Abstract
Vasculature is essential for maintaining the cellular function and balance of organs and tumors. As a key component of the tumor microenvironment (TME), it significantly influences tumor characteristics. Angiogenesis, heavily influenced by the extracellular matrix (ECM), which acts as a structural scaffold and growth factor reservoir, is regulated by various factors. Notably, adipose tissues and adipose-derived stromal cells contribute angiogenic and anti-apoptotic factors that promote angiogenesis. Sustained vasculature is essential for tissue engineering and ex vivo disease modeling. Lack of shear stress from fluid flow leads to vascular instability and regression. Microfluidic models replicate three-dimensional (3D) cultures from original tissues, encapsulate microenvironmental factors, and maintain consistent fluid flow. In our study, we established decellularized adipose ECM (AdECM) derived from bovine sources and engineered a 3D-printed microfluidic device. We observed significant increases in both the length and diameter of vascular networks after coculturing HUVECs and HDFs in a fibrin gel containing 0.5% AdECM. Additionally, gene expression related to ECM remodeling and angiogenesis was significantly enhanced in vasculature cultivated in fibrin gel containing 0.5% AdECM compared to that in fibrin gel alone. The enhanced vasculogenesis was further amplified and sustained by the 3D microfluidic device placed on a rocker during extended cultivation, primarily through the activation of the PI3K and JAK-mediated pathways. Our ex vivo model with vascularized colon tumoroids revealed that integrating AdECM within a microfluidic device correlates with increased tumoroid growth. Therefore, our study underscores the synergistic impact of AdECM and microfluidic device in promoting and sustaining vasculature. This synergy may have significant implications for tissue regeneration and ex vivo disease modeling, facilitating drug testing and efficacy evaluation.
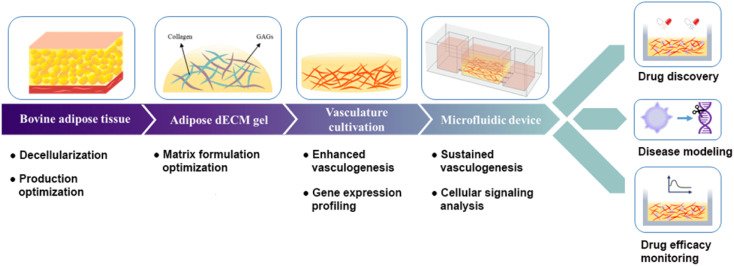
A schematic representation of the synergistic effects of AdECM and the microfluidic device in promoting and sustaining vasculogenesis, along with their applications in drug discovery, disease modeling, and monitoring of drug efficacy.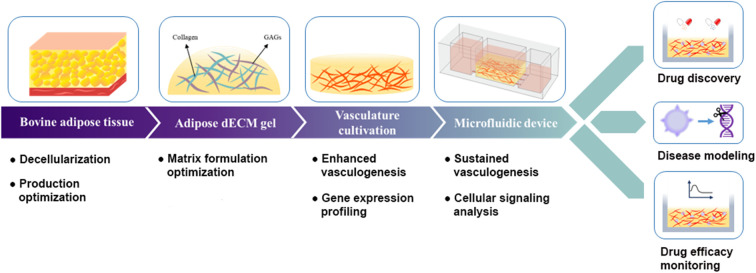
Introduction
The intricate interplay between a cell and its surrounding microenvironment holds paramount significance in the maintenance of physiological equilibrium in normal tissues and the regulation of tumorigenesis. The tumor microenvironment (TME) consists of extracellular matrix (ECM) proteins and diverse stromal cell varieties, including immune cells, endothelial cells, fibroblasts, and adipocytes, all of which wield substantial influence over the progression of tumors. The vascular network within tumors is a pivotal constituent of the TME that profoundly affects tumor behavior, encompassing invasiveness, metastatic potential, and responsiveness to therapeutic interventions. This vascular network diverges structurally, organizationally, and functionally from that observed in normal tissues. Essentially, less convoluted vascular configurations tend to inhibit metastatic expansion and enhance the efficient delivery of chemotherapeutic agents to the tumor.1 In contrast, abnormal vascular configurations can compromise therapeutic outcomes by impeding drug access to the tumor.2 Recent studies have illuminated this issue, revealing a direct correlation between vessel tortuosity and the survival rates of lung cancer patients undergoing checkpoint inhibitor treatments.3 Furthermore, in addition to their role in secreting vascular endothelial growth factors (VEGFs) to stimulate vessel formation, the tumor vasculature, in conjunction with associated pericytes and the extracellular matrix, functions as a barrier, hindering the infiltration of immune cells.4 This barrier can impede the immune system's capacity to effectively target and eradicate cancer cells.
Vasculature is prevalent for supplying essential nutrients and oxygen to maintain cellular homeostasis across diverse organs and within tumor environments.5 The process of angiogenesis, involving the generation of new blood vessels, occurs not only during organ development and maturation but also plays a pivotal role in fundamental physiological processes like wound healing and reproduction. Beyond a distance of 200 μm from a blood supply source, cells are unable to survive, and even tumors are constrained from exceeding a size of 1 mm3, further underscoring the indispensable role of vasculature in regulating cellular growth.6,7
However, angiogenesis is a complex and tightly regulated process, subject to control by a broad spectrum of regulatory factors. Accumulating evidence showed that ECM participates in all stages of angiogenesis, exerting an important role in regulating angiogenic processes in both physiological and pathological conditions.8 ECM fulfills a dual purpose, serving as both a physical scaffold for cells and a reservoir for growth factors. Additionally, the structural and physicochemical attributes of the ECM communicate precise information to cells, significantly impacting their biology through interactions with cell surface receptors referred to as integrins. In the context of angiogenesis, the perivascular ECM plays a pivotal role in shaping the proliferative, invasive, and survival behaviors of local vascular cells in response to angiogenic growth factors. Notably, adipose tissues have been identified as a source of angiogenic factors, such as VEGF, which enhance angiogenesis.9 In addition, adipose-derived stromal cells promote vasculogenesis by secreting angiogenic and anti-apoptotic factors.10 These studies conclusively illustrate that the formation of vasculature is profoundly modulated by its surrounding environment.
The construction of a vascular network is a fundamental requirement for tissue engineering and ex vivo models of disease mimicry, however, ensuring the persistence of this vasculature over the long term remains a significant challenge. Some studies investigate the factors that influencing late-stage angiogenesis from various perspectives. Ets variant 2 (ETV2), a key regulator of hematopoietic and endothelial development, is transiently expressed during angiogenesis.11 Increasing ETV2 expression in mature human umbilical vein endothelial cells (HUVECs) can regain the ability to form embryonic-like vessels, thus enhancing vasculature durability.12 Evidence reveals that a lack of shear stress induced by fluid flow can lead to vascular instability and subsequent regression.13 Increasing evidence suggests supports the use of microfluidic systems to mitigate vasculature degeneration and facilitate extended tissue culture by providing continuous fluid flow.14,15 Furthermore, microfluidic models offer advantages in reconstructing three-dimensional (3D) cultures from original tissues and capturing factors present in the microenvironment. The blood vessels generated within microfluidic devices can exhibit characteristics resembling those of the human circulatory system. Consequently, our investigation aimed to explore strategies for sustaining vascular networks from a multifaceted perspective.
Except for the angiogenic effects of adipose tissue, it existed throughout the human body, underscoring its universal applicability. In this investigation, we sought to assess the synergistic impact of adipose tissue and a microfluidic system on vascular network sustainability. To achieve the goal, we developed a clinically applicable gel of adipose decellularized extracellular matrix (AdECM) from bovine and fabricated a microfluidic device by using a 3D printer. Our findings revealed that AdECM played a pivotal role in promoting angiogenesis by stimulating the expression of angiogenic and extracellular matrix remodeling-related genes. Furthermore, when vascular networks were cultured within the microfluidic system, their integrity was maintained for an extended duration, primarily facilitated by the activation of PI3K/Akt and JAK/STAT3-mediated signaling pathways. The ex vivo model of the vascularized tumoroid demonstrated that vasculature formed in a microfluidic device system combined with a matrix containing AdECM correlated with increased growth of tumoroids. These findings clearly illustrate the synergistic influence of AdECM and the microfluidic model in enhancing and sustaining vasculogenesis (Fig. 1).
Fig. 1. A schematic representation of the synergistic effects of AdECM and the microfluidic device in promoting and sustaining vasculogenesis, along with their applications in drug discovery, disease modeling, and monitoring of drug efficacy.
Experimental section
Decellularization of bovine adipose tissue
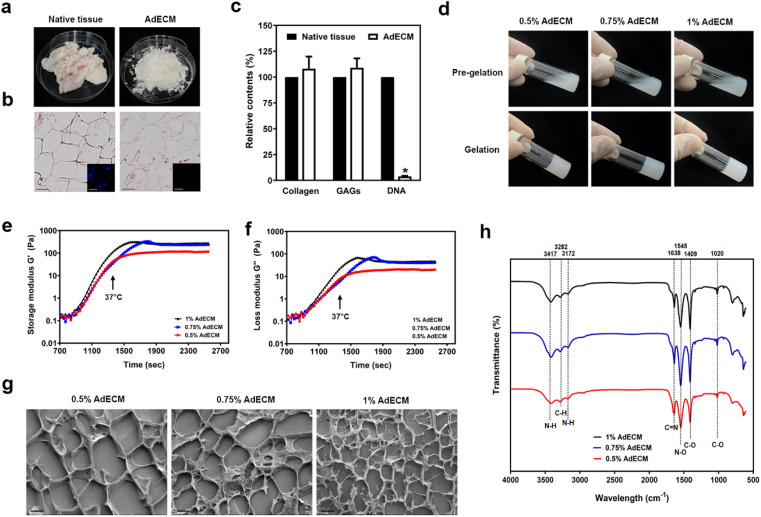
The non-specific connective tissue associated with male bovine adipose tissue was initially removed. The adipose tissue was subsequently sliced into pieces measuring 1–2 cm3. These pieces were then immersed in a PBS solution and stirred at room temperature for 3 hours. Following this, the tissue slices were treated with a 0.5% SDS solution in 1.5 M NaCl, maintained at 37 °C for 24 hours. After the SDS treatment, the slices were incubated in isopropanol at room temperature for a duration of 48 hours, and then treated with 0.1% peracetic acid in 4% ethanol at room temperature for an additional 4 hours. After several washes with water, the decellularized tissues were freeze-dried for 48 hours and then stored at −20 °C until needed. Subsequently, 10 mg of AdECM powder (Fig. 2a) was dissolved in 0.333 ml of 0.5 M acetic acid to form a 3% stock solution, to which 1 mg of pepsin was added. This mixture was incubated at 37 °C for 2 days. The AdECM solution was then neutralized to a physiological pH of 7.4 and stored at 4 °C for use in biological applications.
Fig. 2. Preparation and characterization of AdECM derived from bovine adipose tissue. (a) Structural change of bovine adipose tissues after decellularization. (b) Histological sections of native and decellularized tissue. The inserts showed immunofluorescence images of DAPI staining (nuclei). (c) The content of collagen, GAGs, and DNA were quantified in native adipose tissue (native tissue) and decellularized adipose tissues (AdECM). *P < 0.05, compared to native tissue. (d) The ability of gelation was examined in solution of 0.5, 0.75, and 1% AdECM/0.5 M acetic acid. (e and f) Rheological properties of 0.5, 0.75, and 1% AdECM during gelation. (g) Cryo-SEM images of AdECM exhibited the network of fiber after gelling. Scale bar = 2 μm. Three batches of bovine adipose tissue were processed for the preparation of AdECM.
Biochemical identification of AdECM
Bovine adipose tissues were homogenized in TE buffer to extract DNA. The DNA content in both native and decellularized adipose tissues was quantified using the Quant-iT PicoGreen® dsDNA reagent and kits (Invitrogen). Following tissue hydrolysis, hydroxyproline volume was measured in accordance with the manufacturer's instructions (Abcam) to determine the collagen content in the tissue. GAG levels in the tissues were assessed using the total glycosaminoglycans assay kit (BioVision). Both native and decellularized tissues were fixed in 4% paraformaldehyde and embedded in paraffin. The tissue blocks were sectioned to a thickness of 10 μm and subsequently stained with hematoxylin and eosin.
Oscillatory rheology
The rheological properties of the neutralized AdECM solution at concentrations of 0.5%, 0.75%, and 1% were analyzed using a rheometer (Anton Paar; MCR301). Initially, 230 μL of the AdECM solution was dispensed onto a cone plate. The subsequent analysis was carried out using a parallel plate (25 mm, PP25) set with a gap of 0.5 mm. The temperature was ramped up from 4 °C to 37 °C at a heating rate of 1°C min−1 and was then maintained at a constant 37 °C for 30 minutes. This temperature ramping test was designed to evaluate the gelation kinetics of AdECM by calculating both the storage modulus (G′) and the loss modulus (G′′).
Cryogenic scanning electron microscope (cryo-SEM)
After gelation, the samples underwent pressurization, then moved to a preparation chamber under vacuum. There, they were fractured in liquid nitrogen. A JEOL JSM-7800F Thermal field emission scanning electron microscope (FESEM) was utilized to visualize the microstructure of these samples.
Fourier-transform infrared spectrometer (FTIR)
Gelatinized samples (500 μL) were subjected to −80 °C for 3 hours, followed by overnight freeze drying. Absorbances were measured in the wavenumber spectral range of 500–4000 cm−1, with a spectral resolution of 1 cm−1, as specified by the equipment supplier.
Cell culture
HUVECa (cAP-0001, Angio Proteomie, USA) were grown in EGM™-2 medium (CC-3162, LONZA, Switzerland). HDF (PCS-201-012™, ATCC, USA) were cultured in Fibroblast Medium (#2301, ScienCell, USA). These cells were cultured in an atmosphere humidified with 5% CO2.
Cell viability assay
The proliferation of HUVECs or human dermal fibroblasts (HDFs) were examined when cultivation with 0.5, 0.75, or 1% AdECM at intervals of 1, 3, and 5 days using the PrestoBlue® Cell Viability Reagent (Invitrogen). In brief, cells were exposed to the PrestoBlue solution and kept in the dark at 37 °C for an hour. Absorbance readings were then taken at 570 nm using a microplate reader. Additionally, to evaluate cell toxicity, a live and dead cell assay was conducted on HUVECs or HDFs after they were cultivated in 0.5%, 0.75%, or 1% AdECM for a 5 days span. For this, cells were dyed with calcein-AM for live cells and ethidium homodimer-1 for dead cells. The resulting fluorescence was detected with excitation wavelengths of 494 and 528 nm and emission wavelengths of 517 and 617 nm, respectively.
Fabrication of a microfluidic device
A stereolithographic 3D Printer (MiiCRAFT, Canada) was used to fabricate a microfluidic device using MiiCraft BV007a Clear resin (CADworks3D, Canada), an alternative to PDMS with reduced autofluorescence signals. After printing, 3D-printed device was dried and post-cured with UV for 5 min using the FormCure (FormLabs, USA). A 3D-printed microfluidic device was then mounted to a cover glass (22 × 22 mm2) and wash with distilled water.
Vascular network formation
HUVECs and HDFs were co-cultured in a cultured plate or a microfluidic device at a cell ratio of 9 : 1. They were embedded in a 10 mg ml−1 fibrin with or without AdECM (0.5, 0.75 or 1%) in the presence of thrombin. This mixture was subsequently gelled at 37 °C and incubated in EGM™-2 Endothelial Cell Growth Medium (Lonza, Switzerland). After 5 days of cultivation, a vascular network was formed.
Immunofluorescence staining
Whole vasculature block in microfluidic device or cultured plate was firstly washed with 1X PBS and fixed with 4% paraformaldehyde for 2 h. After washed with 1X PBS and permeabilized with 0.1% Triton X-100, the vasculature block was blocked with 2% BSA for 2 h. Subsequently, the vasculature block was stained with Alexa Fluor™ 647 Phalloidin (Thermo Fisher Scientific, USA) or primary antibodies, anti-pSTAT3 (Abcam, UK), anti-pAkt (Abcam, UK) overnight at 4 °C. Next, the Alexa Fluor 488 Goat Anti-Rabbit (Thermo Fisher Scientific, USA) antibody was added and counterstained with DAPI (Sigma-Aldrich, USA). The fluorescence images were acquired using Dragonfly confocal microscope (ANDOR Dragonfly High Speed confocal system, Germany).
Vessel quantification
Vascular network was stained with F-actin. Vessel area, vessel length, and vessel diameter in different matrices were quantified using ImageJ software with a vessel analysis plugin (National Institutes of Health), based on data from three independent experiments. For each experiment, three measurements of vessel area, 20 measurements of vessel length, and 30 measurements of vessel diameter were quantified from at least three fields of each well. Results are presented as the mean percentage (±SD), and statistical analysis was performed using one-way ANOVA. *P < 0.05.
Measurement of VEGF in medium
Medium were collected from vasculature culture and centrifugated to remove cell debris. The content of VEGF in medium was measured by VEGF ELISA assay (Abcam, United Kingdom). The relative expression of VEGF was analysed from there independent experiments according to one way ANOVA. *P < 0.05.
RNA sequencing and real-time PCR
Total RNA was isolated from vessel block using the TRIzol reagent (Thermo Fisher Scientific, USA). The whole transcriptome was analysed by RNA sequencing. For real-time PCR, the 2 μg of RNA were reversed transcribed to cDNA using 50 ng of random hexamer primers and SuperScript III reverse transcriptase according to the manufacturer's instruction (Thermo Fisher Scientific, USA). The genes were amplified with SYBR Green (Roche, Swiss) with specific primers. Quantitative real-time RT-PCR was carried out to measure mRNA levels of genes using LightCycler 480 Real-Time PCR System (Roche, Swiss).
Colon tumoroid culture
The human colon tumoroid was established using colon tissue samples obtained from patients with colon cancer at China Medical University Hospital in Taiwan according to the procedures as reported previously.16,17 This study was approved by the Research Ethics Committee of China Medical University and Hospital, under project number CMUH111-REC1-083. Colon tumoroids were cultured in Geltrex (Thermo Fisher, United States) with the Advanced DMEM/F12 medium supplemented with B-27, N-2, 50 ng ml−1 EGF, 500 ng ml−1 R-spondin-1, 100 ng ml−1 Noggin, 10 μM SB202190, 500 nM A83-01, 1 mM N-acetylcysteine, 10 μM Y-27632, and 100 μM nicotinamide (colon tumoroid growth medium). The colon tumoroid was formed after 10–14 days, the morphology and specific colon markers have been identified (Fig. S10†). Tumoroids were passaged every 10–14 days according to the protocol described in the previous reports.16,17 Briefly, tumoroids were dissociated with cold Advanced DMEM/F12 medium and TrypLE solution (Thermo Fisher, United States) to small fragments. These fragments were embedded with fresh Geltrex in 24 well plate at 37 °C incubator for 30 min to gelation, and fresh medium were added.
Establishment of vascularized tumoroids
When colon tumoroids were formed, they were dissociated with cold Advanced DMEM/F12 medium. Tumoroids with an area of approximately 0.15 mm2 were then incubated within Geltrex in a microfluidic device at 37 °C for 30 minutes. Subsequently, HUVECs and HDFs were cultured in fibrin gel with or without AdECM and applied to tumoroids cultured in Geltrex at 37 °C for an additional 30 minutes. A mixed medium of colon tumoroid growth medium and EGM™-2 Endothelial Cell Growth Medium at a ratio of 1 : 1 was added. The microfluidic system was placed on a rocker, which was regularly tilted at 30° every 20 minutes. A vascular network formed when HUVECs and HDFs were co-cultured for 5–7 days.
Tumoroid quantification
The size of the tumoroids was determined by measuring the area of F-actin staining in the tumoroids using ImageJ software. Tumoroids grown with vascular networks in various matrices were quantified from three independent experiments for each condition.
Statical analysis
Result was analyzed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA) and are presented as the mean ± SD from at least three independent experiments.
Results
Preparation and gelation of AdECM
Accumulating evidence shows that the ECM participates in all steps of angiogenesis and plays an important role in regulating angiogenic processes.8 The presence of adipocytes in the ECM has been shown to enhance angiogenesis. We are thus attempting to establish adipose ECM from bovine sources to examine the impact of the adipose ECM on angiogenesis. Given the advantages of decellularized tissues in terms of translational potential for clinical applications, we've established a protocol to construct AdECM. Even though the tissue underwent several steps of detergent and enzymatic treatment to ensure the complete removal of nuclei (Fig. S7†), the structure of the decellularized adipose tissue was not significantly changed. As shown in Fig. 2b, the morphology of the decellularized adipose tissue resembles lipid droplet-like structures, similar to that of the native tissue (Fig. 2b). This indicates that the decellularization procedure did not disrupt the adipose tissue's structure. In contrast to the native tissue, abundant proteins expressed in the ECM, such as collagen and glycosaminoglycans (GAGs), are slightly more enriched in the AdECM (Fig. 2c).
Characterization of AdECM gel
The gelation ability of the AdECM solution was confirmed when it successfully formed a hydrogel at 37 °C as illustrated in Fig. 2d. Further, to assess the rheological properties of AdECM hydrogel, 0.5, 0.75, or 1% AdECM solutions were analysed by temperature ramping oscillatory rheology. Temperature from 4 °C to 37 °C, the storage modulus (G′) was higher than the loss modulus (G′′), and all concentrations of AdECM displayed gel-like characteristics (Fig. 2e and f). Comparably, the three formulations of AdECM (0.5%, 0.75%, or 1%) exhibited similar characteristics in terms of gelation ability and rheological properties. Cryo-SEM imaging revealed an interwoven network of fibers in the AdECM gel at concentrations of 0.5%, 0.75%, and 1%. As the concentration of AdECM increased, a more compact and denser structure was evident, as shown in Fig. 2g. Fourier transform infrared (FTIR) spectroscopy images illustrated the biochemical content of the AdECM gel. The characteristic absorption peaks include the O–H stretching vibration at approximately 3417 cm−1, the N–H amides I and II vibrations at around 1638 cm−1 and 1545 cm−1 respectively, the C–N stretching near 1409 cm−1, and the C–C and C–N deformation close to 1020 cm−1 (Fig. 2h).
AdECM exhibited no cytotoxicity
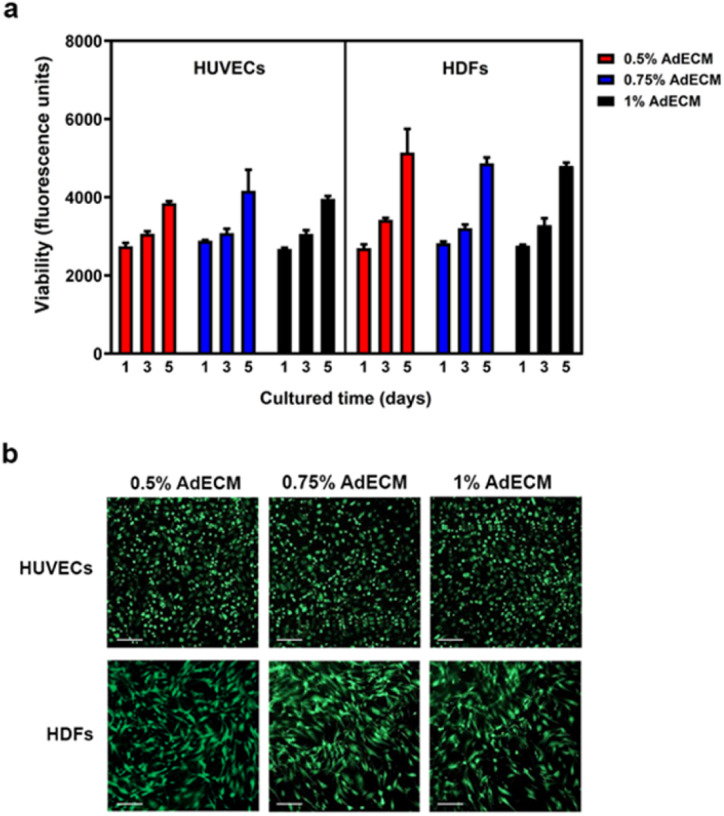
Proteins secreted from fibroblasts, such as growth factors and matrix proteins, have been demonstrated to regulate endothelial cells sprouting and the growth of capillary-like networks in vitro.18,19 Therefore, we co-cultured HUVECs with HDFs to facilitate the formation of vasculature in this study. To assess if AdECM affects cell viability, we conducted a cell toxicity assay using PrestoBlue and the live/dead assays. As shown in Fig. 3, AdECM did not affect the growth of either HUVEC or HDF cells, and no apparent cell death was observed with AdECM at concentrations of 0.5%, 0.75%, or 1%. This indicates that the matrix of AdECM is suitable for cell growth. In contrast, AdECM at any concentration, without the addition of other materials, cannot form vasculature (Fig. S1†).
Fig. 3. AdECM has no toxicity to cells. Cell viability of HUVECs and HDFs was measured by PrestoBlue cell viability assay (a) and live/dead assay (red: dead cells; green: live cells) (b). Scale bar = 200 μm.
AdECM enhanced the vasculogenesis within a fibrin matrix
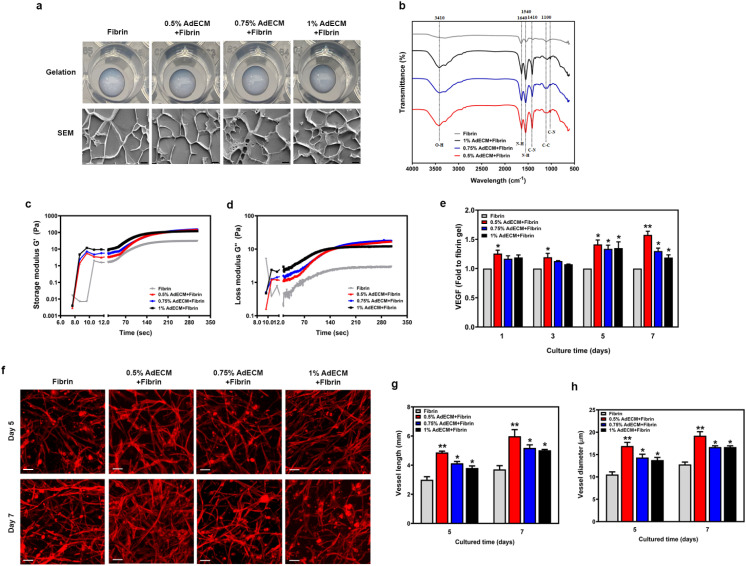
It has been demonstrated that HUVECs embedded in fibrin gel can autonomously form vasculature.20 Additionally, adipocyte extracts derived from human adipose tissue have been shown to promote angiogenesis both in vitro and in vivo.21 Therefore, we aimed to investigate the vascular network formation of AdECM when combined with fibrin. The 10 mg ml−1 fibrin solution was combined with 0.5, 0.75, or 1% AdECM. Cryo-SEM images showed that the addition of AdECM did not significantly affect the structure of the fibrin gel (Fig. 4a). Furthermore, AdECM at any concentration, when mixed with fibrin, achieved a gel-like and stable state more quickly than the fibrin solutions alone when exposed to a temperature of 37 °C. There was no significant difference in rheological properties among the concentrations of 0.5%, 0.75%, and 1% AdECM when combined with fibrin solution (Fig. 4c and d). The biochemical content in the matrix of AdECM and fibrin was characterized by FTIR analysis. The typical peak of O–H vibration around 3410 cm−1 exists only in the fibrin gel mixed with AdECM. The peaks around 1640, 1540, 1410, and 1100 cm−1, corresponding to C–C stretching, N–H deformation, O–H stretching vibration, and C–O stretching respectively, were more pronounced in the fibrin gel containing AdECM, particularly in the 0.5% AdECM/fibrin gel (Fig. 4b). To assess the pro-angiogenic properties of AdECM, we measured the VEGF content in the medium during the co-culture of HUVECs and HDFs in fibrin gel, both with and without AdECM. The inclusion of AdECM at any concentration significantly elevated the VEGF levels when HUVECs were co-cultured with HDFs over periods of 1, 3, 5, and 7 days (Fig. 4e). Profoundly, AdECM combined with fibrin promotes microvessel formation compared to fibrin gel alone, as demonstrated by staining with F-actin (Fig. 4f).
Fig. 4. AdECM-containing fibrin gel enhanced vasculature formation. (a) The ability of gelation was analyzed in matrices of 0.5, 0.75, and 1% AdECM with 10 mg ml−1 fibrin. Cryo-SEM images of AdECM-containing fibrin gel. Scale bar = 4 μm. (b) FTIR spectra of fibrin gel and AdECM-containing fibrin gels. (c and d) Rheological properties of fibrin and AdECM/fibrin solutions during gelation. (e) The level of VEGF in the media was measured after cultivating HUVECs and HDFs on a culture plate with different matrices for the indicated time period. *P < 0.05, compared to fibrin gel alone by one-way ANOVA. All data are shown as the mean (±SD). (f) Fluorescence images of vasculature stained with F-actin. Self-assembled microvascular network on the matrices after cultivation of HUVECs and HDFs for 5 and 7 days. Scale bar = 100 μm. (g and h) Quantitative measurement of average vessel length and vessel diameter of assembled vascular network in different matrices by ImageJ with a vessel analysis plugin from three fields of each well. *P < 0.05, compared to fibrin gel by one-way ANOVA.
The average vessel length and vessel diameter were measured and quantified using ImageJ software with a vessel analysis plugin (National Institutes of Health) after HUVECs and HDFs were embedded within the matrix for 5 and 7 days. For each experiment, at least 20 measurements of vessel length, and 30 measurements of vessel diameter were quantified from three different fields of each well. As shown in Fig. 4g and h, both the vessel length and diameter were significantly increased in the fibrin gel with AdECM especially at 0.5% AdECM. This evidence clearly demonstrates that AdECM promotes vasculogenesis within fibrin gel.
The transcriptomic profile of the AdECM-containing matrix revealed enhanced expression of angiogenesis-related genes
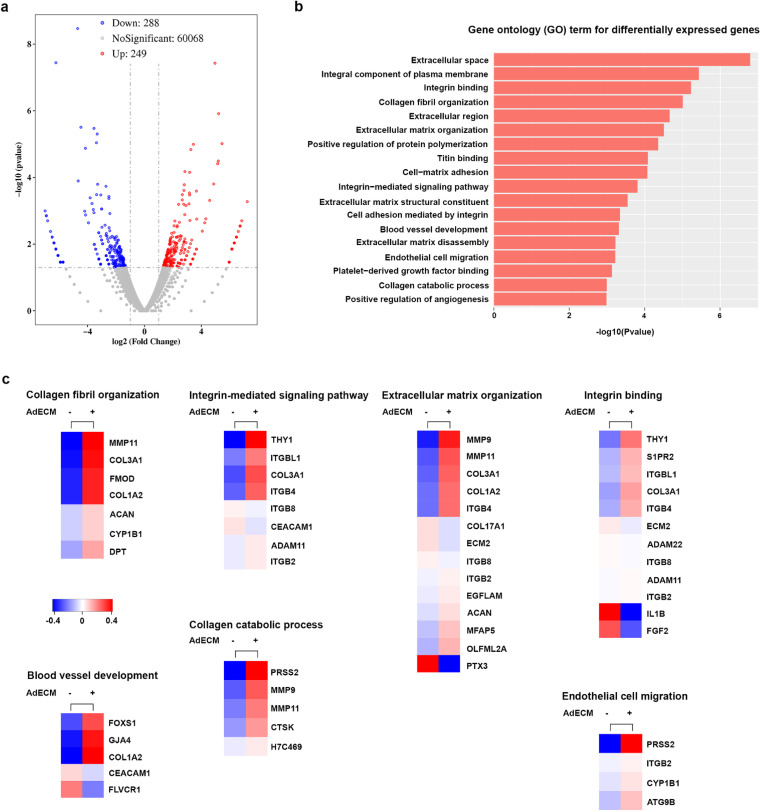
Given the pro-angiogenic properties of AdECM in promoting vasculature formation within fibrin gel, we further investigated the gene regulations underlying the effects elicited by AdECM using RNA-sequencing analysis. Given that 0.5% AdECM more prominently promoted vascular network formation, we sought to understand the transcriptomic profile of the 0.5% AdECM/fibrin matrix compared to fibrin gel alone after co-culturing of HUVECs and HDFs for 7 days. The volcano plot displayed 537 genes that were differentially expressed between these two matrices, in which 288 genes were downregulated and 249 genes were upregulated as shown in Fig. 5a. The differentially expressed genes were further subjected to gene ontology (GO) analysis to discern the biological differences between the two matrices. Protease-mediated remodeling of the extracellular matrix facilitates cell movement, which is essential for creating new blood vessels. Additionally, proteases and angiogenic factors release matrix-bound growth factors, promoting angiogenesis by stimulating the migration and growth of endothelial cells.22 We found the differentially expressed genes were enriched in the GO terms related to extracellular remodeling and angiogenesis, such as collagen fibril organization, integrin-mediated signaling pathway, blood vessel development, and endothelial cell migration (Fig. 5b). For induction of angiogenesis, as anticipated, almost all of the differentially expressed genes associated with blood vessel development and endothelial cell migration were upregulated by supplementation with 0.5% AdECM. In the regulation of extracellular remodeling, the differentially expressed genes were mostly elevated in the GO term of collagen fibril organization, collagen catabolic process, and extracellular matrix organization when fibrin gel mixed with 0.5% AdECM. Given that integrins regulate contacts between cells and between cells and the matrix, they play a crucial role in blood vessel growth by enhancing the migration and survival of endothelial cells.23 Regarding integrin binding and the integrin-mediated signaling pathway, we found that almost all differentially expressed genes were upregulated in the 0.5% AdECM/fibrin gel (Fig. 5c). Collectively, the transcriptomic data clearly show that AdECM promotes angiogenesis by regulating the expression of angiogenesis-related genes.
Fig. 5. Transcriptomic analysis of vasculature formed in the matrix with or without AdECM. (a) A volcano plot. The red dots indicated genes that are significantly upregulated, while the green dots represented genes that have been significantly downregulated, and the gray dots denote genes whose differential expression is not significant. (b) A GO term enrichment analysis of the differentially expressed genes. (c) Heatmap of the genes corresponding to a selection of GO terms derived from (b).
Microfluidic model combined with AdECM-containing gel for maintaining vasculature formation
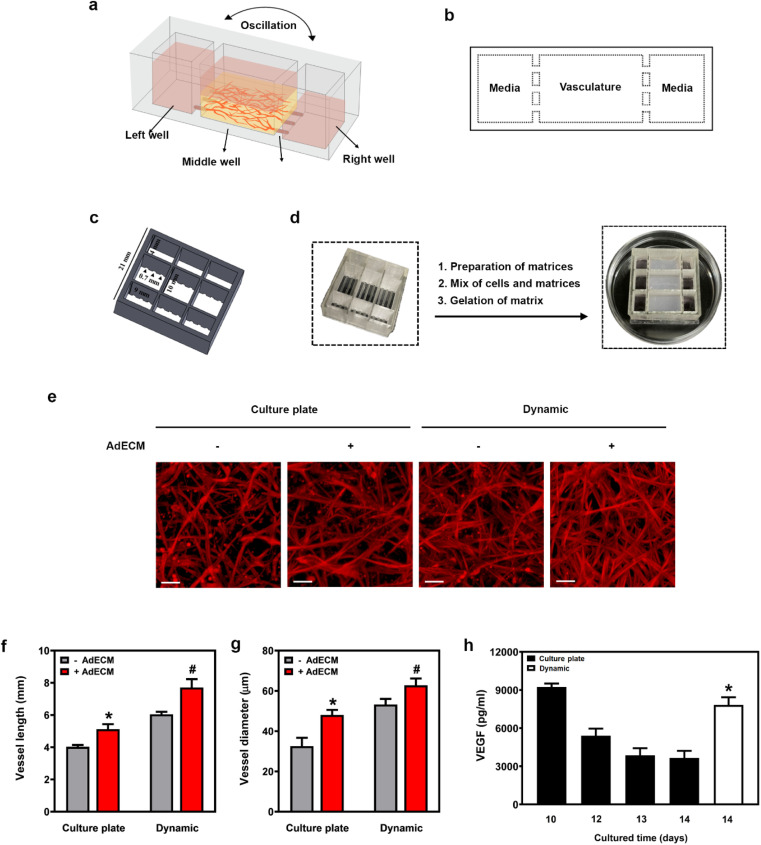
In order to investigate the molecular mechanisms underlying diseases or to evaluate long-term drug efficacy, the persistence of vasculature is required. Both blood flow and growth factors are essential for sustaining active angiogenesis during vascular maturation.24 In our study, we observed that the degree of vascular maturation diminished when the vasculature was cultured on a plate for 14 days (Fig. 6e–g). Similarly, the level of VEGF gradually decreased in the media when the vasculature was cultivated on a culture plate. This underscores the importance of microfluidic flow in sustaining vasculogenesis (Fig. 6h). Consequently, we fabricated a microfluidic device from polydimethylsiloxane (PDMS) using a 3D printer (MiiCraft Ultra). This device, mounted on a 22 mm × 22 mm cover glass slide with a 1 mm thickness in a 9-well format, was placed on a rocker to create fluid flow through the device.
Fig. 6. A 3D-printed microfluidic device sustained vasculogenesis. (a) Illustration of the 3D-printed microfluidic device and experimental setup. (b) Top view of the microfluidic device. (c) Schematic showing the dimensions of the 3D-printed microfluidic device. (d) Experimental procedures for constructing vasculature in a microfluidic device. (e) Vessels cultured in a cultured plate or in a microfluidic device (dynamic) for 14 days, stained with F-actin (red). Scale bar = 100 μm. (f and g) Quantification of vessel length and diameter using ImageJ. *P < 0.05, versus vessels embedded in fibrin matrix without 0.5% AdECM in the cultured plate. #P < 0.05, compared to vessels cultivated in an AdECM-containing fibrin gel on a culture plate by one-way ANOVA. (h) VEGF content in media from the cultivation of vasculature for 10 to 14 days was measured. *P < 0.05, compared to the content of VEGF in media after cultivating vessels on a cultured plate for 14 days by one-way ANOVA. Results are presented as the mean percentage (±SD) of three independent experiments.
As depicted in Fig. 6a and b, an illustration of the microfluidic device with 9 wells shows the details of the device and the experimental setup for the vasculature formation. The device consists of three connected wells that function as a unit. The dimensions of the middle well are 6(L) × 10(W) × 9(H) mm, and it is connected to the left or right well by three channels of 1(L) × 0.7(W) × 0.7(H) mm. In contrast, the dimensions of the left and right chambers are 6(L) × 4(W) × 9(H) mm (Fig. 6c). Vasculature was established in the middle well and connected to both side wells through the connecting channels. By enlarging the middle well's dimensions, specifically its width and height, the volume increased, enabling the media to supply nutrients more effectively to the vasculature. Notably, a vascular network formed more rapidly in a well with a width of 10 mm and a height of 9 mm, compared to one with both a width and height of 6 mm (Fig. S2†).
To emulate human tissues permeated by vasculature, we positioned a 13 mm-long Polyvinyl alcohol (PVA) tube in two distinct connecting channels on both the left and right sides of the middle chamber (Fig. 6d). This allowed the tube to pass through the left, middle, and right chambers, creating microfluidic channels within the cultured tissue. This design aimed to enhance the surface area for more efficient transport of nutrients and oxygen, thereby promoting vasculature formation. When we did not use PVA tube insertion in the connecting channels, there was a dramatic decrease in the extent of vasculature. This further underscores our focus on optimizing the fabrication of our microfluidic device to enhance vasculature formation (Fig. S2†).
The entire microfluidic device will be positioned on a rocker, which will tilt at 30° regularly every 20 minutes. After cultivating HUVECs and HDFs in a 0.5% AdECM/fibrin matrix using a microfluidic device (dynamic) for 14 days, the vascular network developed into a multilayered and extensively branched structure. This was in contrast to the networks formed in a 0.5% AdECM/fibrin matrix within a culture plate over 14 days (Fig. 6e). We found that the vessel length and vessel diameter were noticeably increased when vasculature cultured in the dynamic state for a long-period (14 days) compared to the culture plate (Fig. 6f and g). Comparatively, the pro-angiogenic factor VEGF was markedly elevated when vasculature was formed in the dynamic state for 14 days compared to the culture plate for 14 days, underscoring the crucial role of the microfluidic device in sustaining vasculogenesis. Furthermore, in the dynamic state of using a microfluidic device, the vascular network was notably more complex when HUVECs and HDFs were cultured in AdECM-containing fibrin gel, compared to when the cells were embedded in fibrin gel only. Additionally, the vessel length and vessel diameter were significantly increased, as illustrated in Fig. 6g and f. Thus, the evidence clearly demonstrates the synergistic effects of AdECM and the microfluidic system in promoting vascularization.
Enhanced activity of the PI3K/Akt and JAK/STAT3 pathways in a microfluidic model with an AdECM matrix supports sustained vasculogenesis
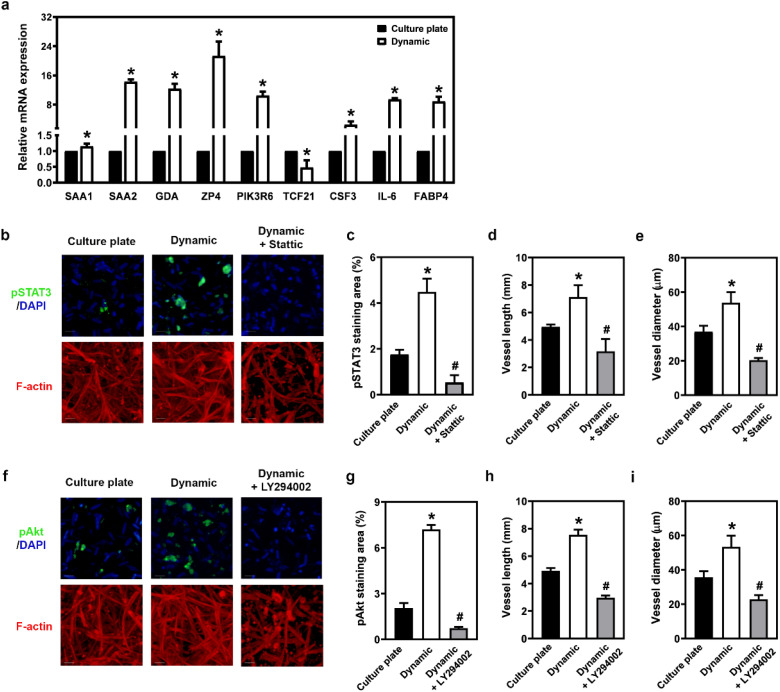
To gain insight into the specific gene regulation during angiogenesis in the microfluidic device, we analysed RNA expression patterns using RNA sequencing assays. Numerous genes exhibited differential expression between the culture plate and dynamic states when vasculature was cultivated in a 0.5% AdECM/fibrin gel over 14 days. qRT-PCR assays further confirmed that the expression of these genes was elevated when HUVECs and HDFs were cultured in a 0.5% AdECM/fibrin matrix in the microfluidic system compared to culture plate condition (Fig. 7a). Many of the genes identified are primarily associated with the PI3K/Akt and JAK/STAT3 signaling pathways. For instance, following endoplasmic reticulum stress, SAA1 transcription is known to be upregulated via JAK/STAT3 signaling.25 Moreover, the IL-6/JAK/STAT3 signaling axis plays a pivotal role in enhancing angiogenesis.26 On another note, PIK3R6 serves as a regulatory subunit in the PI3K/Akt pathway, and inhibiting TCF21 has been shown to activate the PI3K/Akt signaling cascade.27 The transcriptome results were analysed using the KEGG pathway, which also revealed that these genes are primarily involved in the JAK/STAT or PI3K/Akt-related pathways (Fig. S3†). Therefore, we propose that a microfluidic model stimulates vasculogenesis by regulating the PI3K/Akt and JAK/STAT3 pathways. When HUVECs and HDFs were cultured in the dynamic system of a microfluidic device for 14 days, the increased vasculogenesis was concomitant with an observed elevation in the levels of phospho-STAT3 and phospho-Akt (Fig. 7b–i). Based on these findings, it can be concluded that the dynamic system enhanced vasculogenesis by activating the PI3K/Akt and JAK/STAT3 signaling pathways.
Fig. 7. Upregulated PI3K/Akt and JAK/STAT3-mediated signalling are involved in sustaining vasculogenesis in a microfluidic system with an AdECM-containing gel. (a) Relative expression of differentially expressed genes in vasculature cultivated in a culture plate or a microfluidic device (Dynamic) with 0.5% AdECM-containing fibrin gel, as measured by real-time PCR (qPCR). (b) Immunofluorescence staining of the vascular network with pSTAT3 (green) and F-actin (red) after 14 days of cultivation in the presence or absence of static that has inhibitory effects against STAT3 to impede JAK/STAT3 pathway. (c and d) Vessel length and diameter were measured and quantified. (e) Immunofluorescence labeling of the vascular network was performed using pAkt3 (green) and F-actin (red) after cultivating the vasculature for 14 days with or without the addition of LY294002, an inhibitor of PI3K that blocks PI3K/Akt signaling. (f and g) The length and diameter of the vessels were assessed and quantified. Scale bar = 100 μm. *P < 0.05, compared to culture plate condition. #P < 0.05, compared to dynamic condition without inhibitor treatment, by Student's t-test. All data are presented as the mean (±SD) of three independent experiments.
AdECM integration with microfluidic model enhanced vasculature formation and tumoroid growth in the ex vivo vascularized tumoroid model
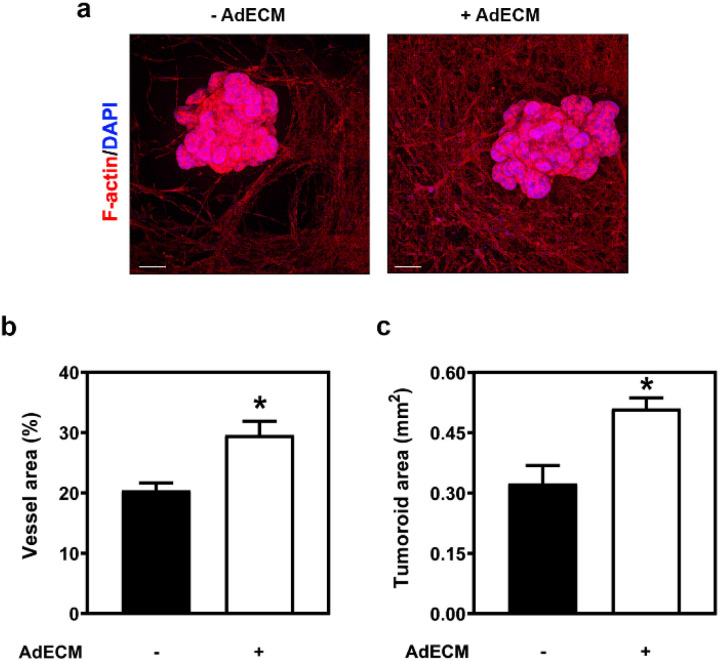
The feasibility of a vascular network cultured in an AdECM-containing gel was evaluated ex vivo using a microfluidic device and a tumoroid model, as the cultivation of HUVECs and HDFs within this gel was shown to accelerate vascular network formation. The tumoroid model recapitulates the genetic characteristics and phenotypic features of tumors within the body (Fig. S10†). To simulate the fluidic environment of tumors, we incubated the tumoroids in the microfluidic device. Colon tumoroids were cultured in Geltrex gel and simultaneously incubated with HUVECs and HDFs in fibrin gel, with and without 0.5% AdECM. After 14 days of cultivation, both the vascular network and tumoroid growth were significantly enhanced in the AdECM-containing gel compared to those in fibrin gel alone (Fig. 8). These data demonstrate that the increased vasculature was associated with enhanced tumoroid growth, revealing the functionality of the vasculature cultured in AdECM-containing gel ex vivo.
Fig. 8. The tumoroid growth was enhanced when the vasculature was cultivated within a gel containing AdECM. (a) Colon tumoroids were co-cultured with vasculature formed in the gel with or without AdECM. (b and c) The vessel area and tumoroid area in the vascularized tumoroids were quantified using ImageJ from three independent samples. Scale bar = 200 μm. *P < 0.05, compared to vasculature cultivated in the gel without AdECM by Student's t-test. All data are shown as the mean (±SD).
Discussion
The human vascular system, a crucial part of the body, supplies nutrients and removes wastes from cells, essential for organ growth and development.28 To study diseases and drug efficacy accurately, researchers aim to recreate organ microenvironments ex vivo. In this study, AdECM from bovine adipose tissue combined with fibrin gel is adapted to enhance angiogenesis. In addition, a 3D-printed microfluidic device is employed to provide microflow during cultivation, resulting in a more robust vascular network in the AdECM-enriched fibrin gel. Traditional 2D culture systems is not sufficient to present and replicate the complexity of the three-dimensional vascular architecture within organs, making 3D culturing systems more realistic.29 Many studies used fibrin gel as a scaffold to create 3D models which promote endothelial self-assembly in the formation of vascular network formation, but its variable ultrastructure (stemming from its formulation and gelation conditions) and lack of tissue specificity pose challenges in providing a consistent and suitable vasculature environment.30
Adipose tissue is a type of connective tissue throughout the body and its extracts stimulate angiogenesis, making them promising for incorporating into an ex vivo scaffold, creating organ-specific environments, and fostering angiogenesis.20 In this study, we decellularized bovine adipose tissue, removed nuclei, leaving collagen and GAGs, creating a scaffold for HUVEC angiogenesis. We found that mixing various percentages of AdECM with fibrin gel significantly enhanced HUVEC angiogenic properties, creating a biomimetic ex vivo disease model.
Additionally, standardizing dECM production is challenging due to variations between batches. To address this, we evaluated key parameters, such as gelation ability, stability during incubation in medium, the constituents of dECM, and cell cytotoxicity for each AdECM batch, ensuring stability and consistency. The high quality controlled AdECM combined with commercial fibrin solution promoted angiogenesis. Our results demonstrated that using a low concentration of AdECM (0.5% AdECM) effectively facilitated the formation of a vascular network. This not only substantially reduced the amount of AdECM needed in the matrix but also minimized batch-to-batch variation, making it a promising step towards clinical application.
To gain a deeper insight into how 0.5% AdECM impacts vasculature formation, we analysed the transcriptomic patterns during its cultivation. The results revealed that the genes present in the 0.5% AdECM/fibrin matrix distinctly aligned with biological pathways associated with angiogenesis and the reshaping of the extracellular matrix. Given that the modification of the extracellular matrix, encompassing both the proteolysis of this matrix and the breakdown of collagen, plays a pivotal role in endothelial sprouting,21 it was significant to observe an upregulation of proteases such as MMP9, MMP11, and PRSS2 in the 0.5% AdECM/fibrin matrix. Moreover, these genes consistently played a role in the collagen catabolic process (Fig. 5c). Therefore, the incorporation of AdECM into fibrin gel significantly promoted vascular network formation by upregulating the genes involved in angiogenic processes.
Furthermore, this study also would like to extend the culture period in a custom microfluidic device for 14 days, aiming to increase vessel length and diameter. Shear stress within blood vessels impacts endothelial cells, affecting gene expression, such as VEGF-A, and angiogenesis, in addition to diversity.31–33 A certain shear stress was identified as the trigger for angiogenic sprouting in endothelial cells, facilitating their penetration of the underlying matrix. Furthermore, blood fluid shear stress has been associated with the upregulation of VEGF-A gene expression.34 In addition, STAT3 is one of the angiogenesis-related transcription factors that has been well-known, and the activation of STAT3 increases the expression of VEGF and MMPs, promoting angiogenesis. In our studies, we found that several genes were differentially expressed during vasculogenesis promoted by a microfluidic system. Among these were SAA1 and SAA2, which have already been reported to stimulate the expression of vascular endothelial growth factor receptor 2 (VEGFR2), the specific receptor for VEGF-A, and to promote angiogenesis.35 SAA1 and SAA2 are activated through a STAT3-mediated pathway.25 Given that STAT3 is a critical factor in the angiogenic pathway,36 our study also revealed its important role in promoting angiogenesis within the microfluidic device environment. Furthermore, we observed that PIK3R6, a component of the PI3K/Akt pathway known for stimulating angiogenesis,37 was upregulated during the dynamic phase of vasculature formation in our research. We observed the upregulation of genes like SAA1, SAA2, and PIK3R6, which promote angiogenesis, emphasizing the role of fluidic flow in vasculogenesis.38 Our 3D-printed microfluidic device can be scaled up, providing dynamic shear stress for better HUVEC angiogenesis. By optimizing AdECM culture and dynamic stress stimuli, we created a functional vasculature system with extended HUVEC culture persistence. In Fig. S4a and the Movie S5,† the formation of an intricate vascular network is observed following the co-culture of HUVECs and HDFs within an AdECM/Fibrin matrix over a 14 days period. This vascular network exhibited clear luminal structures (Fig. S4b and S6†). When integrated with the findings presented in Fig. 8, it is evident that this engineered vascular network closely replicates both the structural and functional characteristics of native vasculature.
This system enables researchers to model ex vivo tissue engineering model or microenvironment with culture various types of organoids or cells; and recapitulate the organs within the human body for further disease or physiological studies.
We have developed a vascularized tumoroid model in a microfluidic device, cultivating vasculature within an AdECM-containing gel to evaluate the feasibility of integrating AdECM with microfluidic technology. After 14 days of co-culturing HUVECs and HDFs in a microfluidic device placed on a rocker, the vessel area was significantly enhanced when these cells were grown in an AdECM-containing matrix. This finding highlights the synergistic effects of AdECM and the microfluidic model in promoting and sustaining vasculogenesis. Additionally, the presence of AdECM in the matrix of the microfluidic system was associated with a notable increase in vasculature, which in turn significantly improved the growth of colon tumoroids. This underscores the functionality of the vasculature within an AdECM-containing gel in a microfluidic device.
Conclusion
The vasculature plays a critical role in the transportation of essential nutrients and oxygen necessary to maintain cellular homeostasis across various organs, including within tumor environments. Angiogenesis, a complex biological process, is regulated by a multitude of factors. This study focuses on the investigation of how the AdECM serves as both a structural framework for cells and a reservoir for growth factors, influencing the regulation of genes associated with angiogenesis and extracellular remodeling. To facilitate this research, a microfluidic device has been designed to induce shear stress, activating PI3K and JAK-mediated pathways, which ultimately promotes the sustainability of the vasculature. The combined utilization of AdECM and the microfluidic device creates optimal conditions for the maintenance of vasculature. Establishing an ideal vasculature system has the potential to lead to the development of biomimetic tissues or disease models suitable for applications such as drug screening and disease investigation.
Data availability
The data supporting this article have been included as part of the ESI.†
Author contributions
Yu-Yin Shih: conceptualization, resources, data curation, formal analysis, funding acquisition, validation, investigation, writing – original draft. Chun-Wei Kao: investigation, visualization, data curation. Yi-Rong Jhong: investigation, visualization. Yi-An Chen: data curation. Yi-Wen Chen: conceptualization, validation, data curation, writing – review & editing, supervision, funding acquisition.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This study was founded by National Science and Technology Council (MOST 111-2314-B-039-049-) and China Medical University Hospital (DMR-111-180).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05573j
References
- Jain R. K. Nat. Med. 2001;7:987–989. doi: 10.1038/nm0901-987. [DOI] [PubMed] [Google Scholar]
- Stylianopoulos T. Munn L. L. Jain R. K. Trends Cancer. 2018;4:292–319. doi: 10.1016/j.trecan.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alilou M. Khorrami M. Prasanna P. Bera K. Gupta A. Viswanathan V. S. Patil P. Velu P. D. Fu P. Velcheti V. Madabhushi A. Sci. Adv. 2022;8:eabq4609. doi: 10.1126/sciadv.abq4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry S. A. Farnsworth R. H. Solomon B. Achen M. G. Stacker S. A. Fox S. B. Front. Immunol. 2016;7:621. doi: 10.3389/fimmu.2016.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. M. Kobayashi H. Rafii S. Nat. Rev. Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman J. N. Engl. J. Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- Auger F. A. Gibot L. Lacroix D. Annu. Rev. Biomed. Eng. 2013;15:177–200. doi: 10.1146/annurev-bioeng-071812-152428. [DOI] [PubMed] [Google Scholar]
- Neve A. Cantatore F. P. Maruotti N. Corrado A. Ribatti D. BioMed Res. Int. 2014;2014:756078. doi: 10.1155/2014/756078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawans P. Behl T. Bhardwaj S. Biomed. Pharmacother. 2020;126:110103. doi: 10.1016/j.biopha.2020.110103. [DOI] [PubMed] [Google Scholar]
- Rubina K. Kalinina N. Efimenko A. Lopatina T. Melikhova V. Tsokolaeva Z. Sysoeva V. Tkachuk V. Parfyonova Y. Tissue Eng., Part A. 2009;15:2039–2050. doi: 10.1089/ten.tea.2008.0359. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N. Garry D. J. Cardiovasc. Res. 2017;113:1294–1306. doi: 10.1093/cvr/cvx133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn J. Mehraban F. Ingle G. Xin X. Bryant J. E. Vehar G. Schoenfeld J. Grimaldi C. J. Peale F. Draksharapu A. Lewin D. A. Gerritsen M. E. Am. J. Pathol. 2000;156:1887–1900. doi: 10.1016/S0002-9440(10)65062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duinen V. Zhu D. Ramakers C. van Zonneveld A. J. Vulto P. Hankemeier T. Angiogenesis. 2019;22:157–165. doi: 10.1007/s10456-018-9647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto Y. Hayashi T. Kunita I. Nakamasu A. Torisawa Y. S. Nakayama M. Takigawa-Imamura H. Kotera H. Nishiyama K. Miura T. Yokokawa R. Integr. Biol. 2017;9:506–518. doi: 10.1039/c7ib00024c. [DOI] [PubMed] [Google Scholar]
- Rajasekar S. Lin D. S. Y. Abdul L. Liu A. Sotra A. Zhang F. Zhang B. Adv. Mater. 2020;32:e2002974. doi: 10.1002/adma.202002974. [DOI] [PubMed] [Google Scholar]
- Driehuis E. Kretzschmar K. Clevers H. Nat. Protoc. 2020;15:3380–3409. doi: 10.1038/s41596-020-0379-4. [DOI] [PubMed] [Google Scholar]
- Pleguezuelos-Manzano C. Puschhof J. van den Brink S. Geurts V. Beumer J. Clevers H. Curr. Protoc. Immunol. 2020;130:e106. doi: 10.1002/cpim.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. C. Nakatsu M. N. Chou W. Gershon P. D. Hughes C. C. Mol. Biol. Cell. 2011;22:3791–3800. doi: 10.1091/mbc.E11-05-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz-Schughart L. A. Schroeder J. A. Wondrak M. van Rey F. Lehle K. Hofstaedter F. Wheatley D. N. Am. J. Physiol.: Cell Physiol. 2006;290:C1385–C1398. doi: 10.1152/ajpcell.00248.2005. [DOI] [PubMed] [Google Scholar]
- Chen M. B. Whisler J. A. Frose J. Yu C. Shin Y. Kamm R. D. Nat. Protoc. 2017;12:865–880. doi: 10.1038/nprot.2017.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. Xia J. Chen H. Wang L. Deng C. Lu F. Stem Cell Res. Ther. 2019;10:252. doi: 10.1186/s13287-019-1356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottile J. Biochim. Biophys. Acta. 2004;1654:13–22. doi: 10.1016/j.bbcan.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Garmy-Susini B. Varner J. A. Lymphatic Res. Biol. 2008;6:155–163. doi: 10.1089/lrb.2008.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhleder S. Fernandez-Chacon M. Garcia-Gonzalez I. Benedito R. Cell. Mol. Life Sci. 2021;78:1329–1354. doi: 10.1007/s00018-020-03664-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. Zhou X. Zou H. Liu L. Li X. Ruan Y. Xie Y. Shi M. Xiao Y. Wang Y. Zhou Y. Wu Y. Guo B. Exp. Cell Res. 2021;408:112856. doi: 10.1016/j.yexcr.2021.112856. [DOI] [PubMed] [Google Scholar]
- Johnson D. E. O'Keefe R. A. Grandis J. R. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y. Duan H. Duan C. Zhu H. Zhou R. Pei H. Shen L. OncoTargets Ther. 2017;10:1603–1611. doi: 10.2147/OTT.S118151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. M. Bissell M. J. Annu. Rev. Cell Dev. Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. Xing Y. Li J. Deng C. Li Y. Ren X. Zhang D. Front. Cell Dev. Biol. 2021;9:639299. doi: 10.3389/fcell.2021.639299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin K. T. Tranquillo R. T. Exp. Cell Res. 2013;319:2409–2417. doi: 10.1016/j.yexcr.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A. Banuth A. M. M. Nader H. B. Dreyfuss J. L. PLoS One. 2020;15:e0241040. doi: 10.1371/journal.pone.0241040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglebert M. Locatelli L. Tsvirkun D. Sinha P. Maier J. A. Misbah C. Bureau L. Biomicrofluidics. 2020;14:024115. doi: 10.1063/1.5143391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douguet D. Patel A. Xu A. Vanhoutte P. M. Honore E. Trends Pharmacol. Sci. 2019;40:956–970. doi: 10.1016/j.tips.2019.10.002. [DOI] [PubMed] [Google Scholar]
- Tsaryk R. Yucel N. Leonard E. V. Diaz N. Bondareva O. Odenthal-Schnittler M. Arany Z. Vaquerizas J. M. Schnittler H. Siekmann A. F. Sci. Rep. 2022;12:4795. doi: 10.1038/s41598-022-08645-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M. Xia Y. F. Li B. Liu H. Pan J. Y. Li B. B. Zhang C. An F. S. J. Physiol. Biochem. 2016;72:71–81. doi: 10.1007/s13105-015-0462-4. [DOI] [PubMed] [Google Scholar]
- Chen Z. Han Z. C. Med. Res. Rev. 2008;28:185–200. doi: 10.1002/med.20101. [DOI] [PubMed] [Google Scholar]
- Wilson L. S. Baillie G. S. Pritchard L. M. Umana B. Terrin A. Zaccolo M. Houslay M. D. Maurice D. H. J. Biol. Chem. 2011;286:16285–16296. doi: 10.1074/jbc.M110.217026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Li J. Zhang X. Zhang X. Zhang X. Zhu Y. Chen C. Liu Z. Wu X. Wang D. Dongye M. Wang J. Lin H. Exp. Cell Res. 2021;398:112362. doi: 10.1016/j.yexcr.2020.112362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this article have been included as part of the ESI.†