Abstract
RNA-1 of peanut clump pecluvirus (PCV) encodes N-terminally overlapping proteins which contain helicase-like (P131) and polymerase-like (P191) domains and is able to replicate in the absence of RNA-2 in protoplasts of tobacco BY-2 cells. RNA-1 also encodes P15, which is expressed via a subgenomic RNA. To investigate the role of P15, we analyzed RNA accumulation in tobacco BY-2 protoplasts inoculated with RNA-1 containing mutations in P15. For all the mutants, the amount of progeny RNA-1 produced was significantly lower than that obtained for wild-type RNA-1. If RNA-2 was included in the inoculum, the accumulation of both progeny RNAs was diminished, but near-normal yields of both could be recovered if the inoculum was supplemented with a small, chimeric viral replicon expressing P15, demonstrating that P15 has an effect on viral RNA accumulation. To further analyze the role of P15, transcripts were produced expressing P15 fused to enhanced green fluorescent protein (EGFP). Following inoculation to protoplasts, epifluorescence microscopy revealed that P15 accumulated as spots around the nucleus and in the cytoplasm. Intracellular sites of viral RNA synthesis were visualized by laser scanning confocal microscopy of infected protoplasts labeled with 5-bromouridine 5′-triphosphate (BrUTP). BrUTP labeling also occured in spots distributed within the cytoplasm and around the nucleus. However, the BrUTP-labeled RNA and EGFP/P15 very rarely colocalized, suggesting that P15 does not act primarily at sites of viral replication but intervenes indirectly to control viral accumulation levels.
After virion uncoating in a cell, the genomic RNAs of positive-strand RNA viruses must first serve as mRNAs. Their translation products, presumably in cooperation with host proteins, then direct synthesis of progeny viral RNA from the infecting templates. For many viruses, two nonstructural proteins with conserved amino acid sequence motifs characteristic of RNA helicases and RNA-dependent RNA polymerases (2, 8, 9, 22) are the only viral proteins required for viral RNA replication. However, in addition to these evolutionarily conserved proteins, other viral proteins without a specific amino acid “signature” sequence for a replication-associated protein may be involved in replication (21). Some are essential, such as the coat protein (CP) of alfalfa mosaic virus, which is needed to activate the replication of genomic RNAs (17, 25), or the proteins encoded by cowpea mosaic virus M-RNA and tomato black ring virus satellite RNA, which are required in cis for trans replication of the RNA (26, 35). Similarly, roles in tobacco etch virus genome amplification have been demonstrated for the HC-Pro protein (19) and for the 6-kDa protein (31), while the protein P1, although not absolutely required, functions as an accessory factor for genome amplification (36).
A number of plant alpha-like viruses, including the carla-, furo-, hordei-, and tobraviruses, possess a gene encoding a small cysteine-rich protein (CRP), which displays sequence similarities to other nucleic acid binding proteins (21) and which has been suggested to act as a regulatory factor during virus replication (10). The CRPs of barley stripe mosaic virus (BSMV) and beet necrotic yellow vein virus (BNYVV) have been shown to affect the accumulation levels of viral RNAs (12, 28, 39), although a direct role for these proteins in viral RNA replication has not been demonstrated.
Here we have studied the function of the peanut clump virus (PCV) RNA-1-encoded 15-kDa protein (P15), which displays homology with the CRPs of BSMV, poa semilatent virus, and soilborne wheat mosaic virus (14). PCV, the type member of the pecluviruses, possesses a messenger sense RNA genome composed of two separately encapsidated RNAs, designated RNA-1 (5,897 nucleotides) and RNA-2 (4,504 nucleotides). RNA-1 is able to replicate independently of RNA-2 in protoplasts, but both RNAs are indispensable for plant infection (15). RNA-2 encodes CP, a 39-kDa protein, and three movement proteins, organized into a triple gene block. RNA-1 encodes 131-kDa (P131) and 191-kDa (P191) proteins which contain the motifs characteristic of proteins involved in viral RNA replication. These two proteins are N-terminally overlapping in the same reading frame, the longer one being produced by readthrough of the shorter protein. RNA-1 also encodes P15, which is expressed via a 3′ proximal subgenomic RNA. P15 has previously been shown to influence viral RNA amplification levels (15).
In the present study, we have confirmed the involvement of P15 in the amplification of the two genomic RNAs and have investigated in more detail the role of P15 in viral RNA replication. In addition an infectious chimeric virus was constructed in which P15 was fused to the enhanced green fluorescent protein (EGFP). The chimera was used to study the subcellular distribution of P15 by epifluorescence and laser scanning confocal microscopy. Specific labeling of newly synthesized viral RNA with 5-bromouridine 5′-triphosphate (BrUTP) has allowed us to investigate whether P15 is present at sites of viral RNA replication.
MATERIALS AND METHODS
Construction of mutant plasmids.
Plasmids pPC1 and pPC2 (Fig. 1A) are full-length cDNA clones of PCV RNA-1 and RNA-2, respectively, and can be used to produce infectious RNA transcripts by in vitro transcription with T7 RNA polymerase (15). Three different P15 mutants of pPC1 were constructed by conventional recombinant DNA techniques or the overlap extension PCR (16).
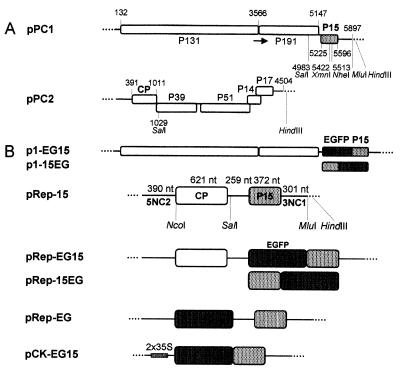
FIG. 1.
Schematic representation of plasmids pPC1 and pPC2 and derived plasmids. (A) pPC1 and pPC2, from which T1 and T2 are obtained. Restriction sites used for the different constructions are indicated. Numbers correspond to the nucleotides' positions in the RNA. Arrow indicates readthrough. (B) pPC1 mutants expressing EGFP fused to either the N or C terminus of P15 and chimeric plasmids expressing P15 or P15 fused to EGFP. 5NC2, 5′ noncoding region from RNA-2; 3NC1, 3′ noncoding region from RNA-1.
Mutant pPC1-15Nh has been previously described (15). Insertion of four nucleotides at the NheI restriction enzyme site (nucleotide 5513) within open reading frame 3 (ORF3) produced a frameshift and a C-terminally truncated P15 of 12 kDa.
Mutant pPC1-15(−), the P15 AUG initiation codon of the P15 cistron, was changed to AAG by replacing the sequence between the SalI site (nucleotide 4983) and MluI site (immediately downstream of the RNA-1 insert sequence) with an equivalent PCR fragment into which the mutation had been introduced by overlap extension mutagenesis.
Mutant pPC1-15LK was obtained by insertion of five 10-mer BglII linkers (Biolabs) between the blunt-ended extremities of XmnI-digested pPC1 (nucleotide 5422) to create an in-frame stop codon. The mutant encodes a 12-kDa protein of which only the N-terminal 9 kDa correspond to the amino acid sequence of P15. The fidelity of all constructs was confirmed by sequencing the region around the mutation or the complete PCR-amplified fragment.
pRep-15 (Fig. 1B) was obtained by replacing the SalI-HindIII fragment of p2MAUGS (a pPC2 derivative with an NcoI site at the beginning of the CP cistron) (13) with the SalI-HindIII fragment (nucleotides 4983 to 5897) from pPC1. The resulting plasmid contains 1,028 nucleotides corresponding to the 5′ region of RNA-2 and 915 nucleotides corresponding to the 3′ end of RNA-1 and carries both the CP and P15 cistrons.
pRep-EG, pRep-EG15, and pRep-15EG (Fig. 1B) were also obtained. To obtain pRep-EG, an NcoI-EcoRI fragment containing the EGFP cistron was excised from pEGFP (Clontech) and was introduced into pRep-15 after elimination of the NcoI-SalI fragment (nucleotides 391 to 1029) containing the CP gene.
To construct plasmids pRep-EG15 and pRep-15EG, which express EGFP/P15 fusion proteins, we used pRep-15′, a derivative of pRep-15 which did not contain the NcoI restriction enzyme site in front of the CP. The construction of pRep-EG15 was performed in two steps. First, a fragment corresponding to the sequence of RNA-1 and overlapping the SalI and MluI sites was amplified and modified by overlap extension PCR so that NcoI and EcoRI sites were introduced in front of the P15 initiation codon. The SalI-MluI-digested PCR fragment was then inserted into pRep-15′ in place of the equivalent wild-type fragment to obtain pRep-NE15. Second, an NcoI-EcoRI fragment, amplified from pEGFP and in which the TAA stop codon of the EGFP cistron had been mutated to TTA, was inserted into NcoI-EcoRI-digested pRep-NE15 to obtain pRep-EG15.
pRep-15EG was constructed similarly. The P15 coding region was amplified and modified by overlap extension PCR so that the P15 termination codon was altered to TAA, and NcoI and EcoRI sites were placed, respectively, 1 and 10 nucleotides further downstream. The PCR product was cleaved with NheI and MluI and inserted into pRep-15′ in place of the corresponding wild-type sequence to obtain pRep-15NE. The NcoI-EcoRI fragment of pEGFP was then introduced between the NcoI and EcoRI sites of this construct to produce pRep-15EG.
p1-EG15 and p1-15EG (Fig. 1B) were constructed by insertion of the SalI-MluI fragment from pRep-EG15 or pRep-15EG into pPC1 in place of the equivalent wild-type fragment.
pCK-EG15 (Fig. 1B) was also obtained. pCK-EGFP, kindly provided by C. Reichel, carries the EGFP coding region placed downstream of a duplicated 35S CaMV promoter (30). To produce pCK-EG15, the NcoI-HindIII fragment of pRep-15EG was inserted into pCK-EGFP in place of the NcoI-BamHI fragment corresponding to the EGFP coding region.
Synthesis of transcripts.
Capped in vitro transcripts were obtained with a Ribomax transcription kit (Promega), following the manufacturer's instructions. pPC1-derived plasmids were linearized with MluI, and pPC2 plasmid was linearized with HindIII before transcription. The resulting transcripts will be referred to as T1 and T2 for the wild-type and T1-15Nh, etc., for the derivatives containing mutant forms of P15.
Protoplast transfection and analysis.
Protoplasts from tobacco BY-2 cells (24) were prepared as previously described (38) with minor modifications (15). Protoplasts (106 in 0.5 ml) were electroporated using a Gene Pulser (Bio-Rad) in a 0.4-cm path-length cuvette, either at 700 V/cm, 100 Ω, and 125 μF using 80 μg of plasmid DNA and 40 μg of sonicated salmon sperm DNA as carrier or at 450 V/cm, 100 Ω, and 125 μF using 5 μg of each transcript. For wild-type transcripts, this concentration was shown in preliminary experiments to maximize the yield of progeny viral RNA. Furthermore, under these conditions the yield was relatively insensitive to small variations in the concentration of the inoculum. For Northern blot analysis, total RNA was isolated from 250,000 protoplasts 48 h postinfection (hpi). The RNAs were separated by electrophoresis on 1% agarose-formaldehyde denaturing gels and were blotted onto Hybond membranes. Viral RNAs were detected by hybridization with specific in vitro-transcribed 32P-labeled RNA probes. Radioactive signals were detected and quantified using a FUJIX BAS1000 phosphorimager and MacBAS image analysis software.
Antibodies.
The DNA sequence corresponding to the P15 gene was amplified by PCR using pPC1 as DNA template and positive- and negative-sense primers containing, respectively, 5′-terminal nonviral BamHI and EcoRI restriction sites. The PCR fragment was digested with BamHI and EcoRI and inserted into BamHI-EcoRI-cleaved pGEX-3X (Pharmacia). Escherichia coli DH5α bacteria were transformed with the resulting plasmid (pGEX-15) and were induced to overexpress a 41-kDa fusion protein containing the glutathione S-transferase (GST) and P15 sequences. The protein was purified from the bacterial extract by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The purified protein was injected into rabbits (5), and serum was collected 2 weeks after the first boost.
Western blotting.
For detection of viral proteins, protoplasts (5 × 105) were collected by centrifugation, and the pellet was resuspended in the equivalent volume of twofold-concentrated gel loading buffer (160 mM Tris-HCl [pH 6.8], 10% SDS, 25% 2-mercaptoethanol). A 10-μl aliquot of each sample was treated at 95°C for 3 min and analyzed by SDS–15% PAGE (23). After electrotransfer for 2 h at 0.8 mA/cm2, the membrane was incubated with 5% powdered milk in phosphate-buffered saline (PBS) for 2 h and then overnight with the CP antiserum diluted 20,000-fold in 5% milk–PBS. The membrane was washed in 1% Tween 20–PBS and incubated for 2 h with a 1:5,000 dilution of alkaline phosphatase-coupled anti-rabbit immunoglobulin G serum in 5% milk–PBS. After washing with 0.5% Tween 20–PBS, bound antibodies were visualized by fluorography with an Immunostar Chemiluminescent kit (Bio-Rad).
Fluorescence microscopy.
Protoplasts were allowed to settle on a multiwell slide, covered with a coverslip, and directly observed with a Nikon Ellipse 800 epifluorescence microscope or processed for immunostaining. For immunofluorescent staining, protoplasts were processed as previously described (33) with the following modifications. Harvested protoplasts (106) were transferred to a microtube and fixed in tobacco BY-2 cell culture medium (38) containing 4% paraformaldehyde for 30 min with gentle agitation. The protoplasts were then centrifuged (2 min at 54 × g) and were washed twice in PBS before suspension in 0.1% NaBH4 in PBS and storage at 4°C. An aliquot of the protoplasts was applied on a poly-l-lysine-coated coverslip and allowed to settle for 1 h at room temperature, followed by incubation in a blocking solution consisting of PBS, 5% bovine serum albumin (BSA), 5% normal goat serum, and 0.1% cold-water fish skin gelatin (Aurion, Wageningen, The Netherlands) for 1 h at room temperature. The protoplasts were then incubated at 4°C overnight with the primary antibody, washed six times in 0.1% Aurion BSA-c in PBS and further incubated in the dark for 1 h at room temperature with goat anti-rabbit (secondary) antibody conjugated to Cy3 (Jackson Laboratories) or a goat anti-mouse antibody conjugated to Alexa 568 (Molecular Probes) diluted 1:500 in 0.1% Aurion BSA-c in PBS. The protoplasts were again washed six times before observation. Photomicrographs were taken with a 3CCD Sony 950DXC videocamera, driven by Visiolab 200 (Biocom, Les Ulis, France). Images were processed using Adobe Photoshop software.
Observations for colocalization experiments were carried out with a Zeiss LSM-510 confocal microscope. For EGFP imaging, excitation at 488 nm was obtained with an argon laser, and for Alexa 568, excitation at 543 nm was obtained with a helium/neon laser. Appropriate emission filters were used to collect the green and red signals simultaneously from the same optical section without overspill of fluorescence.
In vivo RNA labeling.
To label RNA in vivo, protoplasts at 30 hpi were treated for 1 h with 10 μg of actinomycin D/ml and were then further incubated for 15 min in the presence of 10 mM BrUTP (Sigma) (33) or for 6 h with 100 μM BrUTP (7). Incorporation was stopped by addition of the fixation medium (4% paraformaldehyde in tobacco BY-2 cell culture medium) and gentle agitation for 30 min. The protoplasts were then processed as above for immunostaining, and BrUTP incorporation was detected using anti-BrUTP primary antibody (Sigma) and Alexa 568-labeled secondary antibody.
RESULTS
Accumulation of P15 in infected tobacco BY-2 protoplasts.
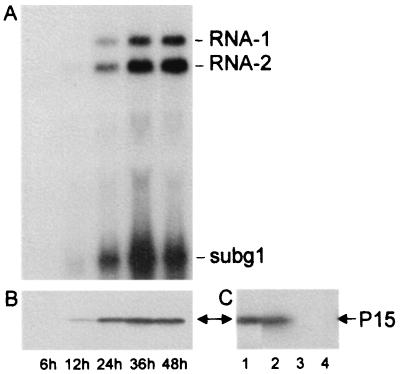
Protoplasts of tobacco BY-2 cells were infected with viral RNA (Fig. 2A). Northern blot analysis of RNA prepared from protoplasts harvested at 24 hpi or later detected progeny RNA-1 and RNA-2, as well as a small RNA of a length corresponding to that predicted for the subgenomic RNA-1 (subg1). Similar results were obtained with a mixture of transcripts corresponding to full-length RNA-1 (T1) and RNA-2 (T2) (not shown). Both genomic RNAs continued to accumulate for at least 48 h, whereas levels of subg1 decreased after 36 h, possibly because the subgenomic RNA is not encapsidated (unpublished observations) and is hence more susceptible to degradation than are the genomic RNAs. A protein of 15 kDa, detected by Western blotting using a GST-P15-specific antiserum (see Materials and Methods), accumulated from 12 hpi and reached a maximum at about 36 hpi (Fig. 2B). This protein (Fig. 2C, lane 2) comigrated with [35S]methionine-labeled P15 translated in vitro from a transcript which encodes P15 (Fig. 2C, lane 1). The band was not detected in extracts from healthy protoplasts (Fig. 2C, lane 3). When the P15-specific antibodies were first allowed to cross-react with purified P15 (obtained from E. coli transformed with a pET vector containing the P15 gene), the putative P15 band was no longer detected (Fig. 2C, lane 4), further confirming that the protein detected in the infected protoplast extracts indeed corresponds to P15.
FIG. 2.
Time course of PCV RNA synthesis and accumulation of P15 in tobacco BY-2 protoplasts infected with PCV RNA. Protoplasts harvested at different times postinfection as indicated below were subjected either to Northern blot analysis of RNA extracts using a riboprobe complementary to the 124 3′-terminal nucleotides of RNA-1 and -2 (A) or Western blot analysis using an antiserum against P15 (B). Protein extracts were separated by SDS–15% PAGE. (C) Analysis by SDS–15% PAGE of the in vitro [35S]methionine-labeled translation product obtained from a transcript encoding the P15 open reading frame of RNA-1 after 2 h of incubation in a wheat germ extract (lane 1) and Western blot analysis of a protein extract from PCV-infected protoplasts at 48 hpi (lanes 2 and 4) or from mock-inoculated protoplasts (lane 3) using P15 antiserum. For lane 4, the P15 antibodies were cross-adsorbed with purified P15 expressed in E. coli before use.
Effect of mutations in the P15 gene on RNA-1 accumulation.
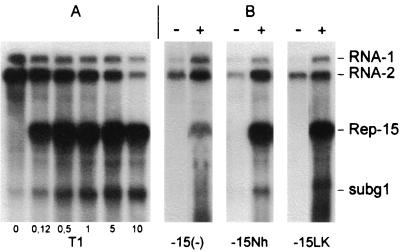
It has previously been shown that the wild-type RNA-1 transcript T1 can replicate without RNA-2 in protoplasts (15) (Fig. 3). To determine if the P15 gene product is required for RNA-1 replication, the RNA-1 transcripts T1-15(−), T1-15Nh, and T1-15LK, each containing a knockout mutation in the P15 gene, were inoculated to BY-2 protoplasts, and total RNA extracts were tested for the presence of progeny RNA-1 by Northern blotting 48 hpi. The experiments revealed that the RNA-1 transcripts containing the different P15 mutations produced progeny RNA-1 and subg1, demonstrating that P15 is not strictly required for viral RNA replication. For mutant T1-15(−), subg1 is not visible in the autoradiogram in Fig. 3, although the band was observed with longer exposure times (not shown). Possibly, the mutation in question has weakened the promoter responsible for synthesis of the subgenomic RNA.
FIG. 3.
Accumulation of genomic RNA 48 hpi of tobacco BY-2 protoplasts inoculated with T1, T1-15(−), T1-15Nh, or T1-15LK, alone (−) or with T2 (+). Northern blot of a representative experiment. 32P riboprobes complementary to nucleotides 5193 to 5515 of RNA-1 and to nucleotides 390 to 1624 of RNA-2 were used for detection.
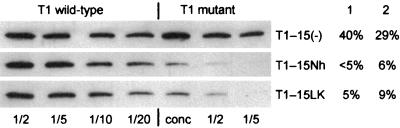
It can be seen in Fig. 3 and Table 1 that the amount of progeny viral RNA produced from the mutants was always significantly lower than for wild-type RNA-1. Thus in four independant experiments, the yield of mutant progeny RNA-1 relative to wild-type RNA-1 levels varied from 21.3 to 34% (average, 27.5%) for T1-15(−), from 17 to 55% (average, 33.1%) for T1-15Nh, and from 3.6 to 26.3% (average, 20.2%) for T1-15LK (Table 1).
TABLE 1.
Amounts of mutant progeny RNA-1 and RNA-2 relative to the wild-type RNAs detecteda in infected protoplasts at 48 hpi
| Inoculum | Progeny | % Progeny detected in expt:
|
Avgc | ||||
|---|---|---|---|---|---|---|---|
| 1b | 2 | 3 | 4 | 5 | |||
| T1-15(−) | RNA-1 | 34.0 | 29.0 | 25.5 | 21.3 | 27.5 ± 4.7 | |
| T1-15(−) + T2 | RNA-1 | 20.0 | 17.5 | 9.5 | 23.0 | 17.5 ± 5.0 | |
| RNA-2 | 27.8 | 24.0 | 22.0 | 24.0 | 24.3 ± 1.8 | ||
| T1-15Nh | RNA-1 | 43.0 | 55.0 | 17.0 | 17.3 | 33.1 ± 16.5 | |
| T1-15Nh + T2 | RNA-1 | 8.5 | 3.8 | 4.9 | 28.0 | 11.3 ± 9.8 | |
| RNA-2 | 32.0 | 7.5 | 15.0 | 23.0 | 19.4 ± 9.8 | ||
| T1-15LK | RNA-1 | 25.0 | 26.0 | 3.6 | 26.3 | 20.2 ± 9.6 | |
| T1-15LK + T2 | RNA-1 | 5.2 | 16.5 | 0.3 | 9.7 | 7.9 ± 5.9 | |
| RNA-2 | 39.0 | 46.0 | 3.7 | 30.0 | 29.7 ± 16.0 | ||
Radioactive signals were detected and quantified using a Fuji phosphorimager (FUJIX BAS1000).
Experiment 1 corresponds to that illustrated in Fig. 3.
Average of experiments ± standard deviation.
Addition of the RNA-2 transcript T2 to the wild-type T1 inoculum provoked an approximately twofold increase in the yield of progeny RNA-1 (Fig. 3). This is probably at least in part a consequence of increased stability of the progeny RNA because of its encapsidation by the CP produced by RNA-2, although we cannot strictly rule out the possibility that other RNA-2-encoded proteins could influence replication as well. Addition of T2 to the inocula containing the RNA-1 P15 mutants, on the other hand, did not enhance accumulation of the progeny RNA-1 (Fig. 3). Instead, both the mutant RNA-1 and the RNA-2 progeny accumulated to lower levels than when the inoculum consisted of wild-type RNA-1 and RNA-2 transcripts (Table 1). Since RNA-2 is dependent on RNA-1-coded proteins for its replication, it is to be expected that the lower levels of RNA-1 progeny provoked by the P15 mutations would diminish RNA-2 replication levels. This could in turn offset any putative stabilizing effect of encapsidation of the progeny RNA by the CP synthesized from RNA-2. Alternatively or additionally, the P15 mutations may interfere directly with CP synthesis from RNA-2 in the protoplasts, a point that we address more fully below.
To determine if the P15 mutations also affect negative-strand RNA synthesis, we probed Northern blots of total RNA from protoplasts infected with T2 plus either wild-type T1 or one of the P15 mutant RNA-1 transcripts with a riboprobe specific for the RNA-1 and -2 negative strands (Table 2). Measurements of the amounts of radioactive probe associated with the negative-strand bands showed that, for the P15 mutants, synthesis of negative-strand viral RNA-1 and -2 was also diminished relative to the levels observed for the wild-type RNA-1 negative strand but that the degree of inhibition was two- to fivefold less than that observed for the positive strands in the same experiment (Table 2). Thus the P15 mutations appear to have a stronger effect on accumulation of positive-sense viral RNA than on the negative-sense RNA.
TABLE 2.
Amounts of positive- and negative-strand RNA-1 and RNA-2 relative to the wild-type RNAs detected in infected protoplasts at 48 hpi
| Inoculum | Progenya | % RNA detected
|
|
|---|---|---|---|
| Positive | Negative | ||
| T1-15(−) | RNA-1 | 21.3 | 47.6 |
| T1-15(−) + T2 | RNA-1 | 23.0 | 52.0 |
| RNA-2 | 24.0 | 49.0 | |
| T1-15Nh | RNA-1 | 17.3 | 41.5 |
| T1-15Nh + T2 | RNA-1 | 28.0 | 79.0 |
| RNA-2 | 23.0 | 66.0 | |
| T1-15LK | RNA-1 | 26.3 | 66.2 |
| T1-15LK + T2 | RNA-1 | 9.7 | 48.0 |
| RNA-2 | 30.0 | 58.0 | |
Progeny RNAs were detected using a probe corresponding to the 3′-terminal 124 nucleotides of RNA-1 for detection of negative strands of both RNA-1 and -2 and to its complement for the detection of the positive strands.
The deficiency of the RNA-1 mutants is complemented by wild-type P15 provided in trans.
The fact that the mutations in P15 interfere with accumulation of RNA-2 as well as of RNA-1 suggests that this effect is due principally to loss of P15 function rather than to hypothetical alteration of putative cis-acting replication signals on RNA-1 by the mutations. To further test this point, experiments were designed to determine if the replication efficiency of the mutants could be recovered when wild-type P15 was expressed via a chimeric transcript in protoplasts infected with T2 plus mutant T1. A chimeric transcript, TRep-15, encoding CP and P15 (Fig. 1B) was first tested for its ability to be replicated. Addition of increasing amounts of TRep-15 to an inoculum consisting of T1 and T2 resulted in production of increasing amounts of Rep-15 and of subg1, presumably derived from both RNA-1 and Rep-15, to the detriment of the genomic RNAs, particularly RNA-2 (Fig. 4A). TRep-15 was also amplified upon coinoculation with T2 plus each of the three T1 mutants (Fig. 4B), but there was a concomitant increase in the amount of progeny T1-15(−), T1-15Nh, and T1-15LK produced. The RNA-1 mutants accumulated to two to five times the level observed in the absence of TRep-15 and thus reached levels nearly equivalent to those obtained with the wild-type RNA. Thus these experiments clearly indicate that the modification introduced in the RNA-1 mutants did not hinder their capacity to be replicated, provided that P15 is furnished, and favor the hypothesis that P15 plays a role in regulating the efficiency of viral RNA replication.
FIG. 4.
Complementation of the replication of P15-deficient mutants in tobacco BY-2 protoplasts by addition of TRep-15 to the inoculum. Northern blot analysis of RNA accumulation in protoplasts inoculated with 5 μg of T1, 5 μg of T2, and the quantity (in micrograms) of TRep-15 indicated below (A) and in protoplasts inoculated with 5 μg of the indicated T1 mutant and 5 μg of T2 without (−) or with (+) 0.5 μg of TRep-15 (B).
P15 does not influence translation of the RNA-2-encoded CP gene.
As mentioned above, addition of T2 to the wild-type T1 inoculum resulted in a significant increase in the yield of progeny RNA-1 (Fig. 3, lane 2). This is in contrast to the situation with the T1 mutants, where addition of T2 had no such stimulating effect (Fig. 3). One possible explanation for this observation could be that P15 not only plays a role in the replication process but also has an effect on viral RNA encapsidation. In the case of BNYVV RNA-2, it was shown that a P14 null mutation inhibited accumulation of RNA-2 (12). Whereas this deficiency could not be complemented by the expression of P14 via a replicon, the translation of CP by RNA-2 was stimulated by the replicon. From these experiments, it was concluded that P14 regulates synthesis of CP in trans. If P15 has a similar role, the amounts of CP produced when the inoculum contains the RNA-1 mutants would be reduced, which could result in deficient packaging of progeny RNA molecules. To test this hypothesis, we investigated whether the amount of CP produced by the mutants was correlated with the amount of progeny RNA-2 neosynthesized in infected protoplasts. Aliquots of different dilutions of a protein extract obtained from protoplasts infected with each T1 mutant and T2 were analyzed by Western blotting, and CP was detected by enhanced chemiluminescence (Fig. 5). The intensity of the bands observed for each combination was compared to the intensity of the bands obtained by serial dilution of an extract from protoplasts infected with wild-type T1 and T2 in a parallel experiment. As shown in Fig. 5, the relative amount of CP detected (Fig. 5, column 1) was in good correlation with the relative amount of progeny RNA-2 for the different mutants as evaluated by Northern blot analysis (Fig. 5, column 2). Further experiments will be required to determine why the presence of RNA-2 does not stimulate accumulation of the P15 mutants, but our findings illustrate that this effect is not related to a specific P15-mediated effect on CP production.
FIG. 5.
Detection of CP in protoplasts inoculated with wild-type or mutant T1 and T2. Protoplasts (106) 48 hpi were sedimented and resuspended in the loading buffer. Then a 10-μl aliquot, diluted as indicated below, was electrophoresed on an SDS–10% polyacrylamide gel. After transfer, CP was detected by enhanced chemiluminescence using a specific CP antiserum. Comparison of the intensity of the bands, evaluated by scanning, was used to estimate the amount of CP present in the different samples. The relative amount of CP present in the samples infected with mutant RNA-1 to that present in the samples infected with wild-type RNA-1 was calculated (column 1). Column 2 gives the relative amounts of progeny RNA-2 present in the same samples analyzed by Northern blot and quantified as in Table 1.
Biological activity of EGFP15 or 15EGFP.
In order to gain a better understanding of the role of P15 in the replication process we attempted to localize P15 fused to EGFP in living cells. RNA-1 transcripts (T1-EG15 and T1-15EG) (Fig. 1B) expressing N- and C-terminal fusion between EGFP and P15 were inoculated along with T2 to tobacco BY-2 protoplasts. Both T1-EG15 (Fig. 6, lane b) and T1-15EG (not shown) replicated, although the yields of mutant RNA-1 were low and the expected subgenomic RNA was difficult to detect in Northern blots. These transcripts thus behave similarly to the mutants in which P15 was knocked out, so, a priori, we cannot exclude the possibility that the fusion of EGFP to P15 perturbs P15 function.
FIG. 6.
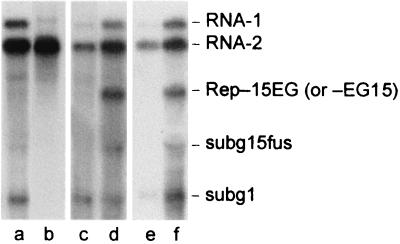
Analysis of the accumulation of RNAs containing the EGFP gene. Protoplasts were inoculated with the following RNA combinations: T1 + T2 (a), T1-EG15 + T2 (b), T1-15Nh + T2 (c), T1-15Nh + T2 + TRep-15EG (d), T1-15Nh + T2 (e), T1-15Nh + T2 + TRep-EG15 (f). Blots were probed with the same 32P riboprobes as given for Fig. 3.
For this reason, the EGFP/P15 fusion proteins were expressed from the replicon (TRep-15EG and TRep-EG15). As a control, a construct expressing a nonfused EGFP was also produced in which the gene encoding the CP in TRep-15 was replaced by EGFP to give TRep-EG (Fig. 1B). Transcripts of all three replicon constructs were amplified in protoplasts when coinoculated with T1 and T2 (not shown). A subgenomic RNA (subg15fus) of the expected size was also detectable in the samples containing the replicons, and Western blot analysis revealed the synthesis of the corresponding fusion protein (not shown). The accumulation of progeny RNA from these transcripts was significantly lower than that from the corresponding transcripts expressing nonfused P15, but addition of each replicon transcript (TRep-15EG or TRep-EG15) to the P15-deficient mutant T1-15Nh resulted in an increased yield of progeny RNA-1 (Fig. 6, compare lanes c and d and lanes e and f). Thus, the EGFP/P15 fusions are still able to stimulate replication of RNA-1 in trans.
Intracellular localization of P15.
To visualize the intracellular distribution of P15, noninfected protoplasts and protoplasts infected for 48 h with the different combinations described above were examined by fluorescence microscopy. Noninfected protoplasts were not fluorescent (Fig. 7A), whereas protoplasts infected with T1 and TRep-EG expressing free EGFP contained diffuse fluorescence characteristic of cytosoluble EGFP throughout the cytoplasm and in the nucleus (Fig. 7B). On the other hand, protoplasts infected with T1-EG15 and T2 (Fig. 7C and D) or T1 and TRep-EG15 (Fig. 7E), in which an EGFP/P15 fusion protein is expressed, contained fluorescent spots of regular size and intensity. These spots were consistently found near the periphery of the nucleus (Fig. 7C and E) but were also dispersed throughout the cytoplasm, as is clearly visible in a cortical view of the protoplasts (Fig. 7D). A similar localization of the EGFP/P15 fusion protein was observed for constructions expressing the C-terminal EGFP-fused P15 (T1-15EG or TRep-15EG) (data not shown).
FIG. 7.
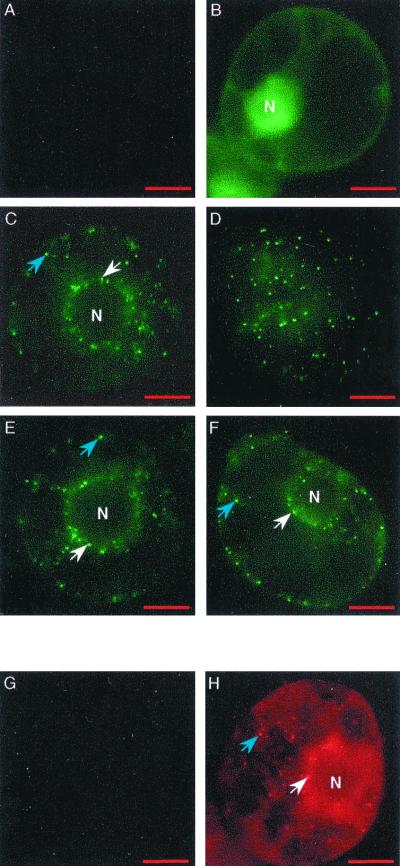
Localization of P15 in tobacco BY-2 protoplasts. Protoplasts were mock inoculated (A and G) or inoculated with T1 + TRep-EG (B), T1-EG15 + T2 (C and D), T1 + TRep-EG15 (E), pCK-EG15 (F), or viral RNA (H). Observations were carried out 48 hpi. EGFP expression (A to F) and immunostaining of P15, using P15 antiserum and Cy3 antibodies (G and H), were detected by epifluorescence microscopy. White arrows indicate a localization of P15 around the nucleus (N), and blue arrows indicate a localization within the cytoplasm. 4′,6′-Diamidino-2-phenylindole (DAPI) staining (not shown) was used to localize the nucleus. For panel F, the number of integrated images was increased to obtain fluorescence comparable to that of panels C to E. Bar, 10 μm.
To prove that the observed distribution of fluorescence was mediated by P15 and was not an artifact due to the fusion of P15 to EGFP, mock-infected and viral RNA-infected protoplasts were harvested at 48 hpi, fixed, and processed for indirect immunofluorescence observation using the anti-P15 serum and an anti-immunoglobulin G Cy3-conjugated secondary antibody. The mock-infected protoplasts (Fig. 7G) were not fluorescent, whereas the infected protoplasts (Fig. 7H) contained well-defined red spots of regular size localized around the nucleus and dispersed in the cytoplasm, exactly as observed with the EGFP/P15 fusion protein.
Localization of P15 is independent of viral infection.
To determine whether the subcellular localization of P15 is dependent on virus infection, protoplasts were transfected with the transient expression vector pCK-EG15, in which the EGF/P15 sequence was under the control of a duplicated 35S promoter (Fig. 1B). Observation of the protoplasts at 48 hpi showed that the EGFP/P15 fusion protein was expressed, although the fluorescence signal was significantly lower than in protoplasts infected with T1-EG15. However, fluorescent spots localized around the nucleus and within the cytoplasm could nevertheless be observed (Fig. 7F), and the distribution of the EGFP/P15 was similar to that obtained previously in infected protoplasts. Thus the observed pattern of accumulation of P15 within the cell occurs in the absence of other viral factors.
Subcellular localization of PCV RNA replication products.
BrUTP incorporation has been used to label active sites of brome mosaic virus RNA synthesis in barley protoplasts (33) and to visualize grapevine fanleaf nepovirus RNA synthesis in tobacco BY-2 protoplasts (7). The procedures used for BrUTP labeling in the two systems were very similar, except that BrUTP was used at a high concentration (10 mM) and incorporated for short periods (15 min) in the first case, whereas a smaller concentration of BrUTP (100 μM) and longer periods of incorporation (6 h) were used in the second case. We employed both conditions to analyze PCV RNA synthesis in T1-EG15-infected protoplasts at 30 hpi. Protoplasts were treated with actinomycin D for 1 h prior to BrUTP labeling to block host DNA-dependent transcription without affecting PCV-directed RNA-dependent RNA synthesis. The protoplasts were then fixed and processed for indirect immunofluorescence using antibodies that recognize bromouridine-containing RNA. The distribution of PCV RNA (red) and EGFP/P15 fusion protein (green) was analyzed using confocal microscopy to image optical sections with a focal depth of only 0.45 μm, thus removing out-of-focus glare and improving resolution. Figure 8 shows representative observations. As expected, no green or red fluorescent signals were observed for the noninfected protoplasts (Fig. 8G and H). In the T1-EG15- and T2-infected protoplasts, green spots corresponding to the EGFP/P15 fusion protein were localized as before around the nucleus and throughout the cytoplasm (Fig. 8A and D). The red spots, corresponding to RNA accumulation sites, were also distributed around the nucleus and throughout the cytoplasm (Fig. 8B and E). However, even when BrUTP incubation was carried out for only 15 min, a time for which only labeling of nascent or freshly completed RNA molecules (32) should have occurred, most of the red spots did not colocalize with green spots (Fig. 8C and F), indicating that P15 is not abundantly present at the sites of RNA replication. While occasionally a green spot was very close to a red spot and in a few (less than 0.2%) cases they apparently colocalized (Fig. 8C), these were extremely rare events.
FIG. 8.
Localization of incorporated BrUTP and EGFP/P15 fusion protein. The two first rows show images from two representative T1-EG15 + T2-infected protoplasts (A to C and D to F) that were harvested 20 hpi, labeled for 6 h with BrUTP, fixed, and processed for immunofluorescence observation using antibodies that detect BrUTP. The last row shows mock-inoculated protoplasts processed identically (G and H). For each protoplast, green fluorescence images of EGFP/P15 (A, D, and G) and red fluorescence images of incorporated BrUTP (B, E, and H) were collected from the same 0.45-μm-thickness optical section with the multitrack confocal microscope and appropriate filters. The two images were digitally superimposed (C and F). Bar, 10 μm.
DISCUSSION
P15 is involved in PCV RNA accumulation.
To address the role of P15 protein in virus replication, mutations affecting the protein were generated and the effects on RNA accumulation were analyzed in tobacco BY-2 protoplasts. The findings reveal that P15 is not an essential protein for replication as the three mutants, including the null mutant T1-15(−), were infectious. We did not analyze whether reversion of the mutations to the wild type has occurred in these experiments, although this seems very unlikely given the short infection times employed (48 h) and our failure to detect expression of P15 in protoplasts infected with the defective mutants.
One possible explanation for the decreased accumulation of the P15 mutants is that the mutations could disrupt cis-acting elements on RNA-1 involved in replication. This is a particular concern for mutant T1-15LK, where the modifications introduced into the RNA were more important. This hypothesis is ruled out, however, by the finding that the mutants can be complemented in trans by addition of a replicon expressing wild-type P15. Therefore, even if certain of the introduced modifications have a slight cis effect on RNA replication, our results nonetheless demonstrate that P15 itself plays an important role in regulating viral RNA accumulation.
The decreased accumulation of RNA in the absence of P15 could be either the result of a lower replication rate, which would suggest a direct role for P15 in the replication process (as a cofactor in the replication complex, for example), or of increased degradation of the progeny RNA, e.g., a reflection of a direct or indirect effect of P15 on the stability of the progeny RNA. This latter result could arise if the replicated RNA is poorly encapsidated. In the case of BSMV, null mutations of γb protein were shown to significantly inhibit the synthesis of α and β BSMV RNAs and to provoke a disproportionate reduction in levels of CP and movement protein (28). As noted earlier, mutation of the BNYVV P14 protein also inhibited the accumulation of RNA-2 and at the same time dramatically reduced the accumulation of CP (12). Therefore, in both cases, the proteins were suggested to be involved primarily in the regulation of gene expression at the translation level. In the case of PCV, on the other hand, the accumulation of both progeny RNA-1 and RNA-2 was reduced significantly when RNA-1 expressed a deficient P15, but the amount of CP produced in the infected protoplasts was approximately proportional to the amount of progeny RNA-2 present. This indicates that the translation of RNA-2 is not directly affected by the P15 mutations and that sufficient amounts of CP were produced to encapsidate progeny RNAs.
P15 rarely colocalized with viral RNA replication sites.
EGFP-fused P15 was detected in spots around the nucleus and within the cytoplasm. Controls showed that P15 could be detected by immunofluorescence at similar sites, confirming that the localization of P15 is not altered by its fusion to EGFP. The aforesaid subcellular localization of P15 occurs in the absence of viral infection, indicating that this behavior is either an intrinsic property of the protein or is mediated by one or more host factors which are present constitutively. Interactions between P15 and cellular factors could, for example, promote changes in the ultrastructure of membranes involved in positive-strand RNA replication (6, 11, 20, 34).
Using confocal microscopy and BrUTP labeling of viral RNA, we observed that viral RNA replication sites were distributed, like sites of P15 accumulation, around the nucleus and within the cytoplasm. This distribution resembles that of brome mosaic virus RNA replication complexes, which colocalized with the replication proteins 1a and 2a and which have been demonstrated to be associated with the endoplasmic reticulum (33). Further analysis is needed to determine with which membrane compartment (3) PCV RNA replication is associated. When EGFP15 and immunofluorescence of BrUTP-labeled RNA were observed in the same protoplasts by confocal microscopy, P15 was sometimes found near RNA accumulation sites but rarely colocalized with these sites.
The fact that P15 did not extensively colocalize with the BrUTP-labeled loci could mean that such loci do not correspond to active RNA replication sites but rather to RNA which has been liberated from the replication complex. This seems unlikely, however, as no differences in localization of BrUTP-labeled RNA were obtained for short and long BrUTP incorporation times. Furthermore, experiments with antibodies specific for 131K and 191K viral replicase proteins showed a perfect colocalization with BrUTP-labeled viral RNA (unpublished data). We conclude that the BrUTP-labeled sites are indeed the sites of replication. Our failure to detect P15 at these sites could have at least two possible explanations. One possibility is that P15 is a direct participant in viral replication but is only very transiently present at the viral RNA replication sites and is hence difficult to detect there. The second possibility, which we favor, is that P15 is never localized at the replication sites and that its effect on viral RNA replication and/or accumulation is indirect. Thus, P15 might localize to the same membranous structures as the RNA replication complex and intervenes in replication by interacting with these structures to create a favorable environment for the replication process and/or to stabilize the progeny RNA prior to encapsidation. Another attractive hypothesis is that P15 could be a suppressor of host defense mechanisms similar or identical to posttranscriptional gene silencing (PTGS) (29). Such activity has recently been attributed to the HC-Pro of tobacco etch virus (18), the 2b protein of cucumber mosaic virus (1), and several other viral proteins (37). The preferential decrease of progeny RNA positive strands relative to negative strands for the P15 mutants is consistent with such a hypothesis, since PTGS-mediated RNA degradation would be expected to preferentially target the more abundant positive-sense RNA (27) in the absence of P15. Experiments are currently underway to determine if P15 plays a role in PTGS suppression during the PCV infection cycle.
ACKNOWLEDGMENTS
We thank T. Dreher for providing plasmids for preparation of probes corresponding to the last 124 nucleotides of RNA-1 and K. Richards for critically reading the manuscript. We are also grateful to D. Scheidecker for technical assistance.
This work was supported by the CNRS and by the Université Louis Pasteur (ULP), Strasbourg. The Zeiss LSM-510 confocal microscope was cofinanced by CNRS, ULP, the “Région Alsace,” the “Association de la Recherche sur le Cancer” (ARC), and the “Ligue de Recherche sur le Cancer.”
REFERENCES
- 1.Brigneti G, Voinnet O, Li W X, Ji L H, Ding S W, Baulcombe D C. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Candresse T, Morch M D, Dunez J. Multiple alignment and hierarchical clustering of conserved amino acid sequences in the replication-associated proteins of plant RNA viruses. Res Virol. 1990;141:315–329. doi: 10.1016/0923-2516(90)90003-2. [DOI] [PubMed] [Google Scholar]
- 3.De Graaff M, Jaspars E M J. Plant viral RNA synthesis in cell-free systems. Annu Rev Phytopathol. 1994;32:311–335. [Google Scholar]
- 4.Donald R G K, Jackson A O. RNA-binding activities of barley stripe mosaic virus gamma b fusion proteins. J Gen Virol. 1996;77:879–888. doi: 10.1099/0022-1317-77-5-879. [DOI] [PubMed] [Google Scholar]
- 5.Erhardt M, Stussi-Garaud C, Guilley H, Richards K E, Jonard G, Bouzoubaa S. The first triple gene block protein of peanut clump virus localizes to the plasmodesmata during virus infection. Virology. 1999;264:220–229. doi: 10.1006/viro.1999.9997. [DOI] [PubMed] [Google Scholar]
- 6.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaire F, Schmitt C, Stussi-Garaud C, Pinck L, Ritzenthaler C. Protein 2A of grapevine fanleaf nepovirus is implicated in RNA2 replication and colocalizes to the replication site. Virology. 1999;264:25–36. doi: 10.1006/viro.1999.9927. [DOI] [PubMed] [Google Scholar]
- 8.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. A conserved NTP-motif in putative helicases. Nature. 1988;333:22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- 9.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gramstat A, Courtpozanis A, Rohde W. The 12 kDa protein of potato virus M displays properties of a nucleic acid-binding regulatory protein. FEBS Lett. 1990;276:34–38. doi: 10.1016/0014-5793(90)80500-i. [DOI] [PubMed] [Google Scholar]
- 11.Hatta T, Francki R I B. Cytopathic structures associated with tonoplasts of plant cells infected with cucumber mosaic and tomato aspermy viruses. J Gen Virol. 1981;53:343–346. [Google Scholar]
- 12.Hehn A, Bouzoubaa S, Bate N, Twell D, Marbach J, Richards K, Guilley H, Jonard G. The small cysteine-rich protein P14 of beet necrotic yellow vein virus regulates accumulation of RNA 2 in cis and coat protein in trans. Virology. 1995;210:73–81. doi: 10.1006/viro.1995.1318. [DOI] [PubMed] [Google Scholar]
- 13.Herzog E, Guilley H, Fritsch C. Translation of the second gene of peanut clump virus RNA-2 occurs by leaky scanning in vitro. Virology. 1995;208:215–225. doi: 10.1006/viro.1995.1145. [DOI] [PubMed] [Google Scholar]
- 14.Herzog E, Guilley H, Manohar S K, Dollet M, Richards K, Fritsch C, Jonard G. Complete nucleotide sequence of peanut clump virus RNA 1 and relationships with other fungus-transmitted rod-shaped viruses. J Gen Virol. 1994;75:3147–3155. doi: 10.1099/0022-1317-75-11-3147. [DOI] [PubMed] [Google Scholar]
- 15.Herzog E, Hemmer O, Hauser S, Meyer G, Bouzoubaa S, Fritsch C. Identification of genes involved in replication and movement of peanut clump virus. Virology. 1998;248:312–322. doi: 10.1006/viro.1998.9287. [DOI] [PubMed] [Google Scholar]
- 16.Ho S N, Junt H D, Horton R M, Puller J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Jaspars E M J. Interaction of alfalfa mosaic virus nucleic acid and protein. In: Davies J W, editor. Molecular plant virology. Boca Raton, Fla: CRC Press; 1985. pp. 151–221. [Google Scholar]
- 18.Kasschau K D, Carrington J C. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 19.Kasschau K D, Carrington J C. Requirement for HC-Pro processing during genome amplification of tobacco etch potyvirus. Virology. 1995;209:268–273. doi: 10.1006/viro.1995.1254. [DOI] [PubMed] [Google Scholar]
- 20.Kim K S. An ultrastructural study of inclusions and disease development in plant cells infected by cowpea chlorotic mottle virus. J Gen Virol. 1977;35:535–543. [Google Scholar]
- 21.Koonin E V, Boyko V P, Dolja V V. Small cysteine-rich proteins of different groups of plant RNA viruses are related to different families of nucleic acid-binding proteins. Virology. 1991;81:395–398. doi: 10.1016/0042-6822(91)90512-a. [DOI] [PubMed] [Google Scholar]
- 22.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- 25.Neeleman L, Bol J F. Cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology. 1999;254:324–333. doi: 10.1006/viro.1998.9568. [DOI] [PubMed] [Google Scholar]
- 26.Oncino C, Hemmer O, Fritsch C. Specificity in the association of tomato black ring virus satellite RNA with helper virus. Virology. 1995;213:87–96. doi: 10.1006/viro.1995.1549. [DOI] [PubMed] [Google Scholar]
- 27.Palauqui J C, Vaucheret H. Transgenes are dispensable for the RNA degradation step of cosuppression. Proc Natl Acad Sci USA. 1998;98:9675–9680. doi: 10.1073/pnas.95.16.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petty I T D, French R, Jones R W, Jackson A O. Identification of barley stripe mosaic virus genes involved in viral RNA replication and systemic movement. EMBO J. 1990;9:3453–3457. doi: 10.1002/j.1460-2075.1990.tb07553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratcliff F, Harrison B D, Baulcombe D C. A similarity between viral defense and gene silencing in plants. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 30.Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C. Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc Natl Acad Sci USA. 1996;93:5888–5893. doi: 10.1073/pnas.93.12.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Restrepo-Hartwig M A, Carrington J C. The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J Virol. 1994;68:2388–2397. doi: 10.1128/jvi.68.4.2388-2397.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol. 1999;73:10303–10309. doi: 10.1128/jvi.73.12.10303-10309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlegel A, Giddings T H, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Bokhoven H, Le Gall O, Kasteel D, Verver J, Wellink J, van Kammen A. Cis- and trans-acting elements in cowpea mosaic virus RNA replication. Virology. 1993;195:377–386. doi: 10.1006/viro.1993.1387. [DOI] [PubMed] [Google Scholar]
- 36.Verchot J, Carrington J C. Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J Virol. 1995;69:3668–3674. doi: 10.1128/jvi.69.6.3668-3674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voinnet O, Pinto Y M, Baulcombe D C. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe Y, Meshi T, Okada Y. Infection of tobacco protoplasts with in vitro transcribed tobacco mosaic virus RNA using an improved electroporation method. FEBS Lett. 1987;219:65–69. [Google Scholar]
- 39.Zhou H, Jackson A O. Analysis of cis-acting elements required for replication of barley stripe mosaic virus RNAs. Virology. 1996;219:150–160. doi: 10.1006/viro.1996.0232. [DOI] [PubMed] [Google Scholar]