Summary
Diffuse pleural mesothelioma (DPM) is a lethal cancer with a poor prognosis and limited treatment options. The Hippo signaling pathway genes, such as NF2 and LATS1/2, are frequently mutated in DPM, indicating a tumor suppressor role in the development of DPM. Here, we show that in DPM cell lines lacking NF2 and in mice with a conditional Nf2 knockout, downregulation of WWC proteins, another family of Hippo pathway regulators, accelerates DPM progression. Conversely, the expression of SuperHippo, a WWC-derived minigene, effectively enhances Hippo signaling and suppresses DPM development. Moreover, the adeno-associated virus serotype 6 (AAV6) has been engineered to deliver both NF2 and SuperHippo genes into mesothelial cells, which substantially impedes tumor growth in xenograft and genetic DPM models and prolongs the median survival of mice. These findings serve as a proof of concept for the potential use of gene therapy targeting the Hippo pathway to treat DPM.
Keywords: diffuse pleural mesothelioma, DPM, WWC proteins, NF2, YAP/TAZ, Hippo, gene therapy
Graphical abstract
Highlights
-
•
NF2 and WWC proteins jointly regulate Hippo signaling and tumorigenesis
-
•
Downregulation of WWC promotes diffuse pleural mesothelioma (DPM) development
-
•
Ectopic expression of a WWC-derived SuperHippo minigene suppresses DPM progression
-
•
AAV6 expressing both NF2 and SuperHippo is a potential gene therapy for DPM
Zhu et al. demonstrate that HPO1 is crucial for maintaining Hippo signaling and preventing rapid tumorigenesis in diffuse pleural mesothelioma (DPM) with defective HPO2. Co-activating HPO1 and HPO2 by AAV6-mediated expression of SuperHippo-P2A-NF2 effectively mitigates tumor progression in DPM mouse models, offering a potential treatment for this aggressive disease.
Introduction
Diffuse pleural mesothelioma (DPM) is an aggressive and treatment-resistant cancer originating from the mesothelial lining surrounding the lungs or other pleural tissues.1,2 Exposure to asbestos or other fibrous minerals is the primary risk factor for DPM.3,4,5 Without treatment, patients with DPM consistently display a life expectancy of less than 1 year.6 Current therapeutic interventions for DPM primarily involve standard cisplatin and pemetrexed chemotherapy.7,8,9 However, the response rate to chemotherapy among patients with DPM only ranges between 30% and 40%, with the median overall survival (OS) slightly exceeding 12 months.7,8,10 A recent phase 3 clinical trial demonstrated that the combined utilization of ipilimumab (a CTLA-4 monoclonal antibody) and nivolumab (a PD-1 monoclonal antibody) can extend the OS by up to 4 months in a subset of patients with DPM.11 However, some patients exhibit primary resistance to immunotherapy, and the overall prognosis for DPM remains discouraging.9,12 Hence, there is an urgent need to explore alternative therapeutic modalities to improve the efficacy of DPM treatment. In this pursuit, acquiring comprehensive insights into the molecular mechanisms, particularly key oncogenic signaling pathways underlying the tumor biology of DPM, is crucial for developing effective therapeutic strategies.
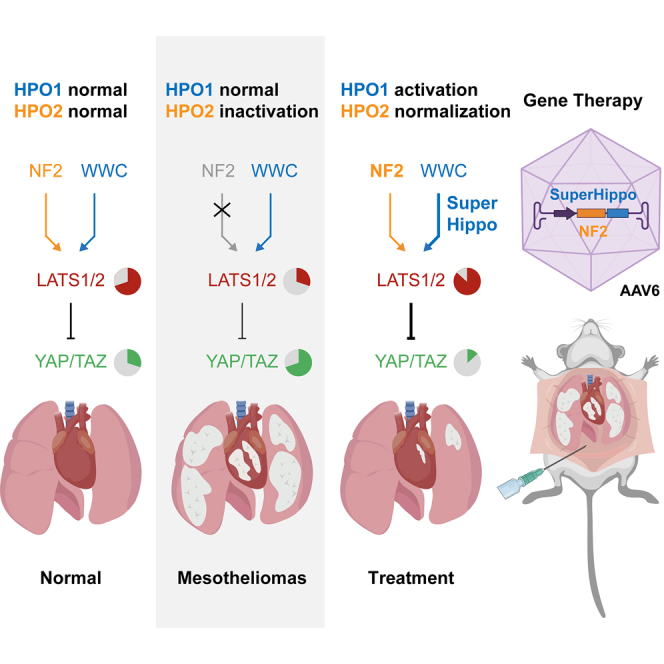
The Hippo signaling pathway is critical in regulating organ size, tissue regeneration, and tumorigenesis.13,14,15,16,17 Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ, also known as WWTR1) are downstream effectors of the Hippo pathway and, as proto-oncoproteins, are frequently activated in diverse cancers.15,16,18,19,20,21,22,23,24,25 The activity of YAP/TAZ is tightly restricted by upstream kinases (MST1/2, MAP4K1–7, and LATS1/2) and scaffolding proteins (SAV1, NF2, WWC1–3, and MOB1).26,27,28,29,30,31,32,33 These upstream regulators of YAP/TAZ are mostly tumor suppressors, and their inactivation leads to tumorigenesis.31,34,35 Recently, we have shown that the Hippo signaling network contains two largely independent modules, HPO1 and HPO2, in which WWC1–3 and NF2, as adaptors, mediate the phosphorylation and activation of LATS1/2 by MST1/2 and MAP4K1–7, respectively31,36,37 (Figure 1A). Concurrent inactivation of HPO1 and HPO2 genes in mouse livers leads to rapid cancer development, suggesting that HPO1 and HPO2 synergistically regulate YAP/TAZ activity and tumorigenesis.31 Moreover, a WWC1-derived SuperHippo minigene can effectively and specifically activate HPO1 signaling and serve as a potent tumor suppressor in liver cancers.30
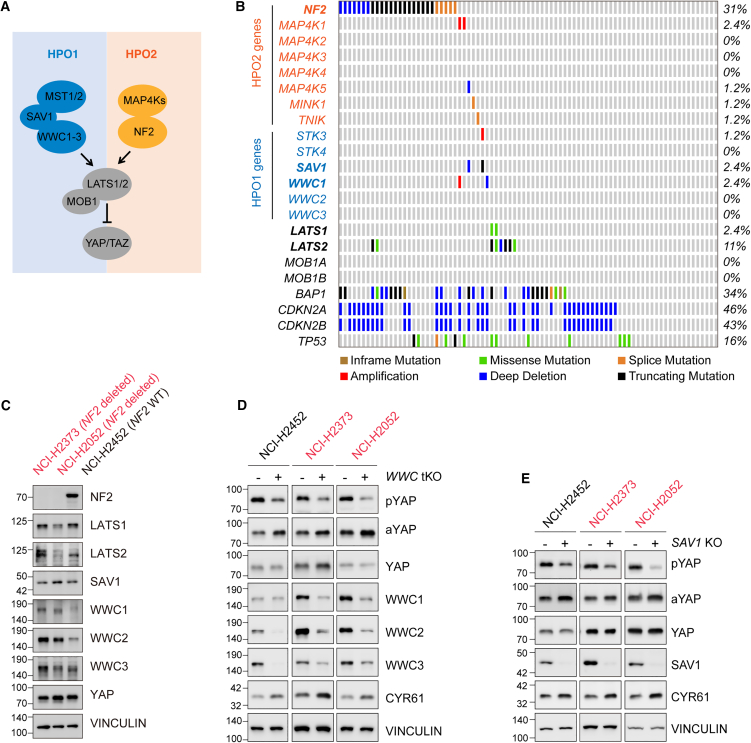
Figure 1.
Deletion of SAV1 or WWC1–3 induces YAP/TAZ activity in DPM cells
(A) Schematic representation of two Hippo signaling modules. There are two modules within the Hippo signaling network: MST1/2-SAV1-WWC1–3-LATS1/2 (HPO1) and MAP4K1–7-NF2-LATS1/2 (HPO2).
(B) Oncoplot depicting the distribution and alteration patterns of Hippo pathway genes in TCGA DPM datasets.
(C) Expression analysis of Hippo pathway component in NCI-H2373, NCI-H2052, and NCI-H2452 mesothelioma cell lines using immunoblotting.
(D and E) Enhanced YAP/TAZ activity in WWC1–3 tKO and SAV1 KO DPM cells. The pYAP and aYAP indicate phosphorylated and active YAP, respectively.
See also Figure S1.
Whole-exome sequencing of human DPM samples has revealed a high prevalence of mutation or deletion in many tumor suppressor genes, such as CDKN2A/B, NF2, BAP1, and TP53.38,39,40,41 In addition, loss-of-function mutations of LATS2 have also been observed in patients with DPM.42,43 Among genes frequently mutated, NF2 and LATS2 are major regulators in the Hippo signaling pathway, indicating that the inactivation of Hippo signaling is involved in DPM pathogenesis. In support of this, genetic inactivation of Nf2 and Trp53 induces tumor formation in the mouse mesothelium.44
In this study, we show that the inactivation of SAV1 or WWC genes in mesothelioma cells results in YAP/TAZ hyperactivation and rapid tumor growth, and expression of SuperHippo has the opposite effect. Hence, HPO1 is crucial for maintaining Hippo signaling and preventing rapid tumorigenesis, especially in HPO2-defective (NF2-mutated) cells, and it may be employed to develop therapeutics for cancers, including DPM. Indeed, activation of both HPO1 and HPO2 signaling modules by concurrent expression of NF2 and SuperHippo leads to complete inactivation of YAP/TAZ and repression of tumorigenesis, regardless of NF2 status. Based on these findings, we have developed a preclinical gene therapy for DPM by using an adeno-associated virus (AAV) to deliver both NF2 and SuperHippo into tumor cells, and this approach has effectively blocked tumor progression in both xenograft and genetic DPM models.
Results
Inactivation of HPO1 induces YAP/TAZ activity in DPM cells
To assess the involvement of the Hippo pathway in the progression of DPM, we examined the status of major Hippo pathway genes in The Cancer Genome Atlas (TCGA) datasets.42 In 87 DPM specimens analyzed, NF2 and LATS2 were altered in 31% and 11% of samples, respectively, whereas SAV1, WWC1, or LATS1 was only altered in 2.4% of samples (Figure 1B). Other Hippo pathway genes were not significantly altered in DPM (Figure 1B). Clearly, there was an enrichment of HPO2 gene (NF2) alterations in DPM. On the other hand, alterations of HPO1 genes (SAV1 and WWC1/2/3) were relatively rare in DPM.
HPO1 and HPO2 are required to regulate LATS1/2 and YAP/TAZ in a synergistic manner.31,36,37 Hence, we sought to test the function of HPO1 in the development of DPM. We first collected three human DPM cell lines: NCI-H2452, NCI-H2373, and NCI-H2052. Among these cell lines, NCI-H2373 has NF2 mutation, NCI-H2052 has both NF2 and LATS2 mutations, and NCI-H2452 has wild-type NF2 and LATS2.35,45 The expression of Hippo pathway components was analyzed by immunoblotting. SAV1 and WWC1–3 were expressed in all three cell lines (Figure 1C). We then ablated SAV1 or WWC1–3 expression in these cells using CRISPR-Cas9 technology and observed a decrease in phosphorylated YAP (pYAP Ser127) and an increase in active/dephosphorylated YAP (aYAP) (Figures 1D and 1E). Furthermore, the mRNA levels of ANKRD1, CTGF, and CYR61, three faithful target genes of YAP/TAZ, were induced in SAV1 knockout (KO) or WWC1–3 triple KO (tKO) cells (Figure S1). The protein level of CYR61 was also induced in SAV1 KO or WWC1–3 tKO cells (Figures 1D and 1E). These results indicate that YAP/TAZ activity is hyperactivated following HPO1 inactivation (SAV1 or WWC1–3 deletion) in all three DPM cell lines tested, regardless of NF2 and LATS2 status.
Inactivation of HPO1 promotes tumorigenesis of DPM cells
YAP/TAZ are proto-oncoproteins, and their activation promotes tumorigenesis.15,16,18,19,20,21,22,23,24,25 We then tested the effect of SAV1 and WWC1–3 deletion on the tumorigenic potential of DPM cells. In a colony formation assay, DPM cells deficient in SAV1 or WWC1–3 formed significantly more colonies, indicating a growth advantage of these cells (Figures 2A, 2B, S2A, and S2B). YAP/TAZ exert their functions mainly by interacting with TEAD transcription factors (TEAD1–4).46,47,48,49,50 The colony formation of SAV1- or WWC1–3-deficient DPM cells was effectively repressed in the presence of VT103 or VT107, two inhibitors of TEAD1 and TEAD1–4, respectively (Figure S2C).51 Hence, downregulation of SAV1 or WWC1–3 can promote the clonogenic potential of DPM cells, which is likely mediated by enhanced YAP/TAZ-TEAD function.
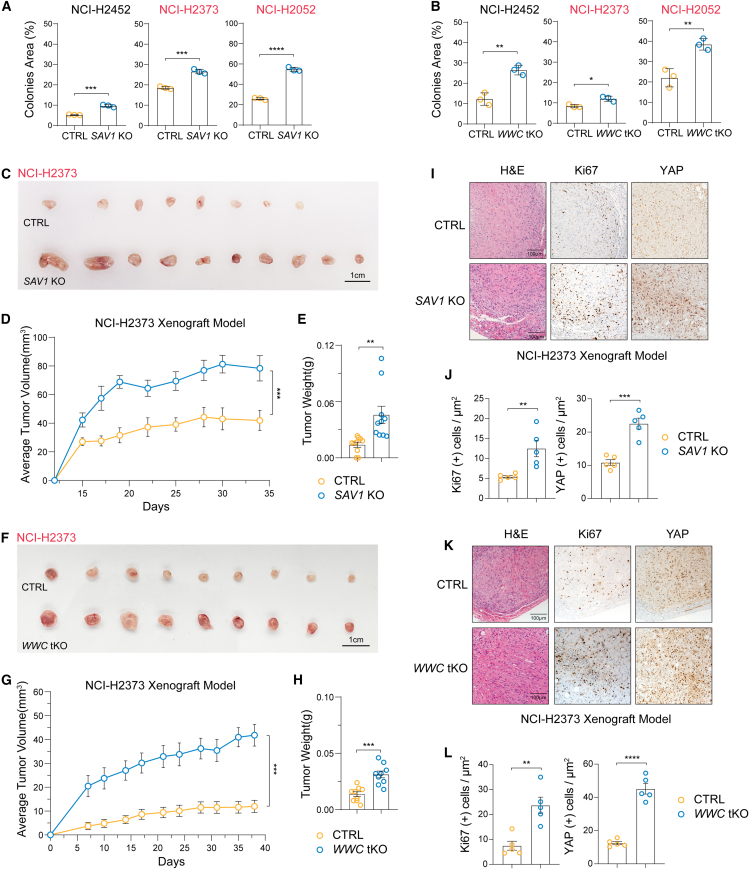
Figure 2.
Downregulation of SAV1 or WWC1–3 expression promotes the tumor-forming capacity of DPM cells
(A) Colony formation of DPM cell lines was enhanced upon SAV1 deletion.
(B) Colony formation of DPM cell lines was enhanced upon WWC1–3 deletion.
(C) Gross tumor images of tumor xenografts of control or SAV1 KO NCI-H2373 cells implanted in nude mice. Scale bar, 1 cm.
(D and E) Growth curve and tumor weight analysis of control and SAV1 KO NCI-H2373 xenografts. Five mice with 10 tumors for each group were analyzed.
(F) Gross tumor images of tumor xenografts of control or WWC1–3 KO NCI-H2373 cells implanted in nude mice. Scale bar, 1 cm.
(G and H) Growth curve and tumor weight analysis of control and WWC1–3 KO NCI-H2373 xenografts. Five mice with 9 tumors for each group were analyzed.
(I and J) Histological assessment of control and SAV1 KO NCI-H2373 tumor xenografts. H&E and immunohistochemistry (IHC) (Ki67 and YAP) staining and quantifications. Scale bar, 100 μm.
(K and L) Histological assessment of control and WWC1–3 KO NCI-H2373 tumor xenografts. H&E and IHC (Ki67 and YAP) staining and quantifications. Scale bar, 100 μm.
Data are presented as mean ± SEM from three independent experiments for (A) and (B). Statistical significance: ∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗, p < 0.001. The two-way ANOVA test was used for tumor growth curves (D, I), and the Student’s t test was used for other data.
See also Figure S2.
SAV1 or WWC1–3 deletion induced YAP/TAZ activity and colony formation of DPM cells, which might enhance tumor growth in vivo. Indeed, DPM cells grew poorly when inoculated subcutaneously into nude mice, with NCI-H2373 cells forming small tumors (<40 mm3) and NCI-H2052 cells failing to establish solid tumors consistently (Figures 2C–2H and S2D–S2I). In contrast, both NCI-H2372 and NCI-H2052 cells deficient in SAV1 or WWC1–3 grew much faster, as indicated by larger tumor size and weight (Figures 2C–2H and S2D–S2I). SAV1- or WWC1–3-deficient tumors also exhibited higher YAP/TAZ and Ki67 expression, as analyzed by immunohistochemistry staining (Figures 2I–2L and S2J–S2M). Hence, by inactivating HPO1 in DPM cells, we have established DPM xenograft models exhibiting rapid tumor onset.
Concurrent inactivation of HPO1 and HPO2 induces rapid mesothelioma onset in mice
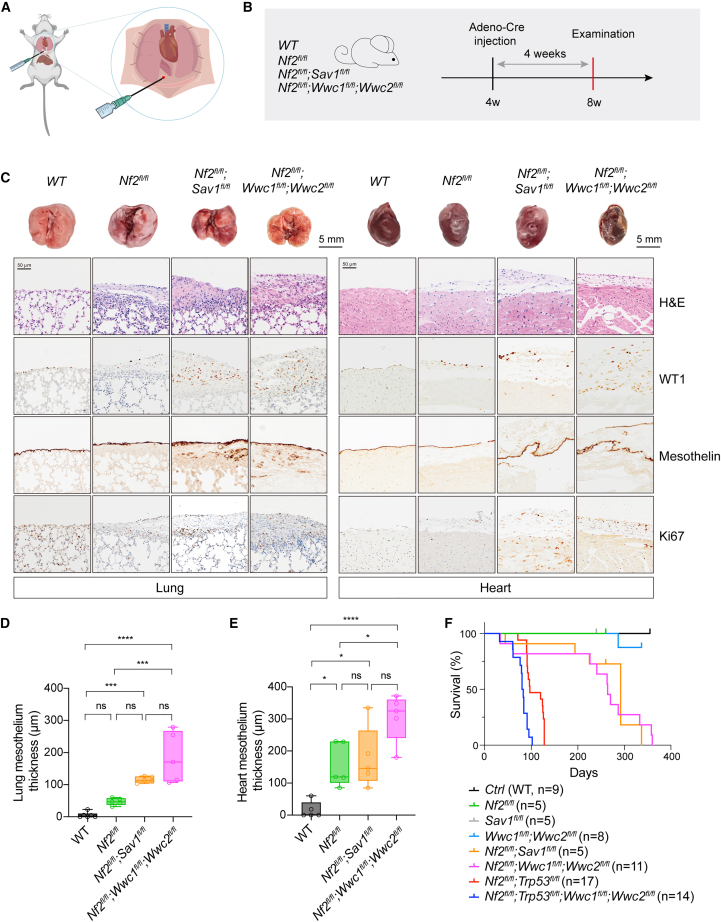
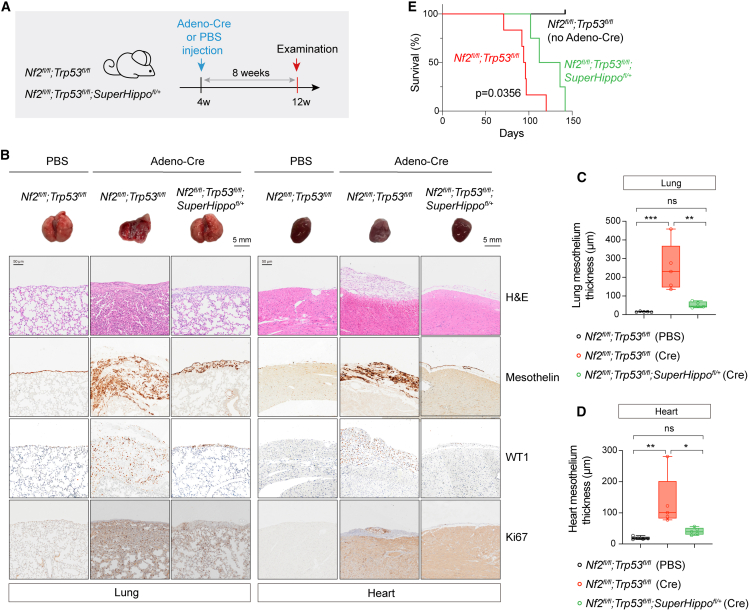
To mimic the initiation and progression of human DPM, genetically engineered mouse models of DPM have been established by selective inactivation of DPM-related tumor suppressor genes, such as Nf2, Trp53, Cdkn2ab, Pten, and Bap1 (Figure 1B).38,39,41,42,52,53 Inactivation of HPO1 further induced YAP/TAZ activity and promoted the growth of NF2-null DPM cells. Hence, deletion of Nf2 simultaneously with Sav1 or Wwc1/2 (no Wwc3 gene in mice), i.e., concurrent inactivation of HPO1 and HPO2, might also elicit rapid DPM development in mice. We then created mice with various genes floxed, such as Nf2fl/fl, Sav1fl/fl, Wwc1fl/fl;Wwc2fl/fl, Nf2fl/fl;Sav1fl/fl, and Nf2fl/fl;Wwc1fl/fl;Wwc2fl/fl mice, and induced gene deletion by administration of an adenovirus carrying Cre recombinase driven by a cytomegalovirus (CMV) promoter (Adeno-Cre) into the pleural cavity of mice (Figure 3A). The visceral pleura of the lung and heart are the most common sites for DPM development. We, therefore, mainly focused on the pathological changes in the lungs and heart. At 4-week after Adeno-Cre administration, the mesothelial lining surrounding the lung or heart thickened markedly in mice with concurrent HPO1 and HPO2 inactivation (Nf2;Sav1 or Nf2;Wwc1;Wwc2 deletion) (Figures 3B–3E). On the other hand, in mice with HPO1 or HPO2 inactivation (Nf2, Sav1, or Wwc1/2 deletion), the expansion of the mesothelium was mild even at 45 weeks after Adeno-Cre administration (Figures S3A and S3B). Cells in the thickened layer expressed Wilm’s tumor-1 (WT1) and Mesothelin, indicating a mesothelium origin (Figure 3C). Moreover, Ki67-positive cells were also increased in the mesothelium of mice deficient in both Nf2 and Sav1 or Wwc1/2, indicating hyperproliferation (Figure 3C). These data demonstrate that the concurrent inactivation of HPO1 and HPO2 in mice accelerates the development of mesothelioma.
Figure 3.
Deletion of Nf2 and Sav1 or WWC1–3 in the mesothelium of pleural cavities in mice led to mesothelioma development
(A) Illustration of intrathoracic injection of Adeno-Cre in mice.
(B) Schematic diagram indicating the experimental procedure. Mice (n = 5 for each group) at 4-week-old were injected with Adeno-Cre to delete corresponding genes. Lungs and hearts were collected after 4 weeks for histological analysis.
(C) Gross images and histological analysis of lungs and hearts. H&E and IHC staining for WT1, Mesothelin, and Ki67 expression were performed. Genotypes of mice were indicated. Scale bars, 5 mm for gross lung and heart images and 50 μm for H&E or IHC images.
(D and E) Quantification of the average thickness of lung (D) and heart (E) mesothelium in mice with different genotypes. Mesothelium thickness is primarily defined by Mesothelin staining signals.
(F) Survival curves of mice with different genotypes following Adeno-Cre injection. The number of mice used was indicated.
Data are presented as mean ± SEM. Statistical significance: ∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗, p < 0.001. The Student’s t test was used for statistical analysis.
See also Figure S3.
TP53 has been reported as one of the highly mutated genes in DPM, of which the frequency of somatic mutation rate is about 16%, as indicated in TCGA DPM datasets (Figure 1B).54 Recently, a mouse mesothelioma model with conditional deletions of Nf2 and Trp53 has been established to mimic human DPM.44 Indeed, Nf2fl/fl;Trp53 fl/fl mice developed mesotheliomas, and, 8 weeks after Adeno-Cre administration, we observed significant thickening of the mesothelium and infiltration of mesothelial cells into the lung and heart (Figures S3C–S3F). To test the effect of Wwc1/2 deletion in the Nf2;Trp53 model, we established Nf2fl/fl;Trp53 fl/fl;Wwc1fl/fl;Wwc2fl/fl mice and observed further enhanced mesothelium thickening and organ infiltration (Figures S3C–S3F). Furthermore, the expression of WT1, Mesothelin, and Ki67 was also significantly increased in the mesothelium of Nf2fl/fl;Trp53 fl/fl;Wwc1fl/fl;Wwc2fl/fl mice (Figure S3D). Hence, inactivation of HPO1 also promotes the development and progression of mesotheliomas in the Nf2;Trp53 mouse model.
The parietal pleura of the diaphragm and thoracic chest wall are also sites for DPM development.44 Diaphragm samples from mice with different genotypes were collected and subjected to histological analysis. Like the mesothelium of the lung and heart, diaphragm mesothelium thickening occurred in all mutant mice, and the phenotype was enhanced in mice with both HPO1 and HPO2 inactivation (Figure S3G). We also compared the life expectancy of mice with various genotypes. While Nf2-, Sav1-, or Wwc1;Wwc2-deficient mice lived for more than 1 year, the median survival of Nf2;Sav1 and Nf2;Wwc1;Wwc2 mice was 292 days and 264 days, respectively (Figure 3F). Trp53 deletion on top of Nf2 deficiency dramatically affected survival, with Nf2;Trp53 mice living for about 125 days, and further KO of Wwc1/2 shortened the medium survival to about 80 days (Figure 3F). This difference in median survival is likely due to both non-aggressive epithelioid or mixed tumors and highly invasive sarcomatoid tumors in Nf2;Trp53 mice, while Nf2;Sav1 or Wwc1;Wwc2 KO mice mainly develop non-aggressive epithelioid tumors (Figures 3C and S3D).44 Hence, HPO1 inactivation effectively promoted mesothelioma progression and reduced the life expectancy in mice with Nf2 deficiency.
HPO1 activation inhibits YAP/TAZ activity and DPM cell proliferation
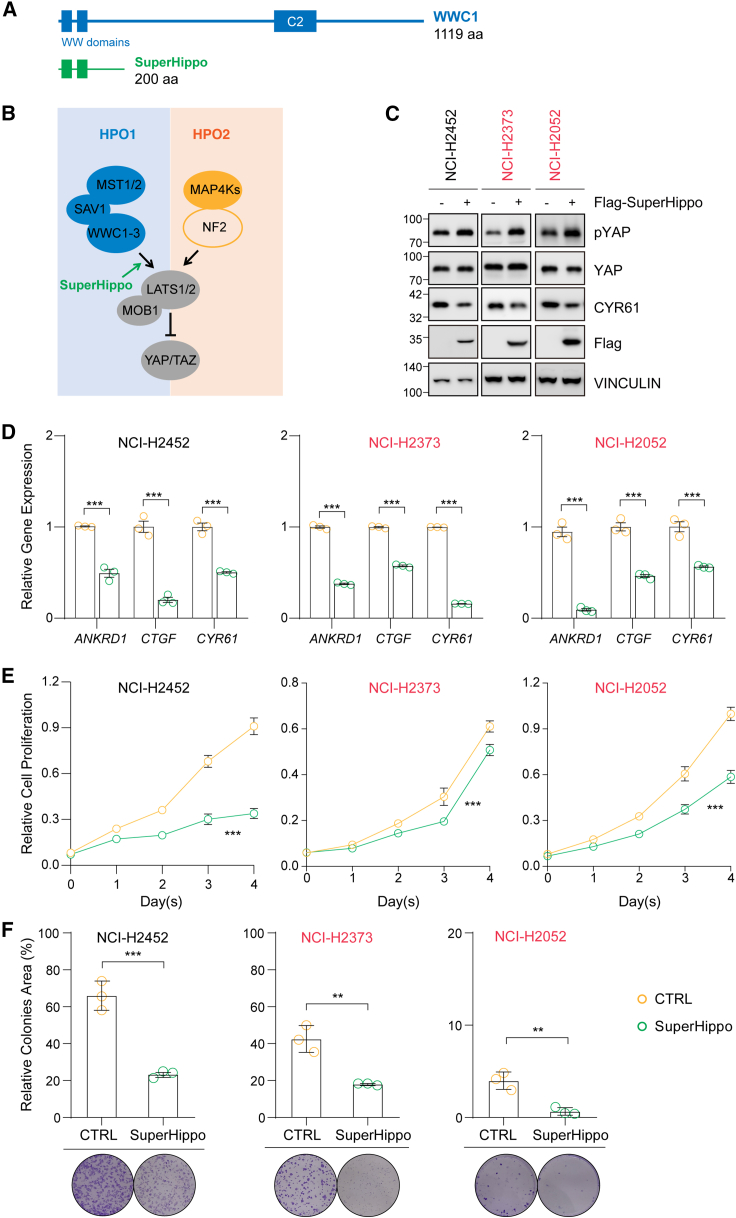
HPO1 inactivation (deletion of Sav1 and Wwc1/2) in Nf2-null cells or tissues significantly induced YAP/TAZ activity and tumorigenesis, indicating that HPO1 and HPO2 signaling may work independently and synergistically, and upregulation of HPO1 signaling in DPM cells may block tumorigenesis. We previously engineered a WWC-derived minigene called SuperHippo to specifically and effectively activate HPO1 signaling, which leads to LATS1/2 activation and YAP/TAZ inhibition30 (Figures 4A and 4B). Ectopic expression of SuperHippo or full-length WWC1 in HEK293A cells resulted in a significant increase in YAP phosphorylation and a decrease in CRY61 expression (Figure S4A). In addition, SuperHippo induced YAP phosphorylation in NF2-null cells, indicating that it works in an NF2-independent manner (Figure S4B).
Figure 4.
Expression of SuperHippo activates Hippo signaling in mesothelioma cell lines
(A) Schematic diagram illustrating full-length WWC1 and SuperHippo (1–200 aa of WWC1).
(B) Schematic diagram illustrating the activation of the Hippo signaling by SuperHippo via its functional engagement in the HPO1 signaling module.
(C) SuperHippo expression in mesothelioma cell lines inactivates YAP.
(D) Decreased mRNA levels of YAP/TAZ target genes ANKRD1, CTGF, and CYR61 upon SuperHippo expression in mesothelioma cell lines.
(E) SuperHippo expression induces cell death in mesothelioma cell lines. Cell counting kit-9 (CCK8) assays were performed.
(F) SuperHippo expression represses colony formation of mesothelioma cell lines. Data are presented as mean ± SEM from three independent experiments. Statistical significance: ∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗, p < 0.001. The Student’s t test was used.
See also Figure S4.
We then tested if SuperHippo can repress YAP/TAZ activity and the proliferation of DPM cells. In three DPM cell lines expressing SuperHippo, phosphorylation of YAP was induced, whereas expression of CRY61 was inhibited (Figure 4C). The mRNA levels of YAP/TAZ target genes ANKRD1, CTGF, and CYR61 were reduced in cells with ectopic SuperHippo expression (Figure 4D). Moreover, the proliferation of DPM cells in the presence of SuperHippo was also significantly slowed down (Figure 4E). In line with these data, the colony-forming ability of DPM cells was markedly repressed by SuperHippo (Figure 4F). Moreover, SuperHippo expression significantly enhanced apoptosis in these DPM cell lines (Figures S4C and S4D). Collectively, these data demonstrate that SuperHippo, by activating the HPO1 signaling, can inhibit the proliferation of DPM cells in vitro.
Activating HPO1 signaling inhibits tumorigenesis in mice
Next, we sought to test the effect of HPO1 activation on tumorigenesis in a mouse DPM model. To this end, we crossed a conditional SuperHippo transgenic mice (Loxp-stop-loxp-3xFlag-SuperHippo driven by the CMV early enhancer/chicken beta-actin/beta-globin intron [CAG] promoter at the Rosa26 locus) with Nf2fl/fl;Trp53 fl/fl mice to generate Nf2fl/fl;Trp53 fl/fl;SuperHippofl/+ mice.30 SuperHippo expression was detected in lung and heart tissues of Nf2fl/fl;Trp53 fl/fl;SuperHippofl/+ mice treated with Adeno-Cre (Figures 5A and S5A). Strikingly, the mesothelium thickening and parenchymal infiltration in both lung and heart resulted from Nf2 and Trp53 deletion was significantly inhibited in mice expressing SuperHippo (Figures 5B–5D). Moreover, the expression of WT1, Mesothelin, and Ki67 all reduced upon SuperHippo expression, indicating a tumor-suppressive role of SuperHippo (Figure 5B). Importantly, SuperHippo expression significantly improved the survival of the mice from 95 to 124 days (Figure 5E). Furthermore, the expression of SuperHippo was well-tolerated by normal mesothelium, because no significant change in the mesothelium thickness or structural integrity was detected upon SuperHippo expression (Figures S5B–S5F). Collectively, these data demonstrate that SuperHippo, by activating HPO1, can inhibit the development of mesothelioma in mice, suggesting a potential of SuperHippo for treating DPM.
Figure 5.
SuperHippo expression represses the development of mesothelioma in a genetic mouse model
(A) Schematic diagram indicating experimental procedure. Adeno-Cre was injected into 4-week-old Nf2fl/fl;Trp53fl/fl or Nf2fl/fl;Trp53fl/fl;SuperHippofl/+ mice (n = 4–6 each); after 8 weeks, lungs and hearts were collected for histological analysis.
(B) Gross image and histological analysis of lungs and hearts. H&E and IHC staining for WT1, Mesothelin, and Ki67 expression was performed. Scale bar, 5 mm for gross images, and 50 μm for H&E or IHC images.
(C and D) Quantification of the average thickness of lung and heart mesothelium. Mesothelium thickness is primarily defined by Mesothelin staining signals.
(E) Survival curves of Nf2fl/fl;Trp53fl/fl and Nf2fl/fl;Trp53fl/fl;SuperHippofl/+ mice (n = 4–6 each) after Adeno-Cre injection.
Data are presented as mean ± SEM. Statistical significance: ∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗, p < 0.001. The log rank (Mantel-Cox) test was used for survival curves (E), and the Student’s t test was used for other data.
See also Figure S5.
Concurrent HPO1 and HPO2 activation inhibits the development of DPM
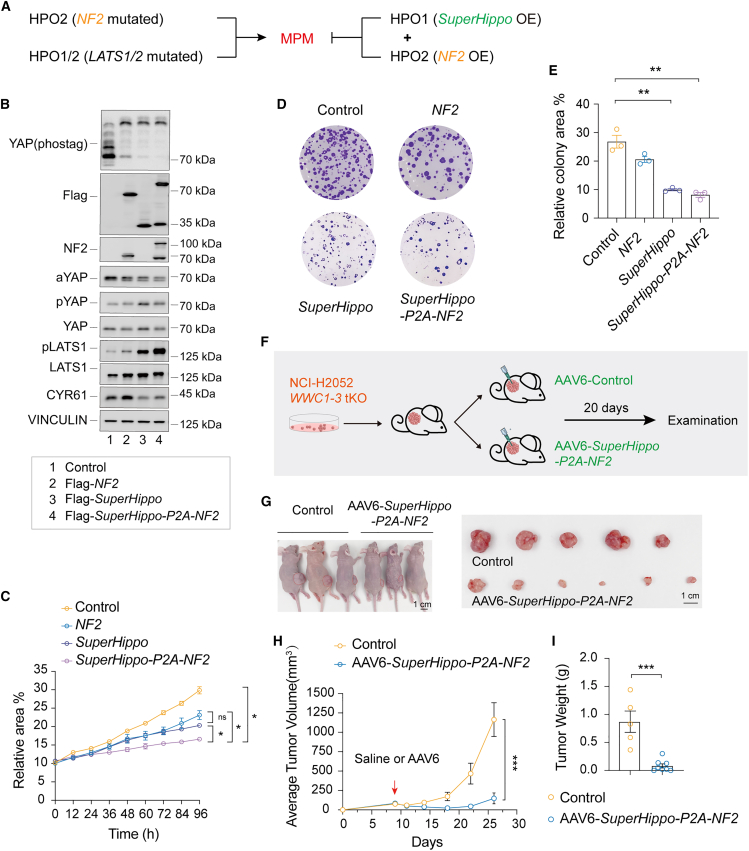
Multiple genes in the Hippo signaling network are mutated in DPMs, such as NF2, LATS1/2, and WWC1 (Figure 1B). Ectopic expression of a functional NF2 gene will, in principle, normalize HPO2 signaling in DPMs with NF2 mutations. However, it is currently unclear if this normalized HPO2 signaling effectively kills DPM cells. Moreover, the expression of NF2 should not work in DPMs with alteration in LATS1/2 and WWC1. In contrast, SuperHippo has a broader application, as its ectopic expression will induce HPO1 signaling in all DPM cells with NF2, LATS1, LATS2, or WWC1 mutations. We reasoned that concurrent activation of HPO1 and HPO2, by expression of both NF2 and SuperHippo, should lead to more complete inactivation of YAP/TAZ oncoproteins and be effective for treating DPMs with distinct gene alterations.
We then assessed the effect of the expression of NF2, SuperHippo, or SuperHippo-P2A-NF2 (a construct expresses a fusion protein that can be specifically cleaved into SuperHippo and NF2) in WWC1–3 tKO NCI-H2052 cells. SuperHippo or NF2 expression alone induced phosphorylation of LATS1/2 and YAP and reduced cell proliferation and colony formation. Interestingly, these changes were significantly amplified in cells expressing SuperHippo-P2A-NF2 (Figures 6A–6E). These results demonstrate an anti-tumor effect of SuperHippo-P2A-NF2 in DPM cells in vitro.
Figure 6.
Expression of SuperHippo-P2A-NF2 represses DPM development
(A) Expression of both SuperHippo and NF2 may effectively repress the development of DPM with NF2 mutations (HPO2) or LATS1/2 mutations (HPO1/2).
(B) Expression of SuperHippo-P2A-NF2 leads to the maximum activation of Hippo signaling in DPM cells. NF2, SuperHippo, or SuperHippo-P2A-NF2 was stably expressed in WWC1–3 tKO NCI-H2052 DPM cells. Cell lysates were subjected to immunoblotting to assess the phosphorylation status of YAP and LATS1/2. These cell lines were also used in (C–E).
(C) SuperHippo-P2A-NF2 expression represses the proliferation of DPM cells. The proliferation of different cells was monitored over 4 days by live cell imaging.
(D and E) SuperHippo-P2A-NF2 expression effectively inhibits the colony formation of DPM cells. Crystal violet staining (D) indicates colony formation capacity, and quantification (E) was shown; data represent mean ± SEM from three independent experiments.
(F) Schematic diagram indicating the experimental procedure. WWC1–3 tKO NCI-H2052 cells were inoculated subcutaneously into nude mice, and, when tumors reached 75 mm3, mice were randomly assigned into two groups. AAV6-control or AAV6-SuperHippo-P2A-NF2 was injected intratumorally, and, 20 days later, tumors were harvested for examination.
(G–I) AAV6-mediated expression of SuperHippo-P2A-NF2 represses the progression of DPM in a xenograft mouse model. n = 5–8 for each group. Tumor gross images, tumor growth curve, and tumor weight were shown (5–6 tumors for each group were analyzed). Scale bars: 1 cm. Data are presented as mean ± SEM. Statistical significance: ∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗, p < 0.001. One-way or two-way ANOVA (C and H) and Student’s t test (E and I) were used.
See also Figure S6.
AAV-based vectors are frequently used for gene therapy.55 Both NF2 and SuperHippo are relatively small and suitable for AAV-based gene delivery. We compared the infection efficiency of multiple AAV serotypes in NCI-H2052 cells and found that AAV6 exhibited a distinct tropism for DPM cells (Figure S6A). Next, we constructed an AAV6-based vector containing a CMV promoter followed by the SuperHippo-P2A-NF2 coding sequence. Subsequently, we evaluated the anti-tumor potential of SuperHippo-P2A-NF2 in the WWC-tKO NCI-H2052 xenograft model. When the average tumor volume reached about 75 mm3, we performed intratumoral injection of AAV6-SuperHippo-P2A-NF2 or AAV6-control. Strikingly, the growth of AAV6-SuperHippo-P2A-NF2-treated tumors was significantly suppressed, as evidenced by the slow increase in tumor volume and a notable reduction in tumor weight (Figures 6F–6I). In addition, the proliferation of tumor cells with SuperHippo and NF2 expression was reduced, as indicated by the decreased Ki67 staining (Figures S6B and S6C). Together, AAV6-mediated expression of SuperHippo-P2A-NF2 is effective in inhibiting DPM tumor growth.
SuperHippo-P2A-NF2 gene therapy mitigates tumor progression in mice
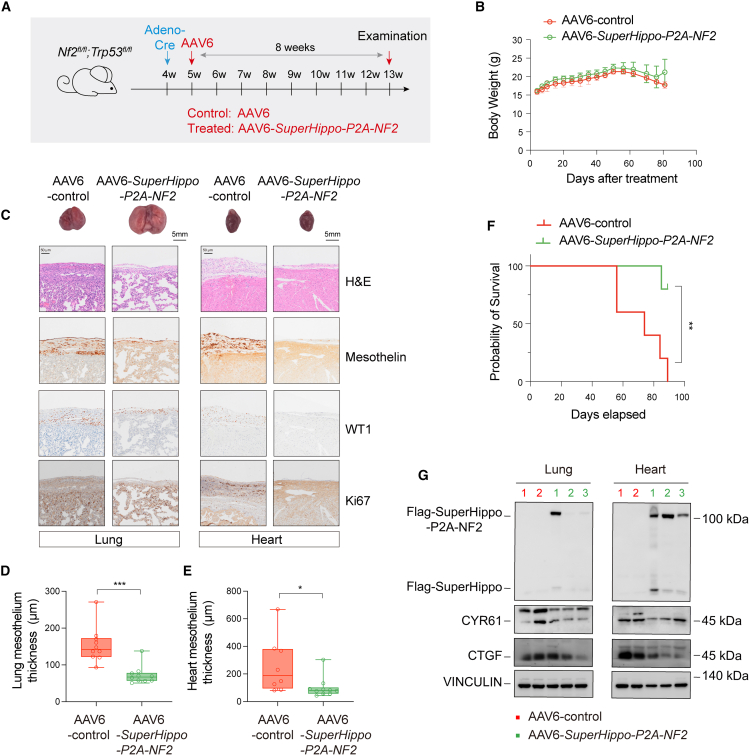
To further assess the in vivo function of AAV6-SuperHippo-P2A-NF2, we tested its effect on the development of mesothelioma in the Nf2;Trp53 mouse model. One week after intrathoracic Adeno-Cre injection in 4-week-old Nf2fl/fl;Trp53 fl/fl mice, AAV6 (control) or AAV6-SuperHippo-P2A-NF2 (treated) was administered (2.3 × 1011 viral genomes/mouse) into the pleural cavity of randomly selected mice (Figure 7A). There was no significant difference in body weight between the control and treated groups within 80 days following treatment (Figure 7B). However, administration of AAV6-SuperHippo-P2A-NF2 resulted in the remission of mesothelioma development, as evidenced by reduced mesothelium thickening and decreased Mesothelin, WT1, and Ki67 expression (Figures 7C–7E). More importantly, the survival of AAV6-SuperHippo-P2A-NF2-treated Nf2;Trp53 mice was significantly prolonged (Figure 7F). Full-length SuperHippo-P2A-NF2 and cleaved SuperHippo were detected in the protein lysates of lungs and hearts of treated mice, and expression of CYR61 and CTGF in these organs was also reduced (Figure 7G). On the other hand, compared to SuperHippo-P2A-NF2, the efficacy of Nf2 or SuperHippo single-agent therapy was lower, as evidenced by a relatively weak reduction in mesothelium thickness (Figures S7A–S7E). These results demonstrate that concurrent activation of HPO1 and HPO2 via AAV6-SuperHippo-P2A-NF2 gene therapy is an effective strategy for DPM management in preclinical tumor models.
Figure 7.
Mitigation of tumor progression by AAV6-SuperHippo-P2A-NF2 in an Nf2;Trp53 dKO mouse DPM model
(A) Schematic diagram indicating the experimental procedure. Adeno-Cre was injected intrathoracically into 4-week-old Nf2fl/fl;Trp53fl/fl mice to initiate DPM, and, one week later, AAV6-control or AAV6-SuperHippo-P2A-NF2 was injected intrathoracically. Lungs and hearts were collected for histological analysis after another 8 weeks. n = 5–7 for each group.
(B) Body weight measurement of mice used.
(C) H&E and IHC staining of lung and heart tissue sections from control and treated mice. IHC for WT1, Mesothelin, YAP, and Ki67 were shown. Scale bar, 5 mm for gross lung and heart images, and 50 μm for H&E or IHC images.
(D and E) Measurement of the average thickness of lung and heart mesothelium in mice from control and treated groups. Mesothelium thickness is primarily defined by Mesothelin staining signals.
(F) Overall survival curve of mice treated with AAV6-control or AAV6-SuperHippo-P2A-NF2.
(G) Immunoblotting analysis of SuperHippo-P2A-NF2 expression and cleavage in lungs and hearts from mice treated with AAV6-SuperHippo-P2A-NF2. Expression of YAP/TAZ targets, CTY61 and CTGF, was also reduced in treated samples.
Data are presented as mean ± SEM. Statistical significance: ∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗, p < 0.001. The log rank (Mantel-Cox) test was used for the overall survival curve (F), and the Student’s t test for other data.
See also Figure S7.
Discussion
We have reported recently that two signaling modules, HPO1 and HPO2, regulate LATS1/2 and YAP/TAZ in a synergistic manner.31,36,37 This study has demonstrated that deleting both HPO1 and HPO2 genes leads to a rapid development of DPM. In contrast, ectopic expression of the SuperHippo minigene effectively enhances HPO1 signaling and suppresses DPM. Furthermore, by using AAV6 to deliver both NF2 and SuperHippo genes, a gene therapy is developed to block the progression of DPM in preclinical models. Collectively, these findings underscore the critical role of the Hippo pathway in the development of DPM and provide a proof of concept for the use of gene therapy in treating this deadly disease.
Inactivation of Hippo pathway genes occurs frequently in patients with DPM and contributes to tumor progression and worse prognosis.35,42,43,56,57,58,59,60 A recent report indicates that alterations of NF2 and LATS2 in DPMs are associated with different molecular signatures.61 Hence, according to the genes altered, various DPMs may have distinct clinical features due to differential YAP/TAZ activation status. Notably, co-occurring alterations of multiple Hippo pathway genes are observed in a small fraction of DPM specimens (Figure 1B). Indeed, a distinct subset of DPMs harboring both LATS2 and NF2 mutations and functional investigations indicate that the inactivation of either NF2 or LATS2 has no apparent effect on the proliferation of DPM cells, whereas the inactivation of both genes effectively enhances the proliferation of DPM cells.58,61 Hence, combined inactivation of multiple genes in the Hippo pathway represents a mechanism underpinning rapid DPM progression.
Targeting the Hippo pathway is an appealing strategy for treating YAP/TAZ-dependent cancer.62,63,64 Hence, therapeutics inactivating YAP/TAZ activity hold a promise for patients with DPM. Directly targeting the YAP/TAZ-TEAD complex, a key downstream effector of the Hippo pathway, is under intensive investigation. Agents have been developed to either directly disrupt YAP/TAZ-TEAD interaction or inhibit TEAD palmitoylation, such as verteporfin, MGH-CP1, TED347, K975, VT103, and VT10749,51,.65,66,67,68,69,70 It would be meaningful to test the anti-cancer effect of these molecules in genetic mouse models of DPM in the future. However, due to the crucial roles of YAP/TAZ in various tissues, systemic delivery of these molecules may cause side effects, which should be carefully assessed in clinical trials. AAV-based gene therapy reported here can achieve localized drug delivery, representing an alternative treatment modality.
DPMs have relatively clear hotspot driver mutations, such as those found in NF2, TP53, CDKN2A/B, and BAP1.38,39,41,42,52,53 In principle, gene therapy is suitable to treat DPMs. Currently, gene therapies, such as oncolytic viruses and virus-carrying suicide genes, are available for patients with DPM in clinical trials.71,72,73 As shown earlier, the Hippo pathway plays a pivotal role in maintaining the homeostasis of the plural mesothelium (Figure 3). Hence, activating the Hippo signaling via gene replacement or functional restoration should repress DPM progression. The mesothelium monolayer and malignant cells provide a relatively large surface area for gene transduction. In addition, the pleural cavity is accessible for gene delivery, and, as a closed space, it may retain viruses with limited diffusion and dilution.73 The AAV6-mediated expression of SuperHippo-P2A-NF2 is efficient and flexible, and this approach may be employed to treat DPMs in the future.
NF2 loss of function, either by mutation or deletion, has been found in different cancers, including DPM, schwannomas, ependymomas, and meningiomas, and these tumors are all derived from monolayer cells covering internal cavities.15,17 We have hypothesized that HPO2 plays a dominant role in two-dimensional monolayer tissues, and the formation of cell-cell junctions in these tissues might be critical for NF2 to activate hippo signaling.31 Conversely, HPO1 appears to function primarily in three-dimensional tissues, exerting a more pronounced effect on organ size.31 In this study, we revealed the roles of HPO1 in mesothelioma, highlighting its importance in sustaining Hippo signaling and preventing rapid tumorigenesis when HPO2 is defective. Based on the common genetic alterations and tissue topological features, it is likely that all these NF2-mutated tumors can be treated with the AAV6-based SuperHippo-P2A-NF2 gene therapy. However, due to specific anatomic features, it remains challenging to deliver AAV-based therapies into different tumors.
Limitations of the study
In this study, gene therapy has been shown to prevent tumor growth in xenograft and genetic DPM models. However, these models may not fully recapitulate the molecular heterogeneity of human DPM. Therefore, it is important to evaluate this gene therapy in patient-derived xenograft (PDX) mesothelioma models. Moreover, since NF2 mutations in DPM often co-occur with CDKN2A/B and BAP1 mutations, the efficacy of this gene therapy in these contexts also warrants further investigation. Furthermore, this gene therapy is designed for YAP/TAZ-driven mesotheliomas; its effect in mesotheliomas without Hippo pathway inactivation should be tested in genetic and PDX models in the future.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Fa-Xing Yu (fxyu@fudan.edu.cn).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Uniform Biological Materials Transfer Agreement.
Data and code availability
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This study was supported by grants from the National Key R&D Program of China (2020YFA0803202, 2018YFA0800304), the National Natural Science Foundation of China (32425017, 32370770, 32200570, 82403053), the Science and Technology Commission of Shanghai Municipality (21S11905000), and the Shanghai Municipal Health Commission (2022XD049). This work was also supported by the Medical Science Data Center and the Core Facility at the Shanghai Medical College of Fudan University.
Author contributions
R.Z., X.L., X.Z., Z.Z., S.Q., R.J., Y.G., and Y.W. performed experiments. C.L., K.C., and D.Y. provided key reagents and comments. R.Z. and F.-X.Y. analyzed data and wrote the manuscript. F.-X.Y. conceived and supervised the project.
Declaration of interests
A patent application about mesothelioma gene therapy has been filed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Merlin (D3S3W) | Cell Signaling Technology | Cat#12888S; RRID: AB_2650551 |
| Rabbit monoclonal anti-LATS1(C66B5) | Cell Signaling Technology | Cat#3477S; RRID: AB_2133513 |
| Rabbit monoclonal anti-LATS2(D83D6) | Cell Signaling Technology | Cat#5888S; RRID: AB_10835233 |
| Rabbit monoclonal anti-SAV1(D6M6X) | Cell Signaling Technology | Cat#13301S; RRID: AB_2798176 |
| Mouse monoclonal anti-WWC1 | Zhang et al.74 | N/A |
| Rabbit polyclonal anti-WWC2 | Abcam | Cat#ab126356; RRID: AB_11140331 |
| Rabbit polyclonal anti-WWC3 | Qi et al.30 | N/A |
| Rabbit monoclonal anti-YAP(D8H1X) | Cell Signaling Technology | Cat#14074S; RRID: AB_2650491 |
| Rabbit monoclonal anti-Vinculin(E1E9V) | Cell Signaling Technology | Cat#13901S; RRID: AB_2728768 |
| Rabbit monoclonal anti-Phospho-YAP(Ser127) | Cell Signaling Technology | Cat#13008; RRID: AB_2650553 |
| Rabbit monoclonal anti-Non-phospho(Active) YAP(Ser127)(E6U8Z) | Cell Signaling Technology | Cat#29495; RRID: AB_2798974 |
| Rabbit polyclonal anti-CYR61(H-78) | Santa Cruz Biotechnology | Cat#sc-13100; RRID: AB_2088733 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat#ab15580; RRID: AB_443209 |
| Mouse monoclonal anti-YAP1(63.7) | Santa Cruz Biotechnology | Cat#sc-101199; RRID: AB_1131430 |
| Rabbit monoclonal anti-WT1 | Cell Signaling Technology | Cat#83535; RRID: AB_2800020 |
| Rabbit polyclonal anti-Mesothelin | Thermo Fisher Scientific | Cat#PA5-79698; RRID: AB_2746813 |
| Rabbit monoclonal anti-Flag tag (D6W5B) | Cell Signaling Technology | Cat#14793S; RRID: AB_2572291 |
| Mouse monoclonal anti-FLAG M2(HRP conjugated) | Sigma-Aldrich | Cat#A8592; RRID: AB_439702 |
| Rabbit monoclonal anti-Phospho-LATS1(Thr1079)(D57D3) | Cell Signaling Technology | Cat#8654S; RRID: AB_10971635 |
| Mouse monoclonal anti-Myc Tag | MBL International | Cat#M192-3; RRID: AB_11160947 |
| Mouse monoclonal anti-GAPDH(HRP conjugated) | Abways Technology | Cat#AB2000 |
| Goat polyclonal anti-CTGF(L-20) | Santa Cruz Biotechnology | Cat#sc-14939; RRID: AB_638805 |
| Goat anti-rabbit IgG antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A11001; RRID: AB_2534069 |
| Bacterial and virus strains | ||
| Trans5α Chemically Competent Cell | TransGen Biotech | Cat#CD201-01 |
| pLVX lentivirus | This paper | N/A |
| LentiCRISPR v2 | Sanjana et al.75 | Addgene 52961 |
| Adeno-Cre | HANBIO | Cat#HBAD-1010 |
| Adeno-associated virus (AAV) | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Fetal bovine serum | Invitrogen | Cat#10091-148 |
| Protease inhibitor cocktail | MCE | Cat#HY-K0010 |
| Phosphatase inhibitor Cocktail I | MCE | Cat#HY-K0021 |
| Phosphatase inhibitor Cocktail II | MCE | Cat#HY-K0022 |
| Phosphatase inhibitor Cocktail II | MCE | Cat#HY-K0022 |
| PEI | Polysciences | Cat#23966-2 |
| PolyJet | Signagen Laboratories | Cat#SL100688 |
| Envision anti-Rabbit | DAKO | Cat#K4002 |
| DAB reagent | GeneTech | GK500705 |
| Prime STAR Max DNA polymerase | Takara | Cat#R045A |
| VT103 | Selleck | Cat#E1598 |
| VT107 | Selleck | Cat#E1599 |
| Critical commercial assays | ||
| TaKaRa MiniBEST Universal RNA Extraction Kit | Takara | Cat#9767 |
| TB Green® Premix Ex Taq™ (Tli RNaseH Plus) | Takara | Cat#RR420A |
| TransScript® First-Strand cDNA Synthesis SuperMix | TransGen Biotech | Cat#AT301-03 |
| ClonExpress MultiS One Step Cloning Kit | Vazyme | Cat#C113-02 |
| High-sig ECL Western Blotting | Tanon | Car#180-501 |
| Cell Counting Kit(CCK-8) | YEASEN | Cat#40203ES60 |
| Deposited data | ||
| TCGA Mesothelioma data | Hmeljak et al.42 | https://portal.gdc.cancer.gov/projects/TCGA-MESO, dbGap Study Accession:phs000178.v11.p8 |
| Experimental models: Cell lines | ||
| Human: HEK293A | Yu et al.76 | N/A |
| Human: HEK293T | National collection of Authenticated Cell Cultures | Cat#SCSP-502 |
| Human: NCI-H2452 | Tang et al.51 | N/A |
| Human: NCI-H2373 | Tang et al.51 | N/A |
| Human: NCI-H2052 | Tang et al.51 | N/A |
| HEK293A NF2 KO | Qi et al.30 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: Nf2fl/fl (C57/BL6J background) | Wang et al.77 | N/A |
| Mouse: Savfl/fl(C57/BL6J background) | Ji et al.78 | N/A |
| Mouse: Wwc1fl/fl;Wwc2fl/fl (C57/BL6J background) | Qi et al.31 | N/A |
| Mouse: Nf2fl/fl;Savfl/fl (C57/BL6J background) | Qi et al.31 | N/A |
| Mouse: Nf2fl/fl;Wwc1fl/fl;Wwc2fl/fl (C57/BL6J background) | Qi et al.31 | N/A |
| Mouse: Nf2fl/fl;Trp53fl/fl (C57/BL6J background) | This paper | N/A |
| Mouse: Nf2fl/fl;Trp53fl/fl;Wwc1fl/fl;Wwc2fl/fl (C57/BL6J background) | This paper | N/A |
| Mouse: SuperHippofl/fl (C57/BL6J background) | Qi et al.30 | N/A |
| Mouse: Nf2fl/fl;Trp53fl/fl;SuperHippofl/+ (C57/BL6J background) | This paper | N/A |
| Oligonucleotides | ||
| WWC1 siRNA: 5′-GGUUGGAGAUUACUUCAUAGA-3′ | This paper | N/A |
| WWC2 siRNA: 5′-GGAUCUUCAUCCAGUACUAAA-3′ | This paper | N/A |
| WWC3 siRNA: 5′-GGAUAUUCAACAAAUACAAAG-3′ | This paper | N/A |
| RT-qPCR primer GAPDH forward 5′-ATGGGGAAGGTGAAGGTCG-3′ | This paper | N/A |
| RT-qPCR primer GAPDH reverse 5′- GGGGTCATTGATGGCAACAATA-3′ | This paper | N/A |
| RT-qPCR primer CTCF forward 5′- CCAATGACAACGCCTCCTG-3′ | Yu et al.76 | N/A |
| RT-qPCR primer CTCF reverse 5′- TGGTGCAGCCAGAAAGCTC-3′ | Yu et al.76 | N/A |
| RT-qPCR primer CYR61 forward 5′- AGCCTCGCATCCTATACAACC-3′ | Yu et al.76 | N/A |
| RT-qPCR primer CYR61 reverse 5′- TTCTTTCACAAGGCGGCACTC-3′ | Yu et al.76 | N/A |
| RT-qPCR primer ANKRD1 forward 5′- CACTTCTAGCCCACCCTGTGA-3′ | Yu et al.76 | N/A |
| RT-qPCR primer ANKRD1 reverse 5′- CCACAGGTTCCGTAATGATTT-3′ | Yu et al.76 | N/A |
| Recombinant DNA | ||
| pLVX-puro vector | Takara | #632164 |
| PsPAX2 | Didier Trono | Addgene #12260 |
| pMD.2g | Didier Trono | Addgene #12259 |
| AAV2/6-CMV-3xFLAG-SuperHippo-P2A-NF2-WPRE | This paper | N/A |
| AAV2/6-CMV-3xFLAG-NF2-WPRE | This paper | N/A |
| AAV2/6-CMV-3xFLAG-SuperHippo-WPRE | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| R (version 3.6.2) | Open Source | https://www.r-project.org/ |
Experimental model and study participant details
Mouse models
All procedures in animal experiments were approved by the Animal Ethics Committee of Shanghai Medical College, Fudan University, and performed in accordance with institutional guidelines. Nf2fl/fl, Wwc1fl/fl, Wwc2fl/fl, and SuperHippo (R26-e(CAG-LSL-3xFlag-SuperHippo)) mice were in-housed generated as described earlier.30,31,77 The Trp53 mice were described previously.79,80 All mice used in this study were maintained in a specific pathogen-free facility under a 12-h light/dark cycle and provided ab libitum access to a standard rodent diet and water. Male and female mice were randomly used in this study unless otherwise indicated. For modeling mesotheliomas in mice, 4-week-old gene-floxed mice were intrathoracically injected with purified adenovirus harboring Cre recombinase driven by a CMV promoter (109 PFU for each mouse).41,81 Briefly, virus particles were slowly released into the pleural space of mice with a 33-gauge needle inserted between the ribs, penetrating the chest wall at a depth of 2–3 mm (Figure 3A). Upon virus injection, mice were monitored daily and euthanized when they had weight loss or breathing abnormalities. Mouse lungs, hearts, and diaphragms were harvested, fixed in 4% formalin, paraffin-embedded, sectioned, and subjected to histological and pathological analysis.
Cell lines and cell culture
Mesothelioma cell lines NCI-H2452, NCI-H2052, and NCI-H2373 were cultured at early passages (less than 10) in RPMI-1640 medium (Gibco), supplemented with 10% fetal bovine serum (FBS) (Gibco or ExCell Bio) and 1% penicillin/streptomycin. Wild-type (WT) HEK293A and NF2 knockout (KO) HEK293A cells were maintained in DMEM (Gibco) medium containing 10% FBS and 1% penicillin/streptomycin.31 Cells were incubated in a 5% CO2 incubator at 37°C. SAV1 KO and WWC1-3 tKO mesothelioma cell lines were generated in this study using the CRISPR/Cas9 system (below). The downregulation of target proteins in each cell line was confirmed by immunoblotting.
Method details
CRISPR/Cas9 gene editing
Gene-specific single-guide RNAs (sgRNAs) were designed using the CRISPR design tool at http://www.genome-engineering.org/crispr. The sgRNA sequences were cloned into the plasmid pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene #62988), which was from Feng Zhang laboratory.82 Cells transfected with CRIPSR/Cas9 plasmids were selected by 2 μg/mL puromycin for 2 days. Pooled cells were used directly for experiments or seeded into 96-well plates (one cell per well) to establish monoclonal KO cells. The guide RNA sequences used were reported previously.31
Transfection of plasmids, RNA interference, and lentivirus production
Plasmids were transfected into HEK293A cells using PolyJet DNA In Vitro transfection Reagent (Signagen, #SL100688) according to the manufacturer’s instructions. For RNA interference, 20 μM siRNA (GenePharma) and 3 μL Lipofectamine RNAiMAX (Life Technologies) were each diluted in 100 μL serum-free medium, mixed gently, and incubated for 10 min at room temperature; the mixture was added dropwise to plates with cells. Cells were incubated for 48–72 h before analysis. The detailed siRNA sequences are as follows: siWWC1:5′-GGUUGGAGAUUACUUCAUAGA-3’; siWWC2:5′-GGAUCUUCAUCCAGUACUAAA-3’; siWWC3:5′-GGAUAUUCAACAAAUACAAAG-3’. For lentivirus production, HEK293A cells were transfected with pLVX-based plasmids together with psPAX2 and pMD.2G packing vectors, and the medium-containing virus was collected and filtered for viral transduction.
Immunoblotting
Proteins in cells or tissue lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Tissues and tumors were collected and lysed with RIPA buffer containing 0.1 mM PMSF. The total protein concentrations were determined using the BCA Protein Assay Kit (Cwbio, #CW0014S). Then proteins were transferred onto a nitrocellulose membrane, blocked with 5% skim milk in PBST, and incubated with primary antibodies in 5% bovine serum albumin (BSA) overnight at 4°C. Membranes were washed and incubated with HRP-coupled secondary antibodies for 1h at room temperature. High-sig ECL Western Blotting Substrate (Tanon, #180–501) was added, and chemiluminescence was detected using a Tanon 5200S imaging system.
RNA extraction, cDNA synthesis, and quantitative RT-PCR (qRT-PCR) analyses
The total RNA was extracted using the RNeasy Plus mini kit (Qiagen). The cDNA was synthesized using the PrimeScript RT kit (TaKaRa) according to the manufacturer’s instructions. qRT-PCR was performed using SYBR Green qPCR Master Mix (TaKaRa) on a 7500 Real-Time PCR system (Applied Biosystems). All experiments were performed in triplicate and for at least three biological repeats. The changes in mRNA levels were determined using the ΔΔCT method and normalized to the housekeeping gene GAPDH. Primers used in PCR were as follows: GAPDH, F:5′- ATGGGGAAGGTGAAGGTCG-3′, R:5′- GGGGTCATTGATGGCAACAATA-3’; CTGF: F:5′-CCAATGACAACGCCTCCTG-3′,R:5′-TGGTGCAGCCAGAAAGCTC-3’;CYR61: F:5′-AGCCTCGCATCCTATACAACC-3′, R:5′-TTCTTTCACAAGGCGGCACTC-3′, ANKRD1: F:5′-CACTTCTAGCCCACCCTGTGA-3′, R:5′-CCACAGGTTCCGTAATGATTT-3’.
Colony formation
Two thousand mesothelioma cells were seeded into each well of 12-well plates. After a 2-week culture, cells were washed with PBS and fixed in 4% PFA for 15 min. The fixed cells were then washed and stained using 0.1% crystal violet (Servicebio, #G104) for 15 min. Pictures were taken using a light microscope and analyzed by ImageJ.
CCK8 assay
Cells cultured in 96-well plates were subjected to CCK8 assay. At indicated time points, 10% CCK8 solution was added to each well of the plate and incubated in the dark at 37°C for 1 h. Then the absorbance of each well at 450 nm was measured using a microplate reader.
Tumor xenograft model
To generate xenograft mouse tumors, mesothelioma cells were resuspended in PBS at a concentration of 1 x 107 cells/ml, and 100 μL of cell suspension was injected subcutaneously into the dorsal flank of nude mice. After implantation, tumor growth was monitored and measured every three days. The tumor volume was calculated using the modified ellipsoidal formula: V = 1/2(Length x Width2). Mice were sacrificed when the average size of mesothelioma xenografts reached 1 cm3.
AAV6 virus production and injection
AAV6 virus was produced by OBiO Technology. pcAAV-3xFlag-SuperHippo-P2A-NF2 was sequenced and packaged into AAV6. The viral titer of AAV6-SuperHippo-P2A-NF2 was 1.50 x 1013 v.g./mL, as determined by quantitative PCR. For intratumoral injections, about 1 x 1011 v.g of the virus was injected slowly into the tumor with a 33-gauge needle. For intrathoracic injection, about 2.5 x 1011 v.g of AAV6-SuperHippo-P2A-NF2 virus was injected into the pleural cavity of mice at a depth of 2–3 mm.
Histology and IHC staining
Tissue or tumor samples were dissected carefully from mice and immediately fixed in 4% formalin, dehydrated through graded alcohols, and embedded in paraffin. Tissue or tumor sections about 5 μm-thick were cut using a microtome (LEICA, RM2235). The sections were then deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E) according to standard protocols. For IHC staining, paraffin-embedded sections were baked at 65°C overnight, followed by deparaffinization and hydration. Then heat-induced antigen retrieval was performed using sodium citrate buffer (Beyotime, P0083) following the manufacturer’s protocol. Endogenous peroxidase activities were inactivated by 3% H2O2 for 30 min. After blocking with 3% BSA-PBST, sections were incubated overnight at 4°C with anti-Mesothelin (PA5-79698, Invitrogen), anti-WT1 (83535, Cell Signaling Technology) and anti-Ki67(ab15580, Abcam) primary antibodies and subsequently with Envision anti-Rabbit (DAKO, K4002) secondary antibody. Sections were counterstained with hematoxylin and mounted with Permount Mounting Media. Images were taken using the Olympus VS200 Microscope.
TEAD inhibitors for cell-based assays
Two TEAD inhibitors, VT103 and VT107, were used to treat mesothelioma cells in this study.51 Briefly, VT103 and VT107 were dissolved in DMSO and added into the culture medium at indicated concentrations. For the colony formation assay, the culture medium containing VT103 and VT107 was refreshed every two days to maintain optimal conditions for the experiment.
Quantification and statistical analysis
All results were presented as mean ± SEM. Statistical analysis was performed using GraphPad Prism 9 software (GraphPad Software, Inc, USA). Comparisons between the two groups were made using unpaired Student’s t-tests. One-way ANOVA was used for multiple comparisons. Data marked with asterisks are significantly different from the control as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and n.s. indicates not significant.
Published: October 4, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101763.
Supplemental information
References
- 1.Craighead J.E. Current pathogenetic concepts of diffuse malignant mesothelioma. Hum. Pathol. 1987;18:544–557. doi: 10.1016/s0046-8177(87)80354-4. [DOI] [PubMed] [Google Scholar]
- 2.Zhou S., Liu L., Li H., Eilers G., Kuang Y., Shi S., Yan Z., Li X., Corson J.M., Meng F., et al. Multipoint targeting of the PI3K/mTOR pathway in mesothelioma. Br. J. Cancer. 2014;110:2479–2488. doi: 10.1038/bjc.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curran D., Sahmoud T., Therasse P., van Meerbeeck J., Postmus P.E., Giaccone G. Prognostic factors in patients with pleural mesothelioma: the European Organization for Research and Treatment of Cancer experience. J. Clin. Oncol. 1998;16:145–152. doi: 10.1200/JCO.1998.16.1.145. [DOI] [PubMed] [Google Scholar]
- 4.Milano M.T., Zhang H. Malignant pleural mesothelioma: a population-based study of survival. J. Thorac. Oncol. 2010;5:1841–1848. doi: 10.1097/JTO.0b013e3181f1cf2b. [DOI] [PubMed] [Google Scholar]
- 5.Zhai Z., Ruan J., Zheng Y., Xiang D., Li N., Hu J., Shen J., Deng Y., Yao J., Zhao P., et al. Assessment of Global Trends in the Diagnosis of Mesothelioma From 1990 to 2017. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Opitz I. Management of malignant pleural mesothelioma-The European experience. J. Thorac. Dis. 2014;6:S238–S252. doi: 10.3978/j.issn.2072-1439.2014.05.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelzang N.J., Rusthoven J.J., Symanowski J., Denham C., Kaukel E., Ruffie P., Gatzemeier U., Boyer M., Emri S., Manegold C., et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 8.Ceresoli G.L., Zucali P.A., Favaretto A.G., Grossi F., Bidoli P., Del Conte G., Ceribelli A., Bearz A., Morenghi E., Cavina R., et al. Phase II study of pemetrexed plus carboplatin in malignant pleural mesothelioma. J. Clin. Oncol. 2006;24:1443–1448. doi: 10.1200/JCO.2005.04.3190. [DOI] [PubMed] [Google Scholar]
- 9.Borea F., Franczak M.A., Garcia M., Perrino M., Cordua N., Smolenski R.T., Peters G.J., Dziadziuszko R., Santoro A., Zucali P.A., Giovannetti E. Target Therapy in Malignant Pleural Mesothelioma: Hope or Mirage? Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24119165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oehl K., Vrugt B., Opitz I., Meerang M. Heterogeneity in Malignant Pleural Mesothelioma. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baas P., Scherpereel A., Nowak A.K., Fujimoto N., Peters S., Tsao A.S., Mansfield A.S., Popat S., Jahan T., Antonia S., et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): a multicentre, randomised, open-label, phase 3 trial. Lancet. 2021;397:375–386. doi: 10.1016/S0140-6736(20)32714-8. [DOI] [PubMed] [Google Scholar]
- 12.Davis A., Ke H., Kao S., Pavlakis N. An Update on Emerging Therapeutic Options for Malignant Pleural Mesothelioma. Lung Cancer. 2022;13:1–12. doi: 10.2147/LCTT.S288535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey K., Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat. Rev. Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 14.Johnson R., Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu F.X., Zhao B., Guan K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanconato F., Cordenonsi M., Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Y., Pan D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell. 2019;50:264–282. doi: 10.1016/j.devcel.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong W., Guan K.L. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin. Cell Dev. Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maugeri-Sacca M., Barba M., Pizzuti L., Vici P., Di Lauro L., Dattilo R., Vitale I., Bartucci M., Mottolese M., De Maria R. The Hippo transducers TAZ and YAP in breast cancer: oncogenic activities and clinical implications. Expet Rev. Mol. Med. 2015;17 doi: 10.1017/erm.2015.12. [DOI] [PubMed] [Google Scholar]
- 20.Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 21.Mo J.S., Park H.W., Guan K.L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan S.W., Lim C.J., Guo K., Ng C.P., Lee I., Hunziker W., Zeng Q., Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 23.Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Z., Hao Y., Liu N., Raptis L., Tsao M.S., Yang X. TAZ is a novel oncogene in non-small cell lung cancer. Oncogene. 2011;30:2181–2186. doi: 10.1038/onc.2010.606. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W., Nandakumar N., Shi Y., Manzano M., Smith A., Graham G., Gupta S., Vietsch E.E., Laughlin S.Z., Wadhwa M., et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q., Li S., Mana-Capelli S., Roth Flach R.J., Danai L.V., Amcheslavsky A., Nie Y., Kaneko S., Yao X., Chen X., et al. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev. Cell. 2014;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S., Cho Y.S., Yue T., Ip Y.T., Jiang J. Overlapping functions of the MAP4K family kinases Hppy and Msn in Hippo signaling. Cell Discov. 2015;1 doi: 10.1038/celldisc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng Z., Moroishi T., Mottier-Pavie V., Plouffe S.W., Hansen C.G., Hong A.W., Park H.W., Mo J.S., Lu W., Lu S., et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y., Wang W., Liu B., Deng H., Uster E., Pan D. Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev. Cell. 2015;34:642–655. doi: 10.1016/j.devcel.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi S., Zhu Y., Liu X., Li P., Wang Y., Zeng Y., Yu A., Wang Y., Sha Z., Zhong Z., et al. WWC proteins mediate LATS1/2 activation by Hippo kinases and imply a tumor suppression strategy. Mol. Cell. 2022;82:1850–1864.e7. doi: 10.1016/j.molcel.2022.03.027. [DOI] [PubMed] [Google Scholar]
- 31.Qi S., Zhong Z., Zhu Y., Wang Y., Ma M., Wang Y., Liu X., Jin R., Jiao Z., Zhu R., et al. Two Hippo signaling modules orchestrate liver size and tumorigenesis. EMBO J. 2023;42 doi: 10.15252/embj.2022112126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai Z.C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 33.Wei X., Shimizu T., Lai Z.C. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang N., Bai H., David K.K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R.A., Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami H., Mizuno T., Taniguchi T., Fujii M., Ishiguro F., Fukui T., Akatsuka S., Horio Y., Hida T., Kondo Y., et al. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 2011;71:873–883. doi: 10.1158/0008-5472.CAN-10-2164. [DOI] [PubMed] [Google Scholar]
- 36.Zhong Z., Jiao Z., Yu F.X. The Hippo signaling pathway in development and regeneration. Cell Rep. 2024;43 doi: 10.1016/j.celrep.2024.113926. [DOI] [PubMed] [Google Scholar]
- 37.Zhong Z., Meng Z., Yu F.-X. Reconstructing the Hippo signaling network. Sci. Bull. 2023;68:2307–2310. doi: 10.1016/j.scib.2023.08.048. [DOI] [PubMed] [Google Scholar]
- 38.Cheng J.Q., Jhanwar S.C., Klein W.M., Bell D.W., Lee W.C., Altomare D.A., Nobori T., Olopade O.I., Buckler A.J., Testa J.R. p16 alterations and deletion mapping of 9p21-p22 in malignant mesothelioma. Cancer Res. 1994;54:5547–5551. [PubMed] [Google Scholar]
- 39.Sekido Y., Pass H.I., Bader S., Mew D.J., Christman M.F., Gazdar A.F., Minna J.D. Neurofibromatosis type 2 (NF2) gene is somatically mutated in mesothelioma but not in lung cancer. Cancer Res. 1995;55:1227–1231. [PubMed] [Google Scholar]
- 40.Bott M., Brevet M., Taylor B.S., Shimizu S., Ito T., Wang L., Creaney J., Lake R.A., Zakowski M.F., Reva B., et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badhai J., Pandey G.K., Song J.Y., Krijgsman O., Bhaskaran R., Chandrasekaran G., Kwon M.C., Bombardelli L., Monkhorst K., Grasso C., et al. Combined deletion of Bap1, Nf2, and Cdkn2ab causes rapid onset of malignant mesothelioma in mice. J. Exp. Med. 2020;217 doi: 10.1084/jem.20191257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hmeljak J., Sanchez-Vega F., Hoadley K.A., Shih J., Stewart C., Heiman D., Tarpey P., Danilova L., Drill E., Gibb E.A., et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018;8:1548–1565. doi: 10.1158/2159-8290.CD-18-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bueno R., Stawiski E.W., Goldstein L.D., Durinck S., De Rienzo A., Modrusan Z., Gnad F., Nguyen T.T., Jaiswal B.S., Chirieac L.R., et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016;48:407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 44.Jongsma J., van Montfort E., Vooijs M., Zevenhoven J., Krimpenfort P., van der Valk M., van de Vijver M., Berns A. A conditional mouse model for malignant mesothelioma. Cancer Cell. 2008;13:261–271. doi: 10.1016/j.ccr.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 45.Hakiri S., Osada H., Ishiguro F., Murakami H., Murakami-Tonami Y., Yokoi K., Sekido Y. Functional differences between wild-type and mutant-type BRCA1-associated protein 1 tumor suppressor against malignant mesothelioma cells. Cancer Sci. 2015;106:990–999. doi: 10.1111/cas.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holden J.K., Cunningham C.N. Targeting the Hippo Pathway and Cancer through the TEAD Family of Transcription Factors. Cancers. 2018;10 doi: 10.3390/cancers10030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin K.C., Park H.W., Guan K.L. Regulation of the Hippo Pathway Transcription Factor TEAD. Trends Biochem. Sci. 2017;42:862–872. doi: 10.1016/j.tibs.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pobbati A.V., Kumar R., Rubin B.P., Hong W. Therapeutic targeting of TEAD transcription factors in cancer. Trends Biochem. Sci. 2023;48:450–462. doi: 10.1016/j.tibs.2022.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Liu-Chittenden Y., Huang B., Shim J.S., Chen Q., Lee S.J., Anders R.A., Liu J.O., Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao B., Kim J., Ye X., Lai Z.C., Guan K.L. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- 51.Tang T.T., Konradi A.W., Feng Y., Peng X., Ma M., Li J., Yu F.X., Guan K.L., Post L. Small Molecule Inhibitors of TEAD Auto-palmitoylation Selectively Inhibit Proliferation and Tumor Growth of NF2-deficient Mesothelioma. Mol. Cancer Therapeut. 2021;20:986–998. doi: 10.1158/1535-7163.MCT-20-0717. [DOI] [PubMed] [Google Scholar]
- 52.Metcalf R.A., Welsh J.A., Bennett W.P., Seddon M.B., Lehman T.A., Pelin K., Linnainmaa K., Tammilehto L., Mattson K., Gerwin B.I., et al. p53 and Kirsten-ras mutations in human mesothelioma cell lines. Cancer Res. 1992;52:2610–2615. [PubMed] [Google Scholar]
- 53.Sementino E., Menges C.W., Kadariya Y., Peri S., Xu J., Liu Z., Wilkes R.G., Cai K.Q., Rauscher F.J., 3rd, Klein-Szanto A.J., Testa J.R. Inactivation of Tp53 and Pten drives rapid development of pleural and peritoneal malignant mesotheliomas. J. Cell. Physiol. 2018;233:8952–8961. doi: 10.1002/jcp.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J.E., Kim D., Hong Y.S., Kim K.P., Yoon Y.K., Lee D.H., Kim S.W., Chun S.M., Jang S.J., Kim T.W. Mutational Profiling of Malignant Mesothelioma Revealed Potential Therapeutic Targets in EGFR and NRAS. Transl. Oncol. 2018;11:268–274. doi: 10.1016/j.tranon.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naso M.F., Tomkowicz B., Perry W.L., 3rd, Strohl W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs. 2017;31:317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunningham R., Jia S., Purohit K., Salem O., Hui N.S., Lin Y., Carragher N.O., Hansen C.G. YAP/TAZ activation predicts clinical outcomes in mesothelioma and is conserved in in vitro model of driver mutations. Clin. Transl. Med. 2023;13 doi: 10.1002/ctm2.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyanaga A., Masuda M., Tsuta K., Kawasaki K., Nakamura Y., Sakuma T., Asamura H., Gemma A., Yamada T. Hippo pathway gene mutations in malignant mesothelioma: revealed by RNA and targeted exon sequencing. J. Thorac. Oncol. 2015;10:844–851. doi: 10.1097/JTO.0000000000000493. [DOI] [PubMed] [Google Scholar]
- 58.Tranchant R., Quetel L., Tallet A., Meiller C., Renier A., de Koning L., de Reynies A., Le Pimpec-Barthes F., Zucman-Rossi J., Jaurand M.C., Jean D. Co-occurring Mutations of Tumor Suppressor Genes, LATS2 and NF2, in Malignant Pleural Mesothelioma. Clin. Cancer Res. 2017;23:3191–3202. doi: 10.1158/1078-0432.CCR-16-1971. [DOI] [PubMed] [Google Scholar]
- 59.Sato T., Sekido Y. NF2/Merlin Inactivation and Potential Therapeutic Targets in Mesothelioma. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19040988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiltbrunner S., Fleischmann Z., Sokol E.S., Zoche M., Felley-Bosco E., Curioni-Fontecedro A. Genomic landscape of pleural and peritoneal mesothelioma tumours. Br. J. Cancer. 2022;127:1997–2005. doi: 10.1038/s41416-022-01979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang H., Hall S.R.R., Sun B., Zhao L., Gao Y., Schmid R.A., Tan S.T., Peng R.W., Yao F. NF2 and Canonical Hippo-YAP Pathway Define Distinct Tumor Subsets Characterized by Different Immune Deficiency and Treatment Implications in Human Pleural Mesothelioma. Cancers. 2021;13 doi: 10.3390/cancers13071561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gong R., Yu F.X. Targeting the Hippo Pathway for Anti-cancer Therapies. Curr. Med. Chem. 2015;22:4104–4117. doi: 10.2174/0929867322666151002112256. [DOI] [PubMed] [Google Scholar]
- 63.Nakatani K., Maehama T., Nishio M., Goto H., Kato W., Omori H., Miyachi Y., Togashi H., Shimono Y., Suzuki A. Targeting the Hippo signalling pathway for cancer treatment. J. Biochem. 2017;161:237–244. doi: 10.1093/jb/mvw074. [DOI] [PubMed] [Google Scholar]
- 64.Dey A., Varelas X., Guan K.L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020;19:480–494. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nouri K., Azad T., Ling M., Janse van Rensburg H.J., Pipchuk A., Shen H., Hao Y., Zhang J., Yang X. Identification of Celastrol as a Novel YAP-TEAD Inhibitor for Cancer Therapy by High Throughput Screening with Ultrasensitive YAP/TAZ-TEAD Biosensors. Cancers. 2019;11 doi: 10.3390/cancers11101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiao S., Wang H., Shi Z., Dong A., Zhang W., Song X., He F., Wang Y., Zhang Z., Wang W., et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 67.Pobbati A.V., Han X., Hung A.W., Weiguang S., Huda N., Chen G.Y., Kang C., Chia C.S.B., Luo X., Hong W., Poulsen A. Targeting the Central Pocket in Human Transcription Factor TEAD as a Potential Cancer Therapeutic Strategy. Structure. 2015;23:2076–2086. doi: 10.1016/j.str.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bum-Erdene K., Zhou D., Gonzalez-Gutierrez G., Ghozayel M.K., Si Y., Xu D., Shannon H.E., Bailey B.J., Corson T.W., Pollok K.E., et al. Small-Molecule Covalent Modification of Conserved Cysteine Leads to Allosteric Inhibition of the TEAD⋅Yap Protein-Protein Interaction. Cell Chem. Biol. 2019;26:378–389.e13. doi: 10.1016/j.chembiol.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Kaneda A., Seike T., Danjo T., Nakajima T., Otsubo N., Yamaguchi D., Tsuji Y., Hamaguchi K., Yasunaga M., Nishiya Y., et al. The novel potent TEAD inhibitor, K-975, inhibits YAP1/TAZ-TEAD protein-protein interactions and exerts an anti-tumor effect on malignant pleural mesothelioma. Am. J. Cancer Res. 2020;10:4399–4415. [PMC free article] [PubMed] [Google Scholar]
- 70.Holden J.K., Crawford J.J., Noland C.L., Schmidt S., Zbieg J.R., Lacap J.A., Zang R., Miller G.M., Zhang Y., Beroza P., et al. Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107809. [DOI] [PubMed] [Google Scholar]
- 71.Pease D.F., Kratzke R.A. Oncolytic Viral Therapy for Mesothelioma. Front. Oncol. 2017;7:179. doi: 10.3389/fonc.2017.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang H., Xu D., Gao Y., Schmid R.A., Peng R.W. Oncolytic Viral Therapy for Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2020;15:e111–e113. doi: 10.1016/j.jtho.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 73.Vachani A., Moon E., Albelda S.M. Gene therapy for mesothelioma. Curr. Treat. Options Oncol. 2011;12:173–180. doi: 10.1007/s11864-011-0153-5. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L., Iyer J., Chowdhury A., Ji M., Xiao L., Yang S., Chen Y., Tsai M.Y., Dong J. KIBRA regulates aurora kinase activity and is required for precise chromosome alignment during mitosis. J. Biol. Chem. 2012;287:34069–34077. doi: 10.1074/jbc.M112.385518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu F.X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H., Zhao J., Yuan H., Tumaneng K., Li H., et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y., Zhu Y., Gu Y., Ma M., Wang Y., Qi S., Zeng Y., Zhu R., Wang X., Yu P., et al. Stabilization of Motin family proteins in NF2-deficient cells prevents full activation of YAP/TAZ and rapid tumorigenesis. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109596. [DOI] [PubMed] [Google Scholar]
- 78.Ji S., Liu Q., Zhang S., Chen Q., Wang C., Zhang W., Xiao C., Li Y., Nian C., Li J., et al. FGF15 Activates Hippo Signaling to Suppress Bile Acid Metabolism and Liver Tumorigenesis. Dev. Cell. 2019;48:460–474.e9. doi: 10.1016/j.devcel.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 79.Li F., Han X., Li F., Wang R., Wang H., Gao Y., Wang X., Fang Z., Zhang W., Yao S., et al. LKB1 Inactivation Elicits a Redox Imbalance to Modulate Non-small Cell Lung Cancer Plasticity and Therapeutic Response. Cancer Cell. 2015;27:698–711. doi: 10.1016/j.ccell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 81.Meuwissen R., Linn S.C., van der Valk M., Mooi W.J., Berns A. Mouse model for lung tumorigenesis through Cre/lox controlled sporadic activation of the K-Ras oncogene. Oncogene. 2001;20:6551–6558. doi: 10.1038/sj.onc.1204837. [DOI] [PubMed] [Google Scholar]
- 82.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.