Abstract
Cell interactions with the extracellular matrix (ECM) influence intracellular signaling pathways related to proliferation, differentiation, and secretion, amongst other functions. Herein, bone-marrow derived mesenchymal stromal cells (MSCs) are encapsulated in a hydrazone crosslinked hyaluronic acid (HA) hydrogel, and the extent of stress relaxation is controlled by systemic introduction of irreversible triazole crosslinks. MSCs form elongated multicellular structures within hydrogels containing RGD peptide and formulated with elastic composition slightly higher than the hydrogel percolation threshold (12 % triazole, 88 % hydrazone). A scaling analysis is presented ( ∼Nα) to quantify cell-material interactions within these structures with the scaling exponent (α) describing either elongated (0.66) or globular (0.33) structures. Cellular interactions with the material were controlled through peptides to present integrin binding ECM cues (RGD) or cadherin binding cell-cell cues (HAVDI) and MSCs were observed to form highly elongated structures in RGD containing hydrogels (), whereases collapsed structures were observed within HAVDI containing hydrogels (). Finally, cytokine secretion was investigated, and a global increase in secreted cytokines was observed for collapsed structures compared to elongated. Taken together, this study presents a novel method to characterize cellular interactions within a stress relaxing hydrogel where altered cluster morphology imparts changes to cluster secretory profiles.
Keywords: Mesenchymal stromal cells, Hydrogels, Stress relaxation, Ligand interactions
Graphical abstract
Highlights
-
•
Stress relaxation and presented peptide epitope guides MSC structure formation.
-
•
Scaling law image analysis enables quantification of MSC multicellular structures.
-
•
Cooperative effect of cell-ECM and secretory properties enable elongated structures.
1. Introduction
Tissue-specific extracellular matrix (ECM) is comprised of an assortment of polysaccharides and structural proteins, such as collagen, fibronectin, and laminin, and defined by its mechanical properties, such as stiffness and stress relaxation [1,2]. Many studies have investigated how ECM composition influences cell migration, fate determination, and secretory properties, all of which influence tissue repair and regeneration [[3], [4], [5]]. For example, in single mammary epithelial cells nuclear accumulation of yes-associated protein-1 (YAP) and subsequent downstream mechanosensing are highly dependent on ECM protein identity, with a decrease in nuclear YAP levels observed for cells cultured on laminin-111 functionalized substrates compared to those functionalized with collagen I or fibronectin [6]. These changes were due to specific integrin activation through β4 integrins which stabilized keratin cytoskeleton networks to shield the nucleus from mechanical sensing. Similarly, ECM composition and the accumulation of fibrillar Collagen I within the colonic epithelial basement membrane, a type of specialized ECM, was shown to induce a mechanosensitive, regenerative cell phenotype leading to repair [7]. Beyond epithelial cells, MSCs have also been utilized to investigate cellular responses to ECM specific cues.
MSCs are adherent cells and require matrix interactions to prevent anoikis, but the type of integrin binding can significantly alter secretory properties [[8], [9], [10], [11]]. For example, Clark and coworkers showed differences in cell-matrix interactions induce changes in MSCs secretion, specifically when MSCs interacted with the collagen I derived peptide GFOGER as opposed to the fibronectin derived peptide RGD. Results showed an upregulation of secreted inflammatory, vascularization, and bone regeneration cytokines, which subsequently led to increased bone regeneration in vivo [8]. Beyond ECM signaling cues, the N-cadherin mimetic peptide, HAVDI, has been used to alter the secretory profile of MSCs. Granular hydrogels modified with HAVDI induced globally higher levels of cytokine secretion by MSCs, even in the smaller clusters (less than 5 cells) or single cells, comparable to that of MSC aggregates and higher than that of RGD alone [10]. Beyond ECM composition, mechanical properties such as stiffness and dynamic cues such as creep compliance, strain stiffening, and stress relaxation are known to influence MSCs.
Hydrogels are water swollen networks comprised of crosslinked, hydrophilic macromolecules that are often functionalized with biological epitopes to capture key biological features of the ECM [12]. Depending on the nature of the molecular interactions between the macromolecules, the networks can be elastic or viscoelastic, the latter imparting unique properties such as stress relaxation and injectability [13,14]. In one example of viscoelastic hydrogels, Chaudhuri and coworkers utilized physical crosslinks within alginate hydrogels where the molecular weight (MW) of alginate macromers was systematically decreased to alter macromer connectivity and even sterically hindered through the coupling of PEG spacers onto the alginate. An increase in stress relaxation, observed through faster relaxation timescales was observed as the MW of the alginate decreased and was maximal when steric hindrances were also introduced. Importantly, faster relaxation timescales resulted in MSC YAP nuclear localization, osteogenic differentiation, and formation of an interconnected mineralized matrix of collagen-1 [15]. This work is particularly interested in covalent adaptable networks (CANs), which once formed can adapt under physiological conditions [13,16]. One of the early examples of a hydrogel CAN used for cell encapsulation was a hydrazone crosslinked poly(ethylene glycol) (PEG) network, achieving relaxation rates spanning multiple orders of magnitude, which allowed spreading and fusion of C2C12 myoblasts [17]. Richardson et al. expanded on this work and encapsulated chondrocytes, demonstrating that the deposition and distribution of type II collagen and glycosaminoglycans depended on the local relaxation properties, where faster relaxing gels led to more robust deposition of cartilaginous tissue after 28 days in culture [18]. Beyond this early work with hydrazone hydrogels, other materials chemistries have been developed that demonstrate the versatility of CANs and their applications for cell encapsulation; these include boronate esters, thioesters, allyl sulfides and dithiolanes [13,[19], [20], [21], [22]].
In this work, we were interested in better understanding the relative role of mechanical signaling via stress relaxation versus biochemical signaling via peptide epitopes on MSC morphology, clustering, and secretory properties. As a platform to study these interactions, we were drawn to hydrogels crosslinked with the covalently adaptable hydrazone bond as they have seen extensive use in tissue engineering applications. In one study, Lou et al. used the hydrazone chemistry to fabricate stress relaxing hyaluronic acid-collagen hydrogels, where the degree of stress relaxation was tuned by the hydrazone-chemistry and relative content of collagen versus hyaluronic acid [23]. MSC morphology and secretion were found to depend upon both the degree of stress relaxation and the composition of the hydrogels (i.e., matrix interactions). Wang and coworkers demonstrated the printability of hydrazone hydrogels by formulating a precursor solution using hyaluronic acid (HA) macromers functionalized with either aldehyde or hydrazide functionalities but adding a norbornene-modified HA to allow for a secondary photoinitiated crosslinking to stabilize the final hydrogel [24]. As an alternative strategy, Borelli et al. tuned the stress relaxation rate and extent of relaxation of hydrazone-crosslinked HA hydrogels by introducing permanent crosslinks via a strain-promoted azide-alkyne cycloaddition (SPAAC) reaction [25]. Nascent protein deposition was found to depend on the extent of stress relaxation (hydrazone: SPAAC crosslinking ratio), but interestingly, multi-cellular, highly extended MSC structures were observed when the hydrazone: SPAAC ratio approached the percolation threshold of the permanent crosslinks in the gel (i.e., permanent gel with a high degree of stress relaxation).
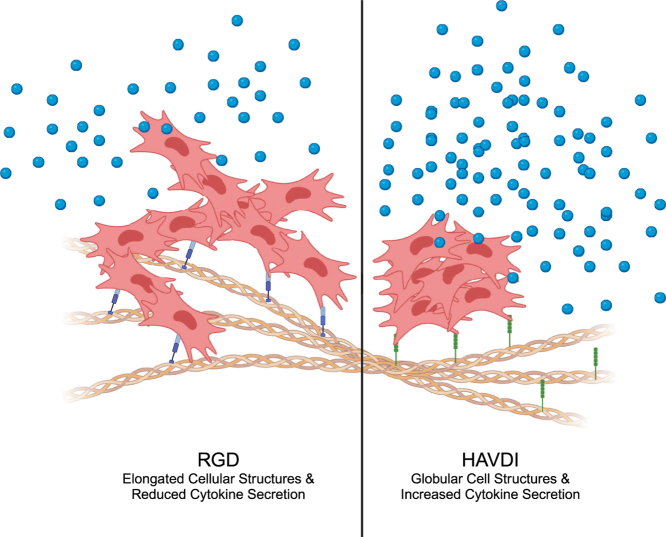
Intrigued by these findings, we set out to better understand the evolution of these highly extended, multi-cellular structures, especially near this critical point (i.e., percolation threshold of permanent crosslinks). MSCs were encapsulated in PEG-HA hybrid hydrogels with varying extents of stress relaxation, specifically 94 %, 88 % and 82 % alkyl-hydrazone bonds conditions and cultured for 7 days. The resulting multi-cellular structures were imaged via a laser scanning confocal or spinning disk confocal microscope and stained for Dapi (nuclei), AF-647 Phalloidin (F-actin), Paxillin and N-Cadherin. To uniformly characterize the complex cell structure, a simple scaling analysis was applied from polymer solution theory to calculate the radius of gyration and how it scales to overall object length. The resulting analysis enabled a better understanding and quantification of the cell-matrix interactions present within the multi-cellular structures. Additionally, experiments were designed to further probe the cell-matrix interactions by tuning the presented peptide epitope within the hydrogel system. Interestingly, upon incorporation of HAVDI, an N-cadherin mimetic peptide, the multi-cellular structures shifted in morphology from elongated, observed in RGD conditions, to globular structures. Upon analysis of the cytokine secretion from these hydrogel systems, broad upregulation of cytokine secretion was seen in the condition containing HAVDI, as compared to RGD. Overall, the results reported herein demonstrate the design of a stress-relaxing hydrogel system that enables control over the biophysical and biochemical cues which the encapsulated MSCs receive, resulting in a better understanding of the cell-matrix interactions required for multi-cellular structure formation, and manipulation of the ensuing secretory profile.
2. Results and discussion
2.1. Designed stress relaxing PEG-HA hydrogels
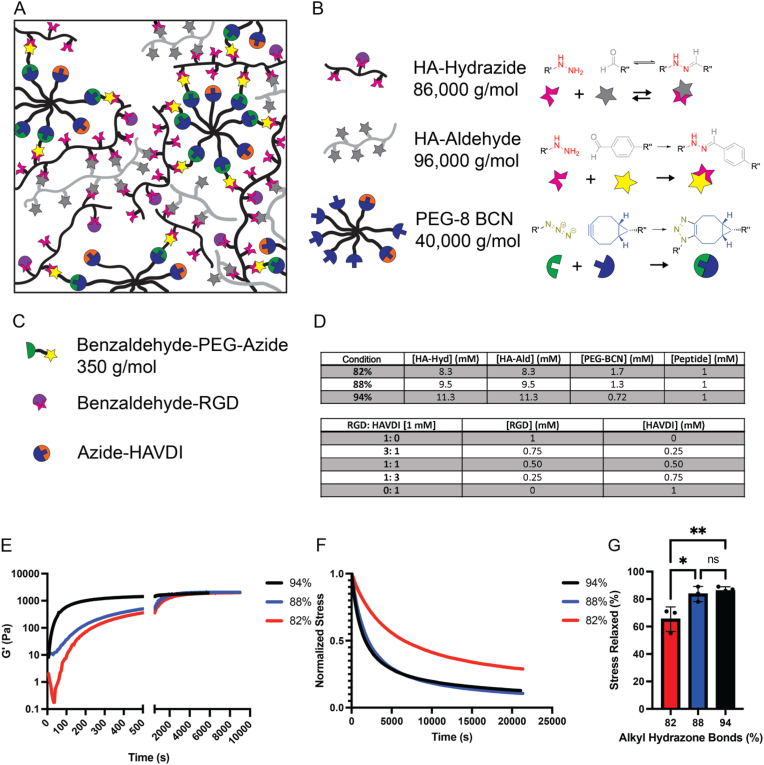
A chemically defined, covalent adaptable hydrogel was formulated to investigate the influence of the extent of stress relaxation and mimetic peptide presentation on MSC cell-ECM and cell-cell interactions. Specifically, a hyaluronic-based (HA) hydrogel containing dynamic hydrazone crosslinks was formed via the reaction of hydrazide and alkyl-aldehyde functionalized hyaluronic acid macromolecules (HA-Hyd and HA-Ald, respectively). To control the extent of stress relaxation, a small dose (1.3–4.2 mM) of benzaldehyde-PEG-azide was first reacted onto the HA-Hyd macromer (4.2–9.5 mM Hyd), converting a fraction of HA-Hyd (1.3–4.2 mM) functional groups to azides. Additionally, RGD or HAVDI (1 mM) was incorporated into the system by reaction of a benzaldehyde-RGD peptide onto the HA-Hyd macromer. Upon subsequent reaction with an 8-arm poly(ethylene glycol)-bicyclononyne (PEG-BCN) macromer (1.3–4.2 mM), stable triazole bonds are formed via a strain promoted azide-alkyne cycloaddition (SPAAC) reaction. In this manner, a percolating network of stable triazole crosslinks can be formed through reaction of the azide functionalized HA-Hyd with the PEG-BCN. For these formulations, the percolation threshold was estimated using Flory-Stockmayer theory and is reached when the triazole crosslinks account for ∼5 % of the total crosslinks [18,26,27]. The remainder of the crosslinks are adaptable and formed through reaction of the HA-Ald with the remainder of HA-Hyd functional groups. Additionally, HAVDI (1 mM) was incorporated into the system by reaction of an azide-HAVDI peptide onto the PEG-BCN macromer. Further, this system enables the control over the ratio of RGD: HAVDI, herein the peptide concentrations will range from 1:0, 3:1, 1:1, 1:3 and 0:1 ([RGD]: [HAVDI]) (Fig. 1A, B, C, D). Notably, the final peptide concentrations within the hydrogels were ∼67 % of the initial peptide loading when swollen to equilibrium, which is ∼150 % the initial volume after formation (Fig. S1).
Fig. 1.
Network structure of hybrid, stress relaxing hydrogel containing mimetic peptides. A) Schematic representation of the dual-network containing both the dynamic hydrazone bond, and the elastic triazole bond and the mimetic peptide's, RGD and HAVDI, attached to the HA-Hyd or PEG-BCN, respectively. B) The schematics of the macromers present within the hydrogel including the hydrazide functionalized HA, aldehyde functionalized HA and BCN functionalized PEG groups. The reactant groups forming the various bonds throughout the hydrogel, including the dynamic hydrazone bonds (either alkyl- or benzyl based), and the triazole bonds. C) The small molecule and peptide epitopes present throughout the hydrogel, enabling control over the formation of the SPAAC network and composition of the hydrogel, respectively. D) Tables containing the formulations of the differing stress relaxing conditions, as well as the concentrations of the varied RGD: HAVDI peptide ratios utilized throughout this paper. E) Hydrogel in situ formation, reaching a final modulus ranging from 1300 Pa to 1600 Pa. F) The stress-relaxation of the hydrogels, over 6 h, after in situ formation with the 94 % and 88 % alkyl hydrazone bonds conditions relaxing the most stress over the timescale. G) The percentage of stress relaxed by the differing alkyl hydrazone compositions. ∗p < 0.05, ∗∗p < 0.01, ns: no significance.
Hydrogels were prepared from 3 wt% polymer solutions using a stoichiometric ratio of aldehyde to hydrazide. Based on prior results that revealed extended multicellular MSC structures in the 88 % alkyl hydrazone condition, which nears the percolation threshold of irreversible SPAAC crosslinks (5 %) in the stress-relaxing hydrazone HA gels, we sought to investigate MSCs and the formation of multicellular structures near this critical point: 94 %, 88 % and 82 % hydrazone crosslinks [25]. Under these conditions, the final shear modulus (G′) upon formation were similar, ranging from 1300 to 1600 Pa after 9000s (2.5 h) of reaction, indicative of a similar network crosslinking density (Fig. 1E–S2). Importantly, however, the ratio of reversible to irreversible crosslinks led to significant differences in the extent of stress relaxation. To measure this, 10 % strain was applied to in situ formed hydrogels, and the stress relaxation (σ/σo) was monitored over a 6-h period. The percent of stress relaxed ranged from 86 ± 2.9 % (94 % hydrazone crosslinks) to 65 ± 8.9 % (82 % hydrazone crosslinks), and the 88 % hydrazone gel's relaxation (84 ± 5.5 %) was not statistically different from the 94 % gel (Fig. 1F and G). The average time constant for stress relaxation (<τ>) was then assessed for each condition and found to be ∼7000 s for both the 94 % and 88 % conditions, which were statistically different from the 82 % condition, with a time constant of ∼19000 s (Fig. S3). Using these formulations with a similar network crosslinking density, we next sought to determine the role of a compositionally defined stress relaxation on the extent of MSC-matrix interactions and its influence on the final multi-cellular morphology.
2.2. The formation and morphology of multi-cellular clusters depend on stress relaxation
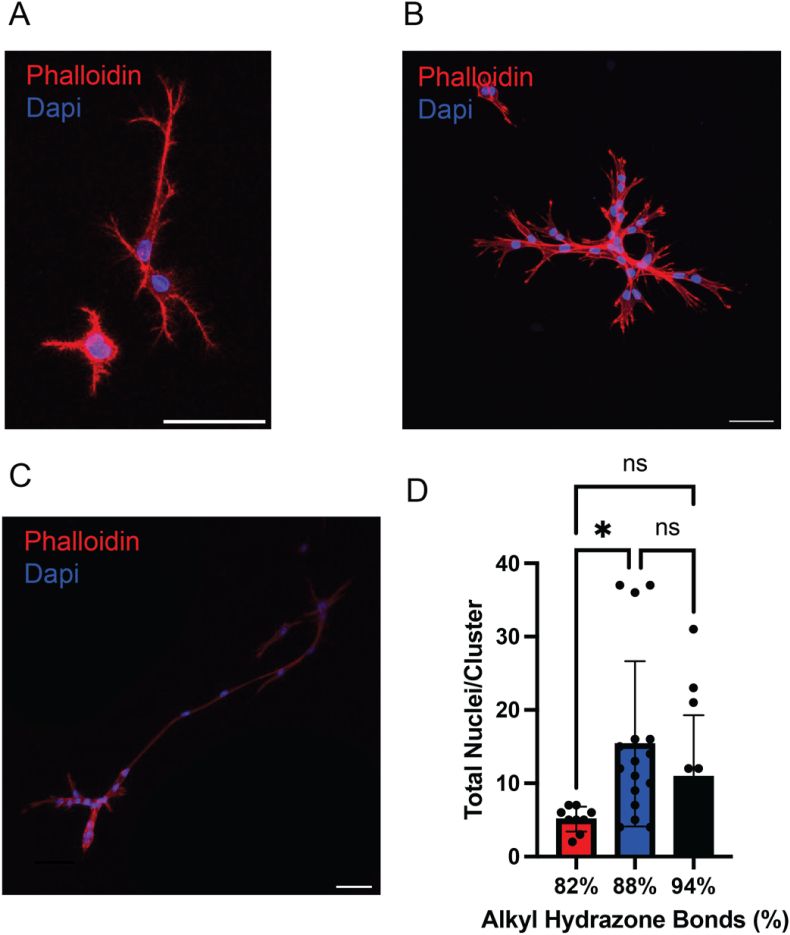
MSCs were encapsulated in HA-hydrazone hydrogels with varying extents of stress relaxation (82, 88, and 94 % hydrazone crosslinks) at a density of 3E6 cells/mL and functionalized with 1 mM RGD to promote MSC-matrix interactions. After 7 days, large multi-cellular structures formed that were imaged using a Zeiss 710 laser scanning confocal microscope (Fig. 2A, B, C). As an initial characterization, the number of nuclei per structure was quantified (DAPI staining), which ranged from 4.9 ± 1.3 nuclei/structure (82 %) to 17 ± 4.5 nuclei/structure (88 %), with the 94 % condition (9.2 ± 3.1 nuclei/structure) being statistically the same as the 88 % (Fig. 2D). To better characterize the cell-matrix interactions in these multi-cellular structures, we immunostained for F-actin (AF-647 Phalloidin), N-cadherin (anti-N-Cadherin antibody) and paxillin (anti-paxillin antibody) expression. N-cadherin was selected as a cell junction protein (cell-cell interactions), paxillin as a marker for mature focal adhesions (cell-ECM interactions), and F-actin to visualize the cell body. Images reveal both extensive paxillin and N-cadherin expression throughout the multi-cellular structures, which was quantified across the stress-relaxing formulations. In general, the intensity of N-cadherin expression was similar across all stress relaxing conditions (Fig. S4); however, the global paxillin intensity (Fig. S5) increased as the hydrazone crosslinking content (88–94 %) and percent of stress relaxation increased. Thus, cell-ECM interactions likely play a role in the formation of these structures. Therefore, we next sought to manipulate the extent of the cell-ECM interactions via controlled compositional variations in our hydrogel formulations. In addition, we introduced a new quantitative analysis of these multicellular structures to better characterize the distinct morphological features that evolve as a result of the extent of stress relaxation, integrin binding, and cellular interactions.
Fig. 2.
Multi-cellular structure formation dependence on stress relaxing hydrogels. A-C) Representative images of the multi-cellular structures observed in the 82 % (A), 88 % (B) and 94 % (C) alkyl hydrazone bonds conditions, scale bar = 50 μm. D) The total number of nuclei/cluster within the multi-cellular clusters for the 82 %, 88 % and 94 % alkyl hydrazone bond conditions. ∗p < 0.05, ns: no significance.
2.3. Analysis of MSC structure morphology using radius of gyration and a power law scaling
As an alternative metric to analyze the size and shape of the MSC multi-cellular structure, the radius of gyration was calculated and a power law scaling developed to describe the structure of macromolecular chains in solution was applied [28,29]. Specifically, the F-actin distribution of the multi-cellular MSC structures was used in a synonymous way to describe polymer chains in solution. To determine the radius of gyration (<Rg, structure2>1/2), the F-actin signal was used to generate a skeleton of the multicellular structure, where the binarized cell-volume was uniformly reduced to render the center-most voxels of a given F-actin signal (FIJI) (Fig. 3A, S6). Then, <Rg, structure2>1/2 was calculated by first finding the structure's center of mass () (eq (1)).
| (1) |
In this equation is calculated as the average position over the total number of voxels (Nv) within the multi-cellular structure for each voxel position (Xi, Yi, Zi). Upon calculating , <Rg, structure2>1/2 was calculated by the root mean squared distance between each voxel element ( and the for the entire multi-cellular structure (eq (2)).
| (2) |
Using < Rg, structure2>1/2, a power law scaling was used to determine how space filling the multi-cellular structure was within the defined radius of gyration (eq (3)).
| (3) |
Fig. 3.
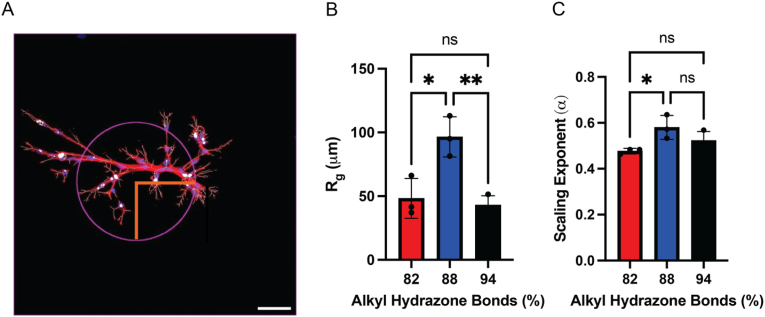
Scaling analysis model to determine cell-cell and cell-ECM interactions. A) Skeletonized image (white), overlaid with AF-647 phalloidin (red) for a representative image within the 88 % alkyl hydrazone bonds condition, including the radius of gyration (magenta circle, with orange radius), yielding an α value of 0.57 for this multi-cellular structure, scale bar = 50 μm. B) The radius of gyration, <Rg, structure2>1/2 for the 82 %, 88 % and 94 % conditions, yielding the largest sized multi-cellular structures in the 88 % alkyl hydrazone bonds condition. C) The scaling exponents for the 82 %, 88 % and 94 % alkyl hydrazone bonds conditions, with the 88 % condition yielding the highest α value, near 0.6, and the 82 % yielding the lowest, significantly different, α-value near 0.5. ∗p < 0.05, ∗∗p < 0.01 ns: no significance.
Here, Nv is as previously defined; Ivoxel is the size of the voxels, based on image acquisition parameters, for this paper defined as 0.6 μm, and α is the scaling exponent. By analogy to polymers in solution, this multi-cellular structure analysis uses the number of voxels (Nv) to represent the total number of monomers within the structure, and the length of the monomers is related to the size of the voxel (lvoxel). In addition, lower values of α (∼1/3) equate to more favorable chain-chain (or weaker cell-ECM) interactions and a more compact, globular state, whereas higher α-values (∼3/5) signify more favorable chain-solvent (or stronger cell-ECM) interactions and an expanded conformation [29].
For the multi-cellular MSC structures within the hydrazone gels, <Rg, structure2>1/2 was 96 ± 15 μm within the 88 % formulation, as compared to 48 ± 16 μm and 36 ± 14 μm in the 82 % and 94 % formulations, respectively, and followed a similar trend to that of the total nuclei/structure and paxillin intensity (Fig. 3B–S2). Using the power law analysis, the scaling exponent (α) varied as a function of matrix stress relaxation from 0.58 ± 0.05 in the 88 % hydrazone formulation, as compared to 0.48 ± 0.01 and 0.52 ± 0.04 for the 82 % and 94 % formulations, respectively, (Fig. 3C). Based on these trends with stress relaxation, we next sought to vary the matrix interactions by altering the matrix adhesivity (i.e, [RGD]) and later weaken cell-ECM interactions by the incorporation of HAVDI.
2.4. Structures of multi-cellular MSC clusters depend on hydrogel adhesivity and peptide identity
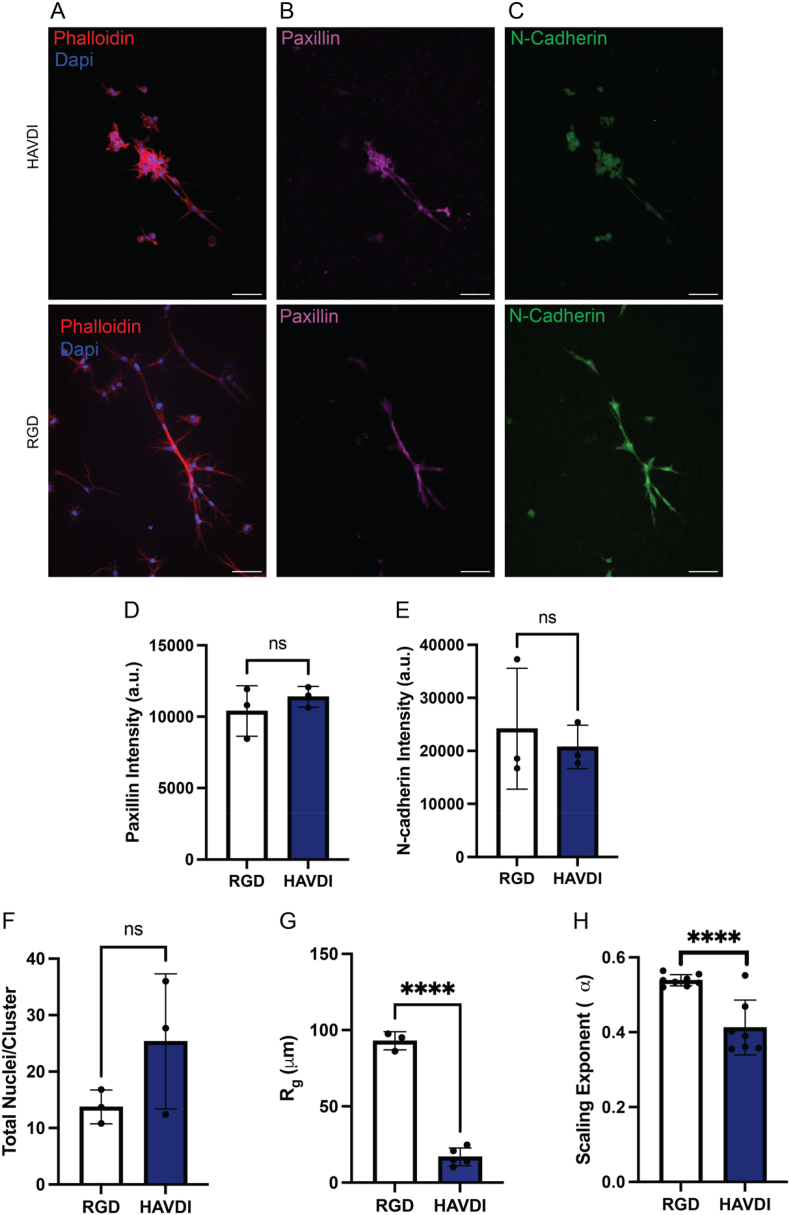
To probe the influence of mimetic peptides on MSC mechanosensing and quantify the impact of its effects on MSC cluster formation, MSCs were encapsulated at a density of 3E6 cell/mL in the 88 % hydrazone formulation but modified with either 1 mM of RGD or 1 mM of HAVDI. After 7 days of culture, the MSCs within the 88 % alkyl-hydrazone condition were immunostained for cell nuclei (DAPI), F-actin (AF-647 Phalloidin), N-cadherin expression (anti-N-Cadherin antibody) and paxillin expression (anti-paxillin antibody), and then imaged on a spinning disk confocal microscope. Notably, the 1 mM HAVDI conditions did not lead to the evolution of large, spread multi-cellular clusters as seen within the 1 mM RGD conditions, but rather, more globular, condensed multi-cellular clusters (Fig. 4A and B,C). Interestingly, there was no significant up or downregulation of either of N-Cadherin or paxillin expression intensity or cell number as compared to the 1 mM RGD condition, with the total number of nuclei within each cluster being similar (Fig. 4D and E,F). While these results are somewhat confounded about the specificity of the peptide interaction, MSC nascent protein deposition is known to increase with higher degrees of adaptability [25,30]. Therefore, the lack of a difference in N-cadherin and Paxillin staining could be explained by the presence of cell-secreted proteins.
Fig. 4.
Influence of RGD and HAVDI integrin-binding on multi-cellular structures. A) Representative image of a multi-cellular structure within the 88 % alkyl hydrazone bonds condition containing 1 mM HAVDI, scale bar = 50 μm. B) Representative image of the 88 % alkyl hydrazone bond condition cell-ECM interactions via visualization of mature focal adhesions, scale bar = 50 μm. C) Representative image of the 88 % alkyl hydrazone bond condition cell-cell interactions via N-cadherin, scale bar = 50 μm. D) The intensity expression of a mature focal adhesion marker, Paxillin, indicating similar expression regardless of presented peptide epitope. E) The intensity expression of a cell-cell junction marker, N-Cadherin, indicating similar expression regardless of presented peptide epitope. F) The total nuclei/structure, again, showing regardless of presented peptide-epitope the number of cell nuclei within each structure is similar. G) The average distance of all the pixels within the structure to the center of mass, or the radius of gyration, for the 1 mM RGD and 1 mM HAVDI conditions within the 88 % alkyl hydrazone bonds condition, with the 1 mM RGD condition leading to a greater radius of gyration, indicating an increase in cell-ECM interactions. H) The derived scaling exponent compared between the 1 mM RGD condition and the 1 mM HAVDI condition within the 88 % alkyl hydrazone bonds condition. The 1 mM RGD condition led to an increased scaling exponent, averaging around 0.56, in comparison to the 1 mM HAVDI condition, averaging around 0.39, indicating increased cell-ECM interactions as compared to cell-cell. ∗∗∗∗p < 0.0001, ns: no significance.
To further quantify the morphological differences observed, <Rg, structure2>1/2 was calculated for the 1 mM HAVDI condition and found to be 17 ± 5.8 μm. For comparison < Rg, structure2>1/2 for the 1 mM RGD condition was significantly larger, 93 ± 5.9 μm (Fig. 4G). In this way, the radius of gyration provides a way to quantify differences observed visually, where the MSC structures are substantially more collapsed in the HAVDI gels compared to the elongated structures in the RGD gels. In addition, the α scaling factor was calculated to be 0.39 ± 0.04 for the HAVDI formulations, as compared to 0.56 ± 0.05 for the RGD, further characterizing the compacted versus elongated structures, respectively (Fig. 4H). Taken together, these results showed that the presented mimetic peptide can significantly influence MSC structure organization and the formation of mature focal adhesions, which we posit is mediated by peptides that promote either weak or strong cell-ECM interactions. To probe this hypothesis, the ratio of RGD:HAVDI was further perturbed and its effects on the multi-cellular MSC structures examined.
2.5. Exploring the balance between cell-ECM and cell-cell interactions in mature focal adhesion and multi-cellular structure formation
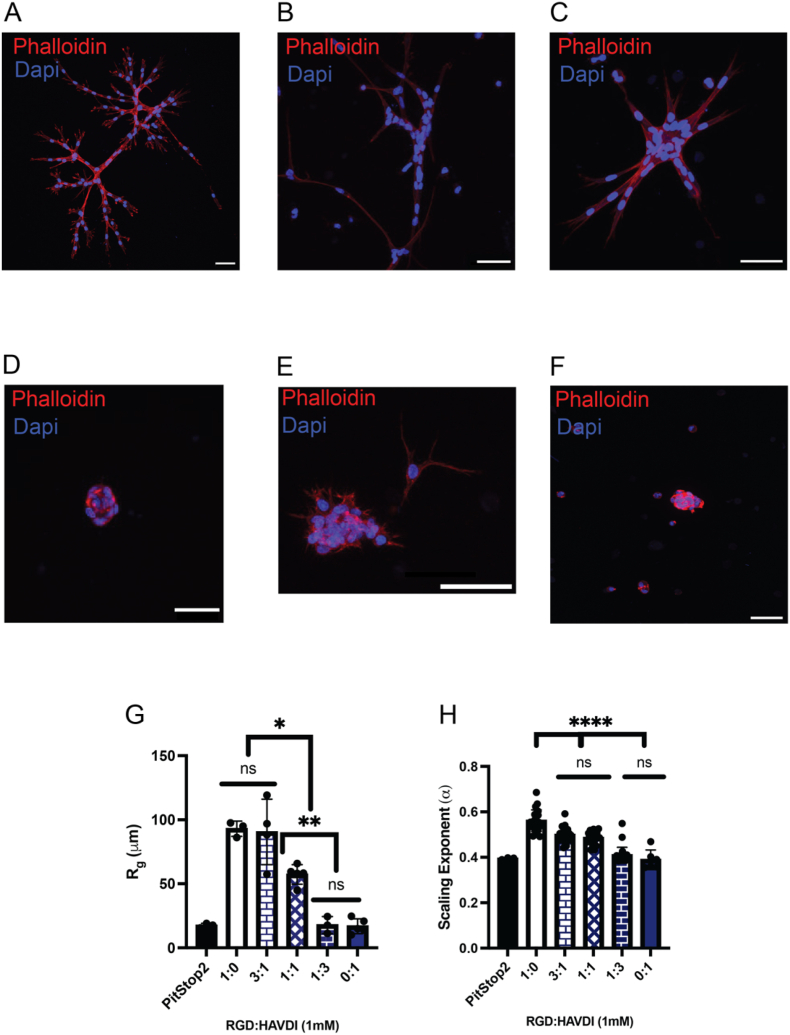
After establishing the differential effects of single peptide epitopes (RGD vs HAVDI) on MSCs and their formation of multi-cellular structures, we next sought to investigate the influence of combinations of RGD:HAVDI (1:0, 3:1, 1:1, 1:3, 0:1), using the same 88 % hydrazone condition, on MSCs. After 7 days in culture, MSCs were again immunostained for cell nuclei (DAPI), F-actin (AF-647 Phalloidin), N-cadherin expression (anti-N-cadherin antibody) and paxillin expression (anti-paxillin antibody), and the multi-cellular structures were visualized within these varying ratio conditions, via confocal microscopy (Fig. 5A–E). Of note, as the ratio of RGD: HAVDI was decreased, the multi-cellular structures became more compact and globular, with fewer extended processes, as visualized with DAPI and AF-647 Phalloidin (Fig. 5D and E). We also stained and confirmed the presence of paxillin marking mature focal adhesions (cell-matrix interactions) and N-Cadherin marking cell-cell interactions (Fig. S7). In general, there was a shift from elongated, extended structures to globular structures at a ratio of 1:3 RGD: HAVDI (Fig. 5A–D).
Fig. 5.
Influence of RGD: HAVDI dual incorporation on MSCs cell-ECM interactions. A-E) Representative images of a multi-cellular structure within the 88 % alkyl hydrazone bonds condition containing 1 mM RGD (A), 3:1 RGD: HAVDI (B), 1:1 RGD: HAVDI (C), 1:3 RGD:HAVDI (D) and 1 mM HAVDI (E) conditions, scale bar = 50 μm. F). The PitStop2, endocytosis inhibitor control representative image, nuclei (blue), and F-actin (red), scale bar = 50 μm. G) The derived radius of gyration compared between PitStop2, 1 mM RGD, 3:1 RGD: HAVDI, 1:1 RGD: HAVDI, 1:3 RGD:HAVDI and the 1 mM HAVDI condition within the 88 % alkyl hydrazone bonds condition. H) The derived scaling exponent compared between PitStop2, 1 mM RGD, 3:1 RGD: HAVDI, 1:1 RGD: HAVDI, 1:3 RGD:HAVDI and the 1 mM HAVDI condition within the 88 % alkyl hydrazone bonds condition. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, ns: no significance.
Further, to explore the necessity of mature stable focal adhesion formations, and better understand the role of cell interactions via paracrine signaling to induce the formation of elongated multi-cellular structures, MSCs were cultured in RGD (1 mM) hydrogels and treated with PitStop2, which inhibits endocytosis of proteins or encapsulated in unmodified formulations. Within the 88 % alkyl hydrazone bond condition containing 1 mM RGD and PitStop2, no significant formation of elongated multi-cellular structures was observed, with few globular structures of more than 5 cells (Fig. 5F). None of the globular multi-cellular structures were seen to the same extent as with the incorporation of either of the mimetic peptides. Upon analysis of the globular structures within the PitStop2 condition, the average size of these structures was found to be <Rg, structure2>1/2 = 17 ± 1.7 μm, with an α-value of 0.39 ± 0.004 (Fig. 5G and H). Furthermore, in the 88 % alkyl hydrazone bond condition containing no RGD or HAVDI, some of the MSCs were observed to have formed small filipodia or protrusions extending from individual cells, although, again not to the same extent as the cells seen within the mimetic peptide containing conditions, and no multi-cellular structures were observed (Fig. S8). Taken together, these results indicate that the RGD peptide is necessary to initiate strong cell-ECM interactions, and in conjunction with cell endocytosis, leads to the formation of stable mature focal adhesions which enables the formation of the elongated multicellular structures.
Upon application of the scaling analysis to the images, with the inclusion of HAVDI, α-values were observed to decrease substantially, even at the 3:1 ([RGD]: [HAVDI]) condition, where α = 0.50 ± 0.03 and <Rg, structure2>1/2 = 90 ± 25 μm, in comparison to the 1 mM RGD only condition (α = 0.56 ± 0.05, <Rg, structure2>1/2 = 93 ± 5.9 μm) (Fig. 5G and H). Additionally, in comparison to the 1 mM HAVDI only condition, the encapsulated MSCs were observed to form more elongated multi-cellular structures up to a ratio of 1:1 (RGD: HAVDI), with an α-value of 0.48 ± 0.03 and <Rg, structure2>1/2 = 57 ± 7.6 μm. These results indicated how the presented peptides influence the strength of the cell-ECM interactions, enabling stable mature focal adhesion formation. Upon the incorporation of RGD: HAVDI at a ratio of 1:3 the multi-cellular structures were noticed to become more globular in structure, similar to those observed within the 1 mM HAVDI condition, with an α value of 0.41 ± 0.04 and <Rg, structure2>1/2 = 18 ± 6.6 μm, which was similar to that of the 1 mM HAVDI condition (α = 0.39 ± 0.04, <Rg, structure2>1/2 = 17 ± 5.8 μm) (Fig. 5G and H). Taken together, these results indicate that stable cell-ECM interactions are necessary to form the elongated multicellular structures, while maintaining cell-cell interactions, whereas the formation of globular multicellular structures occurs strongly through less stable cell-ECM interactions.
2.6. Materials directed multi-cellular structure formation alters MSC cytokine secretory profiles
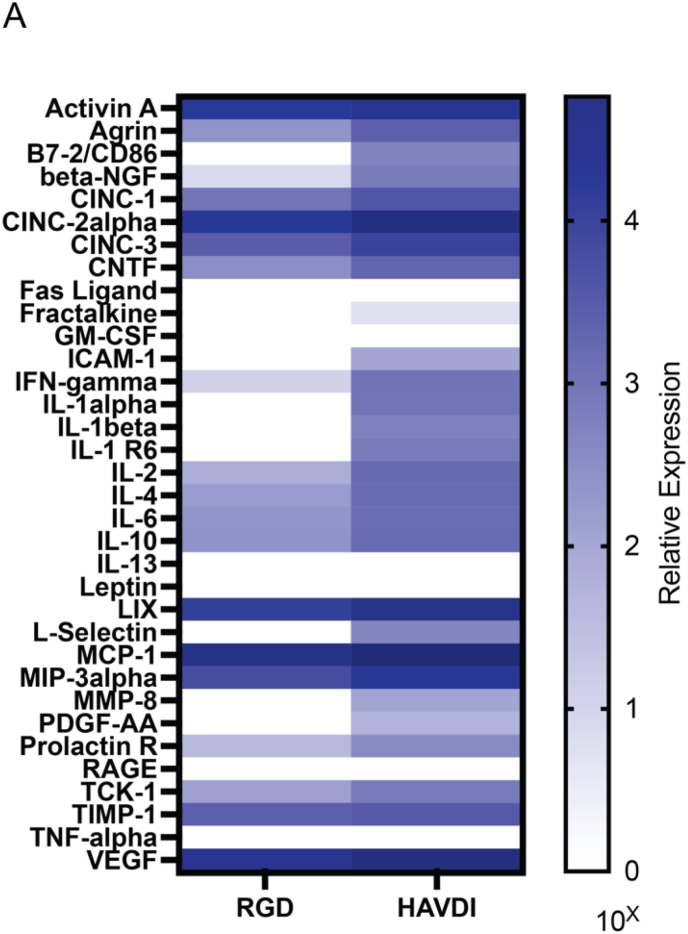
Finally, to understand the functional influence of cell-ECM interactions and the resulting multi-cellular MSC structures on their on their secretory profiles, MSCs were cultured in the 88 % alkyl hydrazone hydrogel modified with either 1 mM RGD (<Rg, structure2>1/2 = 93 ± 5.9 μm, α = 0.56 ± 0.05) or 1 mM HAVDI (<Rg, structure2>1/2 = 17 ± 5.8 μm, α = 0.39 ± 0.04). The MSCs were cultured for 7 days, and the media was collected from day 4 to day 7 to analyze the cytokine content via an array (RayBiotech C2 Array). To normalize the array to cell number, MSC-laden gels were hyaluronidase digested to obtain the total DNA content measured with a PicoGreen assay, and the obtained cytokines values were subsequently normalized to these concentrations (Fig. S10). Notably, MSCs in the HAVDI condition had higher levels of secretion of all cytokines relative to the RGD condition (Fig. 6A). With this in mind, these results indicate cytokine secretion alone is unable to give rise to the elongated multicellular structures, as while the secretion was greatest in the HAVDI condition, the elongated multicellular structures were not observed. This shows a cooperative effect between the MSC secretome and the mechanical microenvironment to form the observed elongated multicellular structures.
Fig. 6.
Integrin presentation influences MSC secretion independent of stable cell-ECM interactions. A) Heatmap of cytokine expression of MSCs encapsulated in the 88 % alkyl hydrazone condition with either 1 mM RGD or 1 mM HAVDI, normalized to total DNA concentration/hydrogel. Blue intensities represent high expression values, and white intensities represent low expression values.
3. Discussion
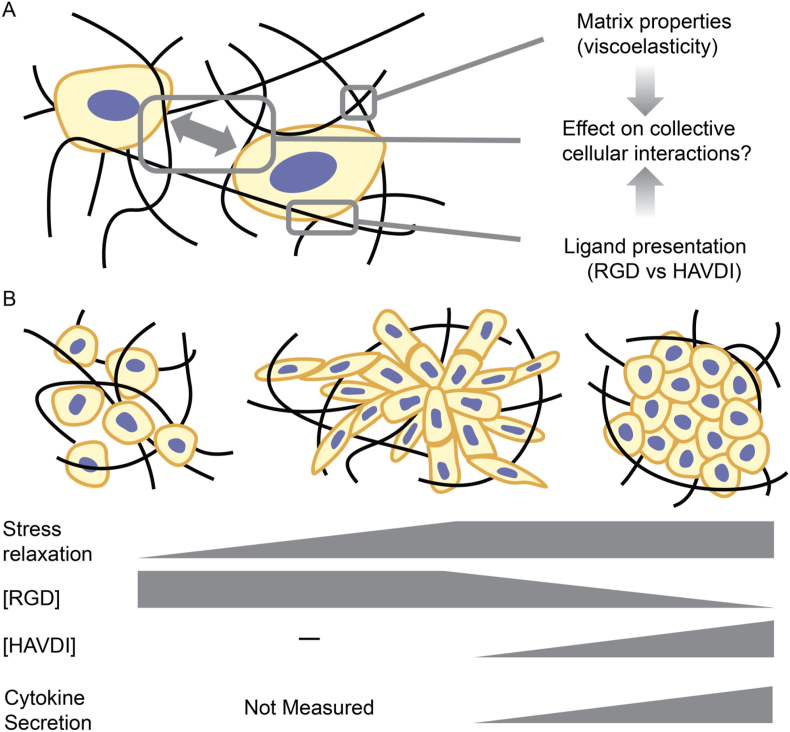
Covalent adaptable networks have been of growing interest to tune the biophysical and biochemical cues which cells receive upon encapsulation into a matrix, to most closely match native ECM [23]. In this work, CANs were formulated by synthetizing HA functionalized with either an alkyl-aldehyde or hydrazide group, forming the dynamic hydrazone chemistry. Additionally, stable triazole bonds were incorporated into the system via a PEG-BCN macromer and a small molecule, benzaldehyde-PEG-azide, which can react with the HA-hydrazide macromer. This enabled the tuning of the hydrazone vs triazole bonds present within the system to control the adaptability and stability of the network. Ultimately, this allowed for the control over the percent of stress relaxed, ranging from 86 ± 2.9 % (94 % alkyl-hydrazone bond condition) to 65 ± 8.9 % (82 % alkyl-hydrazone bond condition). In addition, the presented peptide epitope was varied, containing either 1 mM RGD, 1 mM HAVDI or a ratio of the two within the system. This enabled control over the biochemical cues which the MSCs received leading to changes in their secretory properties. Specifically, using this material system we were able to examine relative effects of stress relaxation versus mimetic peptide presentation on the regulation of the extent of cell-ECM interactions and the ensuing formation of multi-cellular structures of MSCs (Fig. 7A).Scaling theories developed to describe polymer chain structures in solution were applied to quantify how elongated or compacted the multi-cellular structures were, and further elucidated the extent and stability of cell-ECM interactions present within these structures. Here we found, upon utilizing the 88 % alkyl-hydrazone bonds condition, with 1 mM RGD the most elongated structures could be formed, with an α value of 0.56 ± 0.05 and <Rg, structure2>1/2 of 93 ± 5.9 μm, in contrast the conditions containing 1 mM HAVDI led to the most compacted structures with an α value of 0.39 ± 0.04 and a <Rg, structure2>1/2 of 17 ± 5.8 μm. Further, structures which were more compacted than the 1 mM RGD condition, but more elongated than the 1 mM HAVDI condition could be formed by titrating in an equal ratio of [RGD]: [HAVDI] into system (1:1 ratio), where α = 0.48 ± 0.03 and <Rg, structure2>1/2 = 57 ± 7.6 μm.
Fig. 7.
Representative schematic of the influence of varying mimetic peptides and stress relaxation on MSC mechanosensing, specifically the cell-ECM interactions and the ensuing formation of multi-cellular structures.
From the literature, Cosgrove and coworkers showed that HAVDI interactions can induce shielding of MSCs, reducing their mechanosensitivity to the substrate. These results supported the role of inhibition of Rac1-GTP levels, broadly resulting in reduced Yap nuclear localization and reducing mature focal adhesion formation [9]. This further elucidates why, upon incorporation of HAVDI, even at a lower ratio, the stable mature focal adhesion formation and ensuing cell-ECM interactions were decreased as seen by the significantly reduced α-values, and less elongated structures, in comparison to the RGD only condition [9]. Additionally, Caldwell and coworkers showed that upon incorporation of HAVDI into a microgel system, which induced cell clustering, the secretory profile of the MSCs were upregulated in comparison to RGD conditions [31]. Rao and coworkers then expanded upon this work, by using HAVDI to activate the N-Cadherin pathway to induce pro-resorptive secretion by the encapsulated MSCs, for applications towards treating osteoporosis [32]. This illuminates how weaker cell-ECM interactions could enable more dominant cell-cell interactions to induce this global upregulation of secretory properties as seen within the HAVDI containing condition and could be further tuned for therapeutic applications. Simply, these alterations of the cellular microenvironment impacted cellular interactions and attenuated MSCs ability to form stable mature focal adhesions and directed their ensuing cytokine secretion. Collectively, these data establish the isolation of the influence of cell-ECM interactions and the cooperative role between the presented ligand and MSC secretion and provide insight onto the specific role they play in the formation of multi-cellular structures and how these structures change conformation (i.e. elongated versus globular) (Fig. 7b).
Further, this alludes to the conclusion that cells, specifically MSCs, sense differences in stress relaxation through integrin interactions [23,33]. As upon interaction with RGD, and the ensuing formation of focal adhesions, as visualized via paxillin, the MSCs can sense the relaxing matrix, leading to increased cell spreading, and allowing the formation of these elongated structures. However, upon encapsulation with HAVDI, the cells are unable to form stable mature focal adhesion within their environment. In these conditions, MSCs are interacting with the presented ligand, HAVDI, reducing their ability to strongly interact with their ECM, and impacting the cells capacity to spread and form these elongated structures. It should be noted that all of our hydrogels are formed using hyaluronic acid macromers which can facilitate cell-ECM interactions with hyaluronic acid being sensed by CD44, as shown in previous work [34]. This inherently introduced a form of cell-ECM interactions within all of the conditions; however, significant changes to the scaling exponent are seen to be dependent upon the presented peptide epitope, highlighting the sensitivity of this method to infer changes to the cellular interactions.
More generally, this work highlights how MSCs interact in differing microenvironments, be that in a stress relaxing environment, or with differing mimetic peptides, and how these signals influence the complex formation of multi-cellular structures and MSC secretion. Future work could focus on a fundamental understanding of how these cell-ECM interactions impact the stress relaxation of the hydrogel system in a local manner. Beyond the bulk material properties measured using shear rheology, micro-rheological tools, such as optical tweezers, could be employed to more accurately understand local mechanics and influences of cells on matrix stress relaxation [35,36]. Additionally, future work could focus on utilizing this knowledge to further prime the encapsulated MSCs to secrete specific cytokines, such as anti-inflammatory cytokines, for therapeutic applications. For instance, Rao and coworkers found upon clustering of MSCs derived from ovariectomized rats, cytokine factors which support an osteoporotic environment, pro-resorptive factors, were observed. However, upon blocking of N-Cadherin interactions, the secretion of those pro-resorptive factors was decreased [37]. Indicating the role of cell-cell junction interactions. Additionally, this system allows for spontaneous formation of elongated (RGD) or aggregated (HAVDI) MSC multicellular structures within materials of uniform mechanical properties. Beyond functional assays such as the cytokine array used in this study, how this cluster morphology influences cell fate decisions and MSC differentiation is an interesting area of research for future studies. Furthermore, another area which is yet to be investigated is the influence of these cell-ECM interactions on the composition of secreted extracellular vesicles (EVs), as EVs are known to play an important role in cell signaling and immunomodulation through their contents [38,39]. Therefore, the designed system could be used to tune the biochemical and biophysical influences on MSCs and observe the ensuing changes in EV secretion, which is of growing interest within the field of tissue engineering and regenerative medicine. In conclusion, these results suggest a deeper understanding of the mechanism behind the formation of these multi-cellular structures, specifically with regards to the quantification of the cell-ECM interactions and those influences on the conformation of the structures. Further, this knowledge could be utilized in the design of biomaterials to regulate MSC secretion, which has been implicated in many regenerative therapies and tissue engineering applications.
4. Materials and methods
4.1. Synthesis of HA-aldehyde and characterization
HA-Ald was synthesized following previously established protocols [24,27]. Briefly, Ha (500 kDa, 500 mg) was dissolved in dH2O (50 mL), then, sodium periodate (267.5 mg, 1:1 periodate/HA molar ratio) was added into the reaction mixture. The reaction was stirred in the dark for 2 h and successively quenched with the addition of ethylene glycol (70 μL). The reaction was then dialyzed for 3 days against dH2O (8000 MWCO) and lyophilized for 3 days to yield a white powder (471 mg, 94 % yield). HA-Ald was flash frozen in liquid nitrogen and stored at −20 °C until further use. The functionalization of the HA-Ald macromer was quantified using a 2,4,6-Trinitrobenzene Sulfonic Acid (TNBS) assay as previously described [40,41]. Briefly, the HA-Ald was dissolved at 2 wt% (w/v) and then reacted with tert-Butyl carbazate (t-BC, in 1 % trichloroacetic acid) in dH2O. After 24 h, the HA-Ald/t-BC and t-BC standards were reacted with 0.5 mL TNBS (6 mM in 0.1 sodium tetraborate at pH 8) for 1 h. Samples were then reacted with 0.5 N hydrochloric acid and measured at an absorbance of 340 nm on a microplate reader (≈37 % functionalization) (Figs. S10 and 11).
4.2. Synthesis of HA-hydrazide and characterization
HA-Hyd was synthesized following previously established protocols [24,27]. Briefly, Ha (60 kDa, 500 mg) was dissolved in dH2O (100 mL) before adipic acid dihydrazide (≈6.5 mg, >60x molar excess) was added in large molar excess to the reaction and the pH was adjusted to 6.8. 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC, 776 mg, 4 mmol) and hydroxybenzotriazole (HOBt, 765 mg, ≈6 mmol) were separately dissolved in a DMSO/dH2O mixture (1:1) and added dropwise to the reaction. The pH was adjusted to 6.8 every 30 min for 4 h and then allowed to react for 24 h. The solution was dialyzed against dH2O (8000 MWCO) for 3 days and then lyophilized for 3 days. Once dried, the products were weighed, dissolved in a 5 wt% NaCl/dH2O solution, and precipitated into pure ethanol and dialyzed for another 3 days. Products were then lyophilized for 3 days to yield a white powder (430 mg, 86 % yield), and flash frozen in liquid nitrogen and stored at −20 °C until use. HA-Hyd was characterized using 1H NMR (≈40 % functionalization) (Figs. S10 and 12).
4.3. Synthesis of PEG-BCN and characterization
PEG-BCN was synthesized following previously established protocols [42]. Briefly, an 8-arm 40 kDa PEG-amine (1.0 g, 0.2 mmol amine) and BCN-oSu (0.1 g, 0.343 mmol, 1.7x) were added to a 50 mL round bottom (RB) flask (dried overnight at 80 °C). A minimal amount of anhydrous DMF was added to the RB flask to dissolve the contents (10 mL). The flask was then placed under argon and stirred at room temperature. N,N-Diisopropylethyleamine (0.8 mmol, 4x) was added to the solution and the reaction was left to proceed overnight. The following day, the reaction mixture was diluted with dH2O and dialyzed for 3 days (8000 MWCO). The dialyzed solution was lyophilized for 3 days, resulting in a dried white powder as the PEG-BCN product (0.985 g, 98 % yield). End group functionalization was confirmed using 1H NMR (>95 % functionalization) (Fig. S13).
4.4. Hydrogel formation and rheological characterization
Functionalized HAs were dissolved in phosphate buffered saline (PBS, pH 7.4) at 3 wt%. Functionalized PEG-BCN was dissolved in PBS at 10 wt% and the azide-PEG3-phenolaldehyde (BroadPharm) crosslinker was dissolved at a concentration of 20 mm. Hydrogels were formed with 3 w/v% final polymer content based on stoichiometry. In situ rheology measurements were performed using a TA Instruments DHR-3 rheometer equipped with an 8 mm parallel plate geometry. Hydrogel formation was evaluated by time sweeps (1.0 Hz; 0.5 % strain). For stress relaxation experiments, 6 h of measurement followed a 10 % strain applied over 1 s, as dictated by previously published results [27]. Formulations were defined by varying the percentage of alkyl hydrazone bonds present within the hydrogel by modifying the HA-hydrazide with a small molecule, benzaldehyde-PEG-azide, which effectively converts the HA-hydrazide to an azide. The formulations consisted of 94 % (i.e., 6 % of the functional HA-hydrazide arms were functionalized with benzaldehyde-PEG-azide), 88 % alkyl hydrazone, and 82 % alkyl hydrazone bond conditions, incorporating the necessary proportions of PEG-BCN and HA to achieve these functionalities. Mineral oil was applied to the hydrogel perimeter to prevent evaporation during the experiment.
4.5. Cell culture and encapsulation
Sprague–Dawley rMSCs were purchased from Cyagen and were expanded and cultured following the manufacturer's protocol, and as previously described [27]. Briefly, the cells were obtained at passage 2 and expanded to passage 5 in growth medium specifically, Dulbecco's Modified Eagle Medium (ThermoFisher) of low glucose (1 ng mL−1 glucose) supplemented with 10 % FBS (Gibco), 50 μg mL−1 streptomycin (Gibco), and 0.5 μg mL−1 of Amphotericin B (Gibco). The media was changed after 24 h, and then every three days until 80–90 % cell confluency was reached. When the desired cell confluency was reached, the cells were stored at passage 5 in liquid nitrogen. For conditions containing RGD, hydrogels were fabricated by pre-reacting the HA-Hyd with the benzaldehyde-PEG-azide crosslinker, and KGRGDS based on stoichiometry for the desired 3 wt% hydrogel formulation, of 94 %, 88 % and 82 % and allowing them to react overnight. The following day, the HA-Ald, PEG-BCN, and PBS were mixed at stoichiometric ratios in a modified syringe barrel. rMSCs at passage 5 were thawed and placed in growth medium. The rMSC suspension was centrifuged (200 rcf, 5 min) and the resulting pellet was resuspended in the HA-Hyd, benzaldehyde-PEG-azide, KRGDS mixture for a cell density of 3 million cells mL−1. The cell suspension was then mixed into the syringe barrel with the HA-Ald, PEG-BCN, and PBS. The hydrogels were allowed to react until a soft gel formed (5 min or less) and then placed onto benzaldehyde functionalized coverslips and allowed to react for an additional 2 min before 1 mL growth medium was added. This enabled the formation of a 30 μL hydrogel, in a disk shape, with a diameter of ∼4.78 mM and a thickness of ∼1.29 mM, to be reproducibly created for all conditions. For conditions containing HAVDI, hydrogels were fabricated by pre-reacting the HA-Hyd with the benzaldehyde-PEG-azide crosslinker on stoichiometry for the desired 3 wt% hydrogel formulation of 94 %, 88 % and 82 % and allowing them to react overnight. The following day, the HA-Ald, PEG-BCN, HAVDI and PBS were mixed at stoichiometric ratios in a modified syringe barrel. The cell encapsulation continued as previously stated above. For conditions containing both KRGDS and HAVDI, hydrogels were fabricated by pre-reacting the HA-Hyd with the benzaldehyde-PEG-azide crosslinker, and KGRGDS based on stoichiometry for the desired 3 wt% hydrogel formulation of 94 %, 88 % and 82 % and allowing them to react overnight. The following day, the HA-Ald, PEG-BCN, HAVDI and PBS were mixed at stoichiometric ratios in a modified syringe barrel. The cell encapsulation continued as previously stated above.
4.6. Cell morphology, paxillin, and N-cadherin staining
To analyze morphological changes and Paxillin and N-Cadherin signaling, rMSCs grown in the HA-PEG hydrogels, at a concentration of 3 million cells mL−1, were fixed with formalin (30 min, room temperature) following 7 days of growth and then rinsed with PBS. After fixation, the hydrogels were permeabilized with 0.1 % Triton X-100 in PBS (1h, room temperature) and blocked using 5 % BSA (Fisher Scientific) in PBS (1h, room temperature, or overnight, 4 °C). Following blocking, the samples were washed with PBS three times for 5 min intervals at RT. After washing the samples were incubated with the corresponding primary antibodies, Paxillin (Rabbit mAb, Cell Signal 1:400) and N-Cadherin (Mouse mAb, Cell Signal 1:400) in blocking buffer (overnight, 4 °C). The following day the samples were again washed with PBS three times for 5 min intervals at RT, and then incubated with the corresponding secondary antibodies: DAPI (Nuclei, Sigma-Aldrich, 1:1000), Alexa Flour 488 Goat anti-mouse (N-Cadherin, Invitrogen, 1:250), Alexa Flour 555 Goat anti-rabbit (Paxillin, Invitrogen, 1:250) and Alexa Flour 647 Phalloidin (F-actin, Invitrogen,1:300) in PBS with 0.05 % Tween 20 (PBST) (overnight, 4 °C) (morphology). After washing with PBST three times to remove excess stain, the fluorescently labeled rMSCs were imaged using confocal microscopy (Zeiss LSM 710 or Nikon Spinning Disk) with a Plan-Aprochromat 20x (NA -1.0) water objective. ImageJ (NIH) was used to visualize and quantify morphological changes and Paxillin and N-Cadherin intensities using the analyze particles plugin.
4.7. Secretome analysis
Global secretome analysis was measured using a Rat Cytokine Array C2 (RayBiotech) and the manufacturer's protocol was followed. Briefly, media was collected from hydrogel samples between days 4 and day 7. Arrays were blocked for 30 min at room temperature and subsequently incubated with 1 mL of media from each condition and acellular controls overnight at 4 °C. Arrays were washed with the provided washing buffer from the manufacturer for a total of five washes. Next, the arrays were incubated with a biotinylated antibody cocktail overnight at 4 °C. Each array was again washed and the incubated with HRP-streptavidin overnight at 4 °C. After incubation the arrays were again washed and then incubated for 5 min with the detection buffers. Chemiluminescence was detected using a charge-coupled device camera (Image Quant 800). Exposure and incubation times were kept constant between each condition and controls. Raw images were analyzed using the 2D Array feature of ImageQuant (GE Healthcare). The analysis was conducted using the provided excel spreadsheet from the manufacturer's website and the first normalization data was utilized. From here the acellular control signal was subtracted. Intensities from corresponding spots from control arrays were subtracted and each value was normalized to μg DNA as determined by Quant-it Pico Green assay.
Before running the assay, microgels scaffolds were digested in 1 mg mL−1 Papain enzyme (Sigma) in PBE buffer containing 1.77 mg mL−1 L-cysteine and 100 μg mL−1 overnight at 65 °C. DNA concentration per gel was determined using manufacturer's protocol for the Quant-it Pico Green assay.
4.8. Statistical analysis
All data were collected using 3 hydrogel replicates per condition. For each hydrogel condition at least 3 multi-cellular structures per hydrogel were analyzed across all conditions. Data were compared using one-way ANOVAs with Tukey post-hoc comparisons or Student's t-test in Prism 8 (GraphPad Software, Inc). Data were presented as mean ± standard deviation.
Ethics approval and consent to participate
No approval from any ethical committees was required to complete the research presented in this manuscript. No informed consent, animal or human subjects were used in the research of this manuscript.
CRediT authorship contribution statement
Alexandra N. Borelli: Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Courtney L. Schultze: Formal analysis. Mark W. Young: Writing – review & editing, Formal analysis, Conceptualization. Bruce E. Kirkpatrick: Writing – review & editing, Visualization. Kristi S. Anseth: Writing – review & editing, Supervision, Resources, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Kristi Anseth has patent compositions and methods for making and using hybrid network hydrogels pending to the University of Colorado. Alexandra Borelli has patent compositions and methods for making and using hybrid network hydrogels pending to The University of Colorado. Mark Young has patent compositions and methods for making and using hybrid network hydrogels pending to The University of Colorado. Other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge support from the National Institutes of Health, United States (Grant: RO1 DE016523). This work was supported in part by a grant from the United States Defense Advanced Research Projects Agency (DARPA, Grant: W911NF-19-2-0024). The authors thank Dr. Monica Ohnsorg for meaningful discussion of results. The imaging work for Fig. 4, Fig. 5 were performed at the Biofrontiers Institute's Advanced Light Microscopy Core (RRID: SCR_018302). Spinning disc confocal microscopy was performed on Nikon Ti-E microscope supported by the BioFrontiers Institute and the Howard Hughes Medical Institute. Cytokine Arrays (Fig. 6) were imaged using the Amersham ImageQuant 800 biomolecular imager in the Shared Instruments Pool (SIP) core facility (RRID: SCR_018986), Department of Biochemistry, University of Colorado Boulder. Fig. 1, Fig. 7 schematics were created with Adobe Illustrator and Affinity Designer, respectively.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.10.007.
Supporting Information
Supporting Information is available to download or from the author.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Chaudhuri O., Cooper-White J., Janmey P.A., Mooney D.J., Shenoy V.B. Nature. 2020;584:7822. doi: 10.1038/s41586-020-2612-2. 2020, 584, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanton A.E., Tong X., Yang F. Acta Biomater. 2019;96:310. doi: 10.1016/j.actbio.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wechsler M.E., Rao V.V., Borelli A.N., Anseth K.S. Adv. Healthcare Mater. 2021;10 doi: 10.1002/adhm.202001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M G., Ja B. Curr. Opin. Biotechnol. 2013;24:841. doi: 10.1016/j.copbio.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong Z., Jin L., Oliveira J.M., Reis R.L., Zhong Q., Mao Z., Gao C. Bioact. Mater. 2021;6:1375. doi: 10.1016/j.bioactmat.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kechagia Z., Sáez P., Gómez-González M., Canales B., Viswanadha S., Zamarbide M., Andreu I., Koorman T., Beedle A.E.M., Elosegui-Artola A., et al. Nat. Mater. 2023;22:1409. doi: 10.1038/s41563-023-01657-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yui S., Azzolin L., Maimets M., Pedersen M.T., Fordham R.P., Hansen S.L., Larsen H.L., Guiu J., Alves M.R.P., Rundsten C.F., et al. Cell Stem Cell. 2018;22:35. doi: 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark A.Y., Martin K.E., García J.R., Johnson C.T., Theriault H.S., Han W.M., Zhou D.W., Botchwey E.A., García A.J. Nat. Commun. 2020;11:1. doi: 10.1038/s41467-019-14000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosgrove B.D., Mui K.L., Driscoll T.P., Caliari S.R., Mehta K.D., Assoian R.K., Burdick J.A., Mauck R.L., Professor of Orthopaedic Surgery A. N. Mater Author manuscript. Nat. Mater. 2016;15:1297. doi: 10.1038/nmat4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldwell A.S., Rao V.V., Golden A.C., Anseth K.S. Biomaterials. 2020;232 doi: 10.1016/j.biomaterials.2019.119725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García J.R., Clark A.Y., García A.J. J. Biomed. Mater. Res. 2016;104:889. doi: 10.1002/jbm.a.35626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ligorio C., Mata A. Nature Reviews Bioengineering. 2023;1:518. doi: 10.1038/s44222-023-00055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang S., Richardson B.M., Anseth K.S. Prog. Mater. Sci. 2021;120 [Google Scholar]
- 14.Zhou H., Liang C., Wei Z., Bai Y., Bhaduri S.B., Webster T.J., Bian L., Yang L. Mater. Today. 2019;28:81. [Google Scholar]
- 15.Chaudhuri O., Gu L., Klumpers D., Darnell M., Bencherif S.A., Weaver J.C., Huebsch N., Lee H.P., Lippens E., Duda G.N., et al. Nat. Mater. 2016;15:326. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu D.T., Jeffreys N., Diba M., Mooney D.J. Tissue Eng. C Methods. 2022;28:289. doi: 10.1089/ten.tec.2022.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinnon D.D., Domaille D.W., Cha J.N., Anseth K.S. Adv. Mater. 2014;26:865. doi: 10.1002/adma.201303680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson B.M., Wilcox D.G., Randolph M.A., Anseth K.S. Acta Biomater. 2019;83:71. doi: 10.1016/j.actbio.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang S., Ma H., Tu H.C., Wang H.R., Lin P.C., Anseth K.S. Adv. Sci. 2018;5:1. doi: 10.1002/advs.201800638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown T.E., Carberry B.J., Worrell B.T., Dudaryeva O.Y., McBride M.K., Bowman C.N., Anseth K.S. Biomaterials. 2018;178:496. doi: 10.1016/j.biomaterials.2018.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yavitt F.M., Kirkpatrick B.E., Blatchley M.R., Speckl K.F., Mohagheghian E., Moldovan R., Wang N., Dempsey P.J., Anseth K.S. Sci. Adv. 2023;9 doi: 10.1126/sciadv.add5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson B.R., Kirkpatrick B.E., Miksch C.E., Davidson M.D., Skillin N.P., Hach G.K., Khang A., Hummel S.N., Fairbanks B.D., Burdick J.A., et al. Adv. Mater. 2023 doi: 10.1002/adma.202211209. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou J., Stowers R., Nam S., Xia Y., Chaudhuri O. Biomaterials. 2018;154:213. doi: 10.1016/j.biomaterials.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Wang L.L., Highley C.B., Yeh Y.C., Galarraga J.H., Uman S., Burdick J.A. J. Biomed. Mater. Res. 2018;106:865. doi: 10.1002/jbm.a.36323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borelli A.N., Young M.W., Kirkpatrick B.E., Jaeschke M.W., Mellett S., Porter S., Blatchley M.R., Rao V.V., Sridhar B.V., Anseth K.S., et al. Adv. Healthcare Mater. 2022;11 doi: 10.1002/adhm.202200393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice M.A., Anseth K.S. J. Biomed. Mater. Res. 2004;70:560. doi: 10.1002/jbm.a.30106. [DOI] [PubMed] [Google Scholar]
- 27.A. N. Borelli, M. W. Young, B. E. Kirkpatrick, M. W. Jaeschke, S. Mellett, S. Porter, M. R. Blatchley, V.V. Rao, B.V. Sridhar, K. S. Anseth, et al., 2022, DOI 10.1002/adhm.202200393.
- 28.Flory P.J. J. Chem. Phys. 1951;19:1315. [Google Scholar]
- 29.Hofmann H., Soranno A., Borgia A., Gast K., Nettels D., Schuler B. Proc. Natl. Acad. Sci. U.S.A. 2012;109 doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loebel C., Mauck R.L., Burdick J.A. Nat. Mater. 2019;18:883. doi: 10.1038/s41563-019-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldwell A.S., Rao V.V., Golden A.C., Anseth K.S. Biomaterials. 2020;232 doi: 10.1016/j.biomaterials.2019.119725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao V.V., Wechsler M.E., Cravens E., Wojda S.J., Caldwell A.S., Kirkpatrick B.E., Donahue S.W., Anseth K.S. Acta Biomater. 2022;145:77. doi: 10.1016/j.actbio.2022.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam S., Stowers R., Lou J., Xia Y., Chaudhuri O. Biomaterials. 2019:15. doi: 10.1016/j.biomaterials.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Y. Kwon, C. Wang, J. H. Galarraga, E. Puré, L. Han, J. A. Burdick, 2019, DOI 10.1016/j.biomaterials.2019.119451. [DOI] [PMC free article] [PubMed]
- 35.Yavitt F.M., Kirkpatrick B.E., Blatchley M.R., Speckl K.F., Mohagheghian E., Moldovan R., Wang N., Dempsey P.J., Anseth K.S. Sci. Adv. 2023;9 doi: 10.1126/SCIADV.ADD5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson-Anderson R.M. ACS Macro Lett. 2018;7:968. doi: 10.1021/acsmacrolett.8b00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao V.V., Wechsler M.E., Cravens E., Wojda S.J., Caldwell A.S., Kirkpatrick B.E., Donahue S.W., Anseth K.S. Acta Biomater. 2022;145:77. doi: 10.1016/j.actbio.2022.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ju Y., Hu Y., Yang P., Xie X., Fang B. Materials Today Bio. 2023;18 doi: 10.1016/j.mtbio.2022.100522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y., Wu Y., Yang D., Neo S.H., Kadir N.D., Goh D., Tan J.X., Denslin V., Lee E.H., Yang Z. Bioact. Mater. 2023;27:98. doi: 10.1016/j.bioactmat.2023.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell B.A.J., Brendan P., David Lobb, Manoj Charati B., Shauna Dorsey M., Ryan Wade J., Kia Zellers N., Heather Doviak, Sarah Pettaway, Christine Logdon B., James Shuman, Parker Freels D., Joseph Gorman H., Robert Gorman C., Francis Spinale G. Nat. Mater. 2014;13:653. [Google Scholar]
- 41.Su W.Y., Chen Y.C., Lin F.H. Acta Biomater. 2010;6:3044. doi: 10.1016/j.actbio.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 42.Deforest C.A., Tirrell D.A. Nat. Mater. 2015;14:523. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.