Summary
The shortfall in new analgesic agents is a major impediment to reducing reliance on opioid medications for control of severe pain. In both animals and man, attenuating nociceptive transmission from primary afferent neurons with a μ-opioid receptor agonist yields highly effective analgesia. Consequently, deeper molecular characterization of human nociceptive afferents expressing OPRM1, the μ-opioid receptor gene, is a key component for advancing analgesic drug discovery and understanding clinical pain control. A co-expression matrix for the μ-opioid receptor and a variety of nociceptive channels as well as δ- and κ-opioid receptors is established by multiplex in situ hybridization. Our results indicate an OPRM1-positive population with strong molecular resemblance to rodent peptidergic C-nociceptors associated with tissue damage pain and an OPRM1-negative population sharing molecular characteristics of murine non-peptidergic C-nociceptors. The empirical identification of two distinct human nociceptive populations that differ profoundly in their presumed responsiveness to opioids provides an actionable translational framework for human pain control.
Keywords: non-opioid analgesics, μ-opioid receptor, TRPV1, TRPA1, MRGPRD, sodium channels, somatosensory afferent neurons, human nociception, neuropathic pain, translational research
Graphical abstract

Highlights
-
•
OPRM1 expression in the human DRG distinguishes two broad nociceptive populations
-
•
OPRM1-positive nociceptors show molecular resemblance to rodent peptidergic neurons
-
•
Most OPRM1-negative nociceptors express the murine superficial skin marker MRGPRD
-
•
The κ-opioid receptor gene OPRK1 is mainly expressed in satellite glial cells
Staedtler et al. describe a dichotomy of human nociceptors into OPRM1-expressing neurons that share molecular features with rodent peptidergic neurons associated with tissue damage pain and OPRM1-negative neurons that mostly resemble murine non-peptidergic neurons expressing the superficial skin marker MRGPRD. This division provides a cellular-molecular framework for human pain control.
Introduction
Opioids acting at the μ-opioid receptor are mainstays of clinical management of severe tissue damage pain.1,2,3 Their adverse side effect profile and the risk for addiction, however, impose limits on clinical use and drive the search for alternative analgesic targets.4,5,6,7 A crucial element of opioid analgesia is the inhibition of transmission from nociceptive primary afferent neurons to second-order neurons in the dorsal spinal cord,8,9,10 making these afferents critical targets for analgesic drug development. Understanding, identifying, and molecularly distinguishing the most relevant “pain control neuron” are essential steps for focusing analgesic drug development efforts. The idea that a clinically relevant opioid receptor-expressing population is present in the dorsal root ganglion (DRG) is supported by human experimental pain studies that model clinically relevant pain. These models frequently apply sustained experimental noxious stimulation to skin and deep tissues, and significant pain reduction can be achieved by systemic opioids in response to variety of exogenous stimuli including noxious heat, cold, pressure, pinch, and ischemia (Tables S1–S5).11,12,13,14 The variety of stimuli suggests that, in humans, μ-opioid receptors are expressed by heterogeneous and/or multimodal nociceptive afferent populations.
Clinically relevant sustained pain from tissue damage is transmitted mainly by unmyelinated C-fibers,15,16 supporting the idea that C-nociceptors are the major targets of μ-receptor agonists. Based on rodent studies, C-nociceptors have been divided into two major populations, with only one of them having the capacity to transmit sustained pain from tissue damage.17,18,19,20,21 This population has been classically termed “peptidergic” nociceptors due to their production of algogenic peptides such as CGRP (calcitonin gene-related peptide) and substance P. They also express the heat- and inflammation-activated ion channel TRPV1 (transient receptor potential vanilloid receptor 1), the μ-opioid receptor, and the neurotrophic receptor TrkA (tropomyosin receptor kinase A)17,22,23,24,25 and innervate both skin and deep tissues.23,26,27,28 By contrast, the second murine population, termed “non-peptidergic” C-nociceptors, express low levels of neuropeptides and TRPV1, the δ-opioid receptor, and the neurotrophic receptor GFRA2.19,21,29,30,31 The most prevalent non-peptidergic population NP1 is marked by the expression of the itch-related receptor MRGPRD (Mas-related G-protein-coupled receptor D)21 and innervates exclusively the murine superficial epidermis.32 The functional relevance of this division is supported by mouse optogenetic studies that demonstrate guarding behaviors, which are indicative of a sustained pain-like experience, upon stimulation of peptidergic neurons. By contrast, stimulation of non-peptidergic MRGPRD+ neurons causes reflexive paw withdrawal33,34 consistent with a proposal that these neurons form a “biowarning” system that mediates spinal reflex withdrawal prior to tissue damage.35,36 Importantly, these neurons also contribute to pathological pain states such as neuropathic pain.37,38,39
Sequencing studies of human somatosensory afferent transcriptomes have revealed several nociceptive clusters that mostly follow organizational principles of murine DRG neurons, yet a precise delineation into the aforementioned main populations, including an unambiguous expression of low-expressed G-protein-coupled receptors, such as opioid receptors or MRGPRD, has not been achieved.40,41,42 Observations in humans report a high degree of responsiveness to opioids in cases of severe sustained pain, but minimal responsiveness to opioids to short-lasting, threshold-level pain,1 and reduced responsiveness to neuropathic pain (Table S6).43,44,45 This suggests that the basic division of nociceptors is also functionally true in humans. Thus, the aim of the present investigation is to identify the population of DRG nociceptive neurons with the greatest relevance to clinical pain control. Specifically, we hypothesize that this population of human C-fiber neurons is represented by neurons that express the nociresponsive ion channel TRPV1 in conjunction with the μ-opioid receptor. Therefore, this population is sensitive to both opioid agonists and a variety of nociceptive stimuli, making it relevant to tissue damage pain and opioid analgesia. For the empirical identification of this population, we designed a comprehensive set of gene probes for multiplex fluorescence in situ hybridization. This investigation of human nociceptors provides insight into analgesic target validation which is a crucial component for achieving successful translation. Specifically, confirming the expression of putative analgesic targets in the most relevant nociceptive population expressing TRPV1 and OPRM1 is required for peripherally driven analgesia.
Results
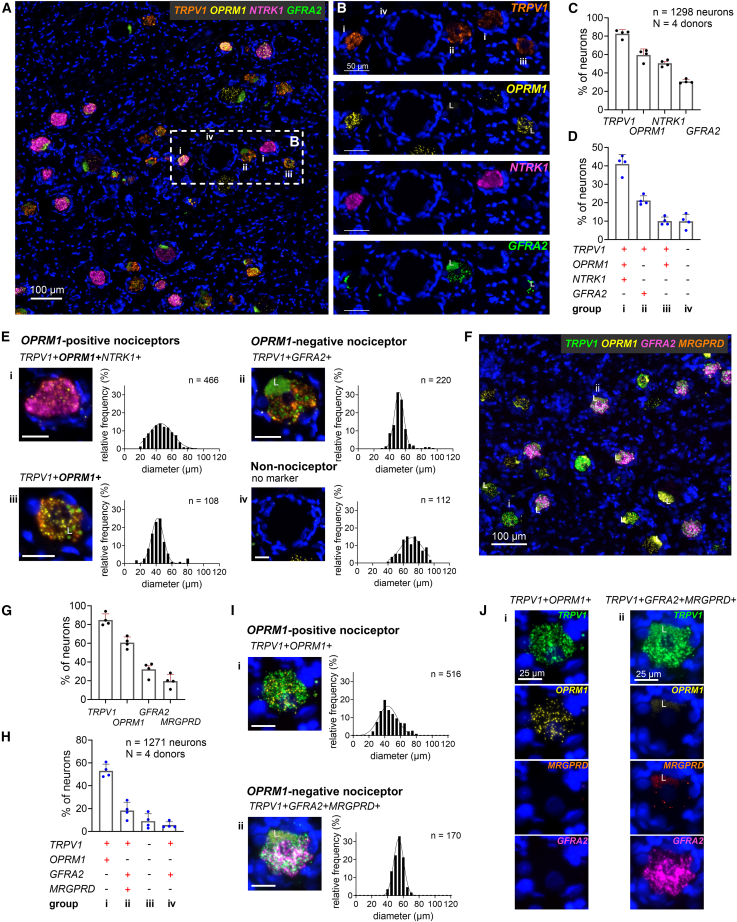
We investigated human DRG neurons from four tissue donors for the expression of TRPV1 and OPRM1. Data from a variety of probe pairs were integrated to obtain a comprehensive picture of the expression of potential analgesic targets (Figure 1A). If we include all experiments and all neurons into the counting analysis, 56.3% ± 2.1% of neurons were characterized as TRPV1+OPRM1+ (Figure S3). We identified a second population of TRPV1+ and OPRM1-negative neurons. Both populations express multiple algesic markers and neurotrophic receptors that provisionally characterize them as nociceptive. A third prominent population of large-diameter neurons did not express any of the algesic markers and was classified as non-nociceptive. These definitions based on transcription can be further substantiated by functional investigations. Additionally, according to their neuronal diameters, 88.7% of TRPV1+OPRM1+ nociceptors could be classified as small- to medium-diameter neurons (see STAR Methods, Figure S7), which is consistent with a nociceptive population.
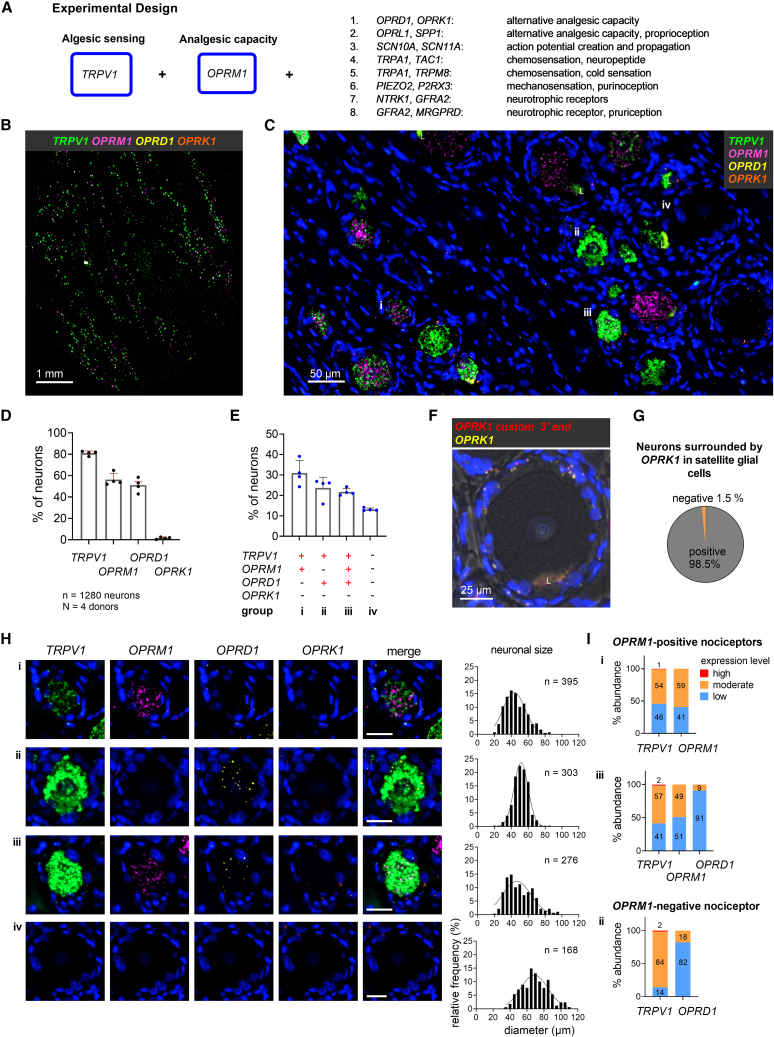
Figure 1.
OPRM1-positive and OPRM1-negative human nociceptors express OPRD1 while OPRK1 is expressed in satellite glial cells
(A) Overall schematic of experimental design for 4-Plex in situ hybridization studies. The major nociceptive ion channel TRPV1 is paired with the major analgesic receptor (μ-opioid, OPRM1) and a series of genes coding for algesic and analgesic mediators.
(B) Scanned image of a complete section from human L3 DRG hybridized for the heat- and inflammation-activated channel TRPV1 (green), the μ-opioid (OPRM1) (magenta), δ-opioid (OPRD1) (yellow), and κ-opioid receptor (OPRK1) (orange). Note the strong expression and high prevalence of neuronal TRPV1 expression which tends to obscure the signal from the other genes at this magnification.
(C) Enlargement showing the multiple neuronal signals. Representative neurons are labeled i–iv and are characterized further in (H).
(D) Percentage of 1,280 DRG neurons expressing each individual transcript. Note the comparatively low neuronal expression of OPRK1.
(E) Percentage of DRG neurons expressing the most common transcript combinations, which defines populations i–iv. Bar graphs in (D) and (E) show mean, standard deviation (SD), and individual values from four independent tissue donors.
(F) Single-neuron example demonstrating the expression of OPRK1 in satellite glial cells surrounding the neuron, as detected by the standard probe (yellow) and, as a technical replicate, the custom probe (red). The large fluorescent patch, “L,” is lipofuscin. See also Figures S4 and S5.
(G) The preponderance of neurons that are surrounded by OPRK1 (κ-opioid receptor) expressing satellite cells.
(H) Individual channel and multi-channel microscopy images of representative neurons for each population (i–iv, as in C) and the corresponding populations’ cell size distribution. Scale bar, 25 μm.
(I) Percentages of nociceptors showing low, medium, or high expression levels for TRPV1 and each opioid receptor transcript averaged across the 4 tissue donors.
OPRM1-positive and OPRM1-negative human nociceptors express OPRD1 while OPRK1 is expressed in satellite glial cells
Both the δ- and κ-opioid receptors (encoded by OPRD1 and OPRK1, respectively) represent potential alternative analgesic targets due to inhibitory effects on neurotransmitter release at synapses in the dorsal horn.2 Whether they are expressed by TRPV1+OPRM1+ nociceptors associated with rodent sustained pain had not been elucidated. We evaluated pooled data of 4 tissue donors (n = 1,280 neurons). TRPV1 was expressed in 81% ± 2.1%, OPRM1 in 56.3% ± 5.9%, OPRD1 in 51.1% ± 6.5%, and OPRK1 in 1.6% ± 1% of human DRG neurons (Figure 1D). The abundance of neurons expressing TRPV1 in the human DRG is shown in the whole DRG section (Figure 1B). When considering the co-expression patterns of all four markers, we observed four prevalent populations (Figures 1E; Table S7), which we characterized for cell size and expression levels of transcripts. Two of them were TRPV1+OPRM1+ nociceptive populations, one was a TRPV1+OPRM1-negative nociceptive population, and one a non-nociceptive population. The most abundant TRPV1+OPRM1+ population (labeled i, detected in 30.9% ± 6.2% of the analyzed neurons) did not express transcripts for any additional opioid receptor subtype, while population iii (21.7% ± 1.8%) expressed OPRD1 in addition to TRPV1 and OPRM1. The OPRM1-negative population (ii) showed positivity for TRPV1 and OPRD1 (23.6% ± 5.3%). A presumably non-nociceptive population did not express any of the four transcripts (iv, 13.1% ± 0.7%). These four main populations represented 89.3% ± 1.5% of sampled neurons. Microscopic images of a representative neuron of each of the four major populations and the cell diameter distributions of each population are shown in Figure 1H. TRPV1+OPRM1+ (i) and TRPV1+OPRM1+OPRD1+ (iii) populations consisted of a heterogeneous group of mostly small- and medium-diameter neurons ( = 45.8 ± 12.7 μm [i], = 50 ± 15.6 μm [iii]). In contrast, OPRM1-negative nociceptors were medium sized with a uniform, homogeneous cell size distribution ( = 51.2 ± 8.7 μm). Neurons that did not express any marker were medium to large in size ( = 69.7 ± 15.4 μm). In order to evaluate the potential of the δ-opioid receptor as a pharmaceutical target for pain relief, including the potential for μ-δ-heterodimers,46 we evaluated the expression level of each transcript in a given population. For this aim, we determined thresholds for each marker in each donor section for low, moderate, and high expression levels. While OPRM1 was expressed similarly both in a low and in a moderate fashion, OPRD1 showed mostly low expression levels, especially in population iii (91%) (Figure 1I). To summarize, the gene encoding the δ-opioid receptor was expressed at low levels in a subpopulation of the relevant TRPV1+OPRM1+ population.
The κ-opioid receptor gene is mainly expressed in satellite glial cells
Transcripts for OPRK1 within sensory neurons were a scarce observation (1.6% of sampled neurons, Figures 1D and S4). Instead, we observed ubiquitous expression of OPRK1 in non-neuronal cells, mostly in subpopulations of satellite glial cells (SGCs) surrounding somatosensory neurons (Figures 1C, 1H, and S5). This was not an expected finding based on our previous investigations in rat47 and the existing literature.48,49,50 To validate our result, we designed a second probe against OPRK1 targeting a different region of the transcript (see STAR Methods section). Co-staining with both probes showed overlapping or closely juxtaposed puncta (Figures 1F and S5). The quantitative results reported in this manuscript are based on the custom-made OPRK1 probe. We quantified that 98.5% ± 0.9% of all characterized neurons (n = 1280) showed OPRK1 transcripts in surrounding SGCs, indicating OPRK1 is likely a ubiquitous transcript in SGCs (Figure 1G). These data indicate that OPRK1 is primarily a non-neuronal receptor in the human DRG.
OPRL1 is expressed by proprioceptors and a subpopulation of OPRM1-positive nociceptors
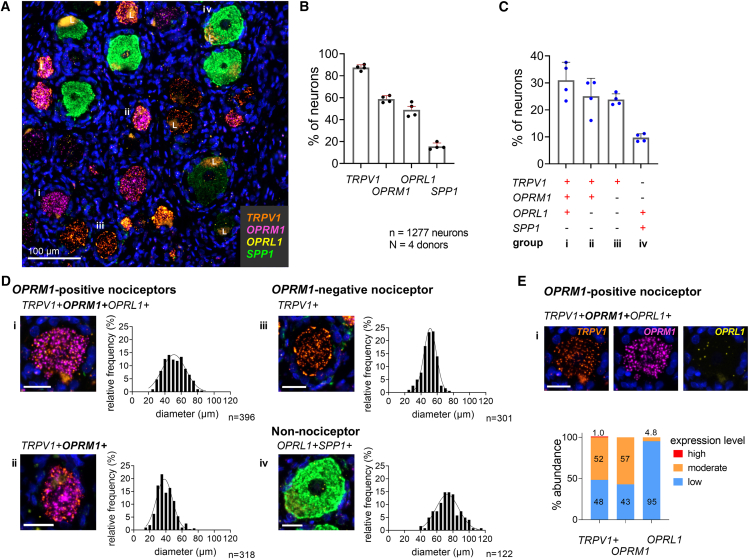
The nociceptin opioid-like receptor (encoded by OPRL1) is a receptor with a wide anatomic distribution in the body, peripheral nervous system (PNS), and CNS that can support a broad spectrum of behavioral and physiological actions.51,52 We previously demonstrated its expression in rat nociceptive and proprioceptive primary afferent neurons.47 Its expression by nociceptive afferents relevant for human pain has not been evaluated. We analyzed human DRGs co-labeled for TRPV1, OPRM1, OPRL1, and the proprioceptive marker osteopontin (SPP1).21,53 We analyzed 1,277 neurons and observed TRPV1 in 87.5% ± 2.6%, OPRM1 in 58.8% ± 3.1%, OPRL1 in 48.9% ± 6.3%, and SPP1 in 15.6% ± 3.1% of neurons (Figure 2B). Analysis of co-expression patterns of all transcripts indicated four prevalent populations (Figure 2C; Table S8) that were representative of 89.6% ± 3.02% of sampled neurons. These included two TRPV1+OPRM1+ nociceptive populations, a TRPV1+OPRM1-negative nociceptive population, and a non-nociceptive population. The largest population consisted of TRPV1+OPRM1+OPRL1+ nociceptors (i, 31% ± 6.7%), which showed a broad cell size distribution ( = 51.8 ± 12.9 μm). The second group (ii, 25.1% ± 6.6%) consisted of small-diameter TRPV1+OPRM1+ neurons ( = 39.6 ± 10.2 μm) that did not express OPRL1. OPRM1-negative TRPV1+ neurons (iii, 23.8% ± 2.1%) did not express OPRL1 and were characterized by a homogeneous medium-sized cell diameter distribution ( = 50.2% ± 8.5) as described before. A relatively small population expressed both SPP1 and OPRL1 (iv, 9.7% ± 1.5%) and consisted of medium- to large-diameter neurons ( = 73.5 ± 15.5 μm) (Figure 2D), indicating that the majority of the previously identified non-nociceptive population expresses both SPP1 and OPRL1. Characterization of OPRL1 expression levels revealed low levels in 95% of the TRPV1+OPRM1+OPRL1+ population (i). The low OPRL1 expression was in contrast to OPRM1 expression levels, which could be classified as moderate in 57% of the same population (Figure 2E). To summarize, OPRL1 was expressed at low expression levels only in a subpopulation of the relevant TRPV1+OPRM1+ population.
Figure 2.
OPRL1 is expressed by proprioceptors and a subpopulation of OPRM1-positive nociceptors
(A) Representative section of human DRG showing positive transcripts for TRPV1, the μ-opioid receptor (OPRM1), the opioid-related nociceptin receptor 1 (OPRL1), and osteopontin (SPP1), a marker for proprioceptive neurons. Lipofuscin is marked with an “L.”
(B) Percentage of somatosensory neurons expressing each individual transcript.
(C) Percentage of 1,277 neurons expressing the most prevalent transcript combinations. Bar graphs in (B) and (C) show mean, SD, and individual values from four independent donors.
(D) Multi-channel microscopy images of a representative individual neuron from each population and the population’s cell size distribution. OPRL1 is expressed at a low level in the neurons illustrated in i and iv. The typical OPRL1 hybridization signal can be seen in (E).
(E) Single-channel images of neuron shown in (Di). Categorized expression levels for each transcript of the TRPV1+OPRM1+OPRL1+ population averaged across 4 independent tissue donors. Scale bars in (D) and (E) represent 25 μm.
The genes encoding NaV1.8 and NaV1.9 show different expression levels in OPRM1-positive and OPRM1-negative nociceptors
Voltage-gated sodium channels (VGSCs) play a crucial role in nociception as they are essential for the initiation and conduction of action potentials from peripheral to central nerve terminals.54,55 The isoforms NaV1.8 (SCN10A) and NaV1.9 (SCN11A) are preferentially expressed in human nociceptive afferents.40,41,42,56 In 1,310 analyzed neurons, we found TRPV1 to be the most expressed of the four markers (87.7% ± 3.3%). More than half of the neurons expressed OPRM1 (57.8% ± 4.1%), consistent with results of earlier probe sets. SCN10A and SCN11A were also expressed by a majority of DRG neurons (83.3% ± 3.5% and 85.5% ± 3.4%, respectively) (Figure 3B). Analysis of co-expression patterns of all four markers revealed three prevalent populations (Figure 3C; Table S9), two nociceptive and a non-nociceptive population. These three populations represented 93.2% ± 1.5% of sampled neurons. A representative cell of each of the three most common neuronal populations with the cell size distribution of that population is shown in Figure 3E. The most abundant population (i) was TRPV1+OPRM1+ nociceptors that expressed transcripts for both VGSCs (53.6% ± 3.5%). This group contained a wide distribution of cell sizes consisting of mostly small- and medium-diameter neurons ( = 50 ± 13 μm), (Figure 3D). The second population (ii) consisted of OPRM1-negative TRPV1+ nociceptors that expressed both VGSCs (28.4% ± 0.9%) and showed a homogeneous cell size distribution ( = 52.5 ± 8.8 μm) as described before. VGSCs showed different expression levels between the two nociceptive populations. SCN10A (NaV1.8) was more highly expressed in the OPRM1-positive population (median intensity 5.3 arbitrary units [a.u.] versus median intensity 4.1 a.u.) (Figure 3F), while SCN11A (NaV1.9) exhibited higher expression in the OPRM1-negative population (median intensity 23.4 a.u. versus median intensity 3.8 a.u.). This population also demonstrated a higher expression of TRPV1 (median intensity 14.8 a.u. versus median intensity 7.8 a.u.). All differences were significant (Mann-Whitney U test, p < 0.001, respectively, after Bonferroni correction). The third population (iii, 11.2% ± 3.6%) expressed none of the four markers and consisted of medium-/large-diameter neurons ( = 70.4 ± 14.8 μm) (Figure 3E). Our results demonstrate that the genes encoding NaV1.8 and NaV1.9 are co-expressed in nociceptive neurons and that NaV1.8 transcripts are enriched in the OPRM1-expressing population.
Figure 3.
The genes encoding NaV1.8 and NaV1.9 show different expression levels in OPRM1-positive and OPRM1-negative nociceptors
(A) Representative section of human DRG showing neurons positive for TRPV1, the μ-opioid receptor (OPRM1), and voltage-gated sodium channels NaV1.8 (SCN10A) and NaV1.9 (SCN11A) transcripts. Representative neurons characterized further in (E) and (F) are labeled with small Roman numerals.
(B) Enlarged field outlined in (A) showing each individual transcript. Overlap of all four transcripts occurs in a substantial subpopulation (i). Lipofuscin is marked with an “L.”
(C) Percentage of 1,310 neurons expressing each individual transcript.
(D) Percentage of neurons expressing the most common transcript combinations. Bar graphs in (C) and (D) show mean, SD, and individual values from four independent donors.
(E) Multi-channel microscopy images of a representative individual neuron of each population and the population’s cell size distribution. Scale bars, 25 μm.
(F) Expression intensity of individual transcripts in OPRM1-positive (i) as compared to OPRM1-negative (ii) nociceptors. Transcript levels for TRPV1 and SCN11A were significantly higher in the OPRM1-negative population. Median and interquartile range indicated. p < 0.001, Mann-Whitney U test.
TAC1 (substance P) is selectively expressed in OPRM1-positive nociceptors
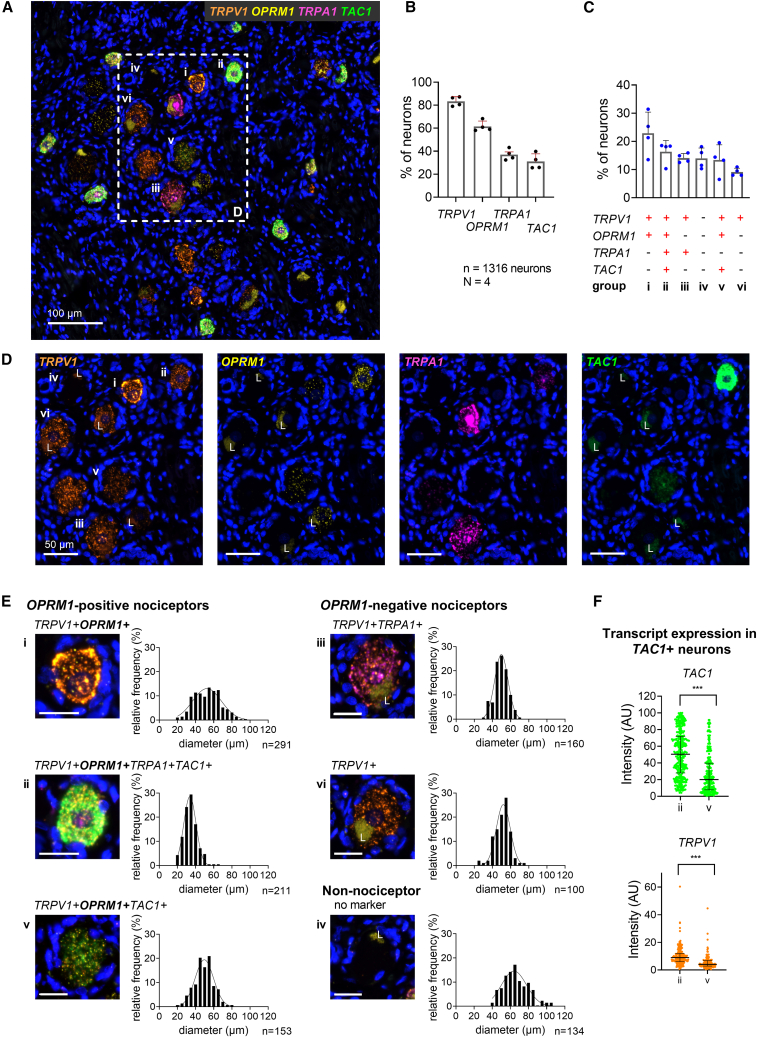
Substance P (encoded by TAC1) is a neuropeptide and a marker for peptidergic nociceptors transmitting sustained pain in rodents.33,34,57 This peptide modulates nociceptive responsiveness of second-order spinal cord neurons,58,59 especially during intense noxious stimulation60 and can include activation of both TRPV1 and TRPA1.61 To investigate the expression of these genes in human nociceptors, we analyzed DRG sections for expression levels of TRPV1, OPRM1, TRPA1, and TAC1. We analyzed 1,316 neurons and observed TRPV1 in 83.3% ± 4.1%, OPRM1 in 61.5% ± 4.6%, TRPA1 in 37.2% ± 4.8%, and TAC1 in 31.2% ± 6.7% of the analyzed neurons (Figure 4B). When we considered the co-expression patterns of all four markers, we detected six prevalent populations: three TRPV1+OPRM1+, two TRPV1+OPRM1-negative nociceptive, and one non-nociceptive population (Figure 4C; Table S10). These six populations represented 89.7% ± 2.8% of the analyzed neurons. TRPV1+OPRM1+ neurons that did not express TRPA1 nor TAC1 were the most common population (i, 22.9% ± 7.5%). They showed a broad cell size distribution of mostly small-/medium-sized neurons ( = 53.2 ± 14.5 μm). Within the TRPV1+OPRM1+ populations, two expressed TAC1: a small-diameter ( = 34.6 ± 7.6 μm) population that also expressed TRPA1 (ii, 16.3% ± 4.0%) and a small-/medium-diameter population ( = 49.4 ± 10.3 μm) that did not express TRPA1 (v, 13.4% ± 5.4%). We observed significantly higher expression levels for TAC1 and TRPV1 in the TRPV1+OPRM1+TRPA1+TAC1+ (i) population than in the TRPV1+OPRM1+TAC1+ (v) population (median intensity for TRPV1 8.9 a.u. versus 4.1 a.u., for TAC1 50.4 a.u. vs. 20 a.u., p < 0.001, Mann-Whitney U test; see Figure 4F). OPRM1-negative populations were characterized by expression of TRPV1 and TRPA1 (iii, 14.1% ± 1.6%) or only TRPV1 (vi, 9.1% ± 1.4%). These neurons were medium sized with a homogeneous cell size distribution (47.2 ± 7.8 μm [iv], = 50.7 ± 8.7 μm [vi]) as described before. Non-nociceptive neurons expressed none of the four markers (iv, 14.0% ± 3.7%) and had medium/large cell sizes ( = 66.4 ± 13.7 μm) (Figures 4C and 4E). In terms of nociception, the OPRM1+TRPV1+TRPA1+TAC1+ neurons are a subpopulation of the aforementioned analyzed TRPV1+OPRM1+SCN10A+SCN11A+ population and are likely associated with sustained tissue damage pain.
Figure 4.
TAC1 (substance P) is expressed in subpopulations of OPRM1-positive nociceptors
(A) Representative section of human DRG showing neurons expressing transcripts for TRPV1, the μ-opioid receptor (OPRM1), the chemo-sensitive receptor TRPA1, and substance P precursor (TAC1).
(B) Percentage of 1,316 neurons expressing each individual transcript.
(C) Percentage of neurons expressing the most common transcript combinations. Bar graphs in (B) and (C) show mean, SD, and individual values from four independent donors.
(D) Enlarged field shown in (A) for each individual transcript.
(E) Multi-channel microscopy images of a representative individual neuron of each population and the population’s cell size distribution. Scale bars, 25 μm. Lipofuscin is marked with an “L.”
(F) Expression intensity for TAC1 and TRPV1 in populations ii and v. The quad+ population ii shows significantly higher expression of TAC1 and TRPV1 and is polyresponsive to algesic mediators. Median and interquartile range indicated. p < 0.001, Mann-Whitney U test.
OPRM1-positive nociceptors express TRPM8
Agonists of the μ-opioid receptor are known to inhibit cold pain induced by sustained stimulation,62,63,64 implicating expression of cold-sensitive channels in OPRM1-expressing nociceptors. The transient receptor potential cation channel subfamily M (melastatin) member 8 (encoded by TRPM8) is activated by compounds such as menthol, mediates cold sensations into the noxious range, and is implicated in cold allodynia.65,66 TRPA1 has been reported to be expressed in human cold-sensing neurons,42 and we examine the colocalization of these two transcripts in this experiment. We analyzed 1,310 DRG neurons for the expression of TRPV1, OPRM1, TRPA1, and TRPM8. We detected TRPV1 in 82.3% ± 4.4%, OPRM1 in 58.3% ± 7.3%, TRPA1 in 44.2% ± 4.6%, and TRPM8 in 39.7% ± 8.0% of neurons (Figure S6B). When we considered the co-expression patterns of all four markers, six prevalent populations were detected (Figure S6C; Table S11), of which three were TRPV1+OPRM1+ nociceptive, two TRPV1+OPRM1-negative nociceptive, and one non-nociceptive population. Neurons of these six populations represented 86.9% ± 3.2% of the analyzed neurons. The three TRPV1+OPRM1+ populations consisted of neurons that also co-expressed TRPA1 and TRPM8 (i, 20.1% ± 4.9%), only TRPM8 (iii, 15.5% ± 3.3%), or neither TRPA1 nor TRPM8 (v, 12.2% ± 3.1%). The TRPV1+OPRM1+TRPA1+TRPM8+ population consisted of small-diameter neurons ( = 39.8 ± 9.2 μm), while the two latter populations showed a broad cell size distribution including mostly small- and medium-diameter neurons ( = 57.0 ± 12.7 μm [iii], = 54.9 ± 13.9 μm [v]). OPRM1-negative nociceptors were either TRPV1+TRPA1+ (ii) (16.8% ± 2.6%) or only TRPV1+ (vi, 9.8% ± 4%). These two prevalent OPRM1-negative nociceptive populations did not express TRPM8. Both groups consisted of medium-sized neurons with homogeneous cell size distributions ( = 53.2 ± 9.5 μm [ii], = 52.5 ± 9.6 μm [vi]) as described in previous paragraphs. Neurons expressing none of the four markers (iv, 12.5% ± 2.4%) were medium- to large-diameter neurons (71.3 ± 14 μm) (Figure S6D). Though we observed a high degree of co-expression of TRPV1, TRPA1, and TRPM8, pairwise analysis of linear correlations between those markers in a pooled sample of all TRPV1+/OPRM1+/TRPA1+/TRPM8+ neurons expressing these markers revealed mostly anticorrelated gene expression of TRPV1 and TRPM8 and TRPM8 and TRPA1, respectively (Figure S6E). A subset of neurons showed significant expression of TRPV1 and TRPM8, indicating potential sensitivity to both heat and cold (Figure S6E). Expression levels of TRPV1 and TRPA1 showed a more complex relationship with a subpopulation of neurons showing high expression levels for both transcripts. Our data demonstrate mostly anticorrelated expression of genes coding for heat- and cold-sensing receptors in TRPV1+OPRM1+ nociceptors, which indicates primarily distinct sensory encoding of noxious heat and cold. Our anatomic evidence supports that TRPM8 is expressed in the TRPV1+OPRM1+ population.
Expression levels of P2RX3 differ between OPRM1-positive and OPRM1-negative nociceptors
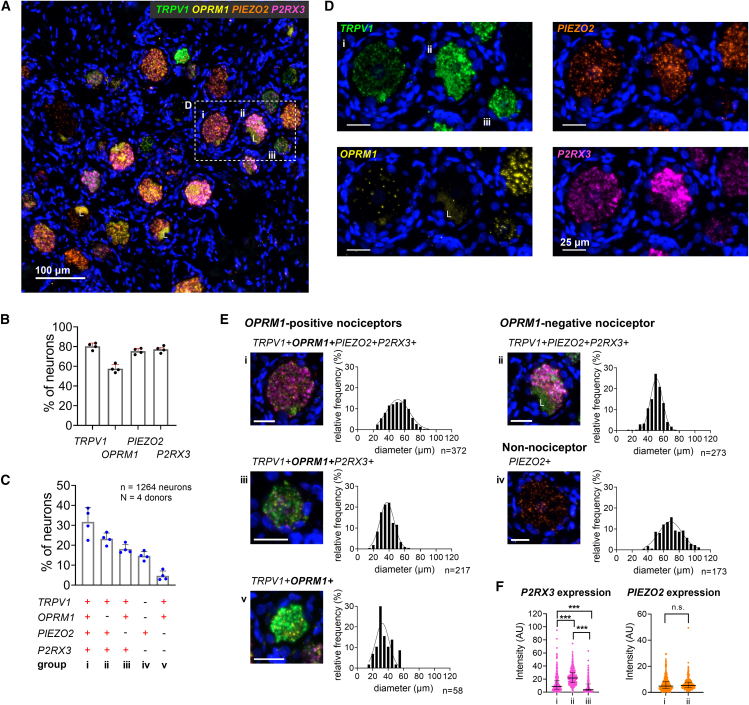
To address the polymodality of human nociceptors including mechanosensation and sensing of indicators of tissue damage such as ATP, we performed an in situ experiment including probes for transcripts of PIEZO2 and P2RX3 (encoding P2X3). PIEZO2 in the somatosensory system plays an essential role in sensing gentle touch, tactile pain, and proprioception.67,68,69 We detected transcripts for TRPV1 in 80.4% ± 3.3%, OPRM1 in 57.6% ± 4.1%, PIEZO2 in 75.4% ± 2.9%, and P2RX3 in 77.3% ± 3.3% of the analyzed neurons (n = 1,264 neurons) (Figure 5B). All molecular markers showed a high degree of co-expression. With this probe set we detected five major populations (Figure 5C; Table S12): three TRPV1+OPRM1+, one TRPV1+OPRM1-negative nociceptive, and a non-nociceptive population. Combined, these five groups represented 87.7% ± 2.7% of the analyzed neurons. Among OPRM1-positive nociceptors, TRPV1+OPRM1+PIEZO2+P2RX3+ neurons (i) were the most common (31.7% ± 7.2%) (Figure 5C), showing a broad cell size distribution ( = 51.3 ± 12.8 μm). A more homogeneous TRPV1+OPRM1+P2RX3+ population (iii, 18.1% ± 2.3%) consisted of small-diameter neurons ( = 38.1 ± 8.6 μm). TRPV1+OPRM1+ nociceptors that did not express PIEZO2 nor P2RX3 represented only a small population of small-diameter cells (v, 4.7% ± 2.4%, = 34.6 ± 9.4 μm). OPRM1-negative TRPV1+ nociceptors expressed both PIEZO2 and P2RX3 (ii) (23.3% ± 2.9%) and showed again a homogeneous cell size distribution peaking at a medium cell diameter ( = 50.1 ± 8.6 μm). In this stain, we found only a minority of cells to not express any of the markers (n = 10, Table S12); instead, we observed a non-nociceptive population of medium-/large-diameter neurons ( = 70.0 ± 14.3 μm) that expressed PIEZO2 (iv, 14.7% ± 2.2%), and presumably the proprioceptive marker SPP1 in a previously described experiment (Figure 2D), which is consistent with the role of PIEZO2 in human proprioception68 (Figure 5E). P2X3 is a purinergic ATP-sensitive receptor selectively expressed in nociceptive afferents70,71 and a marker for rodent non-peptidergic C-fibers.72,73 We noticed a differential expression across neuronal populations. Specifically, P2RX3 showed highest expression (median intensity 21.7 a.u.) in OPRM1-negative nociceptors (ii) (Figure 5F). TRPV1+OPRM1+PIEZO2+P2RX3+ nociceptors (i) showed significantly less P2RX3 expression (median intensity 8.8 a.u.), and TRPV1+OPRM1+P2RX3+ nociceptors (iii) showed the lowest P2RX3 expression level (median intensity 3.9 a.u.). All differences were significant (Mann-Whitney U test, p < 0.001, respectively, after Bonferroni correction). PIEZO2, on the other hand, did not show differences in expression levels between OPRM1-positive and OPRM1-negative nociceptors (median intensity 4.8 a.u. [i], median intensity 5.3 a.u. [ii], p = 0.08, Mann-Whitney U test) (Figure 5F). These data underscore the prevalence of polymodal nociceptors in the human DRG and the high expression of the non-peptidergic marker P2RX3 in OPRM1-negative nociceptors.
Figure 5.
Expression levels of P2RX3 differ between OPRM1-positive and OPRM1-negative nociceptors
(A) Representative section of human DRG showing neurons expressing transcripts for TRPV1, the μ-opioid receptor (OPRM1), the mechano-sensitive receptor PIEZO2, and the purinergic ATP receptor P2X3 (P2RX3).
(B) Percentage of 1,264 neurons expressing each individual transcript.
(C) Percentage of neurons expressing the most common transcript combinations. Group iv expresses PIEZO2 only and very likely represents a population of proprioceptors as shown in Figure 2Div. Bar graphs in (B) and (C) show mean, SD, and individual values from four independent donors.
(D) Enlarged field shown in (A) for each individual transcript.
(E) Multi-channel microscopy images of a representative individual neuron of each population and the corresponding population’s cell size distribution. Scale bars, 25 μm. Lipofuscin is marked with an “L.”
(F) Expression intensities of P2RX3 and PIEZO2 in nociceptive populations. Both transcripts are expressed in OPRM1-positive and -negative nociceptors. While the expression level of PIEZO2 is similar between both populations, P2RX3 shows the highest expression in OPRM1-negative nociceptors. Median and interquartile range indicated. p < 0.001, Mann-Whitney U test, after Bonferroni correction.
Expression of transcripts for neurotrophic and MRGPRD receptors differentiates OPRM1-positive and OPRM1-negative human nociceptors
By labeling for growth factor receptors, we tested the hypothesis that our results, which are indicative of a human nociceptor classification into OPRM1-positive and OPRM1-negative cells, follow the developmental principles of murine DRG neurons. These studies describe a division among nociceptors according to the expression of the neurotrophic receptors TrkA (encoded by NTRK1) for large-diameter A-fiber and peptidergic C-fiber nociceptors, and neurotrophic receptors such as GFRA2 for non-peptidergic C-fiber nociceptors.19,20,74 We analyzed 1,298 neurons and detected TRPV1 in 82.7% ± 4.5%, OPRM1 in 59.6% ± 6.5%, NTRK1 in 50.5% ± 3.5%, and GFRA2 in 30.8% ± 2.2% of neurons (Figure 6C). Classification of neurons according to the co-expression of all markers confirmed our hypothesis: we detected a prevalent TRPV1+OPRM1+NTRK1+ population (i, 41% ± 5.1%) and an OPRM1-negative TRPV1+GFRA2+ population (ii, 21.2% ± 2.6%). Only a small TRPV1+OPRM1+ population did not express NTRK1 (iii, 10% ± 2.2%) (Figure 6D; Table S13). The TRPV1+OPRM1+NTRK1+ population consisted of mostly small- and medium-diameter neurons ( = 46.6 ± 13 μm), while the TRPV1+OPRM1+NTRK1-negative population consisted mainly of small-diameter neurons ( = 42.6 ± 10.5 μm) (Figure 6E). The OPRM1-negative TRPV1+GFRA2+ population, as described for all other experiments, consisted of medium-sized neurons ( = 52.4 ± 8.6 μm). A non-nociceptive population (9.9% ± 3.8%) that did not express any of the markers of this experiment consisted of medium- to large-diameter neurons ( = 70 ± 12.7 μm). These four main populations represented 82.1% ± 5.5% of the analyzed neurons. Only a small fraction of neurons co-expressed both neurotrophic receptors (n = 61, Table S13), which confirms a basic distinction of human nociceptors into NTRK1-expressing “peptidergic” C-nociceptors associated with sustained pain in rodents and GFRA2-expressing “non-peptidergic” nociceptors. These data reinforce our observed dichotomy of the nociceptive neuronal population.
Figure 6.
Expression of transcripts for neurotrophic and MRGPRD receptors differentiates OPRM1-positive and OPRM1-negative human nociceptors
(A) Representative section of human DRG showing neurons expressing transcripts for TRPV1, the μ-opioid receptor (OPRM1), and the neurotrophic receptors TrkA (NTRK1) and GFRA2.
(B) Enlarged window as shown in (A) for each marker individually.
(C) Percentage of 1,298 neurons expressing each individual transcript.
(D) Percentage of neurons expressing the most common transcript combinations. NTRK1 and GFRA2 differentiate OPRM1-positive and -negative nociceptors. Bar graphs in (C) and (D) show mean, SD, and individual values from four independent donors.
(E) Multi-channel microscopy images of a representative individual neuron of each population and the population’s cell size distribution. Scale bars, 25 μm.
(F) Representative section of human DRG showing positive transcripts for TRPV1, the μ-opioid receptor (OPRM1), the neurotrophic receptor GFRA2, and the pruritogenic receptor MRGPRD.
(G) Percentage of neurons showing transcripts for each marker individually.
(H) Percentage of neurons expressing the most common molecular marker combinations. Bar graphs in (G) and (H) show mean, SD, and individual values from each donor.
(I) Multi-channel microscopy images of a representative individual neuron of populations (i) and (ii) and the corresponding cell size distributions. Scale bars, 25 μm.
(J) Individual transcripts of representative neurons shown in (I). Lipofuscin is marked with an “L.”Most OPRM1-positive nociceptors are characterized by expression of NTRK1 (TrkA), while OPRM1-negative nociceptors express transcripts for the neurotrophic receptor GFRA2 and mostly the itch-related receptor MRGPRD, suggesting distinct populations of OPRM1+ “peptidergic” and OPRM1− “non-peptidergic” neurons.
The largest group within the murine non-peptidergic GFRA2+ population consists of nociceptors that express the itch-related receptor MRGPRD.21 In rodents these fibers do not innervate deep tissues but do terminate selectively in the most superficial skin layers.32 Since the human OPRM1-negative population observed in our experiments shares many molecular features with murine non-peptidergic neurons such as high expression levels for P2RX3 and SCN11A,72,73,75,76,77 we hypothesized a human analog to the proposed skin threat detector molecularly defined by co-expression of GFRA2 and MRGPRD.35 In this experiment we found TRPV1 expressed in 86.7% ± 6.5%, OPRM1 in 61.3% ± 6.1%, GFRA2 in 35.8% ± 7.7%, and MRGPRD in 22.6% ± 7.2% of neurons (n = 1,271 neurons) (Figure 6G). We observed a division into two main nociceptive populations: a large TRPV1+OPRM1+ population (i, 53.1% ± 5.8%), encompassing a wide range of mostly small- to medium-sized neurons ( = 47.1 ± 13 μm), and an OPRM1-negative TRPV1+ population co-expressing GFRA2 and MRGPRD (ii, 18.3% ± 7.4%) that consisted of medium-sized neurons with a homogeneous cell size distribution ( = 53.9 ± 6.1 μm) (Figures 6H and 6I; Table S14). Of all OPRM1-negative neurons co-expressing TRPV1 and GFRA2, 74.5% ± 17.2% % also expressed MRGPRD. To summarize, our data support the hypothesis of a human “non-peptidergic” population expressing MRGPRD and define further the molecular distinction between OPRM1-positive and OPRM1-negative nociceptive populations that co-exist in the human DRG.
Discussion
The present study investigates human somatosensory afferent neuronal populations relevant to nociception and opioid analgesia. Based on multiplex combinatorial in situ hybridization experiments, we were able to detect and define two main populations of C-nociceptors. The discriminator between these populations is the expression or lack of expression of OPRM1. They are further delineated by the expression of growth factor receptor genes, which follows the development of murine C-nociceptors. The first population expresses OPRM1 and the gene coding for the nociresponsive channel TRPV1 and shares molecular attributes of murine peptidergic C-nociceptors mediating sustained pain. The second population expresses TRPV1 and other algogenic receptors but not OPRM1. These neurons resemble murine non-peptidergic C-nociceptors. Our observations support the hypothesis of a human “tissue damage” nociceptor that is responsive to clinically used opioids and would be most relevant to analgesic drug development. Multiple experimental opioid administration studies, plus decades of experience with intrathecally administered opioids in human patients, indicate that the first, “peptidergic” population is critical for transmitting clinically relevant nociceptive pain and that this transmission can be controlled by opioids (Table S6). The second, “non-peptidergic” population comprises mainly MRGPRD-positive neurons that are hypothesized and has been shown in mice to terminate superficially in the epidermis and act as a “threat detector.”32 This population does not express OPRM1 and therefore is unlikely to be responsive to opioids. Importantly, MRGPRD+ neurons contribute to pathological pain states including neuropathic pain in rodents.38,39 In humans, neuropathic pain is less responsive to intrathecal opioids than nociceptive pain (Table S6) and less manageable with systemic opioids,43,45 which supports our transcriptionally based findings.
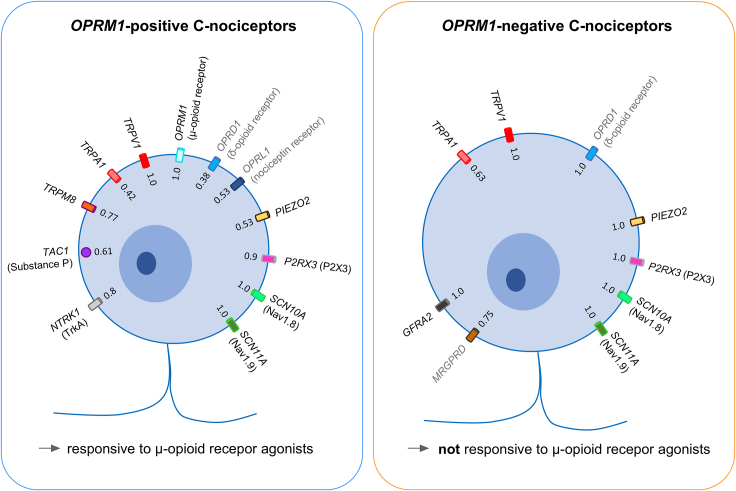
OPRM1-positive nociceptors consist of a heterogeneous group of mostly multimodal neurons expressing markers for cold sensation (TRPM8), chemical sense (TRPA1), inflammation and tissue damage (P2RX3), mechanosensation (PIEZO2), neuropeptides (TAC1), and opioid receptors other than the μ-opioid receptor (OPRD1, OPRL1). OPRM1-negative nociceptors are multimodal neurons expressing transcripts for TRPV1 and PIEZO2, as well as the neurotrophic receptor GFRA2, the itch-related receptor MRGPRD, and the δ-opioid receptor (OPRD1), as well as high expression levels of transcripts for P2X3 and NaV1.9 (Figure 7). An additional finding in this study is that the κ-opioid receptor in humans is expressed in non-neuronal SGCs.
Figure 7.
Expression of transcripts for ion channels, neuropeptide, and receptors in OPRM1-positive and OPRM1-negative C-nociceptors
Transcripts expressed in OPRM1-positive (left) and OPRM1-negative (right) C-nociceptors. Numbers indicate fraction of nociceptors of main populations that do express the individual transcript. Receptors/transcripts in gray indicate genes with low expression levels in the two populations as determined by in situ hybridization. OPRM1-positive nociceptors (left) are highly polymodal and likely consist of several subpopulations. In these neurons the μ-opioid receptor is the main opioid receptor with little contribution from δ-opioid or nociception receptors and nearly no contribution from the κ-opioid receptor (which we show in humans is expressed in satellite glial cells, see Figure 1F). OPRM1-negative neurons are polymodal and typically express TRPV1 and PIEZO2, indicating potential responsiveness to thermal and mechanical stimulation. Most of them express the murine superficial skin marker MRGPRD. In this population the only opioid receptor is the δ-opioid receptor, which is expressed in low levels.
In situ hybridization is a high-fidelity technique that allows for precise identification and localization of gene transcripts expressed in somatosensory neuronal perikarya over a range of expression levels and captures genes with low level transcription, such as opioid receptors and MRGPRD. An unambiguous assignment of these transcripts to human nociceptive populations could not be achieved by sequencing methods due to reasons of sensitivity40,41,42 or spatial resolution.41 Though our results confirm basic organizational principles of human nociceptive afferents of these studies, they formulate some significant differences (Figures S8–S16).
The feasibility of alternative opioid receptors as targets to relieve sustained tissue damage pain
Preclinical data suggest that all opioid receptors including the nociceptin receptor regulate transmission of nociceptive input into the spinal cord,78,79,80 making them potential pharmacological targets for peripheral pain control. Additionally, such efforts were aimed at avoiding adverse side effects of μ-opioid receptor agonists. These considerations generated ongoing efforts to develop agonists to opioid receptors other than the μ-opioid receptor.81,82,83,84 Subsequently, the peripheral κ-opioid agonist difelikefalin was approved for itch, but to date positive results in advanced clinical trials have not been forthcoming for pain (Table S15). Our current results provide a molecular-biological explanation for failures of past efforts and a pathway for future endeavors. The critical parameters are adequate expression of the gene in the correct cell population and that this population is represented by a sufficient number of cells to have a pharmacological impact. Sustained tissue damage pain involves a broad population of nociceptors that support complex transduction mechanisms.85,86 From this frame of reference, OPRD1 and OPRL1 show low amounts of transcript in about half of neurons relevant for analgesia, which implies that peripheral agonist monotherapy would have a marginal analgesic effect and would require a combinatorial approach to fully inhibit relevant primary afferent populations. The fraction of human DRG neurons expressing transcripts for opioid receptors approximately matched previous in situ hybridization (ISH) studies for OPRM1,87 and previous functional studies in human DRG neurons for the μ-opioid (MOR), δ-opioid (DOR), and nociception receptor (NOR) proteins.50 This was different for the κ-opioid receptor (KOR, encoded by OPRK1), which we detected ubiquitously in SGCs and only marginally in neurons. In contrast to this finding, functional studies implied neuronal KOR expression. Though their signal could have been influenced by satellite cell κ-opioid receptors, this discrepancy cannot be resolved without further investigation.49,50 Our data suggest that the potential contribution of KOR to modification of nociception cannot be directly mediated by afferent neurons. To summarize, the low expression levels and small fractions of relevant nociceptors expressing DOR or NOR, plus non-neuronal expression of KOR, make these receptors unlikely candidates for successful peripheral analgesic monotherapy in the context of sustained tissue damage pain.
Nociceptor-selective VGSCs and analgesic efficacy
Our experiments confirm the preferential expression of transcripts for NaV1.8 (SCN10A) and NaV1.9 (SCN11A) in human nociceptors.40,41,42,56 NaV1.8 has gained attention as a most likely source for sustained firing related to tissue injury,88 and conditional knockout of genes on NaV1.8-positive nociceptors has become a surrogate for nociceptor-specific gene modification.89,90 Additionally, interest in these channels comes from human mutations leading to insensitivity to pain.91,92,93 NaV1.8 inhibitors are being currently pursued as analgesics, with VX-548 having entered phase 3 clinical trials for post-surgical pain and painful diabetic neuropathy.94 We detected significantly higher amounts of SCN10A (NaV1.8) in OPRM1-positive than in OPRM1-negative nociceptors. The other channel included in our studies, NaV1.9, is a threshold channel that provides a “window current” which contributes to action potential initiation in response to subthreshold stimuli.95,96,97 The most evident difference in expression among the two sodium channel transcripts was the high expression of SCN11A (NaV1.9) in the OPRM1-negative population, consistent with rodent non-peptidergic C-fibers,75,76,77 and human transcriptomic studies.41,42 A high level of excitability, potentially driven by high NaV1.9 expression in OPRM1-negative nociceptors, supports their hypothesized role as threat detectors and may support altered excitability in pathological states, such as neuropathic pain.37,38 The development of selective NaV1.9 antagonists is at its beginnings98 but seems to be an attractive avenue in controlling pain which is known to be poorly responsive to opioids, such as neuropathic pain (Table S6).44,45
Substance P precursor (TAC1) expression in OPRM1-positive nociceptors and implications for analgesic efficacy
The neuropeptides CGRP and substance P are synthesized by DRG neurons and are modulators of nociceptive transmission at the afferent synapse in the spinal cord.58,59 These neuropeptides also represent molecular markers that identify murine peptidergic C-nociceptors.21,33,34,99,100 In human DRG neurons, CGRP is widely expressed,56 while substance P (encoded by TAC1) displays a more restricted profile in a subpopulation of small-diameter DRG neurons.101,102 Substance P is released during sustained noxious stimulation.60 In line with our hypothesis of a C-nociceptor population that mediates sustained pain and is responsive to μ-opioid receptor agonists, TAC1 expression was selectively detected in two subpopulations of OPRM1-positive nociceptors. One population is of particular interest due to its high expression of TAC1 and co-expression with TRPA1. The presence of TRPA1 in these cells is important because this channel responds to inflammatory conditions, tissue injury, and a wide spectrum of noxious chemicals,103,104 further reinforcing the suggested role of this subpopulation in the transmission of tissue damage pain. Distinguishing the combinatorial expression of nociresponsive genes within distinct cell populations provides key information for evaluating peripheral analgesic strategies and their potential performance in various clinical pain indications. In this regard, nociceptive input of TAC1-expressing neurons is likely sufficient to cause pain; however, blocking transmission from only this population is apparently not sufficient to achieve effective analgesia.105,106 Our data show the presence of an additional population that provides insight into the underlying translational problem. This population (i.e., TAC1-negative, TRPV1+OPRM1+, Figure 4E) is large and highly nociresponsive but transmits nociceptive information in a substance P-independent fashion. The lack of analgesic efficacy of substance P receptor antagonists is consistent with our formulation of incomplete blockade of nociceptive transmission.105,106
Transduction of hot and cold thermosensation
Electrophysiological studies classify most cold-sensitive neurons as C-fiber neurons.107,108 Accordingly, we detected the gene encoding the cold-responsive channel TRPM8 mainly in small-diameter nociceptors, and specifically in OPRM1-positive nociceptors, consistent with human experimental pain studies demonstrating the effect of μ-opioid receptor agonists on sustained noxious cold stimulation.13,63,64 OPRM1-negative C-nociceptors did not express this transcript. Human DRG neurons have been molecularly and electrophysiologically grouped into mostly distinct cold- or heat-sensitive populations.40,41,42,109 We detected a high degree of co-expression between transcripts for TRPV1 and TRPM8. Further analysis revealed that expression levels of transcripts for these two receptors are mostly anticorrelated, as has been shown for rat DRG neurons.47 We also observed a fraction of cells that show moderate/high expression levels of both TRPV1 and TRPM8, implying that they can be activated by both heat and cold stimuli. This is supported by microelectrode recordings in humans that identified heat-cold units with an average heat activation threshold typical for TRPV1.110
The hypothesized cutaneous threat detector
A combinatorial evaluation of all experiments demonstrates two major C-nociceptive populations: the first is a heterogeneous TRPV1+OPRM1+ polymodal population. This population exists alongside a relatively homogeneous TRPV1+OPRM1− population that expressed GFRA2, MRGPRD, and high levels of both SCN11A (NaV1.9) and P2RX3 (P2X3). Based on the molecular profile of the TRPV1+OPRM1− population, we hypothesize a role in first-line cutaneous threat detection. Expression of MRGPRD in non-peptidergic rodent neurons marks nociceptors that exclusively innervate the superficial epidermis.32 In humans, the specific topographical peripheral termination of these neurons is unknown, but experimental pain studies using intradermal injection of the MRGPRD-receptor agonist β-alanine, which causes itch and burning pain, indicate peripheral nerve endings in the skin.111 In contrast to OPRM1-positive nociceptors, TRPV1+OPRM1− nociceptors consistently express PIEZO2, implying responsivity to heat and mechanical stimuli. Many human mechano-heat polymodal skin C-nociceptors start responding early in the stimulus-response function to both heat and mechanical stimulation,112 often with a rapid brief response even to sustained noxious stimulation.113,114 This brief neuronal response triggers withdrawal and escape behaviors that terminate the stimulus suggesting that this population is likely the major population for responding to brief painful stimulation. By contrast, sustained stimulation evokes activity of a second, slow-onset C-population in primates.115 The lack of effect of μ-opioid agonists on threshold-level “sudden and fleeting” skin stimulation is consistent with the absence of OPRM1 in the population we hypothesize to be a threat detector.1,116 Recent studies revealed a role of ATP released from keratinocytes117,118 in response to mechanical stimulation that excites peripheral nociceptive terminals.119 Thus, this purine release stimulus may be quite superficial. The high expression of the ATP-sensing receptor P2X3 in the TRPV1+OPRM1− population is consistent with our hypothesis that this population represents multimodal skin threat detectors.
Implications for analgesic drug development
Beyond providing a combinatorial picture of nociceptive processes, the present dataset leads to several incisive formulations for advancing developmental efforts for new analgesic agents. Our objective is to provide a constructive critique and a framework for progress to determine candidate targets that exhibit more translational potential than others. Additionally, the results highlight the need to query human DRG or spinal cord early in the drug development process to better place animal studies into a stronger translational framework. The present study delineates the most relevant DRG neuron for human clinical analgesia, which we term the tissue damage nociceptor. In particular, peripherally acting analgesics should be directed at these critical cells. Another consideration that can affect peripherally acting analgesics is redundancy. These neurons contain multiple transducers of algesic stimuli,120,121 and antagonism of a single channel is unlikely to result in significant block of nociceptive transmission. Indeed, redundancy was one of the major factors undermining the analgesic actions of TRPV1 antagonists despite clear evidence of target engagement.122 Going forward, it may be a challenge to identify a simplified, single-molecule approach to fully effective peripheral analgesia that provides safety and specificity. However, the approaches outlined provide a template for first-stage evaluation.
Limitations of the study
A limitation of the current interpretation is that we rely on mRNA message to predict functional or pharmacological activity. This implies a correspondence between mRNA and functional receptor protein. In the present study, we have not performed electrophysiological studies in primary cultures of human DRG to elicit responses to TRPV1 stimulation that are differentially responsive to opioids. Such a study implies that opioids would differentially modulate TRPV1 responses in DRG neurons, although opioids have been demonstrated to modulate depolarization elicited by KCl in neurons in mouse in DRG primary cultures.50 In considering communication with post-synaptic spinal cord neurons, we have not conducted recordings of Ca imaging in co-cultures of human DRG and spinal cord to ascertain whether differential actions can be measured on post-synaptic neurons. Such experiments present technical difficulties with respect to sourcing of viable human tissue.123 Nonetheless, the conclusion of distinct functions of the two main OPRM1+ and OPRM1− populations is supported by human clinical trials of opioids in tissue damage and neuropathic pain conditions (see Table S6). Additional functional pharmacologic evidence potentially with in vivo microneurography124,125 or imaging of spinal cord126,127,128 may further validate these predictions.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael Iadarola (michael.iadarola@nih.gov).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this study will be shared by the lead contact upon request. This includes multiplex fluorescence microscopic images and region-of-interest files. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Acknowledgments
We thank Allison Manalo for support with data acquisition and Paige Boyland for support with data analysis. We thank Dr. Misha Bačkonja, Dr. Temugin Berta, and Dr. Wendy Smith for input on the study. This study was supported by the Intramural Research Program of the National Institutes of Health Clinical Center (ZIA CL090033-09 and ZIA CL090033-11 to A.J.M.), and of the National Institute of Neurological Disorders and Stroke (to D.M.). Supplementary funding was provided by the Office of Behavioral and Social Sciences Research, and from a Bench to Bedside grant from the NIH.
Author contributions
Conceptualization, E.S.S. and M.J.I.; methodology, M.R.S., D.M., and M.J.I.; investigation, E.S.S. and D.M.K.; formal analysis, E.S.S.; visualization, E.S.S.; resources, A.J.M., D.M., and A.G.; writing – original draft, E.S.; writing – review and editing, E.S.S., M.R.S., and M.J.I.; funding acquisition, A.J.M.; supervision, D.M., A.J.M., and M.J.I.
Declaration of interests
A.G. is an employee and shareholder of AnaBios Corp.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human DRG Lumbar 3 | AnaBios | 210218DHA |
| Human DRG Lumbar 3 | AnaBios | 210221DHA |
| Human DRG Lumbar 3 | AnaBios | 210325DHA |
| Human DRG Lumbar 3 | AnaBios | 210405DHA |
| Critical commercial assays | ||
| RNAscope® 4-Plex Ancillary kit | Advanced Cell Diagnostics | Cat#323120 |
| RNAscope® Wash Buffer Reagents | Advanced Cell Diagnostics | Cat#310091 |
| RNAscope® Probe Diluent | Advanced Cell Diagnostics | Cat#300041 |
| RNAscope® H2O2 and Protease Reagents | Advanced Cell Diagnostics | Cat#322381 |
| RNAscope® Target Retrieval Reagents | Advanced Cell Diagnostics | Cat#322000 |
| RNAscope® Multiplex Fluorescent Detection Reagents V2 | Advanced Cell Diagnostics | Cat#323110 |
| RNAscope® Multiplex TSA Buffer | Advanced Cell Diagnostics | CAT#322810 |
| Opal 520 Reagent Pack | Akoya Biosciences | SKU: FP1487001KT |
| Opal 570 Reagent Pack | Akoya Biosciences | SKU: FP1488001KT |
| Opal 620 Reagent Pack | Akoya Biosciences | SKU: FP1495001KT |
| Opal 690 Reagent Pack | Akoya Biosciences | SKU: FP1497001KT |
| Oligonucleotides | ||
| RNAscope™ Probe- Hs-GFRA2 (GDNF Family Receptor Alpha 2) | Advanced Cell Diagnostics | Cat#463011 |
| RNAscope™ Probe- Hs-MRGPRD (MAS Related GPR Family Member D) | Advanced Cell Diagnostics | Cat#524871 |
| RNAscope™ Probe- Hs-NTRK1 (Neurotrophic Receptor Tyrosine Kinase 1) | Advanced Cell Diagnostics | Cat#402631 |
| RNAscope™ Probe- Hs-OPRD1 (Opioid Receptor Delta 1) | Advanced Cell Diagnostics | Cat#536061 |
| RNAscope™ Probe- Hs-OPRK1 (Opioid Receptor Kappa 1) | Advanced Cell Diagnostics | Cat#1148211 |
| RNAscope™ Probe- Hs-OPRK1-O1 (Opioid Receptor Kappa 1) | Advanced Cell Diagnostics | Custom made (13 ZZ targeting 1276–2137 bp of NM_000912.5) |
| RNAscope™ Probe- Hs-OPRL1 (Opioid Related Nociceptin Receptor 1) | Advanced Cell Diagnostics | Cat#536071 |
| RNAscope™ Probe- Hs-OPRM1 (Opioid Receptor Mu 1) | Advanced Cell Diagnostics | Cat#410681 |
| RNAscope™ Probe- Hs-PIEZO2 (Piezo Type Mechanosensitive Ion Channel Component 2) | Advanced Cell Diagnostics | Cat#449951 |
| RNAscope™ Probe- Hs-P2RX3 (Purinergic Receptor P2X3) | Advanced Cell Diagnostics | Cat#406301 |
| RNAscope™ Probe- Hs-SCN10A (Sodium Voltage-gated Channel Alpha Subunit 10) | Advanced Cell Diagnostics | Cat#406291 |
| RNAscope™ Probe- Hs-SCN11A (Sodium Voltage-gated Channel Alpha Subunit 11) | Advanced Cell Diagnostics | Cat#404791 |
| RNAscope™ Probe- Hs-SPP1 (Secreted Phosphoprotein 1) | Advanced Cell Diagnostics | Cat#420101 |
| RNAscope™ Probe- Hs-TAC1 (Tachykinin Precursor 1) | Advanced Cell Diagnostics | Cat#310711 |
| RNAscope™ Probe- Hs-TRPA1 (Transient Receptor Potential Cation Channel Subfamily A Member 1) | Advanced Cell Diagnostics | Cat#503741 |
| RNAscope™ Probe- Hs-TRPM8 (Transient Receptor Potential Cation Channel Subfamily M Member 8) | Advanced Cell Diagnostics | Cat#543121 |
| RNAscope™ Probe- Hs-TRPV1 (Transient Receptor Potential Cation Channel Subfamily V Member 1) | Advanced Cell Diagnostics | Cat#415381 |
| Software and algorithms | ||
| Photoshop | Adobe | V25.0.0 |
| Fiji | ImageJ | 14.0/1.54f |
| Prism9 | Graphpad | V9.4.1./9.5.1 |
| Other | ||
| Axio Imager.Z2 microscope | Zeiss | N/A |
Experimental model and study participant details
Patients and ethics statements
Dorsal root ganglia (DRGs) were obtained from organ donors by AnaBios Corporation (San Diego, CA) in partnership with US organ procurement organizations. Legal consent for tissue retrieval and use of that tissue for research in a commercial setting according to US laws and regulations was warranted. The distribution of donor medical information complied with HIPAA regulations regarding donor privacy. All transfers of donor organs to AnaBios are fully traceable and periodically reviewed by US Federal authorities. Upon arriving at AnaBios, each set of DRGs was assigned a unique identifier number that was reproduced on all relevant medical history files, data entry forms, and electronic records. We received only anonymized and coded donor tissue and demographic information with no way to link back to original identifiers. This study did not meet the regulatory definition of human subjects research at NIH and hence did not require IRB approval.
L3 lumbar DRGs from four tissue donors (2 Females, 2 Males, gender as provided by AnaBios Corporation, mean age 22.5 ± 3.1 years, all Caucasian) were used for all analyses in the study. None of the donors suffered from a chronic pain condition or had indications of peripheral nerve damage. Detailed demographic information, cause of death, and tissue retrieval times are available in Table S16.
Due to the small sample size and differences in the causes of death between Female and Male donors, the influence of gender on the results of this study was not systematically assessed (but see Figure S3 for comparison of percentages of TRPV1+OPRM1+ nociceptors between Females and Males). This is a limitation to our research’s generalizability.
Method details
Patients and dorsal root ganglia samples
Human dorsal root ganglia (DRG) were collected from four tissue donors and provided by AnaBios Corporation (San Diego, CA). At the time of tissue harvest, DRGs were flash frozen and stored at −80°C until processing. Immersion fixation was performed by submerging whole DRGs in room temperature 10% neutral buffered formalin, and then refrigerated for 16–24 h for fixation before embedding in paraffin blocks at Histoserv, Inc. (Germantown, MD) and sectioning at 6 μm. For each in situ hybridization experiment, we used one section per individual donor DRG and included all four sections in the analysis.
Fluorescent multiplex in situ hybridization and microscopic imaging
We performed 4-Plex fluorescent RNA in situ hybridizations using the RNAScope Multiplex Fluorescent V2 Assay (Advanced Cell Diagnostics, Newark, CA) following the manufacturer’s instructions for formalin-fixed paraffin-embedded tissue. Target retrieval was performed for 20 min at 100°C. The catalog numbers of the probes used in these experiments are listed in Table S17. After hybridization, slides were imaged using an Axio Imager.Z2 scanning fluorescence microscope (Zeiss, Oberkochen, Germany) as described previously. Filter sets (Semrock, Rochester NY) for detecting DAPI, Opal520, Opal570, Opal620, and Opal690 fluorescent dyes (Opal Reagent Pack; Akoya Biosciences, Marlborough MA) were custom furnished as described previously47,129,130 (Table S18).
Due to the unexpected staining results for OPRK1, a second in situ probe was designed to validate the results. In particular, the original probe was designed against the 3′ end of the transcript. In our redesign, we selected a non-overlapping region 5′ to the original location (base pairs 1276–2137 of NM_000912.5).
Quantification and statistical analysis
Visualization of merged composite images were constructed in Photoshop (v25.0.0, Adobe, San Jose, CA) and Fiji (ImageJ2.14.0/1.54f) in order to analyze the co-expression of transcripts. Cells were identified using a combination of DAPI-labeling of nuclear DNA and differential interference contrast (DIC) imaging. For quantification, cells were counted manually from one section per human tissue donor. In order to capture a representative subset of neurons, multiple windows (1 mm × 1 mm) located in different areas of the DRG were sampled to reach a minimum of 300 neurons per section (range 309–349, mean 323.2 ± 13). Lipofuscin autofluorescence was apparent in the 488 nm, 546 nm, and 594 nm channels, and was excluded from our analyses. This autofluorescence was identified by its simultaneous emission in multiple channels, including the 430 nm channel, which was included to capture autofluorescence. In the representative images, lipofuscin is marked with a capital “L” to distinguish it from real signal. We used the following inclusion criteria for neurons in the quantification. Neurons used for quantification were intact, and in cases where there was substantial lipofuscin, this tissue artifact occupied less than 50% of the cytoplasm. Cells were considered positive for expression of a molecular marker if they showed at least three cytoplasmic puncta. We estimated that three puncta per neuron would be a reasonable threshold to determine whether a neuron actively transcribes the gene of interest. Other groups use a threshold of four puncta,49 but we found that we would miss out on some very small diameter cells (20–30 μm diameter) with low expression levels of some nociceptive markers. In general, neurons with an expression level of 3–5 puncta (mRNA) represented a small percentage of total quantified neurons (see Figure S1), and a change of threshold would have little effect on quantitative measures and qualitative results. In addition, the existence of non-specific signal in DRG tissue sections of all donors was excluded by performing in situ hybridization with negative control probes. Our inclusion criteria of three puncta was definitely above background which allowed to be more inclusive. TRPV1 was usually co-expressed with typical markers for nociception, such as P2RX3, or with the analgesic marker OPRM1 (see Tables S7–S14), which confirms its predominant expression in nociceptive neurons. Given that the sum of all TRPV1+OPRM1+ and TRPV1+OPRM1-neurons for each ISH experiment generally matched known percentages of nociceptive neurons in human56 and mouse19 dorsal root ganglia (Figures 1, 2, 3, 4, 5, and 6), we feel confident that we chose an adequate threshold for positive gene expression.
For each mRNA target and each donor, we determined the percentage of neurons positive for a molecular marker by assessing each neuron as positive or negative for the four mRNAs assessed in each 4-plex combination. This co-expression pattern was used to establish neuronal populations. Each individual (human donor) was assessed for differences before pooling, although no individuals showed notable unique differences in expression patterns. Complete counts of neuronal populations for each experiment can be found in Tables S7–S14. For each of the prevalent neuronal populations (>9% of all counted neurons) we analyzed cell size alongside expression levels of transcripts. We focused on populations comprising 9% or more as this analysis is prone to identifying multiple small subpopulations, and the less prevalent populations can be less reproducible or less biologically relevant.47 One exception to this general rule was that we did characterize some TRPV1+OPRM1+ subpopulations below 9% prevalence as this was a major focus of the study. For cell size analysis, we included only cells that were sectioned through the center of the perikarya to achieve a more accurate circumference.47,56 For calculation of cell size the neuronal cell borders were drawn based on the merged composite of all of the fluorescence channels and DIC using the Fiji freehand selection tool. The neuronal diameter was extrapolated from the area of the drawn region of interest (ROIsize) using the formula for the diameter of a circle (diameter = 2√ (area/π)). Based on existing human DRG literature and our results regarding the cell diameter distribution of TRPV1+GFRA2+MRGPRD+ nociceptors, which represent a molecularly defined C-fiber population21 (see Figure S7), neurons with a diameter smaller than 50 μm were considered small-diameter neurons, and those with a diameter larger than 65 μm were considered as large-diameter cells that likely represent myelinated A-fibers (Figure 7).131,132 For quantification of signal intensity inside individual DRG neurons, ROIs were drawn in the same manner as for ROIsize, but were altered to exclude areas of artifactual autofluorescence, such as that from lipofuscin. This prevented accidental quantification of artifactual signal. We measured the mean gray scale of unmanipulated signal using Fiji (ImageJ2.14.0/1.54f). Due to the TSA amplification, mRNA marked by fluorophore dye visible as puncta can vary in brightness. We found that the mean gray scale as provided by Fiji correlated well with the number of puncta, even when bright and dim puncta were included in the counts (Figure S1). For quantitative graphs, each channel was checked visually for non-specific, “bleed” signal coming from neighboring channels. Signal bleed was detected in some neurons (n = 10) from TRPV1 (488 nm) to OPRD1 (546 nm) in experiment 1 (Figure 1). We corrected for this by subtracting the signal intensity of the 488 nm channel of a region of interest capturing isolated background signal (ROIbleed) from the 546 nm channel in that individual neuron (Figure S2). In order to compare signal intensities of different target genes (and/or detection channels), we determined threshold values for low, moderate, and high expression levels (see Figures 1I and 2E). High expression levels were defined as values larger than 3 standard deviations of the sample mean. For the distinction of low and moderate expression levels we found that a visually based determination of a threshold value was most reliable (manual scoring). Statistical testing was conducted using Prism GraphPad (Version 9.4.1. and 9.5.1.). Representative images were adjusted for brightness and contrast for visibility. Bar graphs in all figures show percentages of neurons expressing individual transcripts or combinations of transcripts for each human subject (mean ± standard deviation) (N = Human subjects; n = cells).
Published: October 15, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101788.
Contributor Information
Ellen S. Staedtler, Email: ellen.staedtler@nih.gov.
Michael J. Iadarola, Email: michael.iadarola@nih.gov.
Supplemental information
References
- 1.Beecher H.K. Pain: one mystery solved. Science. 1966;151:840–841. doi: 10.1126/science.151.3712.840. [DOI] [PubMed] [Google Scholar]
- 2.Stein C., Zöllner C. Opioids and sensory nerves. Sensory Nerves. 2009:495–518. doi: 10.1007/978-3-540-79090-7_14. [DOI] [PubMed] [Google Scholar]
- 3.Inturrisi C.E. Clinical Pharmacology of Opioids for Pain. Clin. J. Pain. 2002;18:S3–S13. doi: 10.1097/00002508-200207001-00002. [DOI] [PubMed] [Google Scholar]
- 4.Eddy N.B., May E.L. The search for a better analgesic. Science. 1973;181:407–414. doi: 10.1126/science.181.4098.407. [DOI] [PubMed] [Google Scholar]
- 5.Benyamin R., Trescot A.M., Datta S., Buenaventura R., Adlaka R., Sehgal N., Glaser S.E., Vallejo R. Opioid complications and side effects. Pain Physician. 2008;11:S105–S120. [PubMed] [Google Scholar]
- 6.Sang C.N., Schmidt W.K. Aligning New Approaches to Accelerate the Development of Non-opioid Analgesic Therapies. Neurotherapeutics. 2020;17:765–769. doi: 10.1007/s13311-020-00935-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Che T., Roth B.L. Molecular basis of opioid receptor signaling. Cell. 2023;186:5203–5219. doi: 10.1016/j.cell.2023.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carstens E., Tulloch I., Zieglgänsberger W., Zimmermann M. Presynaptic excitability changes induced by morphine in single cutaneous afferent C- and A-fibers. Pflugers Arch. 1979;379:143–147. doi: 10.1007/bf00586940. [DOI] [PubMed] [Google Scholar]
- 9.Woolf C.J., Fitzgerald M. Do opioid peptides mediate a presynaptic control of C-fibre transmission in the rat spinal cord? Neurosci. Lett. 1982;29:67–72. doi: 10.1016/0304-3940(82)90366-4. [DOI] [PubMed] [Google Scholar]
- 10.Barpujari A., Ford N., He S.Q., Huang Q., Gaveriaux-Ruff C., Dong X., Guan Y., Raja S. Role of peripheral sensory neuron mu-opioid receptors in nociceptive, inflammatory, and neuropathic pain. Reg. Anesth. Pain Med. 2020;45:907–916. doi: 10.1136/rapm-2020-101779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fillingim R.B., Ness T.J., Glover T.L., Campbell C.M., Hastie B.A., Price D.D., Staud R. Morphine responses and experimental pain: sex differences in side effects and cardiovascular responses but not analgesia. J. Pain. 2005;6:116–124. doi: 10.1016/j.jpain.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Arendt-Nielsen L., Olesen A.E., Staahl C., Menzaghi F., Kell S., Wong G.Y., Drewes A.M. Analgesic efficacy of peripheral kappa-opioid receptor agonist CR665 compared to oxycodone in a multi-modal, multi-tissue experimental human pain model: selective effect on visceral pain. Anesthesiology. 2009;111:616–624. doi: 10.1097/ALN.0b013e3181af6356. [DOI] [PubMed] [Google Scholar]
- 13.Andresen T., Upton R.N., Foster D.J.R., Christrup L.L., Arendt-Nielsen L., Drewes A.M. Pharmacokinetic/pharmacodynamic relationships of transdermal buprenorphine and fentanyl in experimental human pain models. Basic Clin. Pharmacol. Toxicol. 2011;108:274–284. doi: 10.1111/j.1742-7843.2010.00649.x. [DOI] [PubMed] [Google Scholar]
- 14.Olesen A.E., Brock C., Sverrisdóttir E., Larsen I.M., Drewes A.M. Sensitivity of quantitative sensory models to morphine analgesia in humans. J. Pain Res. 2014;7:717–726. doi: 10.2147/jpr.S73044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop G.H., Landau W.M., Jones M.H. Evidence for a double peripheral pathway for pain. Science. 1958;128:712–714. doi: 10.1126/science.128.3326.712. [DOI] [PubMed] [Google Scholar]
- 16.Collins W.R., Jr., Nulsen F.E., Randt C.T. Relation of peripheral nerve fiber size and sensation in man. Arch. Neurol. 1960;3:381–385. doi: 10.1001/archneur.1960.00450040031003. [DOI] [PubMed] [Google Scholar]
- 17.Li J.-L., Ding Y.-Q., Li Y.-Q., Li J.-S., Nomura S., Kaneko T., Mizuno N. Immunocytochemical localization of μ-opioid receptor in primary afferent neurons containing substance P or calcitonin gene-related peptide. A light and electron microscope study in the rat. Brain Res. 1998;794:347–352. doi: 10.1016/S0006-8993(98)00332-1. [DOI] [PubMed] [Google Scholar]
- 18.Scherrer G., Imamachi N., Cao Y.Q., Contet C., Mennicken F., O'Donnell D., Kieffer B.L., Basbaum A.I. Dissociation of the opioid receptor mechanisms that control mechanical and heat pain. Cell. 2009;137:1148–1159. doi: 10.1016/j.cell.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molliver D.C., Wright D.E., Leitner M.L., Parsadanian A.S., Doster K., Wen D., Yan Q., Snider W.D. IB4-binding DRG neurons switch from NGF to GDNF dependence in early postnatal life. Neuron. 1997;19:849–861. doi: 10.1016/s0896-6273(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 20.Marmigère F., Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat. Rev. Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- 21.Usoskin D., Furlan A., Islam S., Abdo H., Lönnerberg P., Lou D., Hjerling-Leffler J., Haeggström J., Kharchenko O., Kharchenko P.V., et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 22.Bennett D.L., Averill S., Clary D.O., Priestley J.V., McMahon S.B. Postnatal changes in the expression of the trkA high-affinity NGF receptor in primary sensory neurons. Eur. J. Neurosci. 1996;8:2204–2208. doi: 10.1111/j.1460-9568.1996.tb00742.x. [DOI] [PubMed] [Google Scholar]
- 23.Cavanaugh D.J., Chesler A.T., Bráz J.M., Shah N.M., Julius D., Basbaum A.I. Restriction of Transient Receptor Potential Vanilloid-1 to the Peptidergic Subset of Primary Afferent Neurons Follows Its Developmental Downregulation in Nonpeptidergic Neurons. J. Neurosci. 2011;31:10119–10127. doi: 10.1523/jneurosci.1299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson S.N., Perry M.J., Prabhakar E., McCarthy P.W. Primary sensory neurones: neurofilament, neuropeptides, and conduction velocity. Brain Res. Bull. 1993;30:239–243. doi: 10.1016/0361-9230(93)90250-f. [DOI] [PubMed] [Google Scholar]
- 25.Price T.J., Flores C.M. Critical evaluation of the colocalization between calcitonin gene-related peptide, substance P, transient receptor potential vanilloid subfamily type 1 immunoreactivities, and isolectin B4 binding in primary afferent neurons of the rat and mouse. J. Pain. 2007;8:263–272. doi: 10.1016/j.jpain.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jimenez-Andrade J.M., Mantyh W.G., Bloom A.P., Xu H., Ferng A.S., Dussor G., Vanderah T.W., Mantyh P.W. A phenotypically restricted set of primary afferent nerve fibers innervate the bone versus skin: therapeutic opportunity for treating skeletal pain. Bone. 2010;46:306–313. doi: 10.1016/j.bone.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malin S., Molliver D., Christianson J.A., Schwartz E.S., Cornuet P., Albers K.M., Davis B.M. TRPV1 and TRPA1 function and modulation are target tissue dependent. J. Neurosci. 2011;31:10516–10528. doi: 10.1523/jneurosci.2992-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F.C., Tan T., Huang T., Christianson J., Samad O.A., Liu Y., Roberson D., Davis B.M., Ma Q. Genetic control of the segregation of pain-related sensory neurons innervating the cutaneous versus deep tissues. Cell Rep. 2013;5:1353–1364. doi: 10.1016/j.celrep.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett D.L., Michael G.J., Ramachandran N., Munson J.B., Averill S., Yan Q., McMahon S.B., Priestley J.V. A distinct subgroup of small DRG cells express GDNF receptor components and GDNF is protective for these neurons after nerve injury. J. Neurosci. 1998;18:3059–3072. doi: 10.1523/jneurosci.18-08-03059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franck M.C.M., Stenqvist A., Li L., Hao J., Usoskin D., Xu X., Wiesenfeld-Hallin Z., Ernfors P. Essential role of Ret for defining non-peptidergic nociceptor phenotypes and functions in the adult mouse. Eur. J. Neurosci. 2011;33:1385–1400. doi: 10.1111/j.1460-9568.2011.07634.x. [DOI] [PubMed] [Google Scholar]
- 31.Bardoni R., Tawfik V.L., Wang D., François A., Solorzano C., Shuster S.A., Choudhury P., Betelli C., Cassidy C., Smith K., et al. Delta opioid receptors presynaptically regulate cutaneous mechanosensory neuron input to the spinal cord dorsal horn. Neuron. 2014;81:1312–1327. doi: 10.1016/j.neuron.2014.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zylka M.J., Rice F.L., Anderson D.J. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Beaudry H., Daou I., Ase A.R., Ribeiro-da-Silva A., Séguéla P. Distinct behavioral responses evoked by selective optogenetic stimulation of the major TRPV1+ and MrgD+ subsets of C-fibers. Pain. 2017;158:2329–2339. doi: 10.1097/j.pain.0000000000001016. [DOI] [PubMed] [Google Scholar]
- 34.Abdus-Saboor I., Fried N.T., Lay M., Burdge J., Swanson K., Fischer R., Jones J., Dong P., Cai W., Guo X., et al. Development of a Mouse Pain Scale Using Sub-second Behavioral Mapping and Statistical Modeling. Cell Rep. 2019;28:1623–1634.e4. doi: 10.1016/j.celrep.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Q. A functional subdivision within the somatosensory system and its implications for pain research. Neuron. 2022;110:749–769. doi: 10.1016/j.neuron.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang T., Lin S.H., Malewicz N.M., Zhang Y., Zhang Y., Goulding M., LaMotte R.H., Ma Q. Identifying the pathways required for coping behaviours associated with sustained pain. Nature. 2019;565:86–90. doi: 10.1038/s41586-018-0793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C., Gu L., Ruan Y., Geng X., Xu M., Yang N., Yu L., Jiang Y., Zhu C., Yang Y., et al. Facilitation of MrgprD by TRP-A1 promotes neuropathic pain. Faseb j. 2019;33:1360–1373. doi: 10.1096/fj.201800615RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warwick C., Cassidy C., Hachisuka J., Wright M.C., Baumbauer K.M., Adelman P.C., Lee K.H., Smith K.M., Sheahan T.D., Ross S.E., Koerber H.R. MrgprdCre lineage neurons mediate optogenetic allodynia through an emergent polysynaptic circuit. Pain. 2021;162:2120–2131. doi: 10.1097/j.pain.0000000000002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Su X., Yan J., Wu Q., Xu X., Wang X., Liu X., Song X., Zhang Z., Hu W., et al. Involvement of Mrgprd-expressing nociceptors-recruited spinal mechanisms in nerve injury-induced mechanical allodynia. iScience. 2023;26:106764. doi: 10.1016/j.isci.2023.106764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen M.Q., von Buchholtz L.J., Reker A.N., Ryba N.J., Davidson S. Single-nucleus transcriptomic analysis of human dorsal root ganglion neurons. Elife. 2021;10:e71752. doi: 10.7554/eLife.71752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tavares-Ferreira D., Shiers S., Ray P.R., Wangzhou A., Jeevakumar V., Sankaranarayanan I., Cervantes A.M., Reese J.C., Chamessian A., Copits B.A., et al. Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors. Sci. Transl. Med. 2022;14:eabj8186. doi: 10.1126/scitranslmed.abj8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung M., Dourado M., Maksymetz J., Jacobson A., Laufer B.I., Baca M., Foreman O., Hackos D.H., Riol-Blanco L., Kaminker J.S. Cross-species transcriptomic atlas of dorsal root ganglia reveals species-specific programs for sensory function. Nat. Commun. 2023;14:366. doi: 10.1038/s41467-023-36014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNicol E.D., Midbari A., Eisenberg E. Opioids for neuropathic pain. Cochrane Database Syst. Rev. 2013;2013:Cd006146. doi: 10.1002/14651858.CD006146.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colloca L., Ludman T., Bouhassira D., Baron R., Dickenson A.H., Yarnitsky D., Freeman R., Truini A., Attal N., Finnerup N.B., et al. Neuropathic pain. Nat. Rev. Dis. Primers. 2017;3:17002. doi: 10.1038/nrdp.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bates D., Schultheis B.C., Hanes M.C., Jolly S.M., Chakravarthy K.V., Deer T.R., Levy R.M., Hunter C.W. A Comprehensive Algorithm for Management of Neuropathic Pain. Pain Med. 2019;20:S2–S12. doi: 10.1093/pm/pnz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomes I., Jordan B.A., Gupta A., Trapaidze N., Nagy V., Devi L.A. Heterodimerization of mu and delta opioid receptors: A role in opiate synergy. J. Neurosci. 2000;20:Rc110. doi: 10.1523/JNEUROSCI.20-22-j0007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma W., Sapio M.R., Manalo A.P., Maric D., Dougherty M.K., Goto T., Mannes A.J., Iadarola M.J. Anatomical Analysis of Transient Potential Vanilloid Receptor 1 (Trpv1+) and Mu-Opioid Receptor (Oprm1+) Co-expression in Rat Dorsal Root Ganglion Neurons. Front. Mol. Neurosci. 2022;15:926596. doi: 10.3389/fnmol.2022.926596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bi J., Tsai N.P., Lin Y.P., Loh H.H., Wei L.N. Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc. Natl. Acad. Sci. USA. 2006;103:19919–19924. doi: 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snyder L.M., Chiang M.C., Loeza-Alcocer E., Omori Y., Hachisuka J., Sheahan T.D., Gale J.R., Adelman P.C., Sypek E.I., Fulton S.A., et al. Kappa Opioid Receptor Distribution and Function in Primary Afferents. Neuron. 2018;99:1274–1288.e6. doi: 10.1016/j.neuron.2018.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moy J.K., Hartung J.E., Duque M.G., Friedman R., Nagarajan V., Loeza-Alcocer E., Koerber H.R., Christoph T., Schröder W., Gold M.S. Distribution of functional opioid receptors in human dorsal root ganglion neurons. Pain. 2020;161:1636–1649. doi: 10.1097/j.pain.0000000000001846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaveri N.T. Nociceptin Opioid Receptor (NOP) as a Therapeutic Target: Progress in Translation from Preclinical Research to Clinical Utility. J. Med. Chem. 2016;59:7011–7028. doi: 10.1021/acs.jmedchem.5b01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El Daibani A., Che T. Spotlight on Nociceptin/Orphanin FQ Receptor in the Treatment of Pain. Molecules. 2022;27:595. doi: 10.3390/molecules27030595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ichikawa H., Itota T., Nishitani Y., Torii Y., Inoue K., Sugimoto T. Osteopontin-immunoreactive primary sensory neurons in the rat spinal and trigeminal nervous systems. Brain Res. 2000;863:276–281. doi: 10.1016/s0006-8993(00)02126-0. [DOI] [PubMed] [Google Scholar]
- 54.Waxman S.G., Dib-Hajj S., Cummins T.R., Black J.A. Sodium channels and pain. Proc. Natl. Acad. Sci. USA. 1999;96:7635–7639. doi: 10.1073/pnas.96.14.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dib-Hajj S.D., Cummins T.R., Black J.A., Waxman S.G. Sodium channels in normal and pathological pain. Annu. Rev. Neurosci. 2010;33:325–347. doi: 10.1146/annurev-neuro-060909-153234. [DOI] [PubMed] [Google Scholar]
- 56.Shiers S., Klein R.M., Price T.J. Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. Pain. 2020;161:2410–2424. doi: 10.1097/j.pain.0000000000001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawson S.N. Phenotype and function of somatic primary afferent nociceptive neurones with C-Adelta- or Aalpha/beta-fibres. Exp. Physiol. 2002;87:239–244. [PubMed] [Google Scholar]
- 58.Pedersen-Bjergaard U., Nielsen L.B., Jensen K., Edvinsson L., Jansen I., Olesen J. Calcitonin gene-related peptide, neurokinin A and substance P: effects on nociception and neurogenic inflammation in human skin and temporal muscle. Peptides. 1991;12:333–337. doi: 10.1016/0196-9781(91)90022-h. [DOI] [PubMed] [Google Scholar]
- 59.Mantyh P.W. Neurobiology of substance P and the NK1 receptor. J. Clin. Psychiatry. 2002;63:6–10. [PubMed] [Google Scholar]
- 60.Duggan A.W., Morton C.R., Zhao Z.Q., Hendry I.A. Noxious heating of the skin releases immunoreactive substance P in the substantia gelatinosa of the cat: a study with antibody microprobes. Brain Res. 1987;403:345–349. doi: 10.1016/0006-8993(87)90073-4. [DOI] [PubMed] [Google Scholar]
- 61.Nakamura Y., Une Y., Miyano K., Abe H., Hisaoka K., Morioka N., Nakata Y. Activation of transient receptor potential ankyrin 1 evokes nociception through substance P release from primary sensory neurons. J. Neurochem. 2012;120:1036–1047. doi: 10.1111/j.1471-4159.2011.07628.x. [DOI] [PubMed] [Google Scholar]
- 62.Jarvik L.F., Simpson J.H., Guthrie D., Liston E.H. Morphine, experimental pain, and psychological reactions. Psychopharmacology (Berl) 1981;75:124–131. doi: 10.1007/bf00432173. [DOI] [PubMed] [Google Scholar]
- 63.Mauermann E., Filitz J., Dolder P., Rentsch K.M., Bandschapp O., Ruppen W. Does Fentanyl Lead to Opioid-induced Hyperalgesia in Healthy Volunteers?: A Double-blind, Randomized, Crossover Trial. Anesthesiology. 2016;124:453–463. doi: 10.1097/aln.0000000000000976. [DOI] [PubMed] [Google Scholar]
- 64.Cleeland C.S., Nakamura Y., Howland E.W., Morgan N.R., Edwards K.R., Backonja M. Effects of oral morphine on cold pressor tolerance time and neuropsychological performance. Neuropsychopharmacology. 1996;15:252–262. doi: 10.1016/0893-133x(95)00205-r. [DOI] [PubMed] [Google Scholar]
- 65.Bautista D.M., Siemens J., Glazer J.M., Tsuruda P.R., Basbaum A.I., Stucky C.L., Jordt S.E., Julius D. The menthol receptor TRPM8 is the principal detector of environmental cold. Nature. 2007;448:204–208. doi: 10.1038/nature05910. [DOI] [PubMed] [Google Scholar]
- 66.Knowlton W.M., Bifolck-Fisher A., Bautista D.M., McKemy D.D. TRPM8, but not TRPA1, is required for neural and behavioral responses to acute noxious cold temperatures and cold-mimetics in vivo. Pain. 2010;150:340–350. doi: 10.1016/j.pain.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coste B., Mathur J., Schmidt M., Earley T.J., Ranade S., Petrus M.J., Dubin A.E., Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chesler A.T., Szczot M., Bharucha-Goebel D., Čeko M., Donkervoort S., Laubacher C., Hayes L.H., Alter K., Zampieri C., Stanley C., et al. The Role of PIEZO2 in Human Mechanosensation. N. Engl. J. Med. 2016;375:1355–1364. doi: 10.1056/NEJMoa1602812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lam R.M., von Buchholtz L.J., Falgairolle M., Osborne J., Frangos E., Servin-Vences M.R., Nagel M., Nguyen M.Q., Jayabalan M., Saade D., et al. PIEZO2 and perineal mechanosensation are essential for sexual function. Science. 2023;381:906–910. doi: 10.1126/science.adg0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen C.C., Akopian A.N., Sivilotti L., Colquhoun D., Burnstock G., Wood J.N. A P2X purinoceptor expressed by a subset of sensory neurons. Nature. 1995;377:428–431. doi: 10.1038/377428a0. [DOI] [PubMed] [Google Scholar]
- 71.Cook S.P., Vulchanova L., Hargreaves K.M., Elde R., McCleskey E.W. Distinct ATP receptors on pain-sensing and stretch-sensing neurons. Nature. 1997;387:505–508. doi: 10.1038/387505a0. [DOI] [PubMed] [Google Scholar]
- 72.Bradbury E.J., Burnstock G., McMahon S.B. The expression of P2X3 purinoreceptors in sensory neurons: effects of axotomy and glial-derived neurotrophic factor. Mol. Cell. Neurosci. 1998;12:256–268. doi: 10.1006/mcne.1998.0719. [DOI] [PubMed] [Google Scholar]
- 73.Vulchanova L., Riedl M.S., Shuster S.J., Stone L.S., Hargreaves K.M., Buell G., Surprenant A., North R.A., Elde R. P2X3 is expressed by DRG neurons that terminate in inner lamina II. Eur. J. Neurosci. 1998;10:3470–3478. doi: 10.1046/j.1460-9568.1998.00355.x. [DOI] [PubMed] [Google Scholar]
- 74.Luo W., Wickramasinghe S.R., Savitt J.M., Griffin J.W., Dawson T.M., Ginty D.D. A hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of nonpeptidergic DRG neurons. Neuron. 2007;54:739–754. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 75.Fjell J., Hjelmström P., Hormuzdiar W., Milenkovic M., Aglieco F., Tyrrell L., Dib-Hajj S., Waxman S.G., Black J.A. Localization of the tetrodotoxin-resistant sodium channel NaN in nociceptors. Neuroreport. 2000;11:199–202. doi: 10.1097/00001756-200001170-00039. [DOI] [PubMed] [Google Scholar]
- 76.Cummins T.R., Black J.A., Dib-Hajj S.D., Waxman S.G. Glial-derived neurotrophic factor upregulates expression of functional SNS and NaN sodium channels and their currents in axotomized dorsal root ganglion neurons. J. Neurosci. 2000;20:8754–8761. doi: 10.1523/jneurosci.20-23-08754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fang X., Djouhri L., McMullan S., Berry C., Waxman S.G., Okuse K., Lawson S.N. Intense isolectin-B4 binding in rat dorsal root ganglion neurons distinguishes C-fiber nociceptors with broad action potentials and high Nav1.9 expression. J. Neurosci. 2006;26:7281–7292. doi: 10.1523/jneurosci.1072-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meunier J.C. Nociceptin/orphanin FQ and the opioid receptor-like ORL1 receptor. Eur. J. Pharmacol. 1997;340:1–15. doi: 10.1016/s0014-2999(97)01411-8. [DOI] [PubMed] [Google Scholar]
- 79.Lambert D.G. The nociceptin/orphanin FQ receptor: a target with broad therapeutic potential. Nat. Rev. Drug Discov. 2008;7:694–710. doi: 10.1038/nrd2572. [DOI] [PubMed] [Google Scholar]
- 80.François A., Scherrer G. Delta Opioid Receptor Expression and Function in Primary Afferent Somatosensory Neurons. Handb. Exp. Pharmacol. 2018;247:87–114. doi: 10.1007/164_2017_58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spahn V., Stein C. Targeting delta opioid receptors for pain treatment: drugs in phase I and II clinical development. Expert Opin. Investig. Drugs. 2017;26:155–160. doi: 10.1080/13543784.2017.1275562. [DOI] [PubMed] [Google Scholar]
- 82.Scholz A., Bothmer J., Kok M., Hoschen K., Daniels S. Cebranopadol: A Novel, First-in-Class, Strong Analgesic: Results from a Randomized Phase IIa Clinical Trial in Postoperative Acute Pain. Pain Physician. 2018;21:E193–E206. [PubMed] [Google Scholar]
- 83.Dalefield M.L., Scouller B., Bibi R., Kivell B.M. The Kappa Opioid Receptor: A Promising Therapeutic Target for Multiple Pathologies. Front. Pharmacol. 2022;13:837671. doi: 10.3389/fphar.2022.837671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Albert-Vartanian A., Boyd M.R., Hall A.L., Morgado S.J., Nguyen E., Nguyen V.P.H., Patel S.P., Russo L.J., Shao A.J., Raffa R.B. Will peripherally restricted kappa-opioid receptor agonists (pKORAs) relieve pain with less opioid adverse effects and abuse potential? J. Clin. Pharm. Ther. 2016;41:371–382. doi: 10.1111/jcpt.12404. [DOI] [PubMed] [Google Scholar]
- 85.Brennan T.J., Vandermeulen E.P., Gebhart G.F. Characterization of a rat model of incisional pain. Pain. 1996;64:493–502. doi: 10.1016/0304-3959(95)01441-1. [DOI] [PubMed] [Google Scholar]
- 86.Luo J., Feng J., Liu S., Walters E.T., Hu H. Molecular and cellular mechanisms that initiate pain and itch. Cell. Mol. Life Sci. 2015;72:3201–3223. doi: 10.1007/s00018-015-1904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mennicken F., Zhang J., Hoffert C., Ahmad S., Beaudet A., O'Donnell D. Phylogenetic changes in the expression of delta opioid receptors in spinal cord and dorsal root ganglia. J. Comp. Neurol. 2003;465:349–360. doi: 10.1002/cne.10839. [DOI] [PubMed] [Google Scholar]
- 88.Akopian A.N., Souslova V., England S., Okuse K., Ogata N., Ure J., Smith A., Kerr B.J., McMahon S.B., Boyce S., et al. The tetrodotoxin-resistant sodium channel SNS has a specialized function in pain pathways. Nat. Neurosci. 1999;2:541–548. doi: 10.1038/9195. [DOI] [PubMed] [Google Scholar]
- 89.Stirling L.C., Forlani G., Baker M.D., Wood J.N., Matthews E.A., Dickenson A.H., Nassar M.A. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain. 2005;113:27–36. doi: 10.1016/j.pain.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 90.Weibel R., Reiss D., Karchewski L., Gardon O., Matifas A., Filliol D., Becker J.A.J., Wood J.N., Kieffer B.L., Gaveriaux-Ruff C. Mu opioid receptors on primary afferent nav1.8 neurons contribute to opiate-induced analgesia: insight from conditional knockout mice. PLoS One. 2013;8:e74706. doi: 10.1371/journal.pone.0074706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Duan G., Sun J., Li N., Zheng H., Guo S., Zhang Y., Wang Q., Ying Y., Zhang M., Huang P., Zhang X. A variant in the SCN10A enhancer may affect human mechanical pain sensitivity. Mol. Pain. 2018;14 doi: 10.1177/1744806918763275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leipold E., Liebmann L., Korenke G.C., Heinrich T., Giesselmann S., Baets J., Ebbinghaus M., Goral R.O., Stödberg T., Hennings J.C., et al. A de novo gain-of-function mutation in SCN11A causes loss of pain perception. Nat. Genet. 2013;45:1399–1404. doi: 10.1038/ng.2767. [DOI] [PubMed] [Google Scholar]
- 93.Woods C.G., Babiker M.O.E., Horrocks I., Tolmie J., Kurth I. The phenotype of congenital insensitivity to pain due to the NaV1.9 variant p.L811P. Eur. J. Hum. Genet. 2015;23:561–563. doi: 10.1038/ejhg.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jones J., Correll D.J., Lechner S.M., Jazic I., Miao X., Shaw D., Simard C., Osteen J.D., Hare B., Beaton A., et al. Selective Inhibition of NaV1.8 with VX-548 for Acute Pain. N. Engl. J. Med. 2023;389:393–405. doi: 10.1056/NEJMoa2209870. [DOI] [PubMed] [Google Scholar]
- 95.Dib-Hajj S.D., Black J.A., Waxman S.G. NaV1.9: a sodium channel linked to human pain. Nat. Rev. Neurosci. 2015;16:511–519. doi: 10.1038/nrn3977. [DOI] [PubMed] [Google Scholar]
- 96.Alsaloum M., Labau J.I.R., Liu S., Estacion M., Zhao P., Dib-Hajj F., Waxman S.G. Contributions of NaV1.8 and NaV1.9 to excitability in human induced pluripotent stem-cell derived somatosensory neurons. Sci. Rep. 2021;11:24283. doi: 10.1038/s41598-021-03608-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cummins T.R., Dib-Hajj S.D., Black J.A., Akopian A.N., Wood J.N., Waxman S.G. A novel persistent tetrodotoxin-resistant sodium current in SNS-null and wild-type small primary sensory neurons. J. Neurosci. 1999;19:Rc43. doi: 10.1523/JNEUROSCI.19-24-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alsaloum M., Higerd G.P., Effraim P.R., Waxman S.G. Status of peripheral sodium channel blockers for non-addictive pain treatment. Nat. Rev. Neurol. 2020;16:689–705. doi: 10.1038/s41582-020-00415-2. [DOI] [PubMed] [Google Scholar]
- 99.Silverman J.D., Kruger L. Lectin and neuropeptide labeling of separate populations of dorsal root ganglion neurons and associated "nociceptor" thin axons in rat testis and cornea whole-mount preparations. Somatosens. Res. 1988;5:259–267. doi: 10.3109/07367228809144630. [DOI] [PubMed] [Google Scholar]
- 100.Segond von Banchet G., Pastor A., Biskup C., Schlegel C., Benndorf K., Schaible H.G. Localization of functional calcitonin gene-related peptide binding sites in a subpopulation of cultured dorsal root ganglion neurons. Neuroscience. 2002;110:131–145. doi: 10.1016/s0306-4522(01)00547-4. [DOI] [PubMed] [Google Scholar]
- 101.Nagao M., Oka N., Kamo H., Akiguchi I., Kimura J. Differential localization of lectin binding sites and neuropeptides in human dorsal root ganglia. Histochemistry. 1994;102:279–286. doi: 10.1007/bf00269164. [DOI] [PubMed] [Google Scholar]
- 102.Landry M., Aman K., Dostrovsky J., Lozano A.M., Carlstedt T., Spenger C., Josephson A., Wiesenfeld-Hallin Z., Hökfelt T. Galanin expression in adult human dorsal root ganglion neurons: initial observations. Neuroscience. 2003;117:795–809. doi: 10.1016/s0306-4522(02)00965-x. [DOI] [PubMed] [Google Scholar]
- 103.Arenas O.M., Zaharieva E.E., Para A., Vásquez-Doorman C., Petersen C.P., Gallio M. Activation of planarian TRPA1 by reactive oxygen species reveals a conserved mechanism for animal nociception. Nat. Neurosci. 2017;20:1686–1693. doi: 10.1038/s41593-017-0005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Viana F. TRPA1 channels: molecular sentinels of cellular stress and tissue damage. J. Physiol. 2016;594:4151–4169. doi: 10.1113/jp270935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Navratilova E., Porreca F. Substance P and Inflammatory Pain: Getting It Wrong and Right Simultaneously. Neuron. 2019;101:353–355. doi: 10.1016/j.neuron.2019.01.034. [DOI] [PubMed] [Google Scholar]
- 106.Borsook D., Upadhyay J., Klimas M., Schwarz A.J., Coimbra A., Baumgartner R., George E., Potter W.Z., Large T., Bleakman D., et al. Decision-making using fMRI in clinical drug development: revisiting NK-1 receptor antagonists for pain. Drug Discov. Today. 2012;17:964–973. doi: 10.1016/j.drudis.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 107.Konietzny F. Peripheral neural correlates of temperature sensations in man. Hum. Neurobiol. 1984;3:21–32. [PubMed] [Google Scholar]
- 108.Campero M., Serra J., Bostock H., Ochoa J.L. Slowly conducting afferents activated by innocuous low temperature in human skin. J. Physiol. 2001;535:855–865. doi: 10.1111/j.1469-7793.2001.t01-1-00855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ackerley R., Watkins R.H. Microneurography as a tool to study the function of individual C-fiber afferents in humans: responses from nociceptors, thermoreceptors, and mechanoreceptors. J. Neurophysiol. 2018;120:2834–2846. doi: 10.1152/jn.00109.2018. [DOI] [PubMed] [Google Scholar]
- 110.Campero M., Serra J., Ochoa J.L. C-polymodal nociceptors activated by noxious low temperature in human skin. J. Physiol. 1996;497:565–572. doi: 10.1113/jphysiol.1996.sp021789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Klein A., Solinski H.J., Malewicz N.M., Ieong H.F.H., Sypek E.I., Shimada S.G., Hartke T.V., Wooten M., Wu G., Dong X., et al. Pruriception and neuronal coding in nociceptor subtypes in human and nonhuman primates. Elife. 2021;10:e64506. doi: 10.7554/eLife.64506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Van Hees J., Gybels J. C nociceptor activity in human nerve during painful and non painful skin stimulation. J. Neurol. Neurosurg. Psychiatry. 1981;44:600–607. doi: 10.1136/jnnp.44.7.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adriaensen H., Gybels J., Handwerker H.O., Van Hees J. Nociceptor discharges and sensations due to prolonged noxious mechanical stimulation--a paradox. Hum. Neurobiol. 1984;3:53–58. [PubMed] [Google Scholar]
- 114.Schmidt R., Schmelz M., Torebjörk H.E., Handwerker H.O. Mechano-insensitive nociceptors encode pain evoked by tonic pressure to human skin. Neuroscience. 2000;98:793–800. doi: 10.1016/s0306-4522(00)00189-5. [DOI] [PubMed] [Google Scholar]
- 115.Wooten M., Weng H.J., Hartke T.V., Borzan J., Klein A.H., Turnquist B., Dong X., Meyer R.A., Ringkamp M. Three functionally distinct classes of C-fibre nociceptors in primates. Nat. Commun. 2014;5:4122. doi: 10.1038/ncomms5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van der Burght M., Rasmussen S.E., Arendt-Nielsen L., Bjerring P. Morphine does not affect laser induced warmth and pin prick pain thresholds. Acta Anaesthesiol. Scand. 1994;38:161–164. doi: 10.1111/j.1399-6576.1994.tb03859.x. [DOI] [PubMed] [Google Scholar]
- 117.Moehring F., Cowie A.M., Menzel A.D., Weyer A.D., Grzybowski M., Arzua T., Geurts A.M., Palygin O., Stucky C.L. Keratinocytes mediate innocuous and noxious touch via ATP-P2X4 signaling. Elife. 2018;7:e31684. doi: 10.7554/eLife.31684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Koizumi S., Fujishita K., Inoue K., Shigemoto-Mogami Y., Tsuda M., Inoue K. Ca2+ waves in keratinocytes are transmitted to sensory neurons: the involvement of extracellular ATP and P2Y2 receptor activation. Biochem. J. 2004;380:329–338. doi: 10.1042/bj20031089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shindo Y., Fujita K., Tanaka M., Fujio H., Hotta K., Oka K. Mechanical stimulus-evoked signal transduction between keratinocytes and sensory neurons via extracellular ATP. Biochem. Biophys. Res. Commun. 2021;582:131–136. doi: 10.1016/j.bbrc.2021.10.046. [DOI] [PubMed] [Google Scholar]
- 120.Woolf C.J., Ma Q. Nociceptors--noxious stimulus detectors. Neuron. 2007;55:353–364. doi: 10.1016/j.neuron.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 121.Middleton S.J., Barry A.M., Comini M., Li Y., Ray P.R., Shiers S., Themistocleous A.C., Uhelski M.L., Yang X., Dougherty P.M., et al. Studying human nociceptors: from fundamentals to clinic. Brain. 2021;144:1312–1335. doi: 10.1093/brain/awab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rowbotham M.C., Nothaft W., Duan R.W., Wang Y., Faltynek C., McGaraughty S., Chu K.L., Svensson P. Oral and cutaneous thermosensory profile of selective TRPV1 inhibition by ABT-102 in a randomized healthy volunteer trial. Pain. 2011;152:1192–1200. doi: 10.1016/j.pain.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 123.Iadarola M.J., Sapio M.R., Mannes A.J. Be in it for the Long Haul: A Commentary on Human Tissue Recovery Initiatives. J. Pain. 2022;23:1646–1650. doi: 10.1016/j.jpain.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fiebig A., Leibl V., Oostendorf D., Lukaschek S., Frömbgen J., Masoudi M., Kremer A.E., Strupf M., Reeh P., Düll M., Namer B. Peripheral signaling pathways contributing to non-histaminergic itch in humans. J. Transl. Med. 2023;21:908. doi: 10.1186/s12967-023-04698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Löken L.S., Backlund Wasling H., Olausson H., McGlone F., Wessberg J. A topographical and physiological exploration of C-tactile afferents and their response to menthol and histamine. J. Neurophysiol. 2022;127:463–473. doi: 10.1152/jn.00310.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eippert F., Finsterbusch J., Bingel U., Büchel C. Direct evidence for spinal cord involvement in placebo analgesia. Science. 2009;326:404. doi: 10.1126/science.1180142. [DOI] [PubMed] [Google Scholar]
- 127.Tinnermann A., Büchel C., Cohen-Adad J. Cortico-spinal imaging to study pain. Neuroimage. 2021;224:117439. doi: 10.1016/j.neuroimage.2020.117439. [DOI] [PubMed] [Google Scholar]
- 128.Li H., Badawi R.D., Cherry S.R., Fontaine K., He L., Henry S., Hillmer A.T., Hu L., Khattar N., Leung E.K., et al. Performance Characteristics of the NeuroEXPLORER, a Next-Generation Human Brain PET/CT Imager. J. Nucl. Med. 2024;65:1320–1326. doi: 10.2967/jnumed.124.267767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maric D., Jahanipour J., Li X.R., Singh A., Mobiny A., Van Nguyen H., Sedlock A., Grama K., Roysam B. Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks. Nat. Commun. 2021;12:1550. doi: 10.1038/s41467-021-21735-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sapio M.R., King D.M., Staedtler E.S., Maric D., Jahanipour J., Kurochkina N.A., Manalo A.P., Ghetti A., Mannes A.J., Iadarola M.J. Expression pattern analysis and characterization of the hereditary sensory and autonomic neuropathy 2 A (HSAN2A) gene with no lysine kinase (WNK1) in human dorsal root ganglion. Exp. Neurol. 2023;370:114552. doi: 10.1016/j.expneurol.2023.114552. [DOI] [PubMed] [Google Scholar]
- 131.Davidson S., Copits B.A., Zhang J., Page G., Ghetti A., Gereau R.W., 4th Human sensory neurons: Membrane properties and sensitization by inflammatory mediators. Pain. 2014;155:1861–1870. doi: 10.1016/j.pain.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chang W., Berta T., Kim Y.H., Lee S., Lee S.Y., Ji R.R. Expression and Role of Voltage-Gated Sodium Channels in Human Dorsal Root Ganglion Neurons with Special Focus on Nav1.7, Species Differences, and Regulation by Paclitaxel. Neurosci. Bull. 2018;34:4–12. doi: 10.1007/s12264-017-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this study will be shared by the lead contact upon request. This includes multiplex fluorescence microscopic images and region-of-interest files. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.