Abstract
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) open reading frame 50 (ORF50) encodes a viral transcriptional activator, which binds to the KSHV promoter and stimulates the transcription of viral early and late genes, thus activating the lytic cycle of KSHV. We report here that KSHV ORF50 binds to the cellular proteins CREB-binding protein (CBP) and histone deacetylase (HDAC) and these binding events modulate ORF50-activated viral transcription. Binding of ORF50 to CBP and HDAC activates and represses, respectively, ORF50-mediated viral transcription. KSHV ORF50 was shown to bind to the C/H3 domain and the C-terminal transcriptional activation domain of CBP, while CBP bound to the amino-terminal basic domain and the carboxyl-terminal transactivation domain of ORF50. The LXXLL motif within the transcriptional activation domain of ORF50 is reminiscent of the CBP-binding sequence found in nuclear receptor proteins. The adenovirus E1A protein, which also binds to the C/H3 domain of CBP, repressed the transcriptional activation activity of ORF50. The cellular protein c-Jun, which binds to the kinase-induced activation domain of ORF50, stimulated ORF50-mediated viral transcription. The HDAC1-interacting domain of ORF50 was shown to be a central proline-rich sequence. Our data provide a framework for delineating the regulatory mechanisms used by KSHV to modulate its transcription and replication through interaction with both histone acetyltransferases and HDACs.
The acetylation states of histones have been known to correlate with transcriptional status. The level of histone acetylation is determined by histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities (14, 19, 32). The transcriptional status of a gene may thus be determined in part by an intrinsic balance of HAT and HDAC activities (14, 19, 32). The best-characterized cellular proteins that are positive regulators of histone acetylation are the cyclic AMP (cAMP)-responsive element binding protein (CREB)-binding protein (CBP) and p300. p300 and CBP are transcriptional coactivators, and p300 was first identified as an adenovirus E1A-interacting protein. p300 and CBP are highly homologous both structurally and functionally (14), and both have intrinsic HAT activities that contribute directly to the acetylation of histones (5, 25).
Well-defined, highly conserved regions of these coactivators interact with a large number of cellular proteins. Cellular transcriptional regulatory proteins, cAMP-activated CREB, and c-Jun bind to the kinase-induced activation (KIX) domain of CBP and p300 and use CBP and p300 as coactivators for their transcription (2, 10, 18, 20). Members of a class of cellular transcription factors known as the nuclear receptors bind to the N-terminal region of CBP (18). The cellular bZIP protein c-Fos (4) and adenovirus E1A (1, 13, 23) bind to the C/H3 region of CBP. In addition to the transcription factors mentioned above, a group of proteins that participate in transcriptional coactivation, such as the p300/CBP-associated factor (P/CAF) and SRC-1, also form transcriptional coactivator complexes with CBP and p300 (18, 35). The tumor suppressor protein p53 binds to several conserved regions (C/H1, C/H3, and KIX domains) of CBP (3, 15, 30). Interestingly, the presence of CBP and p300 is correlated with the differentiation and proliferation states of cells, as these coactivators interact with a number of cell cycle regulatory proteins.
HDACs, on the other hand, are known as repressors of transcription. Deacetylation reverses the acetylation process and causes the formation of tightly packaged nucleosomes, which are inaccessible to transcription factors (34). Transforming growth factor β receptor-activated Smad 2 and 4 signaling proteins move into the nucleus and associate with CBP to activate transcription, while activated Smads can also be induced by tumor growth inhibitory factor to recruit the inhibitory HDAC complex to the site of transcription (33). Spl, another well-characterized cellular transcription factor, binds to the CBP-p300 coactivator complex, and Sp1 activity is repressed by direct interaction with HDAC1 (12). Similarly, cellular transcription factor YY1 also binds to CBP and HDAC and uses both HAT and HDAC activities to regulate transcription (34). Several proteins from animal viruses use HDAC to repress their own viral promoters and to modify host cell growth. For example, the E7 oncoprotein from human papillomavirus interacts with Mi2 and the HDAC complex to promote cell growth (8), and the Epstein-Barr virus (EBV) nuclear antigen 3C interacts with HDAC to repress transcription of the latency-associated viral Cp promoter (26).
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) has been identified as an important pathogen in KS (6). In KSHV infection, the viral DNA is found principally in B cells, and KSHV is in fact associated with abnormal lymphoproliferation (9). In a KS tumor, KSHV DNA is found in spindle cells, and viral lytic replication is important in KS tumor development (7). The most important step in the KSHV life cycle may be the switch from latency to lytic replication. Upon chemical induction, KSHV produces immediate-early viral transcripts. These transcripts encode viral transcriptional activator proteins such as open reading frames (ORFs) 50 and K8, which are necessary to induce the lytic phase (36). ORF50 is a homolog of the EBV immediate-early gene product Rta. It has been reported that ORF50 could activate the lytic cycle of KSHV and is expressed earlier than K8, a homolog of EBV Zta protein, which induces the lytic cycle of EBV (22, 28). ORF50 is a viral transcriptional activator, which activates the early and late genes in the KSHV lytic cycle (21). We speculate that KSHV ORF50 uses CBP/p300 as a coactivator for transcriptional activation.
Here, we show that CBP participates in transcriptional activation by ORF50 and that CBP-binding proteins such as E1A and c-Jun also influence the transcriptional activation activity of ORF50. We also identified the CBP-binding domains in ORF50, which include the N-terminal basic domain and the conserved LXXLL motif of ORF50. In addition, we show that HDAC suppressed ORF50 activity and directly binds to the proline-rich sequences in the middle region of ORF50. Our data suggest that both HAT and HDAC activities may play important roles in controlling latent and lytic cycles of KSHV by acting as sensors of the conditions for lytic replication in infected cells.
MATERIALS AND METHODS
Plasmids.
The expression plasmids containing hemagglutinin (HA)-tagged CBP and HDAC1 are kind gifts from Didier Trouche. The vector containing p300 was obtained from Steve Grossman. The c-Jun-containing vector was provided by Dennis J. McCance. The pFR-Luc reporter was obtained from Stratagene (San Diego, Calif.). pM and pVP16 used in the mammalian two-hybrid assay were from Clontech (Palo Alto, Calif.). ORF50 and the ORF50 deletion mutants were subcloned into the pcDNA3 (Invitrogen, Carlsbad, Calif.) (the EcoRI and XhoI sites), pM (the EcoRI and XbaI sites), and pVP16 (the EcoRI and XbaI sites) vectors using PCR. Flag/ORF50 was derived from pFLAG-CMV-2. Green fluorescent protein (GFP) fusion vectors were constructed using pEGFP-C1 (Clontech). The CBP fragments were introduced into pGEX4T-1 (Amersham Pharmacia Biotech, Uppsala, Sweden), and the GST fusion proteins were expressed and purified according to the manufacturer's instruction.
Transient-transfection assays.
Transfection assays were performed in 293T cells using the calcium phosphate method. In all assays, the luciferase activity derived from the reporter plasmids was determined after normalizing to β-galactosidase activity from a cotransfected Rous sarcoma virus–β-galactosidase (RSV-βgal) control plasmid. All experiments were performed in triplicate. Equivalent expression of each plasmid was verified by Western blot assay (data not shown). 293T cells were transfected with 1 μg of reporter plasmid, 20 ng of RSV-βgal control plasmid, and the amounts of the expression plasmids indicated in the figures. The total amount of each expression vector was kept constant by adding empty cytomegalovirus expression plasmid. BJAB cells were transfected by electroporation as described previously (21).
GST pull-down assays.
All ORF50 and deletion mutants were in vitro transcribed and translated using a T7-coupled transcription-translation system (Promega, Madison, WI). The labeled proteins were incubated with 1 μg of a glutathione S-transferase (GST) fusion protein in binding buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 0.5% NP-40 supplemented with protease inhibitors). Glutathione beads were then added, and the reaction mixture was incubated at 4°C overnight. The beads were then washed four times with binding buffer, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added, and the proteins were analyzed by SDS-PAGE and visualized by autoradiography or phosphorimaging (BAS-1500, Fuji Film Co., Tokyo, Japan).
Coimmunoprecipitation assays.
293T cells were transfected with 7 μg of each expression plasmid using the calcium phosphate method. The cells were harvested 48 h after transfection and lysed in the binding buffer (containing 1% Triton X-100) used in the GST pull-down assays. The cell lysates were rotated in the buffer for 1 h at 4°C before the cell debris was removed via centrifugation. The appropriate lysates were immunoprecipitated with the addition of antibodies to HA or Flag (Sigma, St. Louis, Mo.) and protein G resin (Santa Cruz Biotechnology, Santa Cruz, Calif.). The beads were washed four times, and the proteins were analyzed by SDS-PAGE, transferred to a nitrocellulose membrane, and visualized with ECL reagent according to the manufacturer's instructions (Amersham Pharmacia Biotech).
RT-PCR.
BCBL-1 cells were transfected by electroporation. RNA was prepared from the transfected cell lysates, and reverse transcription (RT)-PCR was performed as described previously (27). The first PCR product (1 μl) was obtained, and PCR (20 cycles) was performed again. The final PCR product was analyzed by agarose gel electrophoresis and ethidium bromide staining.
Immunofluorescence.
pGFP/ORF50 (1 μg) and expression vectors containing HA-tagged CBP and HDAC were transfected into 293T cells (1 μg). Cells were fixed and immunostained 48 h after transfection. HA-tagged CBP and HA-tagged HDAC1 were detected using a rhodamine-conjugated secondary antibody.
RESULTS
CBP, p300, and HDAC1 modulate the transcriptional activation function of ORF50.
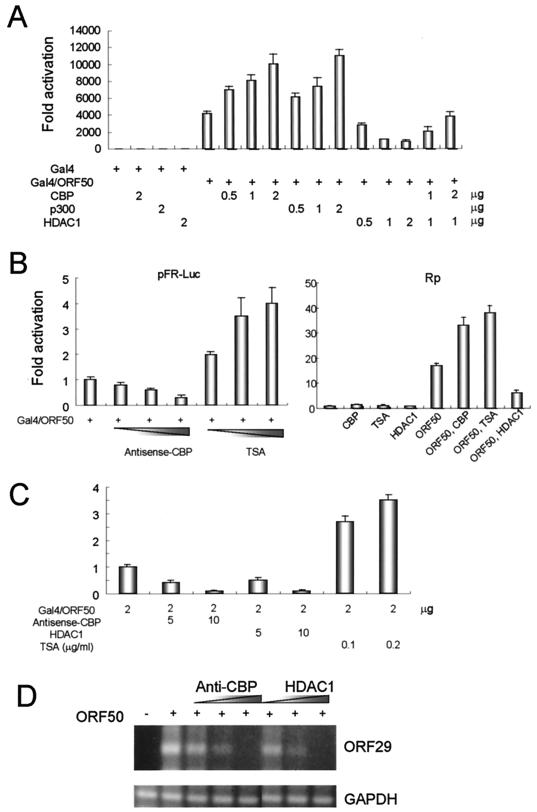
To investigate whether KSHV ORF50 interacts functionally with HATs and HDACs, we fused ORF50 to the Gal4 DNA-binding domain (Gal4/ORF50) and tested its ability to activate transcription of a GAL4 reporter plasmid (pFR-Luc) in transfected 293T cells. Transcriptional activation by Gal4/ORF50 was ∼4,000 times that of the Gal4 DNA-binding domain alone (Fig. 1A). We then measured Gal4/ORF50 activity in the presence of CBP and p300 to assess the effect of HAT activity on ORF50 function. We next tested whether HDAC activity could also modulate ORF50 activity. Cotransfection of an HDAC1 expression vector resulted in the repression of ORF50 activity (Fig. 1A), and this repression was relieved by cotransfection of the CBP.
FIG. 1.
CBP, p300, and HDAC1 modulate the transcriptional activation function of ORF50. (A) 293T cells were transiently cotransfected with a reporter gene construct (pFR-Luc, which contains Gal4 DNA-binding sites) and one or more of the following expression vectors: Gal4, a pM vector containing a Gal4 DNA-binding domain alone (10 ng), Gla4/ORF50, a pM vector containing the Gal4-ORF50 fusion protein with the Gal4 DNA-binding domain fused to ORF50 (10 ng); CBP (amounts shown in the figure); p300 (amounts shown in the figure); and HDAC1 (amounts shown in the figure). The effects of CBP, p300, and HDAC1 on the transcriptional activation function by ORF50 were determined by measuring the luciferase activity of the reporter gene (pFR-Luc) in the transiently transfected 293T cells. The luciferase activity of Gal4 in the absence of the other expression vectors is normalized to 1. (B) To confirm the results shown in panel A, antisense CBP DNA and the HDAC inhibitor TSA were introduced into the 293T cells along with the transfected DNAs indicated (left panel). The effects of the two inhibitors were verified using the pGL2-basic promoter gene under the control of the promoter region of KSHV ORF50 (nucleotides 70501 to 71595) (Rp) (right panel). The basal luciferase activity of Rp in the absence of the other expression vectors is normalized to 1. (C) antisense CBP DNA, HDAC1, and the HDAC inhibitor TSA were introduced into the B-cell lymphoma cell line BJAB along with the transfected DNAs indicated. (D) The RT-PCR product (300 bp) of ORF29, the KSHV capsid protein, was analyzed with RNA prepared from KSHV-containing BCBL-1 cells transfected by electroporation with pcDNA3/ORF50 alone or with an HDAC1 expression vector or antisense CBP DNA. The RT-PCR product of the cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene served as a loading control and was unchanged in all experiments.
In order to investigate the functional interactions among CBP, p300, HDAC1, and ORF50, the effects of antisense CBP DNA and an inhibitor of HDAC, trichostatin A (TSA), were measured in our transfection assay. The cotransfection of antisense CBP DNA reduced transcriptional activation by ORF50 to 0.2-fold of the control value (Fig. 1B). Conversely, increasing concentrations of TSA stimulated the activity of ORF50 up to fourfold. These results confirm that HDAC1 and CBP interact functionally with ORF50 in vivo to repress and activate, respectively, its transcriptional activation activity. The above experiments were repeated with the natural KSHV promoter (Rp). ORF50 stimulated this transcription from the natural promoter up to 18-fold. ORF50 activity was increased by cotransfection of CBP or addition of TSA and decreased by cotransfection of HDAC1 (Fig. 1B). Cotransfection of antisense CBP DNA and HDAC1 expression vectors reduced transcriptional activation by ORF50 to 0.1-fold in the B-cell lymphoma BJAB cell line (Fig. 1C). Conversely, increasing concentrations of TSA stimulated the activity of ORF50 up to fourfold in the BJAB cells. We also tested the effects of CBP and HDAC1 on the ORF50-induced lytic phase of KSHV in the BCBL-1 cell line, which carries a latent KSHV genome. KSHV ORF29 (encodes a capsid protein) expression occurs late in the lytic phase and is followed by viral DNA replication. Therefore, in order to detect entry into the lytic phase, we performed RT-PCR with oligonucleotides 29A and 29B, which spanned both ends of ORF29A and ORF29B (27). An RT-PCR product of ∼300 bp was observed when ORF50 was introduced into the BCBL-1 cells. Antisense CBP DNA and HDAC1 reduced the amount of ORF29 RT-PCR product produced in a dose-dependent manner (Fig. 1D). We conclude that CBP and HDAC1 have stimulatory and inhibitory effects, respectively, on the lytic cycle-inducing functions of ORF50.
In vivo and in vitro interaction of ORF50 with CBP.
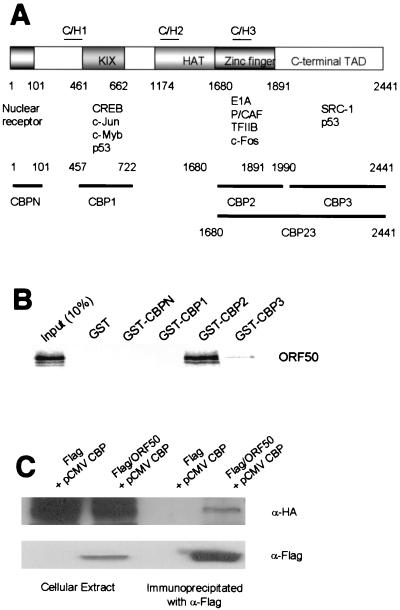
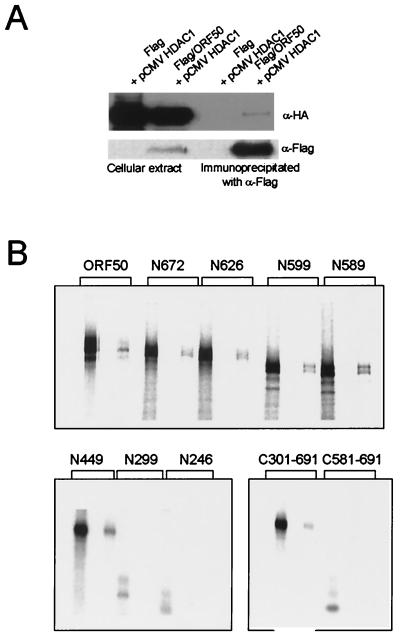
We next sought to determine whether ORF50 interacts directly with CBP and HDAC. To detect CBP-ORF50 interactions in vitro, we performed GST pull-down assays using in vitro-translated ORF50 and GST-CBP fusion proteins (made with various CBP deletion mutants) (Fig. 2A). ORF50 bound with high affinity to the CBP2 fragment (C/H3 region) and weakly to the CBP3 fragment (C-terminal transcriptional activation domain) (Fig. 2B). The CBP-ORF50 interaction was also observed in vivo when plasmids expressing Flag-tagged ORF50 and HA-tagged CBP were transfected into 293T cells. The cell extracts were subjected to immunoprecipitation with anti-Flag antibodies and Western blotting with anti-HA antibodies or anti-Flag (Fig. 2C).
FIG. 2.
In vivo and in vitro interaction of ORF50 with CBP. (A) The domains of CBP that interact with various cellular factors and the GST-fused CBP fragments are indicated. CBP contains the KIX, HAT, and zinc finger motifs and carboxyl-terminal transcriptional activation domain (TAD). (B) The GST-fused CBP fragments were purified and used in GST pull-down assays performed with glutathione-Sepharose beads. ORF50 was translated in vitro and labeled with [35S]methionine. (C) The coimmunoprecipitaion of in vivo-synthesized ORF50 and CBP was analyzed by Western blotting. 293T cells were transfected with an HA-tagged CBP expression vector (pCMVCBP) and either a blank (control) vector or an expression vector carrying Flag-tagged ORF50 (Flag/ORF50). Proteins in the cell lysates were precipitated with anti-Flag antibody, subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and visualized with either anti-Flag (α-Flag) or anti-HA (α-HA) antibodies.
Modulation of the transcriptional activation function of ORF50 by CBP-interacting proteins.
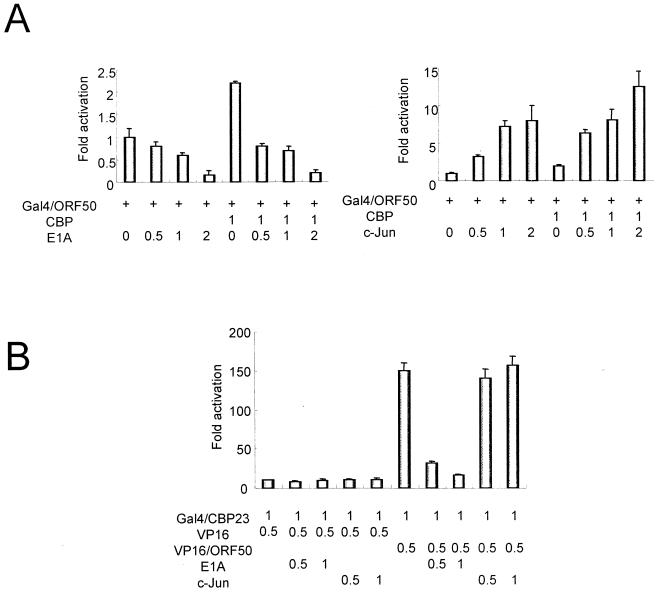
Dozens of cellular transcription factors bind to CBP (Fig. 2A). We thus selected several CBP binding proteins and tested their effects on the transcriptional activation activity of ORF50 in transiently transfected 293T cells. The adenovirus E1A protein, which binds to the C/H3 conserved region of CBP (amino acids 1680 to 1891) (1, 13, 23), repressed ORF50 activity in a dose-dependent manner. ORF50 repression by E1A was greater when a CBP expression vector was cotransfected into the 293T cells (Fig. 3A). These results imply that E1A represses ORF50 transcriptional activation activity by interacting with CBP and that ORF50 uses CBP as a transcriptional coactivator.
FIG. 3.
Modulation of the transcriptional activation function of ORF50 by c-Jun and E1A. (A) Luciferase activity from pFR-Luc was measured in 293T cells that had been transiently cotransfected with Gal4/ORF50 and either E1A or c-Jun expression vectors. The luciferase activity generated from Gal4/ORF50 activation of the reporter in the absence of the other expression vectors is normalized to 1. (B) The mammalian two-hybrid assay was performed to test the effects of E1A and c-Jun on CBP23 activation of VP16/ORF50. The luciferase activity generated from Gal4/CBP23 activation of the reporter in the absence of the other expression vectors is normalized to 1.
We next tested the effects of several cellular proteins that associate with the KIX domain (amino acids 457 to 722) of CBP. c-Jun protein stimulated ORF50 activity in a dose-dependent manner (Fig. 3A), and the addition of CBP yielded an incremental increase in ORF50 activity. We also used the mammalian two-hybrid assay to perform a competition experiment with E1A and c-Jun and measure their effects on interaction in vivo of ORF50 with CBP fragments 2 and 3 (CBP23; amino acids 1680 to 2441) fused to the Gal4 DNA-binding domain (Gal4/CBP23). ORF50 was fused to the transcriptional activation domain of VP16 (VP16/ORF50), and a luciferase gene fused to Gal4 DNA-binding sites served as the reporter. The VP16/ORF50 fusion protein caused an increase in luciferase production over that achieved with Gal4/CBP23 alone. The E1A protein inhibited the VP16/ORF50-Gal4/CBP23 interaction in a dose-dependent manner, while c-Jun had no effect (Fig. 3B). These results imply that E1A competes with ORF50 for binding to the same region of CBP.
CBP-interacting domains within ORF50.
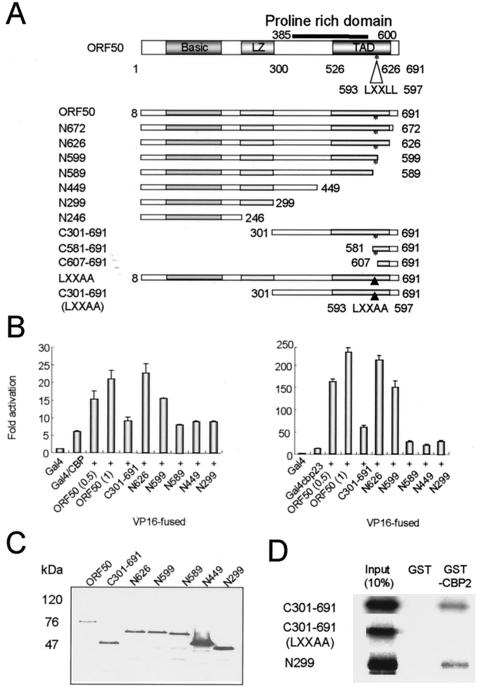
To determine the CBP-binding domain of ORF50, we constructed various ORF50 deletion mutants (Fig. 4A). The LXXLL motif was found in the transcriptional activation domain of ORF50. This motif constitutes a CBP-interacting motif in several proteins, including SRC-1 and p/CIP (16, 24, 29, 31). We hypothesized that the LXXLL motif of ORF50 constitutes a domain for binding to CBP and constructed several deletion mutants that did not contain this motif (N589, N449, N299, N246, C607-691, and LXXAA) (Fig. 4A) for use in ORF50-CBP binding assays. We first performed the mammalian two-hybrid assay using vectors expressing the various ORF50 deletion mutants fused to the VP16 transcriptional activation domain and CBP fused to the Gal4 DNA-binding domain (Gal4/CBP). Gal4/CBP activated transcription of Gal4-Luc more than did the Gal4 DNA-binding domain alone. This activity was stimulated a further threefold in the presence of the ORF50-VP16 fusion (Fig. 4B). The ORF50 mutants N626 and N599-VP16 displayed activity similar to that of wild-type ORF50-VP16, but the ORF50 mutant N589-VP16 lost the ability to transactivate the Gal4-Luc reporter gene. Unexpectedly, another CBP-interacting domain was identified in the mammalian two-hybrid assay. The ORF50 mutant C301-691 fusion protein, which has a deletion of 300 amino acids from the N terminus of ORF50, lost most of its ability to activate transcription of the reporter gene. We then repeated the mammalian two-hybrid assay with Gal4/CBP23. As expected, the CBP23 fragment showed the same pattern as the wild-type CBP (Fig. 4B). More C301-691-VP16 than of ORF50-VP16 was expressed, as shown in Fig. 4C. The expressed amounts of N589, N449, and N299-VP16 deletion mutants were at least not smaller than the amount of ORF50-VP16, although some variations were detected (lanes 1, 5, 6, and 7). Since these deletion mutants did not activate the reporter activity as did the wild-type ORF50, even though the deletion mutants were expressed in greater amounts than the wild type, we can conclude that the decreased interaction of C301-691, N589, N449, and N299 with CBP was not due to lower expression of the deletion mutants.
FIG. 4.
CBP-interacting domains within ORF50. (A) The domains within ORF50 and the ORF deletion mutants are shown. ORF50 contains the basic domain, leucine zipper motif (LZ), and the transcriptional activation domain (TAD). The proline-rich domain is located between amino acids 385 and 600. The LXXLL motif, which is conserved in SRC-1 and P/CAF, is located between amino acids 593 and 597 of ORF50. (B) The mammalian two-hybrid assay was performed to define the CBP interaction domain of ORF50. CBP was introduced into the pM vector to generate the Gal4 DNA-binding domain-fused CBP construct (left graph). ORF50 and its deletion mutants were introduced into the VP16 vector, and the luciferase reporter gene was fused to Gal4 DNA-binding sites. Identical assays were performed with pM/CBP23, which contained the CBP2 and CBP3 fragments (right graph). In each case, the luciferase activity generated by Gal4 activation of the reporter in the absence of the other expression vectors is normalized to 1. (C) Expression of VP16 fusion proteins. 293T cells were transfected with the indicated vectors, and extracts were analyzed by blotting with anti-VP16 antibody. (D) GST pull-down assays were performed with in vitro-translated, 35S-labeled amino- or carboxyl-terminally deleted forms of ORF50 and GST-CBP2.
In order to determine ORF50 domain binding to CBP, we performed the GST pull-down assay using a GST-CBP2 fragment and in vitro-translated C301-691 and N299 (Fig. 4D). Unexpectedly, both of these two domains interact with CBP in the in vitro binding assay. To define the role of the LXXLL motif in the interaction with CBP more clearly, we constructed point-mutated proteins LXXAA and C301-691 (LXXAA). C301-691 (LXXAA) lost its ability to bind to CBP (Fig. 4D). This mutation reduced the transcriptional activity of Gal4 DNA-binding domain-fused ORF50 to 0.3-fold of the wild-type level (Table 1). These facts imply that this LXXLL motif should have a role as a CBP-interacting domain in vivo and in vitro. From these experiments, we concluded that ORF50 has two CBP-binding domains, the basic domain and the LXXLL motif, and that these two domains are necessary for stable binding to CBP in vivo.
TABLE 1.
The effects of ORF50 and HDAC1 cotransfection and TSA treatment of 293T cells on the transcriptional activation function of ORF50 and ORF50 deletion mutantsa
| Expression plasmid transfected | Relative luciferase activity
|
||||
|---|---|---|---|---|---|
| No addition | + TSA | + TSA/no addition | + HDAC1 | + HDAC1/no addition | |
| Gal4 | 1 | 0.94 | 0.94 | 1.5 | 1.50 |
| Gal4/ORF50 | 4,192 | 8,562 | 2.04 | 1,090 | 0.26 |
| Gal4/N672 | 4,548 | 9,333 | 2.05 | 1,240 | 0.27 |
| Gal4/N626 | 2,962 | 5,933 | 2.00 | 607 | 0.20 |
| Gal4/N599 | 7.8 | 30 | 3.84 | 3 | 0.38 |
| Gal4/N589 | 5.54 | 41.2 | 5.77 | 2.5 | 0.45 |
| Gal4/N449 | 8.42 | 42 | 4.99 | 3 | 0.36 |
| Gal4/N299 | 3.54 | 2.72 | 0.77 | 3.98 | 1.12 |
| Gal4/N246 | 2.54 | 3.1 | 1.22 | 3.32 | 1.31 |
| Gal4/C301-691 | 2,500 | 6,250 | 2.5 | 750 | 0.3 |
| Gal4/C581-691 | 546 | 500 | 0.92 | 539 | 0.99 |
| Gal4/C607-691 | 240 | 205 | 0.85 | 255 | 1.06 |
| Gal4/LXXAA | 1,240 | —b | — | — | — |
A luciferase reporter gene with Gal4 binding sites was cotransfected with a control expression plasmid (Gal4) or an expression plasmid encoding various versions of ORF50 fused to the Gal4 DNA binding domain (10 ng). In some experiments, an expression plasmid encoding HDAC1 (1 μg) was cotransfected (+ HDAC1). Some of the transfected cells were treated with TSA (0.1 μg/ml) (+ TSA). The luciferase activity of each ORF50 mutant was determined. Three independent experiments were performed for each condition, and a representative result is shown for each experiment. The outcomes of all three experiments were very similar. In each case, the activity of Gal4 in the absence of HDAC1 and TSA is normalized to a value of 1.
—, not determined.
HDAC1-interacting domain within ORF50.
To determine whether ORF50 and HDAC1 associate in vivo, we transiently transfected plasmids expressing Flag-tagged ORF50 and HA-tagged HDAC1 into 293T cells. Cell extracts were prepared and immunoprecipitated with a monoclonal antibody against the Flag epitope. Anti-Flag immunoprecipitates were separated on SDS-PAGE and blotted with anti-HA antibody. HDAC1 was detected in the immunoprecipitates from cells transfected with Flag-tagged ORF50, but not from cells transfected with the blank vector (Fig. 5A). We then performed the GST pull-down assay using the various ORF50 deletion mutants and GST-HDAC1 to determine the ORF50 binding domain of HDAC1 (Fig. 5B). ORF50 N299 and N246 did not show binding to GST-HDAC1, while N449 and all of the larger ORF50 mutants bound to HDAC1. With respect to the N terminal ORF50 deletion mutants, C301-691 bound to the HDAC, while C581-691 did not.
FIG. 5.
In vivo interaction of ORF50 with HDAC1 and HDAC1-interacting domain within ORF50. (A) Immunoprecipitation of extracts from 293T cells transfected with Flag-tagged ORF50 and HA-tagged HDAC1. (B) The HDAC interaction domain within ORF50 was identified using GST-fused HDAC and GST pull-down assays.
To characterize the ORF50-HDAC interaction in vivo, we tested the effects of HDAC1 and TSA treatment on the ability of the ORF50 deletion mutants to activate transcription (Table 1). 293T cells were cotransfected with Gal4-Luc reporter plasmid, expression plasmids encoding HDAC1 and ORF50, and the various ORF50 deletion mutants fused to the Gal4 DNA-binding domain. In some experiments, the cells were treated with TSA. The ORF50 N449 fragment was responsive to HDAC1 cotransfection and TSA treatment, but further C-terminal ORF50 deletions abolished this responsiveness. C301-691 was sensitive to HDAC1 and stimulated by TSA, but further N-terminal ORF50 deletions abolished these effects. From the above results, we conclude that the HDAC1-binding domain in ORF50 is located between amino acids 301 and 449, which contains several proline residues (Fig. 4A).
CBP and HDAC1 colocalize with ORF50 in vivo.
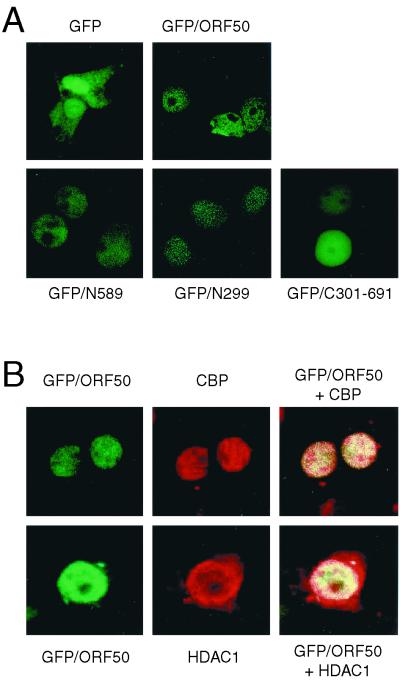
We next assessed whether ORF50, HDAC, and CBP complexes were colocalized in 293T cells. GFP-tagged ORF50 was located mainly in the nucleus, while GFP alone showed a diffuse pattern throughout the cytoplasm and nucleus (Fig. 6A). All of the ORF50 deletion mutants tested in this study localized to the nucleus, and thus the difference in the transcriptional activities of the mutants did not result from changes in their subcellular localization (Fig. 6A). Cotransfection of HA-tagged CBP and HA-tagged HDAC1 with GFP-ORF50 yielded a yellow color, indicative of colocalization in the nucleus (Fig. 6B).
FIG. 6.
ORF50 colocalizes with CBP and HDAC1 in 293T cells. (A) pGFP/ORF50 and GFP-fused ORF50 deletion mutants (1 μg) were transfected into 293T cells. (B) pGFP/ORF50 (1 μg) and an HA-tagged CBP or HA-tagged HDAC expression vector (1 μg) were transfected with 293T cells. Cells were fixed and immunostained 48 h after transfection. HA-tagged CBP and HDAC were detected using a rhodamine-conjugated secondary antibody against a monoclonal HA antibody.
DISCUSSION
Here we showed that CBP and HDAC1 interact with KSHV ORF50. CBP bound to the amino-terminal basic domain and the carboxyl-terminal transactivation domain of ORF50. The Tat transactivator from the human immunodeficiency virus also interacts with CBP/p300 through its basic domain (17). p160 factors such as NCoA-1/SRC-1 and other transcriptional coactivators capable of interacting with liganded nuclear receptors have a common motif that contains a core consensus LXXLL sequence which is also found in ORF50 (16, 24, 29, 31). SRC-1, TIF-2/GRIP-1/NcoA-2, and p/CIP contain LXXLL motifs in a conserved central domain, which has been identified as the nuclear receptor interaction domain. SRC-1 also has LXXLL motifs in a carboxyl-terminal domain that interacts with the hydrophobic binding pocket of CBP/p300 (24). In this study, we showed that the LXXLL motif in the transcriptional activation domain of ORF50 interacts with CBP. It is the first report that the viral protein uses the LXXLL motif as a CBP-interacting domain in addition to the cellular proteins. It is interesting that the basic domain and the LXXLL motifs of ORF50 interact independently with CBP and that these two domains are necessary for ORF50 transcriptional activation function.
Transcriptional repression domains have been reported to exist in transcriptional factors. Several transcription factors contain alanine-rich, glutamine-rich, glycine-rich, and proline-rich sequences in their repression domains, although the precise consensus sequences and the mechanism by which these domains repress transcription have yet to be determined (11, 34). Comparative analysis of the amino acid sequence of the ORF50 repression domain (amino acids 301 to 449) has failed to reveal any homology to the primary sequence motifs of the repression domains described above. Repression of transcription by ORF50 was mediated through interaction of the proline-rich central region of ORF50 with HDAC1. Whether the interaction of a proline-rich domain in a transcription factor with HDAC is common mechanism of transcriptional repression by other transcription-regulatory proteins remains to be determined.
In this study, we demonstrated that KSHV ORF50 bound to CBP/p300 and HDAC1 and these transcriptional cofactors modulated the transcriptional activation function of ORF50. It is possible that the interaction of ORF50 with CBP and HDAC1 is determined by the relative concentration of each protein in KSHV-infected cells; however, we are not able to test whether the interaction of ORF50 with CBP and HDAC1 is mutually exclusive. Using the results herein, we propose a model for the regulation of ORF50 transcriptional activation. The starting point of the lytic cycle of KSHV is the induction of ORF50. CBP functions as a positive regulator of the ORF50 transactivator, while HDAC1 acts as a negative regulator. The repression of ORF50 through interaction with HDAC1 could set a threshold level for activation of the KSHV lytic cycle. If the endogenous concentration of HDAC1 is higher than that of CBP, depending on the status of the infected cells, the lytic phase is slowed, and cells return to the latency phase. HDAC1 may play an important role in controlling latent and lytic cycles of KSHV by acting as a sensor of the conditions for lytic replication in the infected cells. KSHV could use this inhibitory regulation by cellular proteins to replicate at the proper time, according to cellular status. Alternatively, the repression of ORF50 transcriptional activity by HDAC1 may function as a defense mechanism to protect the cell from replication of the virus.
In the present work, we focus our attention on the transcriptional activity of ORF50 modulated by CBP. However, it is possible that ORF50 could modulate the transcriptional activities of the cellular transcription factors through interaction with CBP, as in the case of several CBP-interacting viral proteins. If CBP is a limiting factor in the cells, the interaction of ORF50 with CBP could repress the transcriptional activities of other transcription factors through depletion of CBP. Experiments designed to assess the role of ORF50 in regulation of the cellular transcription factors are under way.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Research Laboratory Program of the Korea Institute of Science & Technology Evaluation and Planning (KISTEP), the Korea Science and Engineering Foundation (KOSEF) through the Protein Network Research Center at Yonsei University, and BK21 Program of the Ministry of Education, Korea.
REFERENCES
- 1.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 2.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff C, Weiss R A. Kaposi's sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 7.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 8.Brehm A, Nielsen S J, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 10.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 11.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 12.Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C. Histone deacetylase 1 can repress transcription by binding to Spl. Mol Cell Biol. 1999;19:5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 14.Giles R H, Peters D J, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Shi X-L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–822. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Hottiger M O, Nabel G J. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–8256. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 19.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 20.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 21.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 23.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 24.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogryzko V, Schiltz R, Russanova V, Howard B, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 26.Radkov S A, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday M J. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 30.Van Orden K, Giebler H A, Lemasson I, Gonzales M, Nyborg J K. Binding of p53 to the KIX domain of CREB binding protein: a potential link to human T-cell leukemia virus, type I-associated leukemogenesis. J Biol Chem. 1999;274:26321–26328. doi: 10.1074/jbc.274.37.26321. [DOI] [PubMed] [Google Scholar]
- 31.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 32.Wolffe A P. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 33.Wotton D, Lo R S, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 34.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 36.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]