10 |
| 15 | 80 | 10 | 10 |
UV detection was carried out at 226, 270, 340, and 424 nm for BER and 226, 270, 384 for NR16. Chromatograms were recorded using Xcalibur, a chromatography software for LC systems (Version 4.2). The sample injection volume was 2 μL, the method run time was 15 min and the column temperature was 40 °C.
Preparation of standard and sample solutions
A BER stock standard of 0.6 mg mL−1 was prepared in CH3OH and subsequent dilutions were carried out to obtain five standard solutions (17.8, 35.5, 44.9, 89.5, and 177 μM, respectively). Additionally, six standard solutions (2, 4, 6, 8, 10, and 12 μM, respectively) were obtained to determine the limit of quantification (LOQ) and the limit of detection (LOD).
A NR16 stock standard of 0.35 mg mL−1 was prepared in CH3OH and subsequent dilutions were carried out to obtain five standard solutions (33.0, 69.4, 138.8, 288.3, and 577.5 μM, respectively). Additionally, six standard solutions (1, 2, 4, 6, 8, and 10 μM, respectively) were obtained to determine the limit of quantification (LOQ) and the limit of detection (LOD).
All the standards and samples were filtered through a 0.22 μm pore-size filter (Millipore, Bedford, USA).
Method validation
The HPLC method was validated in terms of specificity, linearity, precision (intra-day), accuracy, LOD, and LOQ (according to the International Conference on Harmonization (ICH) guidelines). The specificity was evaluated by comparing the representative chromatograms of samples containing possible interfering substances (excipients used in nanoparticle composition) and BER standard. The identification of BER was based on comparison of retention times and UV spectra of peak of BER standard with the corresponding peak in the sample chromatogram. The same criteria were applied for the analysis of NR16. The linearity was determined by calculating a regression line from the plot of the peak area versus concentration for the five standard solutions in CH3OH using a linear least squares regression. Precision was assessed as repeatability. The repeatability of the measurements was assessed by testing three different standard solutions (35, 45, and 60 μM, respectively, n = 10) during the same day. The results were reported as the standard deviation (SD) and relative standard deviation (RSD). The accuracy was determined by calculating the percent recovery of the BER at three concentration levels and then determining the RSD. The mean concentration value obtained for each level was compared to the theoretical value, which was considered to be 100%. The LOD and LOQ were determined from the calibration curve obtained from standard solutions. The following equations were used according to ICH:LOD = 3.3 × σ/S,LOQ = 10 × σ/S,where σ is the standard deviation of the response and S is the slope of the calibration curve.
Linearity
BER linearity was evaluated at five concentration levels (17.8, 35.5, 44.9, 89.5, 177 μM, respectively) by calculating the following regression equation and the correlation coefficient (r) using the least squares method:
y = 8E−08x − 0.0011; r = 0.9999, while NR16 linearity was evaluated at five concentration levels (33.0, 69.4, 138.8, 288.3, 577.5 μM, respectively) by calculating the following regression equation and the correlation coefficient (r) using the least squares method:
y = 9E−05x − 3.4884; r = 0.9999, where y is the peak area and x is the standard solution concentration in μM.
Accuracy
Accuracy was evaluated by calculating the percent recovery of the average analyte concentration at three different concentrations. Three standard solutions (35, 45, and 60 μM, respectively) were carefully prepared in triplicate and analysed using the previously proposed method. The average percent recovery of BER from the samples was 74% (RSD = 2.43%, n = 9) and for NR16 72% (RSD = 2.49%, n = 9).
Precision
Precision expressed as RSD for repeatability was evaluated over three concentrations for both BER and NR16 (35, 45, and 60 μM, respectively); for each analyte, solutions were prepared in triplicate and analyzed over one day to assess intra-day variation. The RSDs of the responses were calculated for each case; the results indicate that precision was achieved because the maximum RSD value obtained was 0.63%.
Limit of quantification and limit of detection
The lowest concentration at which an analyte can be detected (LOD) or quantified (LOQ) with acceptable precision and accuracy was calculated from the SD of the response and the slope obtained from linear regression of the calibration curve. The LOD and LOQ for BER were found to be 1.2 and 3.5 μM, respectively. The LOD and LOQ for NR16 were found to be 0.18 and 0.54 μM, respectively.
Specificity
To evaluate the specificity of the method, chromatograms of the standards and samples of BER were compared with those of potential interfering components of the formulation. The supernatant obtained after centrifugation in the blank nanoparticle preparation (nondrug-loaded nanoparticles) was diluted in CH3OH and analyzed by the described HPLC method. The representative chromatogram of the BER sample (Fig. S1,† red line) showed the BER peak at about 7.3 min, in agreement with that obtained for the BER standard (Fig. S2,† blue line). No peaks were observed in the chromatogram of the supernatant of the blank test at the same retention time, indicating that there was no interference from the formulation components in the quantitative determination of BER.
Spectroscopic characterization of BER and SMA micelles
BER, NR16, SMA-BER, and SMA-NR16 UV-Vis absorption spectra were acquired from 190 nm to 800 nm. BER was dissolved in pure DMSO, while NR16, SMA-BER, and SMA-NR16 were dissolved in DDW. To have a direct comparison between free drug and encapsulated drug, dilutions were prepared considering the loading. For BER, the following concentrations were analyzed: 20, and 10 μg mL−1. For SMA-BER, were analyzed the following dilutions: 151, and 75 μg mL−1. On the other hand, for NR16 were analyzed the following concentrations: 20, 10, and 5 μg mL−1. Respectively, for SMA-NR16 were prepared and analyzed the following concentrations: 159, 79.5, and 39.74 μg mL−1.
1H-NMR analysis
The experiments were performed in D2O at 27 °C and the chemical shifts are reported as δ (ppm) referenced to the resonance of residual HOD.
FTIR and ATR analysis
The spectra of copolymers were recorded using an attenuated total reflection (ATR) technique in the spectral range 450–4000 cm−1 with a resolution of 2 cm−1 and accumulations of 10 scans which were combined to average out random absorption artifacts. Spectra of polystyrene (transmittance) was recorded and used as a reference for band assignments.
Stability studies
UV-Vis stability studies on SMA-BER and SMA-NR16
Stability studies were conducted upon SMA-BER and SMA-NR16 by UV-Vis spectrophotometry, analyzing their behavior in three different solvents: DDW, NaCl 0.9% w/v and 10 mM phosphate buffer, containing NaCl (137 mM) and KCl (2.7 mM) at pH 7.4 (PBS). SMA-BER stock solutions of 400 μg mL−1, and SMA-NR16 stock solutions of 100 μg mL−1 were achieved in different solvents, and samples were stored at 4 °C, r.t., and 37 °C until use. UV-Vis spectra were recorded for each sample at different time points, t = 0, 1, 3, 5, 7, and 14 days, respectively.
DLS stability studies on SMA-BER and SMA-NR16
A 50 μg mL−1 of SMA-BER and SMA-NR16 dispersions in DDW were prepared and stored at 4 °C, r.t., and 37 °C for 7 days. At prefixed timepoints (0, 4, and 7 days) the stored dispersions were analyzed with a Malvern Zetasizer Pro (Malvern Panalytical, United Kingdom) to detect eventual changes in the particle size, PDI, and zeta potential during times at different storage conditions.
Release studies
The release rate was measured by the dialysis bag method, as previously reported for other SMA nanoparticles.21 Briefly, 4 mg of SMA-BER or SMA-NR16 were dissolved in 4 mL of DDW and inserted into a sealed visking dialysis bag (MWCO 12–14 kDa; Medicell Technology – London, UK). Subsequently, the dialysis bag was fully immersed into a sealed container filled with 40 mL of PBS pH 7.4, which was kept at 37 °C and 100 osc min−1 for the entire duration of the experiment, 3 days. At prefixed timepoints (0, 2, 4, 6, 8, 24, 48, and 72 h) 1 mL of solution was taken from the outside of the dialysis bag and the absorbance was read with an Agilent 8453 UV-Vis spectrophotometer (Agilent, Waldbronn, Germany) at 422 nm for BER and 282 nm for NR16. The percentage of drug released was calculated by means of a calibration curve of the drug, plotting the obtained absorbance values and considering the initial amount of drug into the nanomicelles. All experiments were performed in triplicate. Percentage release was reported as mean percentage ± standard error.
Antibacterial studies
Microorganisms and growth conditions
The antibacterial and antibiofilm activity of the compounds was tested on nine ATCC bacterial strains representative of the ESKAPE group, one biofilm producer S. epidermidis, and one fungal ATCC strain.22 Selected microorganisms were E. coli ATCC 25922, K. pneumoniae ATCC BAA-2814, A. baumannii ATCC 179878, P. aeruginosa ATCC 27853, vancomycin susceptible (VS) E. faecalis ATCC 29212, vancomycin resistant (VR) E. faecalis ATCC 51299, methicillin susceptible (MS) S. aureus ATCC 12598 and MS S. aureus ATCC 25923, methicillin resistant (MR) S. aureus USA300, S. epidermidis ATCC 35984, and C. albicans ATCC 10231. Gram-negative and Gram-positive bacteria were grown at 37 (±1) °C overnight on MacConkey NO.3 and MSA, respectively. C. albicans strain was grown on Sabouraud dextrose agar at 37 (±1) °C for 24–48 hours. All strains were stored at −80 °C until use.
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)
MIC evaluation was performed according to EUCAST [The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 14.0, 2024. http://www.eucast.org] and CLSI [NCCLS. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard—Second Edition. NCCLS document M27-A2 [ISBN 1-56238-469-4]. NCCLS, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898 USA, 2002] guidelines for bacteria and Candida albicans, respectively, using Cation adjusted Mueller Hinton broth or RPMI with 34.5% (w/v) of MOPS (cat. no. M3183, Sigma-Aldrich/Merck, Darmstadt – Germany).
For each compound, a suspension equal to 20 000 mg L−1 of active principle was prepared in sterile H2O or DMSO for BER, diluted in the appropriate media to a concentration of 1024 mg L−1 and then transferred to 96-well microplates performing serial two-fold dilutions (higher DMSO concentration of 2.5%). Empty SMA was tested together with the other compounds.
To determinate MBC or minimum fungicidal concentration (MFC), the whole content (100 μL) from the wells equal to the MIC values and higher concentrations was streaked on agar plates.
MIC value was determined at the lowest concentrations of the antimicrobial agent inhibiting bacterial growth, MBC as the lowest concentration of each antimicrobial agent resulting in microbial death, as defined by a decrease of 103 CFU mL−1 in comparison to the starting inoculum. The MIC and MBC values were expressed in mg L−1. All assays were carried out in triplicate with independently grown cultures. The results are geometric means of three different experiments.23,24
Biofilm inhibition
The ability of the tested compounds to inhibit biofilm formation and to disaggregate mature biofilm was evaluated as previously published.25 Briefly, an overnight culture of S. epidermidis ATCC 35984 was diluted 1 : 100 in fresh TSB with glucose 0.25%, then transferred to the wells of a 96 multiwell plate and incubated overnight at 37 (±1) °C.
For the biofilm inhibition test, two-fold serial dilutions of the compounds were added to the plate at the same time of the bacterial inoculum. For biofilm disaggregation test, two-fold serial dilutions of the compounds were added to the plate after the overnight formation of the biofilm.
Subsequently, the broth was carefully removed from the wells and the plate was washed thrice with sterile physiologic solution. Wells were stained with a 0.4% crystal violet solution and washed again. The amount of biofilm produced in the well was evaluated by reading the optical density at 490 nm (OD490).26
Each test was performed in the presence of a positive biofilm formation control (inoculum without compounds) and a blank control (TSB with 0.25% glucose).
Biocompatibility screening
Hemolysis test on SMA-BER and BER
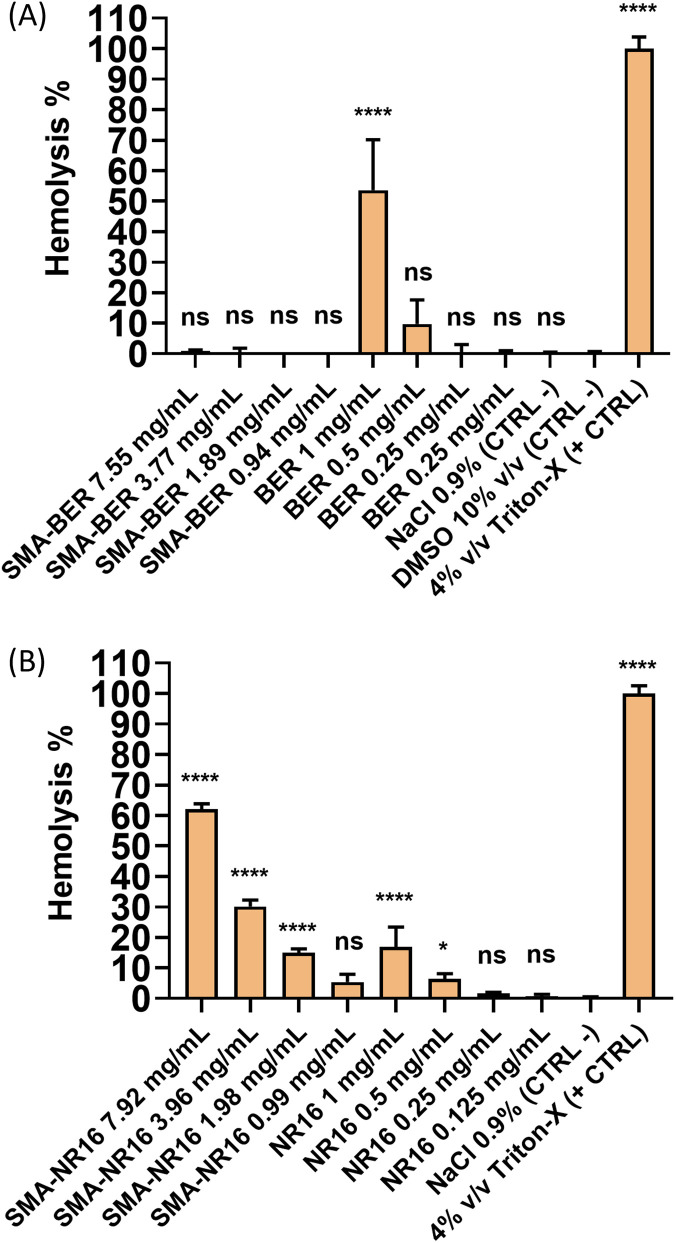
To evaluate the hemocompatibility of the SMA-BER and SMA-NR16 nanomicelles, as well as the free-drug BER and NR16, a hemolysis test was carried out according to previously reported protocols, with some modifications.27,28 Human blood anticoagulated with ethylene diamine tetraacetic acid (EDTA) was from Centro de Transfusión de Galicia (Santiago de Compostela, Spain). The blood was diluted with a NaCl 0.9% w/v solution to reach final 3.5% v/v diluted blood solution. A 10 mg mL−1 BER stock solution was prepared in DMSO and added to 900 μL of diluted blood to obtain final BER test concentrations of 1, 0.5, 0.25, and 0.125 mg mL−1 (DMSO max. conc. 10%). On the other hand, a 10 mg mL−1 NR16 stock solution in 50% DMSO/NaCl 0.9% w/v solvent mixture was prepared and added to 900 μL of diluted blood to obtain final concentrations of 1, 0.5, 0.25, and 0.125 mg mL−1 (DMSO max. conc. 5%). Conversely, SMA-BER and SMA-NR16 were dissolved in NaCl 0.9% w/v.
Thus, considering the loading of 13.24%, a SMA-BER stock solution of 75.5 mg mL−1 was achieved, and aliquots were added to 900 μL of diluted blood to obtain final SMA-BER test concentrations of 7.55, 3.77, 1.89, and 0.94 mg mL−1. For SMA-NR16, taking into account the loading of 12.63%, a stock solution of 79.2 mg mL−1 was prepared, and aliquots were added to 900 μL of blood to obtain final test concentrations of 7.92, 3.96, 1.98, and 0.99 mg mL−1. All test dilutions were prepared into 1.5 mL plastic tubes, and all the concentrations were prepared in order to directly compare nanomicelles to the free drug (same drug concentration). The samples were incubated for 1 h at 37 °C, being gently shaken at 100 osc min−1. Then, the sample tubes were centrifuged at 10 000 rpm for 10 min and the supernatant was transferred in a 96 well plate. The released hemoglobin, due to hemolytic activity, was measured by recording absorbance (Abs) of the supernatant using a plate reader (Fluostar Optima, BMG Labtech, Germany) at 550 nm. Triton X-100 4% v/v in DDW was used as the positive control (PC) (100% lysis), while DMSO 10% v/v, DMSO 5% in NaCl 0.9% w/v, and NaCl 0.9% w/v were used as negative controls (NC). Results, expressed as hemolysis percentage were obtained by the following equation: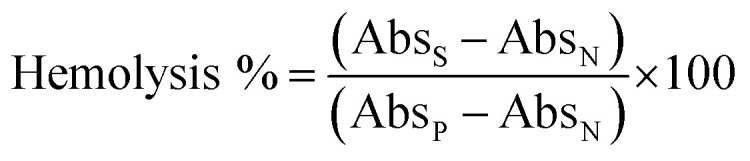 where, AbsS represents the absorbance derived from the tested concentrations hemolytic effect, AbsN is the absorbance of the negative control, while AbsP is the absorbance of the positive control.
where, AbsS represents the absorbance derived from the tested concentrations hemolytic effect, AbsN is the absorbance of the negative control, while AbsP is the absorbance of the positive control.
Toxicity evaluation of SMA-BER, SMA-NR16, BER, and NR16 in mammalian HEK-293 cell culture
Human HEK-293 embryonic kidney cell line was grown at 37 °C in 4.5% CO2 in Advanced RPMI 1640 (1×) growth medium supplemented with 5% (v/v) fetal bovine serum (FBS), penicillin-streptomycin antibiotics, and l-Glutamine 200 mM (100×). Cell passage was performed every 3–4 days and suspensions of HEK-293 cells were produced from confluent cultures using scraping method. Cell concentration was then determined using a hemocytometer where equal quantities of HEK-293 cells (6 × 103 cells per well) were seeded directly into two 96 well-plates. Drugs for testing were prepared as follows. Both free BER and free NR16 were dissolved in 20% (v/v) DMSO to a final concentration of 5.1 mg mL−1. Conversely, SMA-BER and SMA-NR16 were dissolved in DDW and brought to the same final concentration of the active principle (BER or NR16) of 5.1 mg mL−1. All drugs were then diluted with DDW to prepare lower doses, 2.55, 1.275, and 0.638 mg mL−1, respectively.
In the 96 well-plates, cells were allowed to settle and adhere for 24 h. Then, cells on the first plate were treated either with BER dissolved in 20% DMSO or SMA-BER, at the same doses (5.1, 2.55, 1.275, and 0.638 mg mL−1, respectively). On the other hand, cells on the second plate were treated with the same concentrations of either free NR16 dissolved in 20% DMSO or SMA-NR16. Subsequently, the two plates were incubated for 72 h at 37 °C in 4.5% CO2. For free drugs, DMSO was used as the relative control. For SMA-BER and SMA-NR16, unmodified cells were used as the relevant control. The Sulforhodamine B (SRB) assay was utilized to ascertain the cell density at each well. After the 72 h incubation, cells were fixed with 10% (w/v) trichloroacetic acid (TCA) and incubated for 1 h at 4 °C. After that, TCA was eliminated, and the wells were washed three times using DDW. After that, plates were left at 54 °C for 10 min to dry thoroughly. Cells were then stained with SRB dye and allowed to settle for 1 h at r.t. Excess of dye was then eliminated by washing three times with 1% (v/v) CH3COOH. Afterwards, plates were dried for 10 min at 54 °C. After dissolving the cells-bound dye in 10 mM Tris-based solution, the mixture was shaken for 15 min.
OD of the cells-bound dye at each well was determined at 570 nm using a plate reader. OD means ± SD were calculated, and cell viability percentages were then determined according to the OD results.
Statistical analysis
The statistical analysis was carried out by means of GraphPad Prism (GraphPad software, La Jolla, CA, USA) software, using One-way Analysis of Variance (ANOVA) and Tukey's multiple comparison post-tests and considering significant differences for p < 0.05. 0.1234 (ns), 0.0332 (*), 0.0021 (**), 0.0002 (***), <0.0001 (****).
Results and discussions
Formulation and characterization of SMA-BER and SMA-NR16
Micelle formation has been accomplished by means of a well-established method developed by our research group.19 As reported in Table 1, the recovery of SMA-BER and SMA-NR16 was 79% and 74%, respectively. DLS analysis of the micelles showed a size distribution of 123.3 ± 2.63 and 137.2 ± 2.29 for SMA-BER and SMA-NR16, respectively (Fig. S1†). The polydispersity index (PDI) of SMA-BER and SMA-NR16 micelles indicates a homogeneous distribution of size population (Table 1). Zeta (ζ) potential of the SMA-BER and SMA-NR16 micelles proved to be highly negative, being indicative of a stable system.
Recovery, loading, DLS analysis and Zeta potential value of SMA-BER and SMA-NR16 micellesa.
| Recoveryb | Loadingc (w/w) | Size (nm) | PDId | ζ potential (mV) | |
|---|---|---|---|---|---|
| SMA-BER | 79% | 13.24% | 123.3 ± 2.63 | 0.24 ± 0.015 | −46.36 ± 1.86 |
| SMA-NR16 | 74% | 12.63% | 137.2 ± 2.29 | 0.15 ± 0.023 | −49.23 ± 0.13 |
Data are shown as mean values ± standard deviation (SD). Values are the mean of triplicate experiments.
Starting from 1 g of SMA and 200 mg of free BER, 947 mg of SMA-BER have been obtained (79% recovery); starting from 1 g of SMA and 200 mg of NR16, 0.891 mg of SMA-NR16 have been obtained (74% recovery).
Loading as measured by HPLC-MS.
PDI = polydispersity index.
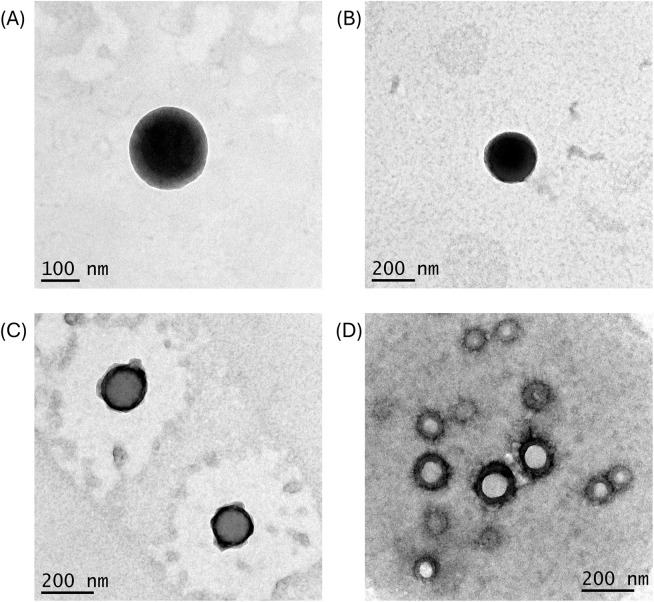
TEM analysis was conducted to visualize SMA-BER and SMA-NR16 structure and morphology and to confirm the DLS size data (Fig. 2). The collected images showed a monodisperse size distribution for both nanoparticles. Size analysis conducted with ImageJ software reported a mean size of 134.26 ± 22.69 nm for SMA-NR16 and 128.58 ± 47.83 nm for SMA-BER, further validating the data obtained by DLS (Fig. S3†). Furthermore, the nanoparticles showed a spherical shape, with a relatively smooth surface, which is the result of a correct polymer assembly, and nanomicelles formation (Fig. 2). The collected data suggested that both nanomicelles possess an optimal size range for cellular uptake in inflamed tissues subjected to bacterial infection (Table 1 and Fig. S3 and S4†). Indeed, size-dependent accumulation of nanoparticles, mostly medium-sized ranging from 20 to 200 nm, has been reported in several inflamed tissues, including infected areas and tumour tissue.29 In addition, the abovementioned size makes them less susceptible to opsonization processes and reticuloendothelial system (RES) uptake, potentially increasing circulation time.30
Fig. 2. TEM images of SMA-NR16 and SMA-BER: (A) image of SMA-NR16 taken with magnification set at 15 000×; (B) image of SMA-BER taken with magnification set at 8000×; (C) image of SMA-NR16 taken with magnification set at 10 000×; image of SMA-BER taken with magnification set at 10 000×.
The drug loading value was obtained by HPLC comparing the concentration with a previously constructed analytical curve for each of the two analytes. The amount of BER trapped in the nanoparticles was determined by treatment of SMA-BER with 0.5 M HCl followed by an ultracentrifugation procedure with Vivaspin® centrifugal concentrators. The amount of NR16 was determined by the same procedure but using 0.1 M HCl. For each of the analytes, the analyses were performed in triplicate. As a result, the loading in SMA-BER was found 13.24% (w/w) while the one of SMA-NR16 was 12.63% (w/w) confirming the correct encapsulation of the two drugs.
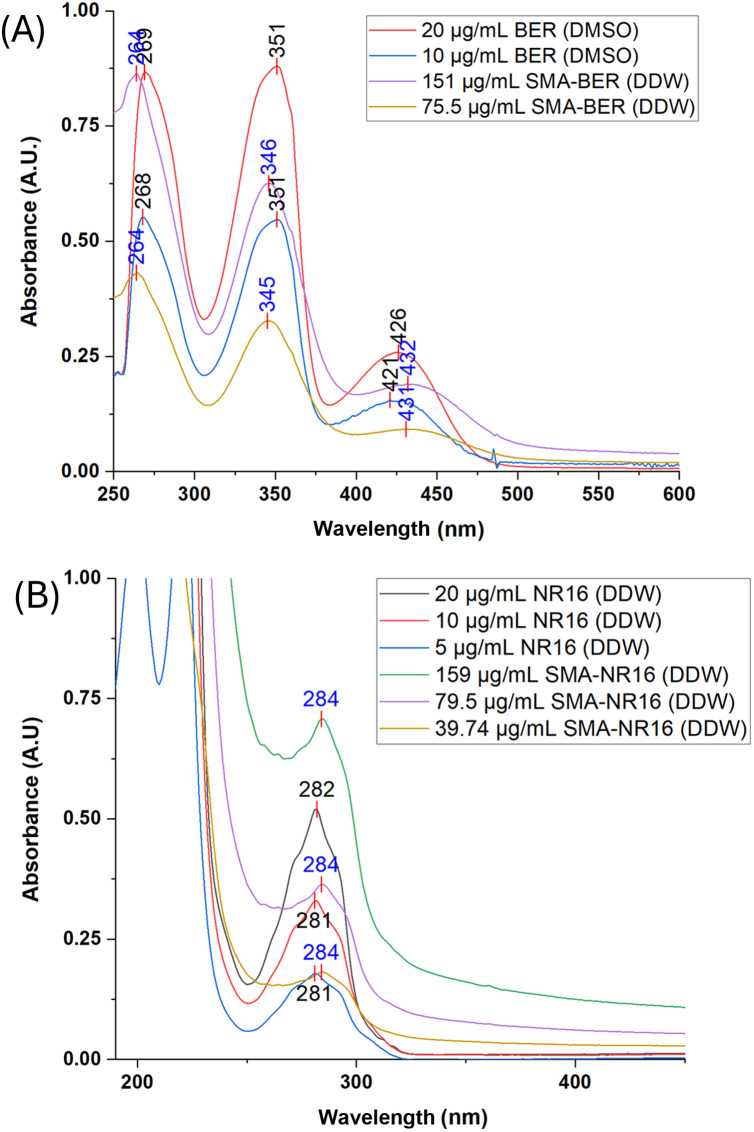
The interactions of SMA polymer with BER and NR16 were studied by UV/Vis absorption spectroscopy. SMA-BER and SMA-NR16 exhibited different spectral profiles compared to the free drug as they are influenced by the novel interactions established with the polymer (Fig. 3A and B).
Fig. 3. (A) UV-Vis spectra of BER (dissolved in DMSO) and SMA-BER (dissolved in DDW), spectra were recorded scanning from 200 to 800 nm, using a 1 cm path length quartz cells; (B) UV-Vis spectra of NR16 and SMA-NR16 (dissolved in DDW), spectra were recorded scanning from 200 to 800 nm, using a 1 cm path length quartz cells.
Considering the loading of BER of 13.24% w/w into the nanomicellar system, solutions of BER and SMA-BER were prepared in DMSO and DDW, respectively, to achieve solutions containing equal concentration in BER. A hypochromic effect can be observed for all the peaks of absolute/relative maximum, other than a hypsochromic shift for the two peaks at lower wavelengths, and a bathochromic shift for the peak at 420–430 nm (Fig. 3A). The observed variations may also be attributable to influence of the different used solvents.31–33
For NR16 and SMA-NR16 UV-Vis analysis, solutions of NR16 and SMA-NR16 were prepared in DDW achieving equal concentrations in NR16 (Fig. 3B). The results showed that the spectra profile of NR16 differs from the encapsulated ones, showing a bathochromic shift from 282 nm to 284 nm. Moreover, at increased concentrations the difference in absorbance became more evident, with SMA-NR16 showing higher absorbance values than the free drug. The present phenomena can be reconducted to the concomitant absorbance of NR16 and SMA polymer, which absorbs in frequency <300 nm.34
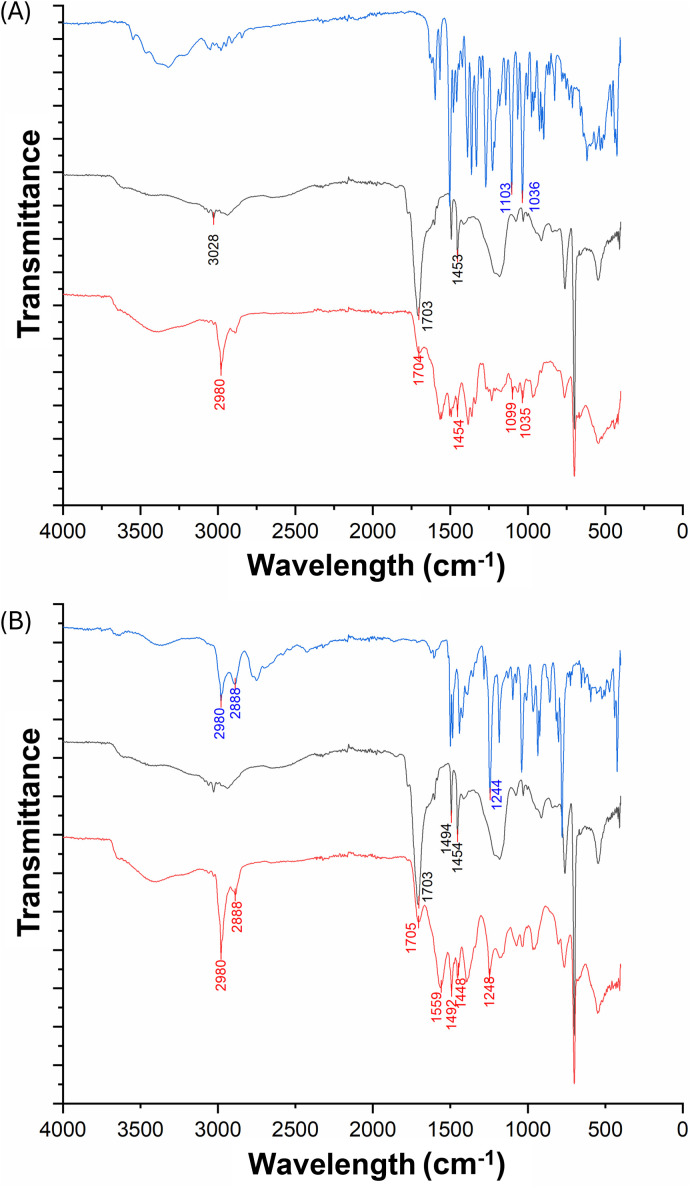
In addition, FT-IR and NMR studies have been performed on both nanomicelle preparations (Fig. 4).
Fig. 4. (A) Comparison between FTIR-ATR spectra of: BER (blue line); H-SMA (black line); H-SMA-BER (red line); (B) comparison between FTIR-ATR spectra of: NR16 (blue line); H-SMA (black line); H-SMA-NR16 (red line); the starting materials used to form the encapsulated systems (H-SMA and BER), subjected to FTIR-ATR analysis, showed spectroscopic characteristics identical to those reported in the literature.35,36 In the H-SMA copolymer, the carboxyl band appeared at 1703 cm−1 and a strong absorption band was observed at 1454 cm−1 (COO− symmetric vibrations), while a weak band (C–H stretching of the aromatic rings) was present at 3028 cm−1 (Fig. 4A).
In parallel, the FTIR-ATR analysis performed on BER showed two intense absorption bands at 1103 and 1036 cm−1 (CH in-plane bending and C–H vibrations). The spectrum of the nanomicellar system shows the characteristic bands of both constituent substances, confirming their presence. Moreover, the variations in the aromatic C–H signals (from 3028 to 2980 cm−1 in the polymer and from 1103 to 1099 cm−1 in the BER) would seem to indicate the presence of a hydrophobic interaction between the two molecular entities. The synthetic analogue of BER, the NR16, was also co-formulated with H-SMA co-polymer. As in the case of the system consisting of BER and H-SMA, the characteristic signals shown in the IR spectrum of the nanomicelles (red line, Fig. 4B), indicate the presence of both substances. Furthermore, the presence of a band at 1559 cm−1 suggests the formation of an ionic residue –NH2+ providing clear evidence of an ionic-type interaction between the two compounds.37 Variations observed in both the carboxylic (1703 cm−1 and 1448 cm−1, COO− symmetric vibrations) and the aromatic CH-related signals (1248 cm−1, CH in-plane bending) further support the existence of hydrophobic interactions within the system.38
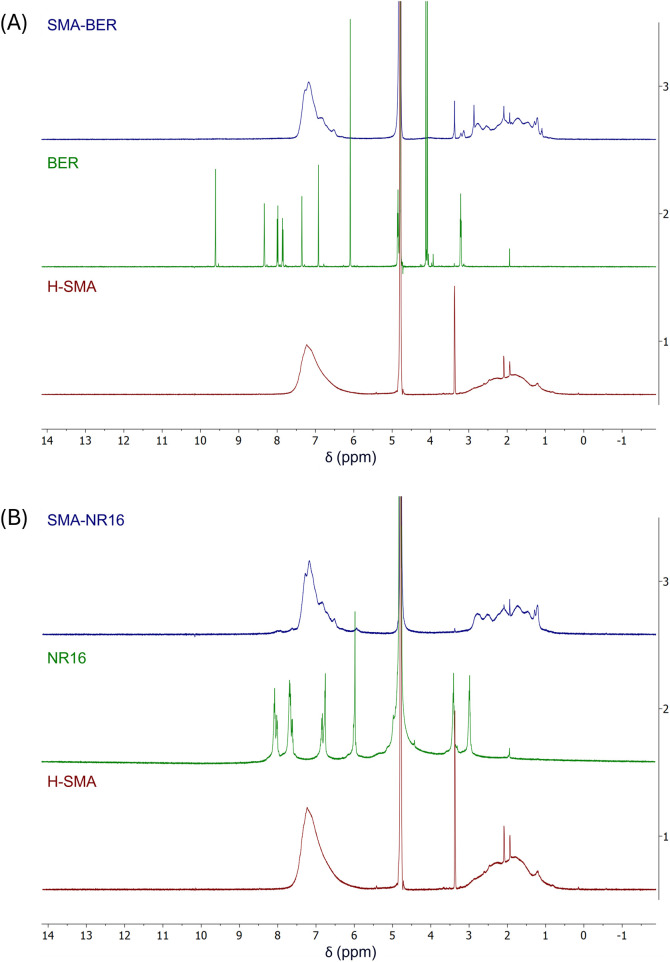
1H-NMR spectra of both the reference compounds and the nanostructured systems were recorded in D2O. The solvent was chosen to assess the real structure of the nanostructured systems in a biological environment39,40 (Fig. 5).
Fig. 5. (A) Superimposed 1H-NMR spectra of: BER (green line); H-SMA (red line); H-SMA-BER (blue line); (B) superimposed 1H-NMR spectra of: NR16 (green line); H-SMA (red line); H-SMA-NR16 (blue line).
The NMR spectrum of the H-SMA (Fig. 5A and B, red line) showed a very broad signal attributable to all the protons of the styrenic ring in the aromatic region, while the very broad signals of the polymeric backbone were displayed in the 1–3 ppm range. The NMR spectrum of BER (Fig. 5A, green line) showed signals in agreement with the previously reported ones.41 The synthesized nanomicellar system was subjected to the same experimental procedure under the same conditions. The obtained spectrum (Fig. 5A, blue line) showed the same signals as those obtained for the polymer (typical broadened peaks, between 6.5–8 and 1–3 ppm). However, the presence of BER was evidenced by the superimposed signals, which due to hydrophobic and ionic interactions, clearly vary their chemical shift value, also assuming the typical broadened appearance of the signals that constitute polymers.42 Results obtained by NR16 analysis (Fig. 5B, green line) and SMA-NR16 (Fig. 5B, blue line) showed a behavior mostly similar to the previously described system, confirming the presence of hydrophobic interactions between the constituents.
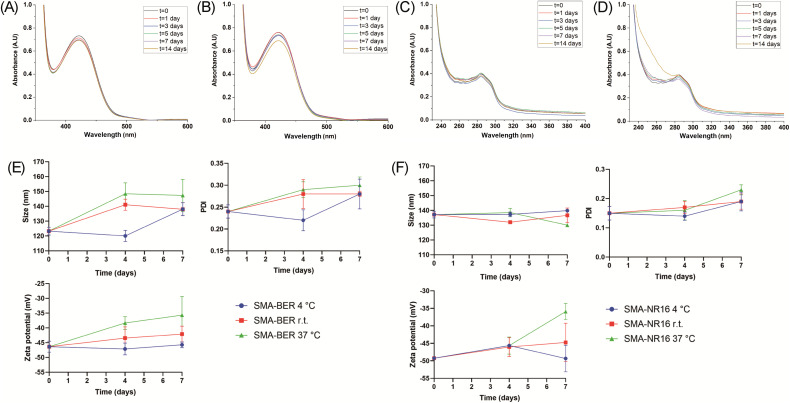
Stability studies were performed by UV/Vis analysis in DDW, NaCl, and PBS at three different temperatures (4 °C, room temperature (r.t.), and 37 °C), for 14 days. SMA-BER, in DDW, showed only slight changes in the UV/Vis spectra starting from day 7 at 4 °C, and from day 5 at r.t. and 37 °C as reported in Fig. S5–S7 (ESI†). All measurements in NaCl and PBS showed no evident changes in the UV/Vis spectra (Fig. S8–S11†, 6A and 6B). These results highlighted an excellent stability of the nanomicelles in the used media and at different storage temperatures. SMA-NR16 showed optimal stability in DDW at 4 °C and r.t. at all the timepoints (Fig. S12† and Fig. 6C). Good stability was also seen in DDW at 37 °C for about 7 days, while increased scattering phenomena were visible after 14 days at 37 °C (Fig. 6D). In NaCl, SMA-NR16 showed good stability when stored at 4 °C (Fig. S13†) and r.t. (Fig. S14†) along all the timepoints, showing only a slight increase in the absorbance spectra when compared to t = 0. On the other hand, the nanomicellar system was unstable when stored at 37 °C (Fig. S15†), presenting a massive increase of the absorbance values in the zone of 240–290 and a shift of the peak related to the drug at lower frequency. Similar results were seen in PBS for SMA-NR16, in which it maintained good stability when stored at 4 °C (Fig. S16†) and r.t. (Fig. S17†), while showing degradation phenomena when stored at 37 °C (Fig. S18†), starting from day 1. Indeed, it was possible to notice a lower absorbance at the frequency relative to the drug peak (∼284 nm) and an increase of the absorbance in the zone between 240-265 nm.
Fig. 6. UV-Vis spectra stability studies of: (A) SMA-BER dissolved in PBS aqueous solution and stored at 37 °C for 14 days (SMA-BER = 400 μg mL−1); (B) SMA-BER dissolved in NaCl aqueous solution and stored at 37 °C for 14 days (SMA-BER = 400 μg mL−1); (C) SMA-NR16 dissolved in DDW and stored at r.t. for 14 days (SMA-RN16 = 100 μg mL−1); (D) SMA-NR16 dissolved in DDW and stored at 37 °C for 14 days (SMA-RN16 = 100 μg mL−1). All the spectra were recorded scanning from 200 to 800 nm, using a 1 cm path length quartz cells; (E) DLS stability study of SMA-BER in DDW (conc. 50 μg mL−1) stored at 4 °C, r.t., and 37 °C for 7 days; (F) DLS stability study of SMA-NR16 in DDW (conc. 50 μg mL−1) stored at 4 °C, r.t., and 37 °C for 7 days.
To better highlight and understand from a nanoscale point of view the behaviour of nanomicelles at different storage temperature and conditions over time, the size, PDI, and zeta potential of SMA-BER and SMA-NR16 were also analyzed throughout 7 days by DLS. As previously mentioned, SMA-BER dissolved in DDW and stored at different temperatures showed only slight changes in the UV/Vis spectra starting from day 7 at 4 °C and from day 5 at r.t. and 37 °C. The same tendency was confirmed by DLS, in which nanoparticles stored at 4 °C (Fig. S19†) maintained similar features until day 4; a slight increase in particle size and PDI was seen at day 7 due to the formation of aggregates (3.4 + 2.16%) and presence of sub-population (may be due to micelles opening). The analysis of SMA-BER stored at r.t. (Fig. S20†) and 37 °C (Fig. S21†) showed that the tendency to form sub-populations and aggregates was accelerated by the increased temperature. When SMA-BER was stored at 37 °C for 7 days, the percentage of other populations amount increased to 10.32 ± 7.63%, showing higher PDI and zeta potential values of the nanoparticle system. This was most probably due to drug-release, disassembly of the nanosystem, and higher tendency in forming aggregates/floccule (Fig. 6E, and Fig. S21†).
Nevertheless, a high percentage of the initial nanoparticle population was seen in all the conditions, indicating good stability. The same DLS stability study protocol was performed on SMA-NR16. Even in this case, obtained data agreed with the UV-Vis stability study analysis. SMA-NR16 proved to maintain high stability in DDW in all the conditions (Fig. S22–S24†), showing only signs of slightly lower stability when stored at 37 °C for 7 days. Indeed, a small presence of lower size sub-population was detected (2.49 ± 1.66%), lower zeta potential, as well as slight aggregates formation. Nevertheless, high stability of the system was detected during all the experiments, maintaining high percentage of the initial nanoparticle population (Fig. 6F).
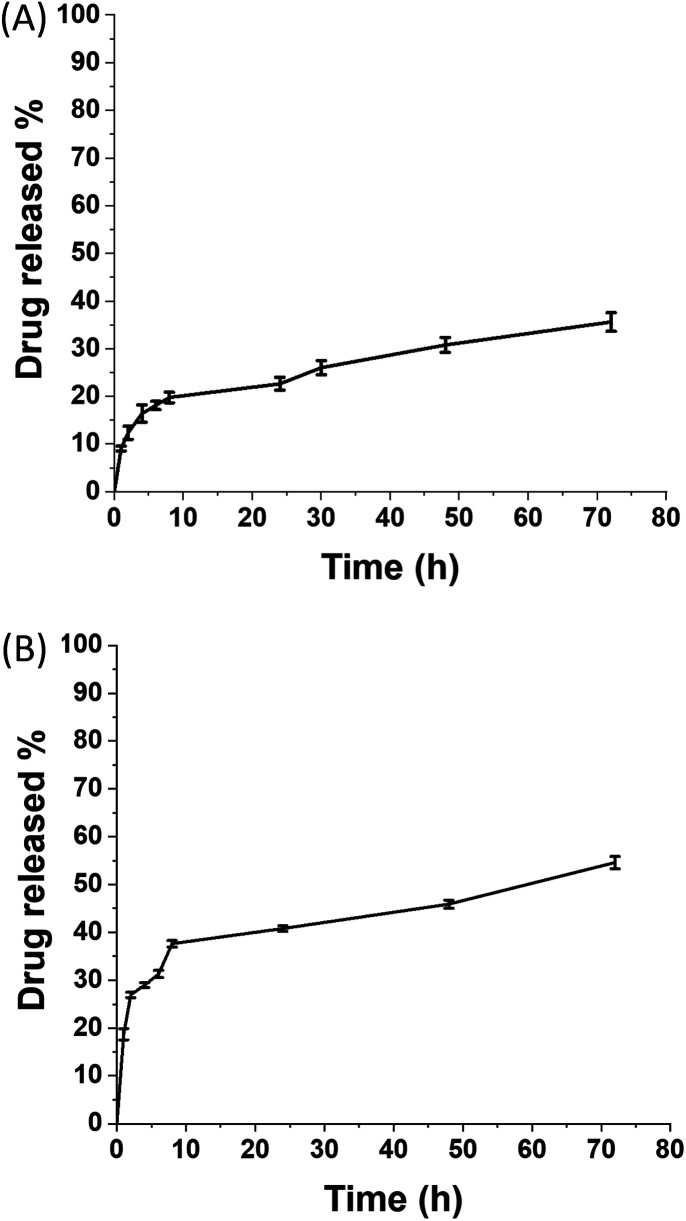
Finally, release studies have been performed in PBS pH 7.4 to characterize and evaluate the release profile of the drug from the nanomicellar systems. The release profiles have been explored by means of dialysis bag method. The quantification of drugs released over time has been performed by UV-Vis spectroscopy, using calibration curves of the free drugs (Fig. 7).
Fig. 7. Release rate at 37 °C of: (A) BER from SMA-BER; (B) RN16 from SMA-NR16.
SMA-BER nanomicelle system possessed an initial release of ∼9% after 1 hour, attributed to BER associated to the micelles surface, followed by a steady release of up to 35% until 72 h which can be ascribed to the drug encapsulated in the core (Fig. 7A). These results align with those previously reported for SMA nanoparticles encapsulating lipophilic drugs.16,21 SMA-NR16 showed a burst release with ∼40% of the drug released during the first 8 h, followed by a steady release up to 55% over 3 days (Fig. 7B). The different release profiles can be explained by the affinity and solubility of the two drugs for the water phase, which is sensibly higher for NR16 than for BER. Moreover, obtained results for SMA-NR16 agreed with the stability study performed in PBS at 37 °C, confirming that the recorded instability was also related to drug release during the first day.
SMA-BER and SMA-NR16 antibacterial studies
Once ascertained SMA-BER and SMA-NR16 have been fully characterized in terms of stability and drug release profile, they were screened for their antimicrobial properties. To this extent, both nanomicelle systems were tested against a panel of bacterial strains of greatest concern belonging to the ESKAPE group and a strain of Candida albicans, in comparison with the free drugs. The results of their antibacterial and antifungal activity, expressed as their minimum inhibitory concentration (MIC) and minimum bactericidal (or fungicidal) concentration (MBC), are reported in Table 2 and described below. The raw data from the assays conducted as biological triplicates are reported in the SI. MIC values were obtained by broth microdilution in three different biological replicates. Interestingly, BER, SMA-BER, and NR16 (but not SMA-NR16) inhibited the growth of E. faecalis, S. aureus, and S. epidermidis as well as the C. albicans strain. BER was able to inhibit VS E. faecalis growth at 512 mg L−1, MR and MS S. aureus as well as VR E. faecalis at 64–128 mg L−1, S. epidermidis at 32 mg L−1, and C. albicans at 256 mg L−1. The observed MIC values were in agreement with those reported in the literature.43–45 Several studies highlighted that BER is more active against Gram-positive bacteria (including S. aureus and S. epidermidis) than Gram-negative bacteria.45 Indeed, Mangiaterra et al. described no inhibitory activity of BER up to 320 μg mL−1 towards P. aeruginosa strains.46 No inhibitory activity against E. coli and K. pneumoniae was also reported at concentration ≥500 μg mL−1.47 Conversely, the obtained activity against Gram-positive bacteria, such as S. aureus (included MRSA and periprosthetic joint infections (PJI) associated isolates), S. epidermidis, and E. faecalis underlined the inhibitory activity of BER against these strains, similarly as reported in literature.43–45 Also in agreement were the data obtained over C. albicans, showing similar results as described by Xie et al.48 MBC evaluation revealed that BER was unable to kill bacteria not even at the highest tested concentration of 512 mg L−1, with the exception of S. epidermidis ATCC 35984 and S. aureus ATCC 25923 having an MBC of 64 mg L−1. BER seemed to exhibit a fungicidal effect at 256 mg L−1. The encapsulation of BER inside SMA nanomicelles promoted its activity against E. faecalis regardless of their vancomycin susceptibility profile, with the best antimicrobial performance achieved on VR E. faecalis. The inhibitory effect was similar to that of BER for all the Staphylococcus strains as well as for C. albicans. However, SMA-BER showed bactericidal activity against sensitive and resistant S. aureus and S. epidermidis, which are the most common and threatening bacteria in implant-related infections and infected wounds.49 On the contrary, SMA-BER was only able to inhibit Enterococcus growth. These results indicate that the encapsulation within SMA nanomicelles notably enhanced BER efficacy against Gram-positive bacterial strains. Moreover, other than antibacterial effect, SMA-BER also presented antifungal activity against C. albicans, showcasing its potential as a versatile antimicrobial agent. The BER synthetic derivate NR16 had MIC values akin to BER and SMA-BER, resulting in being a little more efficient than BER in inhibiting E. faecalis growth but less efficient than SMA-BER. Interestingly, MBC values obtained for NR16 unveiled a greater bactericidal effect when compared to the MBC of BER. NR16 was more efficient than SMA-BER in killing the tested E. faecalis strains. Instead, the encapsulation of NR16 into SMA resulted in being detrimental for NR16 efficacy, showing MIC values >512 mg L−1 for all the strains. Results also highlighted lack of antibacterial activity at the tested concentrations of BER, SMA-BER, and SMA-NR16 against representatives of Gram-negative strains at the highest tested concentrations (512 mg L−1). NR16 showed inhibitory and bactericidal effect on E. coli, K. pneumoniae and A. baumannii, but not against P. aeruginosa, in agreement with what was previously reported.13
MIC and MBC of BER, SMA-BER, NR16, SMA-NR16, H-SMA.
| BER | SMA-BERb | NR16 | SMA-NR16b | H-SMA | ||||
|---|---|---|---|---|---|---|---|---|
| Strain | MICa | MBCa | MIC | MBC | MIC | MBC | MIC | MIC |
| E. coli ATCC 25922 | >512 | — | >512 | — | 256 | 512 | >512 | >512 |
| K. pneumoniae ATCC BAA-2814 | >512 | — | >512 | — | 256 | 512 | >512 | >512 |
| A. baumannii ATCC 179878 | >512 | — | >512 | — | 256 | 512 | >512 | >512 |
| P. aeruginosa ATCC 27853 | >512 | — | >512 | — | >512 | — | >512 | >512 |
| VS E. faecalis ATCC 29212 | 512 | >512 | 128 | >512 | 256 | 256 | >512 | >512 |
| VR E. faecalis ATCC 51299 | 128 | >512 | 16 | >512 | 64 | 256 | >512 | >512 |
| MS S. aureus ATCC 12598 | 128 | >512 | 128 | 128 | 256 | 256 | >512 | >512 |
| MS S. aureus ATCC 25923 | 64 | 64 | 64 | 64 | 128 | 256 | >512 | >512 |
| MR S. aureus USA300 | 128 | >512 | 128 | 128 | 256 | 256 | >512 | >512 |
| S. epidermidis ATCC 35984 | 32 | 64 | 32 | 64 | 128 | 256 | >512 | >512 |
| C. albicans ATCC 10231 | 128 | 256c | 128 | 256c | 256 | 256c | >512 | >512 |
MIC and MBC values calculated as the geometric mean of three different tests and they are expressed as mg L−1.
SMA-BER and SMA-NR16 concentrations are referred to the amount (mg) of encapsulated drug.
For C. albicans the MBC is the minimum fungicidal concentration.
Finally, the uncharged SMA, as expected, did not have any antibacterial effect at tested concentrations.
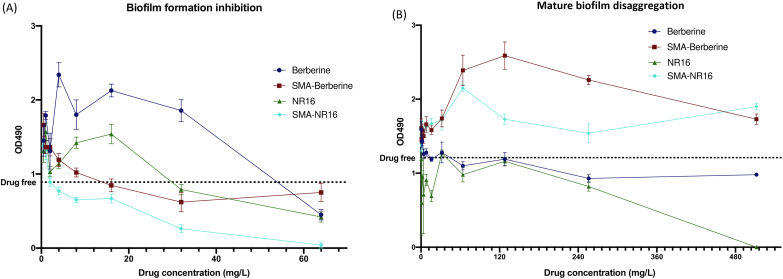
The effect of the formulations and free drugs on S. epidermidis ATCC 35984 biofilm formation and eradication was also studied. Indeed, S. epidermidis has become one of the most common causes of nosocomial infection associated with medical device implants (e.g., prosthesis, catheter, or other biomaterial) due to its tendency to form biofilm and colonize biomaterials.50 Among the compounds, SMA-NR16 showed the highest activity against biofilm formation, lowering its production at 4 mg L−1 and almost totally inhibiting biofilm production at 64 mg L−1. SMA-BER and NR16 had a similar effect on biofilm formation, lowering its growth at concentrations ≥32 mg L−1, but not reaching the complete inhibition of biofilm formation neither at 64 mg L−1. BER seemed to promote biofilm formation even at high concentrations, resulting active in lowering biofilm production only at 64 mg L−1 (Fig. 8A). The results evidenced that SMA-BER and, especially, SMA-NR16 were able to significantly reduce the S. epidermidis biofilm formation, opening to novel possible strategies in preventing biofilm formation and subsequent life-threatening infections.51 Regarding mature biofilm disaggregation, the encapsulated SMA-BER and SMA-NR16 did not have any anti-biofilm activity, while BER slightly reduced biofilm presence at concentrations ≥256 mg L−1. NR16 exerted a stronger anti-biofilm activity at concentrations equal to BER ones, resulting in a complete biofilm disaggregation at 512 mg L−1 (Fig. 8B).
Fig. 8. Effect of the compounds in study on (A) biofilm formation and (B) mature biofilm disaggregation. Biofilm producing strain S. epidermidis ATCC 35984 (the dot line indicates the mean production of biofilm in the absence of any compound).
Biocompatibility screening
Formulations that can be administered by endovenous route or used in an environment rich in blood (e.g. wounds) ought to not produce toxic effects on blood cells.52 To assay the biocompatibility of SMA-BER and SMA-NR16, a hemocompatibility and cytocompatibility screening was performed, in comparison with the free drugs. Hemocompatibility of BER, SMA-BER, NR16, and SMA-NR16 was assessed by means of a hemolysis test. Considering the calculated drug loading of the nanomicellar systems, concentrations corresponding to 1, 0.5, and 0.25 mg mL−1 of BER and NR16 were tested for SMA-BER and SMA-NR16, respectively. As shown by the results, 10% v/v and 5% v/v DMSO showed no hemolytic effect, being suitable for solubilizing the drugs. On the other hand, 1 mg mL−1 of BER showed a high hemolytic effect. A concentration of 0.5 mg mL−1 of BER was only mildly hemolytic, while it proved to be safe at 0.25 mg mL−1 and 0.125 mg mL−1 (Fig. 9A). Conversely, SMA-BER was no hemolytic at all concentrations, proving to be safer than the free drug. Thus, the encapsulation of the compound into the nanomicellar system could protect from the hemolytic effect that BER can produce at high concentrations. An inverted tendency was seen for NR16 and SMA-NR16, in which the nanoparticle system produced higher hemolytic effect than the free drug. Nevertheless, the hemolytic effect showed by SMA-NR16 at 0.99 mg mL−1, corresponding to 0.125 μg mL−1 in NR16 content, proved to produce only low hemolytic effect (Fig. 9B). However, since SMA-NR16 was able to strongly inhibit the biofilm formation of S. epidermidis at 64 mg L−1, the nanoparticle system may be used for therapeutic use without causing toxicity against blood cells.
Fig. 9. (A) BER and SMA-BER hemolytic activity. (B) NR16 and SMA-NR16. One way ANOVA: p < 0.05. 0.1234 (ns), 0.0332 (*), 0.0021 (**), 0.0002 (***), <0.0001 (****).
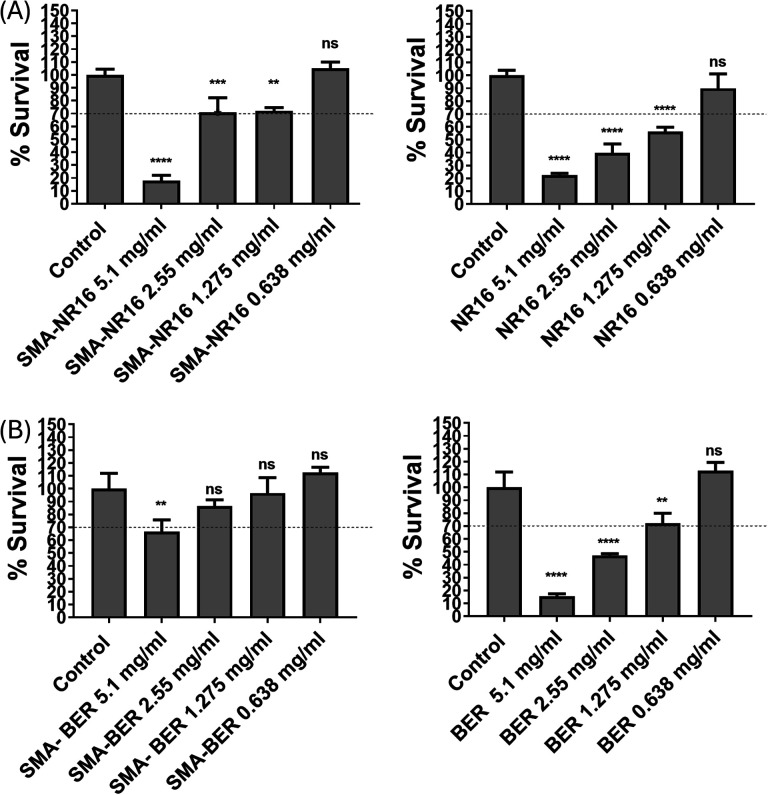
In vitro safety assessment of both nanomicelle formulations, free BER, and NR16 was performed on human embryonic kidney 293 (HEK-293) cells. A 10-fold higher dose range than the highest dose that demonstrated antimicrobial activity was applied to evaluate the potential safety in normal cells compared to bacteria. Moreover, the safety profiles of free drugs and their corresponding SMA nanomicelle formulations were compared within the defined dose range. As shown in Fig. 10A, NR16 displayed a dose-dependent cytotoxic effect against HEK-293 cells. However, at the lowest dose tested (0.638 mg mL−1)—close to the highest antibacterial concentration tested—neither the free NR16 nor the SMA-formulated drug demonstrated any significant reduction in cell number compared to the control group. However, the results indicated that SMA-NR16 significantly outperforms free NR16 in safety at a concentration of 2.55 mg mL−1. This statistical difference suggests that the nanoformulation of NR16 enhances its safeness. Similar results were observed for BER and SMA-BER, as shown in Fig. 10B, where no reduction in cell number was observed at the lowest dose tested compared to the control group. SMA-BER demonstrated significant improvements over free BER at concentrations of 5.1, 2.55, and 1.27 mg mL−1. This statistical evidence strongly supports our hypothesis that the nanoformulation of BER could enhance safety and efficacy when applied to whole-body systems. The cytocompatibility study results confirmed a general absence of toxicity towards normal cells of nanomicelles and corresponding free drugs at the concentration used for antibacterial activity. The lower toxicity trend observed for the SMA-formulated drugs compared to their free counterparts can be potentially attributed to different cellular uptake mechanisms. While free drugs readily diffuse across the plasma membrane, SMA-formulated drugs would require active internalization through endocytosis, a cellular activity that is dependent on both energy and time. These findings suggest that the produced SMA nanomicelles can be a promising approach for enhancing the safety profile of BER and NR16, by mitigating eventual toxic effects on normal cells. Moreover, the encapsulation into SMA highly increased the water solubility of the free drugs, thereby avoiding scarce bioavailability and high toxicity due to drug precipitation. In addition, the structural features showed by the nanoparticle system can be suitable for a passive targeting of the infected and inflamed site. The enhanced antimicrobial and bactericidal activity of SMA-BER against sensitive and resistant S. aureus, S. epidermidis, and C. albicans, jointed with high biocompatibility, make the nanoparticle system an important, effective, and safe alternative to antibiotics. Nonetheless, even though it did not show any efficacy in reducing microbial growth, SMA-NR16 showed that it can be a promising candidate as a strong S. epidermidis biofilm inhibitor.
Fig. 10. In vitro safety of BER and NR16 formulations on normal kidney cells HEK- 293: (A) HEK-293 cell survival after being treated with SMA-NR16 and NR16; (B) HEK-293 cell survival after being treated with SMA-BER and BER. One way ANOVA: p < 0.05. 0.1234 (ns), 0.0332 (*), 0.0021 (**), 0.0002 (***), <0.0001 (****).
Conclusions
The rapid spread of MDR bacteria has become a public health issue which requires innovative approaches and alternatives to antibiotics. Targeting microbial virulence factors may be an alternative winning approach, which can result in a minor evolutionary pressure for the development of bacterial resistance.53 In this context, natural substances represent an endless source of bioactive secondary metabolites and novel chemotypes active against MDR bacteria.54 However, one of the most important drawbacks of natural compounds is their unfavorable pharmacokinetic properties. BER, long known for its plethora of beneficial effects, including antibacterial activities, has no clinical use due to its scarce water solubility and consequent bioavailability.12,14 Bearing in mind the increasing seriousness of antibacterial infections caused by MDR and non-resistant bacteria, we reported the preparation of two different nanoparticle systems, SMA-BER and SMA-NR16, with the aim of increasing drug water solubility, stability, and antimicrobial properties.
Overall, the new nanomicelle formulations with their unique physicochemical properties showed several advantages compared to the free drugs. These include enhanced water solubility, high stability over time and temperature, adequate size distribution indicative of targeting to diseased tissues, and sustained release over at least three days. Antibacterial and antifungal activities against a panel of pathogens of great concern were observed, with the SMA-BER nanomicelles showing the greatest activity against MRSA, MSSA, VR, and VS E. faecalis. Both SMA-BER and SMA-NR16 nanoparticles showed greater activity against S. epidermidis biofilm formation in comparison with the corresponding free molecules, thus underlining the effectiveness of the developed nanocarrier-based delivery approach in preventing bacterial infections derived from biofilm formation.
Overall, these findings emphasize the significant role of SMA in enhancing the antimicrobial activity of BER and NR16, thereby offering promising and safe strategies in the fight against bacterial and fungal infections, including those associated with antibiotic resistance.
Data availability
All data are available in the main text and as part of ESI.†
Author contributions
Nicola F. Virzì: conceptualization, methodology, investigation, software, formal analysis, data curation, writing – original draft; Valentina Greco: methodology, software, validation, formal analysis, data curation, writing – original draft; Stefano Stracquadanio: methodology, software, validation, formal analysis, data curation, writing – original draft; Anfal Jasim: methodology, software, validation, formal analysis, data curation; Khaled Greish: investigation, writing – review & editing, visualization, supervision, project administration; Patricia Diaz-Rodriguez: methodology, software, validation, formal analysis, data curation; Natalie P. Rotondo: methodology; Stefania Stefani: supervision, project administration, funding acquisition; Valeria Pittalà: supervision, visualization, writing – review & editing, project administration, funding acquisition; Alessandro Giuffrida: supervision, project administration, funding acquisition.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This research was funded by: (1) Italian PON Project BONE++ Development of Micro and Nanotechnologies for Predictivity, Diagnosis, Therapy and Regenerative Treatments of Pathological Bone and Osteo-Articular Alterations. Project number ARS01_00693; (2) EU funding within the NextGeneration EU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT). We would like to thank Prof. Carmen Alvarez Lorenzo's group, I+D FARMA, R+D in Drug Dosage Form and Drug Delivery Systems, for support received in hemolysis study. Facultad de Farmacia, Departamento de Farmacia y Tecnología Farmacéutica, 15782 Santiago de Compostela (España).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04457f
Notes and references
- Di Lodovico S. Fasciana T. Di Giulio M. Cellini L. Giammanco A. Rossolini G. M. Antonelli A. Antibiotics (Basel) 2022;11(7):832. doi: 10.3390/antibiotics11070832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M. Hadjifrangiskou M. Hultgren S. J. Cold Spring Harbor Perspect. Med. 2013;3(4):a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L. Costerton J. W. Stoodley P. Nat. Rev. Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Hall C. W. Mah T. F. FEMS Microbiol. Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- Cangui-Panchi S. P. Ñacato-Toapanta A. L. Enríquez-Martínez L. J. Reyes J. Garzon-Chavez D. Machado A. Curr. Res. Microb. Sci. 2022;3:100175. doi: 10.1016/j.crmicr.2022.100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G. Midiri A. Gerace E. Biondo C. Pathogens. 2021;10(10):1310. doi: 10.3390/pathogens10101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer P. Paulin S. Bull. W. H. O. 2020;98(3):151. doi: 10.2471/BLT.20.251751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Chen L. Mao C. Jin L. Wu S. Zheng Y. Cui Z. Li Z. Zhang Y. Zhu S. Jiang H. Liu X. Small. 2024;20(9):e2306553. doi: 10.1002/smll.202306553. [DOI] [PubMed] [Google Scholar]
- Dorman H. J. Deans S. G. J. Appl. Microbiol. 2000;88(2):308–316. doi: 10.1046/j.1365-2672.2000.00969.x. [DOI] [PubMed] [Google Scholar]
- Silva L. N. Zimmer K. R. Macedo A. J. Trentin D. S. Chem. Rev. 2016;116(16):9162–9236. doi: 10.1021/acs.chemrev.6b00184. [DOI] [PubMed] [Google Scholar]
- Franco C. M. Vazquez B. I. Antibiotics (Basel) 2020;9(5):217. doi: 10.3390/antibiotics9050217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana T. Devi O. B. Singh Y. D. Curr. Pharmacol. Rep. 2023;9:329–340. [Google Scholar]
- Milani G. Cavalluzzi M. M. Solidoro R. Salvagno L. Quintieri L. Di Somma A. Rosato A. Corbo F. Franchini C. Duilio A. Caputo L. Habtemariam S. Lentini G. Biomedicines. 2021;9(5):452. doi: 10.3390/biomedicines9050452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Li W. Zhang H. Wang X. Huang Y. Li Y. Pan G. Phytomedicine. 2022;104:154288. doi: 10.1016/j.phymed.2022.154288. [DOI] [PubMed] [Google Scholar]
- Feng R. Shou J.-W. Zhao Z.-X. He C.-Y. Ma C. Huang M. Fu J. Tan X.-S. Li X.-Y. Wen B.-Y. Chen X. Yang X.-Y. Ren G. Lin Y. Chen Y. You X.-F. Wang Y. Jiang J.-D. Sci. Rep. 2015;5:12155. doi: 10.1038/srep12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virzì N. F. Fallica A. N. Romeo G. Greish K. Alghamdi M. A. Patanè S. Mazzaglia A. Shahid M. Pittalà V. RSC Adv. 2023;13(44):31059–31066. doi: 10.1039/d3ra04885c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezike T. C. Okpala U. S. Onoja U. L. Nwike C. P. Ezeako E. C. Okpara O. J. Okoroafor C. C. Eze S. C. Kalu O. L. Odoh E. C. Nwadike U. G. Ogbodo J. O. Umeh B. U. Ossai E. C. Nwanguma B. C. Heliyon. 2023;9(6):e17488. doi: 10.1016/j.heliyon.2023.e17488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta V. Giuffrida A. Grasso G. Maccarrone G. Mazzaglia A. Messina M. Vecchio G. J. Sep. Sci. 2011;34(1):70–76. doi: 10.1002/jssc.201000470. [DOI] [PubMed] [Google Scholar]
- Greish K. Pittala V. Taurin S. Taha S. Bahman F. Mathur A. Jasim A. Mohammed F. El-Deeb I. M. Fredericks S. Rashid-Doubell F. Nanomaterials (Basel) 2018;8(11):884. doi: 10.3390/nano8110884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasrija A. Singh R. Katiyar C. K. Int. J. Ayurveda Res. 2010;1(4):243–246. doi: 10.4103/0974-7788.76789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greish K. Mathur A. Al Zahrani R. Elkaissi S. Al Jishi M. Nazzal O. Taha S. Pittalà V. Taurin S. J. Controlled Release. 2018;291:184–195. doi: 10.1016/j.jconrel.2018.10.030. [DOI] [PubMed] [Google Scholar]
- Rice L. B. J. Infect. Dis. 2008;197(8):1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- Granata G. Stracquadanio S. Leonardi M. Napoli E. Consoli G. M. L. Cafiso V. Stefani S. Geraci C. Food Chem. 2018;269:286–292. doi: 10.1016/j.foodchem.2018.06.140. [DOI] [PubMed] [Google Scholar]
- Granata G. Stracquadanio S. Leonardi M. Napoli E. Malandrino G. Cafiso V. Stefani S. Geraci C. Molecules. 2021;26:4055. doi: 10.3390/molecules26134055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata G. Stracquadanio S. Consoli G. M. L. Cafiso V. Stefani S. Geraci C. Carbohydr. Res. 2019;476:60–64. doi: 10.1016/j.carres.2019.03.005. [DOI] [PubMed] [Google Scholar]
- O'Toole G. A. J. Visualized Exp. 2011;(47):2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rodriguez P. Gonzalez P. Serra J. Landin M. Mater. Sci. Eng., C. 2014;41:232–239. doi: 10.1016/j.msec.2014.04.058. [DOI] [PubMed] [Google Scholar]
- Farto-Vaamonde X. Diaz-Gomez L. Parga A. Otero A. Concheiro A. Alvarez-Lorenzo C. J. Controlled Release. 2022;352:776–792. doi: 10.1016/j.jconrel.2022.10.060. [DOI] [PubMed] [Google Scholar]
- Tu Z. Zhong Y. Hu H. Shao D. Haag R. Schirner M. Lee J. Sullenger B. Leong K. W. Nat. Rev. Mater. 2022;7(7):557–574. doi: 10.1038/s41578-022-00426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S. Nanomedicine (London) 2010;5(4):523–528. doi: 10.2217/nnm.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty G. Pillai V. S. Chittela R. K. J. Photochem. Photobiol., A. 2021;419:113454. [Google Scholar]
- Soulie M. Carayon C. Saffon N. Blanc S. Fery-Forgues S. Phys. Chem. Chem. Phys. 2016;18:29999–30008. doi: 10.1039/c6cp05848e. [DOI] [PubMed] [Google Scholar]
- Diaz M. S. Freile M. L. Gutierrez M. I. Photochem. Photobiol. Sci. 2009;8:970–974. doi: 10.1039/b822363g. [DOI] [PubMed] [Google Scholar]
- Yamamoto S. Kaneo Y. Maeda H. J. Drug Delivery Sci. Technol. 2013;23:231–237. [Google Scholar]
- Kopf A. H. Koorengevel M. C. van Walree C. A. Dafforn T. R. Killian J. A. Chem. Phys. Lipids. 2019;218:85–90. doi: 10.1016/j.chemphyslip.2018.11.011. [DOI] [PubMed] [Google Scholar]
- Hu Z. Zhao K. Chen X. Zhou M. Chen Y. Ye X. Zhou F. Ding Z. Zhu B. Int. J. Mol. Sci. 2023;24(22):16286. doi: 10.3390/ijms242216286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock R. A. Marion L. Can. J. Chem. 1956;34(12):1782–1795. [Google Scholar]
- Ali E. A. Eweis M. Elkholy S. Ismail M. N. Elsabee M. Macromol. Res. 2018;26(5):418–425. [Google Scholar]
- Kobchikova P. P. Efimov S. V. Klochkov V. V. Membranes (Basel) 2023;13(2):196. doi: 10.3390/membranes13020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D. Rajan R. Yusa S.-i. Nakada M. Matsumura K. Mater. Adv. 2022;3:4252–4261. [Google Scholar]
- Hasada K. Yoshida T. Yamazaki T. Sugimoto N. Nishimura T. Nagatsu A. Mizukami H. J. Nat. Med. 2011;65(2):262–267. doi: 10.1007/s11418-010-0490-x. [DOI] [PubMed] [Google Scholar]
- Lamch L. Gancarz R. Tsirigotis-Maniecka M. Moszynska I. M. Ciejka J. Wilk K. A. Langmuir. 2021;37(14):4316–4330. doi: 10.1021/acs.langmuir.1c00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. Wang J. Yang C. Zhu C. Guo G. Tang J. Shen H. BMC Complementary Altern. Med. 2019;19(1):218. doi: 10.1186/s12906-019-2558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. H. Kim K. J. Cha J. D. Kim H. K. Lee Y. E. Choi N. Y. You Y. O. J. Med. Food. 2005;8(4):454–461. doi: 10.1089/jmf.2005.8.454. [DOI] [PubMed] [Google Scholar]
- Wojtyczka R. D. Dziedzic A. Kępa M. Kubina R. Kabała-Dzik A. Mularz T. Idzik D. Molecules. 2014;19(5):6583–6596. doi: 10.3390/molecules19056583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiaterra G. Cedraro N. Laudadio E. Minnelli C. Citterio B. Andreoni F. Mobbili G. Galeazzi R. Biavasco F. J. Nat. Prod. 2021;84(4):993–1001. doi: 10.1021/acs.jnatprod.0c01151. [DOI] [PubMed] [Google Scholar]
- Sun N. Chan F. Y. Lu Y. J. Neves M. A. Lui H. K. Wang Y. Chow K. Y. Chan K. F. Yan S. C. Leung Y. C. Abagyan R. Chan T. Wong K. Y. PLoS One. 2014;9:e97514. doi: 10.1371/journal.pone.0097514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y. Liu X. Zhou P. Drug Des., Dev. Ther. 2020;14:87–101. doi: 10.2147/DDDT.S230857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira W. F. Silva P. M. S. Silva R. C. S. Silva G. M. M. Machado G. Coelho L. Correia M. T. S. J. Hosp. Infect. 2018;98(2):111–117. doi: 10.1016/j.jhin.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Fey P. D. Olson M. E. Future Microbiol. 2010;5(6):917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visperas A. Santana D. Klika A. K. Higuera-Rueda C. A. Piuzzi N. S. J. Orthop. Res. 2022;40(7):1477–1491. doi: 10.1002/jor.25345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S. Shi Y. Liang Y. Liu L. Sun K. Li Y. Asian J. Pharm. Sci. 2021;16(5):551–576. doi: 10.1016/j.ajps.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A. Sperandio V. Nat. Rev. Drug Discovery. 2010;9(2):117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- Saleem M. Nazir M. Ali M. S. Hussain H. Lee Y. S. Riaz N. Jabbar A. Nat. Prod. Rep. 2010;27(2):238–254. doi: 10.1039/b916096e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and as part of ESI.†