Abstract
Meloidogyne incognita is one of the globally serious plant parasitic nematodes. New control measure is urgently needed to replace the common chemical control method. Ascarosides are pheromones regulating the nematodes’ aggregation, avoidance, mating, dispersal and dauer recovery and formation. Ascr#9, one of the ascarosides, exhibits the potential to repel M. incognita. However, the nematode genes involved in the perception of ascr# 9 signal are totally unknown. In this study, the transcriptome of ascr#9-treated second stage M. incognita juveniles (J2s) was analyzed, 44 pathways were significantly affected, multiple ligand-receptor and mucin type O-glycan were induced, and olfactory transduction was disturbed. A total of 11 highly differentially expressed genes involved in neuroactive ligand-receptor interaction and FMRFamide-like peptide related process were identified and knocked down by RNAi. The dispersal rates of M. incognita with three knocked-down genes (flp-14, mgl-1 and ADOR-1) significantly decreased, respectively, when ascr#9 was present. The results demonstrate that flp-14, mgl-1, and ADOR-1 are involved in the dispersal behavior of M. incognita nematodes responding to ascr#9, which promotes the interaction study between ascarosides and M. incognita, and provides new ideas for the prevention and control of M. incognita by using pheromone ascarosides.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76370-5.
Keywords: Meloidogyne incognita, Ascaroside, Dispersal, RNAi
Subject terms: RNAi, Conservation biology, Animal behaviour, Entomology
Introduction
Plant parasitic nematodes (PPN), which live in soil and harm plants, cause global economic losses of $157 billion annually1,2. Meloidogyne incognita is one of the globally important plant parasitic nematodes. The second stage infective juveniles (J2s) of M. incognita penetrate the elongated region of the plant root tip and establish a permanent feeding site in the root, inducing giant cell formation and resulting in hypertrophy and enlargement of plant cells3,4. This plant nematode species possesses the ability to exploit plant-derived photosynthates and nutrients, leading to mortality in tender plant, infection in mature crops and subsequent reduction in crop yield5,6. Their extensive host range makes them one of the most difficult agricultural pests to control7. Cultural practices, crop rotation and resistant crop plantation, together with chemical nematicides, are generally employed to control plant parasitic nematodes. However, many toxic nematicides are removed from the market because of their negative impacts on the environment and public health8–10. Therefore, novel and environmentally friendly techniques for the control of these harmful plant nematodes are urgently needed.
PPNs undergo their life cycle by means of a robust chemical sensing system, which allows them to perceive and respond to various chemical signals from soil and plants. Usually, the root secretions by plants strongly attract the second stage larvae of M. incognita11. Like many organisms, nematodes secrete and use pheromones for interindividual conspecifics, which trigger neuronal stimulation related to various behavioral responses and modulate developmental or physiological responses to adapt to new environments12. Ascarosides are widely believed to be the pheromones of nematodes and play a critical role in the regulation of nematodes’ aggregation, avoidance, mating, dispersal and dauer recovery and formation13–20. Ascr#9 is a common small molecule in the dispersal behavior of entomopathogenic nematodes17,19,21, Caenorhabditis elegans21,22. and M. incognita23. Ascr#9 also has a significant impact on the dispersal behavior of M. incognita20. However, the genes involved in the dispersal behavior of M. incognita under the stress of ascr#9 need to be identified.
J2s of all plant parasitic nematodes do not exhibit feeding behavior before entering the plant, and RNA is not easily introduced into J2s by microinjection. Soaking has been demonstrated as an efficient approach for achieving RNA interference (RNAi) in plant parasitic nematodes, especially when utilizing the neurostimulator octopamine to enhance dsRNA absorption24. Subsequently, RNAi technology was quickly used to study and control plant parasitic nematodes including M. incognita and H. glycines25–28.
In this study, based on the effects of ascr#9 on the dispersal of M. incognita, the transcriptome of ascr#9-treated M. incognita J2s was analyzed and 11 highly differentially expressed genes in response to ascr#9 were knocked down by RNAi. Three genes (flp-14, mgl-1 and ADOR-1) were found to have a significant reduction in the dispersal rate of M. incognita when ascr#9 was present. The results demonstrated that flp-14, mgl-1 and ADOR-1 were involved in the dispersal behavior of M. incognita nematodes responding to ascr#9, and may provide useful clues for the establishment of alternative technologies by using ascarosides as repellents for the safe and effective control of M. incognita.
Materials and methods
Nematodes
M. incognita J2s were collected from infected tomato (Lycopersicon esculentum) roots at Baiyun District, Guangzhou, Guangdong Province (113.27°N, 23.16°E), according to the previous description20. The roots were cut into 1–2 cm pieces, immersed and dramatically shaken in 1% (v/v) NaClO (Tianjin Best Chemical Co., Ltd.) for 3 min. The resulting mixture was filtered by 100 (150 μm), 300 (48 μm) and 500 mesh (25 μm) screens(Shanghai Machinery Equipment Factory, Shanghai, China), respectively, and the collecting nematode eggs on the 500 mesh were washed with sterile ultrapure water and placed in an incubator at 25℃ for 5 days29. The hatching second infective juveniles were collected, washed with sterile ultrapure water three times, sterilized with streptomycin sulfate solution (100 µg/mL) overnight at a 100 rpm shaker at 25℃ and washed three times with sterile ultrapure water before use. The nematodes were identified based on the random amplified polymorphic DNA (RAPD) fragments specific for Meloidogyne incognita, M. javanica and M. arenaria, respectively. by using 1000 J2s30.
Transcriptome analysis of M. Incognita J2s in response to ascr#9
Based on the results of Dai et al. (2022)20, the synthesized ascr#9 by Wuxi Pharmaceutical (Wu Xi App Tec), with a purity of over 95%, was selected to determine its effect on the dispersal of M. incognita, for the transcriptome analysis. 5000 M. incognita J2s in 200 µL sterile ultrapure water were placed onto the center (diameter of 3 cm) of a 9 cm plate containing 20 mL 2% (w/v) agar at 25℃, then 125 µL of ascr#9 (0.5 µM) was introduced into the plate center (Fig. 1). Sterile ultrapure water was used as a control. The assay started when the liquid was absorbed by the agar medium and the nematodes were free to move from the deployment site18,21. Four replicate plates were set up for each treatment. The nematodes remaining in the 3 cm circle and on the circle line (regarded as non-dispersal IJs), and moving out of the circle (regarded as dispersal IJs) were collected and counted in each plate after 1 h. The experiment was independently performed two times.
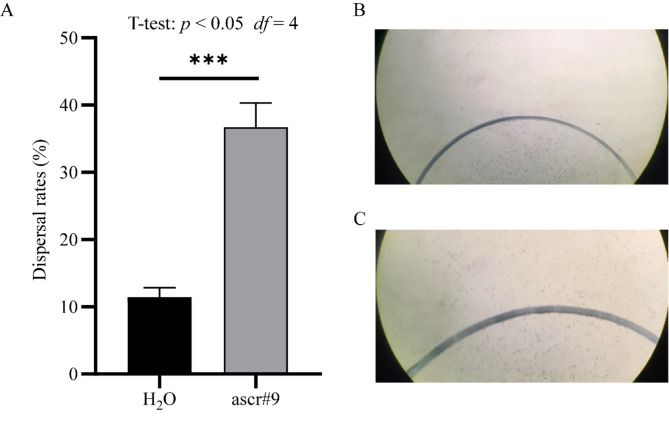
Fig. 1.
Effect of ascr#9 on the dispersal ofM. incognita. M. incognita J2s were placed in the center of a 9 cm diameter agar plate containing 20 mL 2% agar. The dashed lines represent a 1.1 cm diameter circle as the deployment boundary. The juveniles that moved outside the boundary were considered dispersed. (A) The dispersal rates of the treated M. incognita with 0.5 µM ascr#9 for 1 h. The asterisks on the horizontal line indicated extremely significant (T-test, p < 0.001, df = 4). (B) Dispersal of M. incognita J2s treated with sterile extrapure water at 6.75x; (C) Dispersal of M. incognita J2s treated with 0.5 µM ascr#9 under a 10× microscope.
After 1 h treatment, the ascr#9 treated dispersal and control non-dispersal M. incognita J2s were washed off the agar with sterile ultrapure water, respectively, and immediately treated in liquid nitrogen, and stored in a -80 °C refrigerator for RNA extraction.
RNA preparation and sequencing
Total RNA was extracted using the EASYspin RNA micro Kit (Aidlab, Beijing, China), according to the product instruction. The Agilent 2100 RNA Nano 6000 Assay Kit (Agilent Technologies, Santa Clara, CA, USA) and non-denaturing agarose gel electrophoresis were used to determine the quantity and quality of RNA in the samples. A total of 8 cDNA libraries were constructed from Ascr#9 treated samples and sterile ultrapure water treated (control) samples. Each sample with four biological replicates and 5000 individuals was used for each biological replicate. The quality check of the libraries were analyzed on Qubit 2.0, Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA), and ABI StepOnePlus Real-Time PCR system (ABI, Waltham, MA, USA). An Illumina HiSeq2000 platform (Illumina, San Diego, CA, USA) was used for sequencing. All of the raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1010386.
De novo assembly and annotation of transcriptomes
The reads contained adaptor, low-quality (average quality < Q20), and single end with N bases more than 3 were removed from the raw data to obtain the clean reads. Clean reads were assembled using Trinity de novo assembler v.2.8.531 with default option to obtain transcripts. Subsequently, the transcripts were clustered, and the longest transcripts in the same cluster were selected as unigenes. Protein coding capacity of unigenes were identified by TransDecoder v5.5.0 (https://github.com/TransDecoder). Overall unigenes were annotated against NCBI non-redundant protein database (nr) and Swiss-prot database using BlastX (E-value < 1e-5). In addition, possible gene functions were assessed for the unigenes based on Gene Ontology (GO)32. Pathway annotation was assigned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) online service33.
Analysis of differentially expressed genes (DEGs)
Clean reads of each library were aligned to the unigenes using Bowtie software (https://bowtie-bio.sourceforge.net/index.shtml) with default parameters. The abundance of transcripts and the fragments per kilobase per million mapped read (FPKM) were estimated using RSEM from the utility package of the Trinity software34. DEGs were calculated using edgeR35. p-values were corrected for multiple hypothesis tests, and the threshold p-value by False Discovery Rate (FDR) was determined. Genes in different samples with FDR < 0.05 and |log2FC| > 1 were considered as DEGs. Pearson correlation-base heatmap was conducted by R package gplots heatmap.2 function, and PCA analysis were performed by using the base R princomp function and redraw by ggplot2.
GO annotations were performed with Blast2GO program36 to determine each putative protein’s role in biological process, molecular function and cellular component, with default setting. The pathway enrichment analysis of DEGs was conducted by KOBAS 3.037 with a threshold p-value set to ≤ 0.05.
Quantitative real-time PCR (qRT-PCR)
Total RNA from 10,000 M. incognita J2s treated or untreated with ascr#9 using the EASYspin RNA micro Kit (Aidlab, Beijing, China) was used to validate the RNA-seq experiment, and reverse transcription and gDNA removing were followed. A total of 1 µg of RNA from each sample was used for cDNA synthesis according to the manufacturer’s protocol (TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix, TRAN, Beijing, China). The primers for the genes of interest were designed using the Primers designing tool online (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) (Table S1), and synthesized by Sangon (Shanghai, China). qRT-PCR was performed on the CFX Connect™ Optics Module (Bio-Rad Laboratories, Hercules, CA, USA)38. Specificity of amplification was assessed by disassociation or melt curve analysis at 60–95 °C after 40 cycles, followed by a melting curve analysis. Each reaction mix contained 1.0 µL previously diluted cDNA (1:10), 5.0 µL 2×TransStart®Top /Tip Green qPCR SuperMix (TRAN, Beijing, China) and 10.0 mM of each primer, for filling a final volume of 10 µL using 3.6 µL RNase-free water. The amplification reactions were carried out at a hot start of 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min in a qRT-PCR high profile non skirted white 96-well plate (Bio-Rad Laboratories, Hercules, CA, USA). Four biological replicates were conducted for each treatment, with three technical replicates performed per biological replicate in the qRT-PCR analysis. Quantitative measurements were normalized by the reference gene Mi-actin, and relative expression levels were calculated using 2−ΔΔCt method39,40.
RNA interference (RNAi) validation of DEGs
M. incognita cDNA was used as a template to synthesize double-stranded RNA (dsRNA) with primers targeting the key gene sequences (Table S2) according to the T7 RiboMAX™ Express RNAi System (Promega, Madison, USA), and the synthesized dsEgfp was used as a control.
RNAi of the key genes from the transcriptome analysis was conducted by soaking method41. Fluorescein isothiocyanate (FITC) was used as a tracer to assess uptake efficiency. Freshly harvested 10,000 J2s were immersed in a 50 µL soaking buffer containing 5 µL FITC (1 mg/mL), 5 µL octopamine (300 mM) and 10 µL of dsRNA (5 mg/mL)42. Soaked J2s were incubated for 6 h in the dark at room temperature on an 100 rpm rotator. As controls, J2s were incubated in the soaking buffer alone or in the soaking buffer with dsEgfp. After incubation, J2s were washed five times with nuclease free water by centrifugation at 6000 rpm, 10 °C for 3 min. FITC in the treated nematodes were observed under an Olympus BX51 fluorescence microscope. qRT-PCR was performed to evaluate the RNAi efficiency, following the above method.
In brief, 200 J2s from dsRNA treatments (soaking buffer with 1 mg dsRNA/mL) and three controls (soaking buffer with dsEgfp, soaking buffer alone, and sterile ultrapure water) in 10 µL sterile ultrapure water were placed onto the center (diameter = 1.1 cm) of a 9 cm plate containing 20 mL 2% (w/v) agar at 25℃, then 5 µL of ascr#9 at 0.5 µM was introduced into the plate center. The experiment began when the water was absorbed by the medium and the nematodes were free to move from the deployment site18,21. Three replicate plates were set up for each treatment or control. The nematodes remaining in the center circle and on the circle line (regarded as non-dispersal IJs), and moving out of the circle (regarded as dispersal IJs) were collected and counted in each plate after 30 min. The experiment was independently performed two times.
Data analysis
The data was expressed as Mean ± SE. All percentage data were arcsin square root transformed prior to statistical analysis by ANOVA, SPSS 17.0 (SPSS, Chicago, IL). Tukey multiple comparison method was employed for significant difference test (p < 0.05). GraphPad Prism 8 was used for graphing and statistics of the data.
Results
Ascr#9 influence on the dispersal behavior of M. Incognita J2s
The dispersal rate (36.7 ± 2.06%) of M. incognita J2s treated by ascr#9 was significantly higher (T-test, P < 0.001, df = 4, R2 = 0.97) than that in the control (11.4 ± 0.81%) (Fig. 1). It appeared that most of the non-treated nematodes were still retained in the circle (Fig. 1B), however, much more ascr#9-treated J2s apparently moved out of the circle (Fig. 1C), indicating the significant influence of the ascr#9 on M. incognita J2s dispersal.
RNA sequencing analysis
Based on the dispersal assay, eight cDNA libraries were constructed from the J2s outside the ascr#9-treated circles and inside the water-treated circles (four replicates for the treatment or control).
After removal of adaptor sequences and low-quality reads, eight libraries for ascr#9 treated samples (T1-T4) and control samples (CK1-CK4) yielded 41.7 (T1), 43.7 (T2), 41.1 (T3), 39.6 (T4), 47.1 (CK1), 46.8 (CK2), 45.6 (CK3), and 44.4 (CK4) million high quality reads (Q20 > 96.8%; Q30 > 91.73), respectively (Table S3). A total of 132,125 unigenes were generated by Trinity software, and 114,509 unigenes with FPKM > 1 in at least one sample were used for further analysis (Table S4).
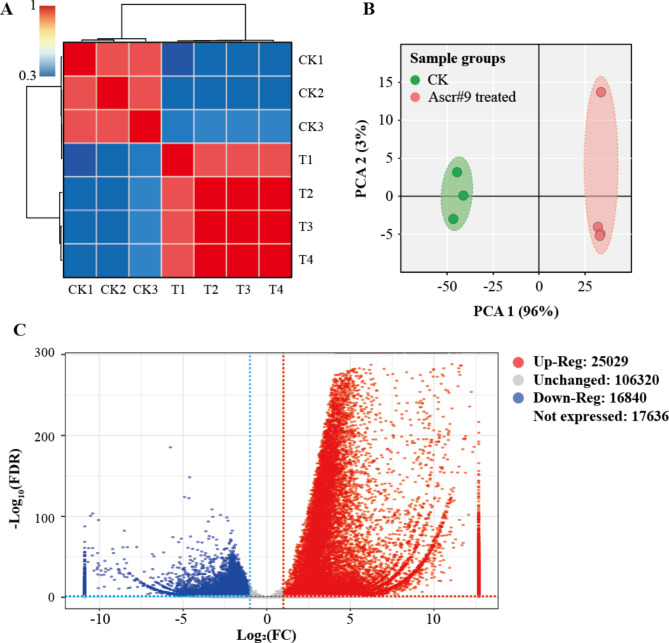
The FPKM value of expressed genes was shown in Table S5. The correlation coefficient of the intragroup samples always exceeded 0.99, and were significantly higher than that of the intergroup samples (Fig. 2A). A similar result was obtained in the principal components analysis (PCA) (Fig. 2B). A total of 41,932 DEGs (|log2FC| > 1 and FDR < 0.05) were detected in M. incognita under ascr#9 induction, of which 25,029 were up-regulated and 16,840 down-regulated (Fig. 2C).
Fig. 2.
Correlation and differential gene expression analysis. (A) Heatmap of duplicate samples of ascr#9 treated and control group, with the color spectrum ranging from blue through yellow to red, representing Pearson correlation coefficient from 0.3 to 1. CK, blank control; T, treat with ascr#9. (B) Principal components analysis (PCA) of the transcriptome for ascr#9 treated and control samples of M. incognita. Treatment and control were indicated by different colors. (C) Gene expression pattern between ascr#9 treatment and control. The number of up- and down-regulated DEGs in M. incognita were shown. The logarithms with FDR values larger than 300 were all plotted at 300. The regions delimited by red and blue lines represented up-regulated (log2FC > 1, FDR < 0.05) and down-regulated (log2FC < − 1, FDR < 0.05) genes, respectively.
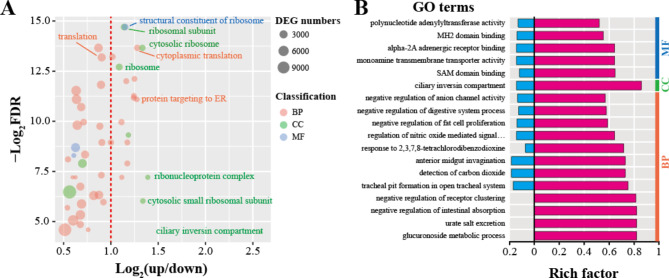
In order to better understand the putative function of the DEGs, GO enrichment analysis was carried out (Table S6). Totally 50 enriched GO processes were identified (FDR < 0.05). Up-regulated genes in 20 GO processes were at least 2 times more than that of down-regulated genes, including cytoplasmic translation (GO:0002181), ribosome assembly (GO:0042255), and protein targeting to ER (GO:0045047) in biological processes; cytosolic ribosome (GO:0022626) and ciliary inversin compartment (GO:0097543) in cellular component; structural constituent of ribosome (GO:0003735) in molecular function (Fig. 3A and Table S6). These processes were mainly participated in ribosomal construction and translation. Notably, some GO processes exhibiting significantly changes between up- and down-regulated genes but with higher FDR value (FDR > 0.05, p-value < 0.05) should also be worthy of attention. For example, urate salt excretion (GO:0097744), response to 2,3,7,8-tetrachlorodibenzodioxine (GO:1904612), and glucuronoside metabolic process (GO:0019389), the up-regulated genes in these biological processes are ten times more abundant than the down-regulated genes (Fig. 3B and Table S6).
Fig. 3.
GO enrichment analysis forM. incognitain response to ascr#9. (A) The X-axis represented the log2 ratio of up-regulated genes and down-regulated genes. The gene numbers were indicated by the size of the bubble, and different colors represented different classifications. (B) Significantly biased GO term in M. incognita after ascr#9 treatment. The X-axis was divided by “0”, and the rich factor of the up-regulated and down-regulated genes were shown in the right (magenta color) and left (blue color) parts, respectively. BP, biological process; CC, cellular component; MF, molecular function.
To better identify the significantly responsive pathways after the treatment of ascr#9, DEGs with FDR < 0.001 and |log2FC| > 4 was used as input data. A total of 12,470 up-regulated genes and 2419 down-regulated genes were enriched in 132 and 55 pathways, respectively (Table S7). 44 pathways showed significantly positive effects (up-regulated genes were 6 times more than down-regulated genes in the corresponding pathway) after ascr#9 was treated, such as apoptosis in cellular processes; Notch, MAPK, and ErbB signaling pathways, neuroactive ligand-receptor interaction in environmental information processing. Melanogenesis, neurotrophin signaling pathway, and olfactory transduction in organismal systems; Histidine, galactose, fructose, mannose, sulfur, nitrogen in metabolism (Fig. 4A).
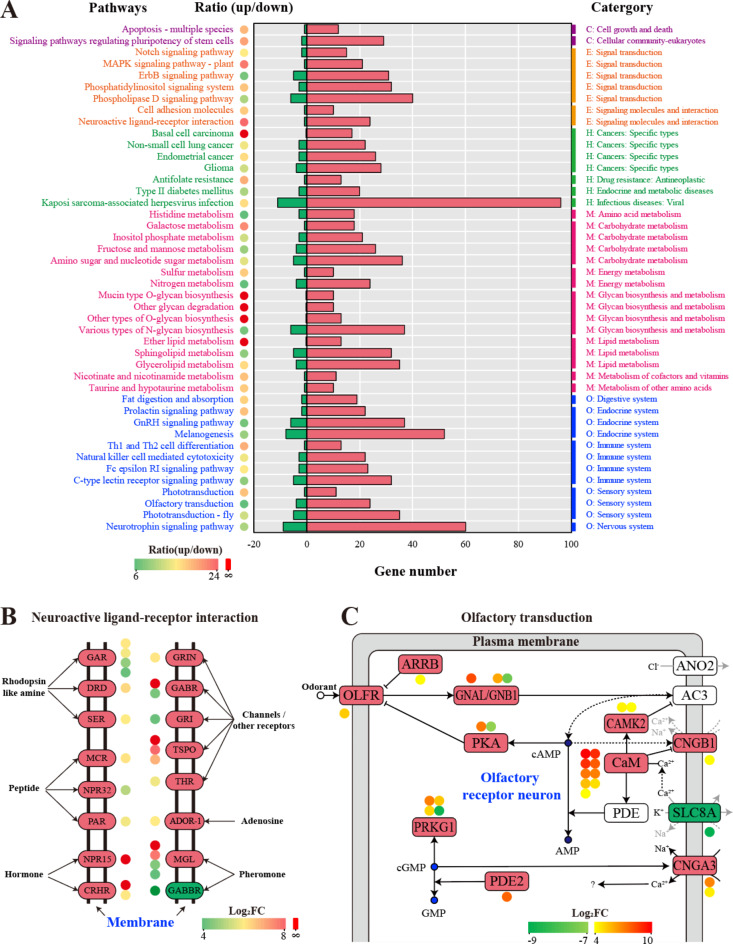
Fig. 4.
Pathway analysis of the DEGs inM. incognitainduced by ascr#9. (A) 44 significantly responsive pathways in M. incognita after the treatment of ascr#9. Up- or down-regulated genes under ascr#9 treatment were indicated by different colors, and the right (red color) and left (green color) parts of the X-axis (divided by “0”) represent the numbers of up- and down-regulated genes, respectively. The pathways with |up/down| > 6 times were shown, and the circle color from green through yellow to light red represented the log2(up/down) value from low to high, and the red circle denoted the pathways without down-regulated genes. The pathways and corresponding categories (level 2) are shown on the left and right, respectively. The level 1 categories were denoted by different colors, including cellular processes (purple), environmental information processing (orange), human diseases (green), metabolism (dark red), and organismal systems (blue). (B) Neuroactive ligand-receptor interaction33 and (C) olfactory transduction pathway33 under ascr#9 treatment. The genes in the red and green boxes represented the gene’s up- and down-regulation after being treated with ascr#9, respectively. The number and color of the bubbles near the gene exhibited the corresponding gene numbers and the log2FC value, respectively.
Neuroactive ligand-receptor interaction is a biological function that involves the interaction between various neuroactive ligands and their corresponding receptors in the nervous system. It refers to the binding of specific ligands, such as neurotransmitters, hormones, or drugs, to their respective receptors, which triggers a series of signaling events and influences neuronal activity and function. The interaction between neuroactive ligands and receptors is highly specific, with each ligand typically binding to its corresponding receptor with high affinity, leading to the activation or modulation of downstream signaling pathways37. Interestingly, after ascr#9 treatment, neuroactive ligand-receptor interaction pathway in M. incognita showed significantly biased between up-regulated genes and down-regulated genes (24 times). A total of 24 receptors were significantly up-regulated (Table S7), including muscarinic acetylcholine receptor (gar-1, gar-2), 5-hydroxytryptamine receptor 1 (ser-2), somatostatin receptor 2 (npr-32), adenosine receptor A2a (ADOR-1), thyrotropin-releasing hormone receptor (npr-15), dopamine receptor D1-like (drd1), metabotropic glutamate receptor (mgl-1) and melanocortin 2 receptor (mc2r) (Fig. 4B), indicating that the ascr#9 affected neuronal activity and behavior of M. incognita by inducing multiple ligands and receptors.
In addition, ascr#9 could also significantly affect the olfactory transduction (Table S7). Up-regulation of nine calmodulin (CAM) genes in olfactory receptor neuron (ORN) may directly suppress cyclic nucleotide gated channel beta 1 (CNGB1), which allowing cations, particularly Na+ and Ca2+, to flow down their electrochemical gradients into the cell. Meanwhile the CAM could induce CaM kinase II (CAMK2) to suppress the adenylate cyclase 3 (AC3), then suppress the intracellular cAMP levels, this may also suppress the CNGB1 expression and block the Na+ and Ca2+ transferring into cell. In addition, the exchanger solute carrier family 8 (SLC8A) was significantly suppressed, which function in transferring the Na+ into the cell, and the anoctamin-2 (ANO2) was lowly expressed, this might block the Cl- to flow out of the cell. Consequently, Na+ and Ca2+ intracellular transferring and Cl- extracellular transferring were impeded, the depolarization of ORN was disturbed, and the olfactory transduction might be disorder.
Furthermore, four metabolic processes only contain up-regulated genes, including mucin-type and other types of O-glycan biosynthesis, other O-glycan degradation, and ether lipid metabolism, indicating the special role of these pathways after ascr#9 treatment. In addition, several human disease pathways were enriched, most of which involved in the subfunction of cell growth, cell proliferation, and cell survival (Table S8).
qRT-PCR validation of transcriptome data
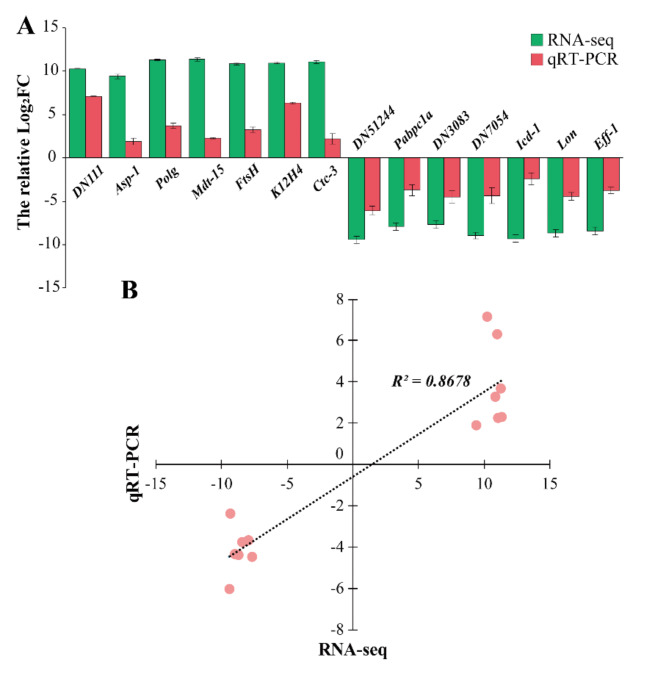
To validate the veracity and reliability of the DEGs identified by RNA-seq, 14 DEGs were selected for qRT-PCR validation from those with differential expression patterns based on fold changes and functional enrichment results. qRT-PCR data were consistent with the expression trend of the RNA-Seq results, demonstrating the credibility of the transcriptome results by Illumina sequencing (Fig. 5).
Fig. 5.
Validation of the selected DEGs from RNA-seq inM. incognitaby qRT-PCR. (A) RNA-seq and qRT-PCR results of selected genes. The gene information is present in Table S1. (B) Comparison of the log2 of gene expression ratios between RNA-seq data and qRT-PCR results. Correlation analysis result was also shown on the figure.
RNAi validation of key DEGs in response to ascr#9
To verify whether key genes of M. incognita play a critical role in the dispersal response to ascr#9, 11 highly expressed genes (Table S2) mainly involved in neuroactive ligand-receptor interaction were used for RNAi validation.
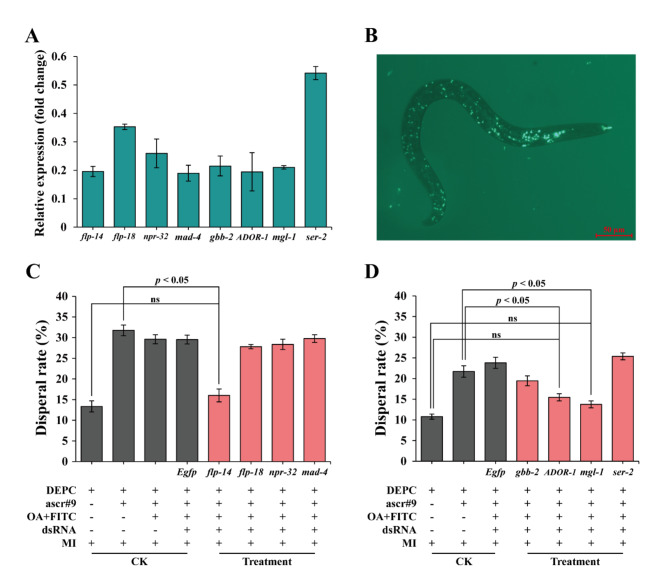
The efficiency of gene knockdown was assessed by performing qRT-PCR analysis on the transcription levels of 11 genes, out of which 8 genes (flp-14, flp-18, npr-32, mad-4, gbb-2, ADOR-1, mgl-1 and ser-2) were successfully knocked down. Remarkably reduced expression levels (p < 0.001) were observed for these genes (Fig. 6A), with knockdown efficiencies ranging from 45.2 to 80.6%. these efficiency 8 genes were utilized in subsequent RNAi experiment. The luminescence inside M. incognita body (mouthparts, intestine, and head) indicated the uptake of dsRNA by M. incognita (Fig. 6B). As shown in Fig. 6C, J2s exhibited no dispersal in response to DEPC-treated water (dispersal rate of 13.36 ± 1.32%), but showed significant dispersion in the presence of ascr#9 (31.76 ± 1.13%; Tukey test; F = 35.086, dfB = 7, dfW = 24, p < 0.05). After treated by different dsRNAs (dsflp-14, dsflp-18, dsnpr-32 and dsmad-4; dsEgfp as a control), there were no differences observed in the dispersal rates of J2s upon knockdown of flp-18, npr-32 and mad-4; however, a significantly decreased dispersal rate of J2s was found when flp-14 gene expression was partially silenced, from 31.76 ± 1.13% to 16.01 ± 1.56% (Fig. 6C). In another experiment, the dispersal rates of M. incognita J2s treated with the remaining dsRNAs exhibited variations. Treatment with the dsRNA of gbb-2, ser-2, and Egfp did not results in a significant difference in J2 dispersal rate; however, treatment with ADOR-1 or mgl-1 dsRNA led to a significant reduction in dispersal rate (Tukey test; F = 34.782, dfB = 9, dfW = 30, p < 0.001) (Fig. 6D). All these results indicated that the genes flp-14, ADOR-1, and mgl-1 were involved in regulating dispersal behavior of M. incognita.
Fig. 6.
RNAi validation of key DEGs in response to ascr#9. (A) qRT-PCR verification of the gene expression changes in M. incognita J2s treated with corresponding dsRNA. Actin was employed as an internal reference gene. (B) the luminescence inside M. incognita body indicated the uptake of FITC by M. incognita. Dispersal rates of M. incognita J2s treated with ascr#9 and (C) dsRNAs (dsflp-14, dsflp-18, dsnpr-32 and dsmad-4; dsEgfp as a control), and (D) dsRNAs (dsgbb-2, dsADOR-1, dsmgl-1, and dsser-2; dsEgfp as a control). ns indicated no significant differences; “+” means addition of reagents or DEPC water or J2s; “-” means no addition of reagents or DEPC water or J2s. OA, octopamine; FITC, fluorescein isothiocyanate; MI, M. incognita. The whole experiments were repeated 4 times.
Discussion
In this study, the results demonstrate that flp-14, ADOR-1, and mgl-1 are involved in the dispersal behavior of M. incognita nematodes responding to the pheromone signal ascr#9, which promotes the interaction study between ascarosides and M. incognita, and provides new ideas for the prevention and control of M. incognita by using ascarosides.
The ability to perceive and respond to the environment is critical to the survival of nematodes43,44. Transduction of pheromone signals requires the TAX-4 cGMP-gated channel and the OCR-2 and OSM-9 TRPV channels in ASK (sensory neuron) and ADL (sensory neuron), respectively44–46. The GPA-3 Ga protein is also involved in ascaroside avoidance behaviors47. Osas molecules are comprised of ascarosides connected to succinylated octopamine. Osas#9 (derived from ascr#9) induces robust avoidance behaviors by starved C. elegans larvae and adults, suggesting that this molecule acts as a signal promoting dispersal from unfavorable conditions48,49, which is mediated by the TYRA-2 tyramine/octopamine receptor and the GPA-6 Ga protein in the ASH nociceptive neurons49,50. M. incognita juveniles were repelled, to various degrees, by most of the tested ascarosides (especially ascr#9)20,21. Ascr#9 is also a common small molecule inducing the dispersal behavior of entomopathogenic nematodes19,22,51. However, the genes or receptors sensing ascarosides in M. incognita juveniles are largely unknown. In this study, the transcriptome data showed that differentially expressed genes in M. incognita under ascr#9 stress were mainly up-regulated, including up-regulated 25,029 and down-regulated 16,840 genes, in multiple pathways, such as multiple neuroactive ligand-receptor interaction, mucin type O-glycan, and olfactory transduction. To validate the functions of the expressed genes that sense ascr#9 in M. incognita, eleven genes mainly from neuroactive ligand-receptor interaction and FMRF amide-like peptide related process were selected for RNA interference. Significant decrease in ascr#9-induced nematode dispersal rates was observed by knocking down the flp-14, mgl-1 and ADOR-1 genes.
flp-14 encodes a secreted FMRF amide-like neuropeptide that disrupts the migration of M. incognita in response to root exudates52,53 and towards host roots, as well as subsequent invasion into the roots. This discovery presents a potential tool for controlling M. incognita and inducing C. elegans specific motor states, such as forward movement54, while also acting as a context-specific driver for avoidance behavior in C. elegans55. The reduced dispersal rate of flp-14 knocked down M. incognita J2s induced by ascr#9 further demonstrates that this nematode species senses ascr#9 at least by flp-14 for dispersal.
As one of the metabotropic glutamate receptors in C. elegans, mgl-1 is important for normal embryonic growth rate and size56, for fat regulation57,58, for systemic starvation response59, for feeding behavior60, and for governing behavioral response to food availability61. Mgl-1 mediates foraging behavior and reproductive plasticity in adult C. elegans62,63. Mgl-1 is also up-regulated under the stress of neurotoxicity64 and involved in FLP-2 neuropeptide signaling65. But mgl-1 role in locomotion is not clearly known. The results from this study indicated that this metabotropic glutamate receptor is involved in the dispersal of M. incognita J2s induced by ascr#9, which reflects the broad functions of mgl-1 in different nematode taxa.
Adenosine, a purine nucleoside with neuromodulatory actions, is part of the purinergic signaling system (PSS). The adenosine A2A receptor is an adenosine receptor that regulates the concentration of adenosine and has a key role in the activation of several receptors that affect neurotransmitter release and synaptic transmission, particularly the neuropeptide-related receptor, metabotropic glutamate receptor66. Adenosine can activate the FOXO signaling pathway, the human homolog of DAF-16, via the adenosine A2A receptor67. In C. elegans, only one adenosine receptor has been identified so far, adenosine receptor-1 (ADOR-1), which belongs to the superfamily of G-protein coupled receptors and is homologous to adenosine A2A receptors in humans, but the function of this receptor is sparsely known68,69. Based on the adenosine receptor, adenosine signaling induced by caffeine70 and Guarana (Paullinia cupana)71 has been shown to partially extend the lifespan of C. elegans. ADOR-1 is involved in the modulation of fat metabolism in C. elegans by an extract of a drink plant Ilex paraguariensis72. It is reported that the protective effect of adenosine in terms of the paraquat-induced oxidative stress in C. elegans was mainly regulated by ADOR-168. ADOR-1 also mediates to ameliorate C. elegans locomotion by caffeine73. Surprisingly, in this study, to the best of our knowledge, for the first time, the gene homologous to ADOR-1 in M. incognita senses pheromone ascr#9 and contributes to dispersal behavior.
The present study demonstrate that flp-14, mgl-1 and ADOR-1 are involved in the dispersal behavior of M. incognita nematodes responding to ascr#9, based on the analysis of the transcriptome of ascr#9-treated second stage M. incognita juveniles and knock down of 11 highly differentially expressed genes by RNAi. The results promote the interaction study between ascarosides and M. incognita, and provides new ideas for the prevention and control of M. incognita by using pheromone ascarosides.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The work was supported by the National Key R & D Program of China (2024YFE0109600) Guangzhou Science and Technology Project (202206010120), GDAS Special Project of Science and Technology Development (2022GDASZH2022010101).
Author contributions
Conceptualization, L.C and R.H; Data curation, K.D; Funding acquisition, L.C; Methodology, K.D; Project administration, R.H and L.C; Resources, L.C and C.X; Software, Z.R; Validation, K.D; Writing - original draft, K.D; Writing - review & editing, R.H, Z.R, C.X and L.C.
Data availability
All of the raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1010386.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chengti Xu, Email: xchti@163.com.
Li Cao, Email: caol@giz.gd.cn.
References
- 1.Jones, J. T. et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol.14(9), 946–961. 10.1111/mpp.12057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyne, D. L. et al. Plant-parasitic nematodes and food security in sub-saharan africa. Annu. Rev. Phytopathol.56, 381–403. 10.1146/annurev-phyto-080417-045833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Favery, B. et al. Gall-forming root-knot nematodes hijack key plant cellular functions to induce multinucleate and hypertrophied feeding cells. J. Insect. Physiol.84, 60–69. 10.1016/j.jinsphys.2015.07.013 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Juvale, P. S. & Baum, T. J. Cyst-ained research into Heterodera parasitism. PLoS Pathogens 14(2), e1006791 (2018). 10.1371/journal.ppat.1006791 [DOI] [PMC free article] [PubMed]
- 5.Castagnone-Sereno, P. Genetic variability in parthenogenetic root-knot nematodes, Meloidogyne spp. and their ability to overcome plant resistance genes. Nematology. 4(5), 605–608. 10.1163/15685410260438872 (2002). [Google Scholar]
- 6.Khanal, C. et al. Identification and haplotype designation of Meloidogyne spp. of Arkansas using molecular diagnostics. Nematropica. 46(2), 261–270 (2016). https://www.researchgate.net/publication/314089197 [Google Scholar]
- 7.Kepenekci, I. et al. Application methods of Steinernema feltiae, Xenorhabdus bovienii and purpureocillium lilacinum to control root-knot nematodes in greenhouse tomato systems. Crop Prot.108, 31–38. 10.1016/j.cropro.2018.02.009 (2018). [Google Scholar]
- 8.Ntalli, N. G. & Caboni, P. Botanical nematicides: A review. J. Agric. Food Chem.60, 9929–9940. 10.1021/jf303107j (2012). [DOI] [PubMed] [Google Scholar]
- 9.Abdel-Rahman, F. H. et al. Nematicidal activity of terpenoids. J. Environ. Sci. Health Part. B. 48(1), 16–22. 10.1080/03601234.2012.716686 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Maleita, C. et al. Naphthoquinones from walnut husk residues show strong nematicidal activities against the root-knot nematode Meloidogyne hispanica. ACS Sustain. Chem. Eng.5(4), 3390–3398. 10.1021/acssuschemeng.7b00039 (2017). [Google Scholar]
- 11.Oota, M. et al. Identification of naturally occurring polyamines as root-knot nematode attractants. Mol. Plant. 13(4), 658–665. 10.1016/j.molp.2019.12.010 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Brennan, P. A. & Zufall, F. Pheromonal communication in vertebrates. Nature. 444(7117), 308–315. 10.1038/nature05404 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Jeong, P. Y. et al. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature. 433(7025), 541–545. 10.1038/nature03201 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Kim, B. et al. Root exudation by aphid leaf infestation recruits root-associated Paenibacillus spp. to lead plant insect susceptibility. J. Microbiol. Biotechnol.26(3), 549–557. 10.4014/jmb.1511.11058 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Butcher, R. A. Decoding chemical communication in nematodes. Nat. Prod. Rep.34(5), 472–477. 10.1039/c7np00007c (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Reuss, S. H. Exploring modular glycolipids involved in nematode chemical communication. Chimia. 72, 297–303. 10.2533/chimia.2018.297 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Hartley, C. J. et al. Infective juveniles of entomopathogenic nematodes (Steinernema and Heterorhabditis) secrete ascarosides and respond to interspecific dispersal signals. J. Invertebr. Pathol.16810.1016/j.jip.2019.107257 (2019). [DOI] [PubMed]
- 18.Oliveira-Hofman, C. et al. Pheromone extracts act as boosters for entomopathogenic nematodes efficacy. J. Invertebr. Pathol.164, 38–42. 10.1016/j.jip.2019.04.008 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Wang, J. et al. Influence of the ascarosides on the recovery, yield and dispersal of entomopathogenic nematodes. J. Invertebr. Pathol.18810.1016/j.jip.2022.107717 (2022). [DOI] [PubMed]
- 20.Dai, K. et al. Influence of entomopathogenic nematodes, symbiotic bacteriaand ascarosides on the dispersal behaviour of Meloidogyne incognita. Nematology. 0, 1–11. 10.1163/15685411-bja10184 (2022). [Google Scholar]
- 21.Kaplan, F. et al. Interspecific nematode signals regulate dispersal behavior. PloS One. 7(6). e38735 (2012). [DOI] [PMC free article] [PubMed]
- 22.Choe, A. et al. Ascaroside signaling is widely conserved among nematodes. Curr. Biol.22(9), 772–780. 10.1016/j.cub.2012.03.024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manohar, M. et al. Plant metabolism of nematode pheromones mediates plant-nematode interactions. Nat. Commun.11(1), 208. 10.1038/s41467-019-14104-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urwin, P. E., Lilley, C. J. & Atkinson, H. J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant-Microbe Int.15, 747–752. 10.1094/MPMI.2002.15.8.747 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Chen, Q., Rehman, S., Smant, G. & Jones, J. T. Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Mol. Plant. Microbe Int.18, 621–625. 10.1094/mpmi-18-0621 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Dong, L. et al. Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts Mi-flp-18 expression to block infection. J. Exp. Bot.65(1), 131–141. 10.1093/jxb/ert356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fosu-Nyarko, J., Iqbal, S. & Jones, M. G. K. Targeting nematode genes by RNA silencing. Plant. Gene Silencing Mech. Appl.5, 176–192. 10.1079/9781780647678.0176 (2017). [Google Scholar]
- 28.Iqbal, S., Fosu-Nyarko, J. & Jones, M. G. K. Attempt to silence genes of the RNAi pathways of the Root-Knot Nematode, Meloidogyne incognita results in diverse responses including increase and no change in expression of some genes. Front. Plant. Sci.11, 328 (2020). Published 2020 Mar 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi, I. et al. RNA-Seq of plant-parasitic nematode Meloidogyne incognita at various stages of its development. Front. Genet.810.3389/fgene.2017.00190 (2017). [DOI] [PMC free article] [PubMed]
- 30.Meng, Q. P. et al. PCR assays for rapid and sensitive identification of three major root-knot nematodes, Meloidogyne incognita, M. Javanica and M. arenaria. Acta Phytopathologica Sinica. 34(3), 204–210 (2004). [Google Scholar]
- 31.Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc.8, 1494–1512. 10.1038/nprot.2013.084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner, M. et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet.25, 25–29. 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa, M. & Goto, S. K. E. G. G. Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30. 10.1093/nar/28.1.27 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, B., Dewey, C. N. RSEM accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform.12, 323. 10.1186/1471-2105-12-323 (2011). [DOI] [PMC free article] [PubMed]
- 35.Robinson, M. D. et al. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26, 139–140. 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatic. 21(18), 3674–3676. 10.1093/bioinformatics/bti610 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Xie, C. et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res.39, W316–W322. 10.1093/nar/gkr483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papolu, P. K. et al. Utility of host delivered RNAi of two FMRF amide like peptides, flp-14 and flp-18, for the management of root knot nematode, Meloidogyne incognita. PLoS ONE. 8(11), e80603. 10.1371/journal.pone.0080603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25, 402–408. 10.1006/meth.2001.1262 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol.3, 7, RESEARCH0034. 10.1186/gb-2002-3-7-research0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong, L. et al. Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts Mi-flp-18 expression to block infection. J. Exp. Bot.65(1), 131–141. 10.1093/jxb/ert356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierce, K. L. et al. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol.3(9), 639–650. 10.1038/nrm908 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Chen, S. A. et al. Prey sensing and response in a nematode-trapping fungus is governed by the MAPK pheromone response pathway. Genetics. 217(2), iyaa008. 10.1093/genetics/iyaa008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferkey, D. M. et al. Chemosensory signal transduction in Caenorhabditis elegans. Genetics. 217(3), iyab004 (2021). [DOI] [PMC free article] [PubMed]
- 45.Macosko, E. Z. et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. Elegans. Nature. 458, 1171–1175. 10.1038/nature07886 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jang, H. et al. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. Elegans. Neuron. 75, 585–592. 10.1016/j.neuron.2012.06.034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, D. et al. A conserved neuronal DAF-16/FoxO plays an important role in conveying pheromone signals to elicit repulsion behavior in Caenorhabditis elegans. Sci. Rep.7(1), 7260. 10.1038/s41598-017-07313-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Artyukhin, A. B. et al. Succinylated octopamine ascarosides and a new pathway of biogenic amine metabolism in Caenorhabditis elegans. J. Biol. Chem.288(26), 18778–18783. 10.1074/jbc.C113.477000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chute, C. D. et al. Co-option of neurotransmitter signaling for inter-organismal communication in C. Elegans. Nat. Commun.10(1), 3186. 10.1038/s41467-019-11240-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rex, E. et al. TYRA-2 (F01E11.5): A Caenorhabditis elegans tyramine receptor expressed in the MC and NSM pharyngeal neurons. J. Neurochem.94(1), 181–191. 10.1111/j.1471-4159.2005.03180.x (2005). [DOI] [PubMed] [Google Scholar]
- 51.Roderick, H. et al. Rational design of biosafe crop resistance to a range of nematodes using RNA interference. Plant Biotechnol. J.16(2), 520–529. 10.1111/pbi.12792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalzell, J. J. et al. Non-nematode-derived double-stranded RNAs induce profound phenotypic changes in Meloidogyne incognita and Globodera pallida infective juveniles. Int. J. Parasitol.39, 1503–1516. 10.1016/j.ijpara.2009.05.006 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Dalzell, J. J. et al. Short interfering RNA-mediated gene silencing in Globodera Pallida and Meloidogyne incognita infective stage juveniles. Int. J. Parasitol.40, 91–100. 10.1016/j.ijpara.2009.07.003 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Lim, M. A. et al. Neuroendocrine modulation sustains the C. Elegans forward motor state. Elife. 5, e19887. 10.7554/eLife.19887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gershkovich, M. M. et al. Pharmacological and functional similarities of the human neuropeptide Y system in C. Elegans challenges phylogenetic views on the FLP/NPR system. Cell. Communication Signal.17(1), 123. 10.1186/s12964-019-0436-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamath, R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421, 231–237. 10.1038/nature01278 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Greer, E. R. et al. Neural and molecular dissection of a C. Elegans sensory circuit that regulates fat and feeding. Cell Metabol.8, 118–131. 10.1016/j.cmet.2008.06.005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo, Z. et al. Obesogenic effect of erythromycin on Caenorhabditis elegans through over-eating and lipid metabolism disturbances. Environ. Pollut.294, 118615. 10.1016/j.envpol.2021.118615 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Kang, C. & Avery, L. Systemic regulation of starvation response in Caenorhabditis elegans. Genes Dev.23(1), 12–17. 10.1101/gad.1723409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dillon, J. et al. Metabotropic glutamate receptors: modulators of context-dependent feeding behaviour in C. Elegans. J. Biol. Chem.290(24), 15052–15065. 10.1074/jbc.M114.606608 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahmadi, M. & Roy, R. AMPK acts as a molecular trigger to coordinate glutamatergic signals and adaptive behaviours during acute starvation. Elife. 5, e16349. 10.7554/eLife.16349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeong, H. & Paik, Y. K. MGL-1 on AIY neurons translates starvation to reproductive plasticity via neuropeptide signaling in Caenorhabditis elegans. Dev. Biol.430(1), 80–89. 10.1016/j.ydbio.2017.08.014 (2017). [DOI] [PubMed] [Google Scholar]
- 63.López-Cruz, A. et al. Parallel multimodal circuits control an innate foraging behavior. Neuron. 102(2), 407–419. 10.1016/j.neuron.2019.01.053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De la Parra-Guerra, A. et al. Intergenerational toxicity of nonylphenol ethoxylate (NP-9) in Caenorhabditis elegans. Ecotoxicol. Environ. Saf.19710.1016/j.ecoenv.2020.110588 (2020). [DOI] [PubMed]
- 65.Chai, C. M. et al. Interneuron control of C. Elegans developmental decision-making. Curr. Biol.32(10), 2316–2324. 10.1016/j.cub.2022.03.077 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ribeiro, J. A. Adenosine A2A receptor interactions with receptors for other neurotransmitters and neuromodulators. Eur. J. Pharmacol.375(1–3), 101–113. 10.1016/s0014-2999(99)00230-7 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Friedman, B. et al. Adenosine A2A receptor signaling promotes FoxO associated autophagy in chondrocytes. Sci. Rep.11(1), 968. 10.1038/s41598-020-80244-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ling, C. et al. AdoR-1 (Adenosine Receptor) contributes to protection against paraquat-induced oxidative stress in Caenorhabditis elegans. Oxidative Med. Cell. Longev.175900910.1155/2022/1759009 (2022). [DOI] [PMC free article] [PubMed]
- 69.da Silva, T. C. et al. Exogenous adenosine modulates behaviors and stress response in Caenorhabditis elegans. Neurochem. Res.48(1), 117–130. 10.1007/s11064-022-03727-5 (2023). [DOI] [PubMed] [Google Scholar]
- 70.Bridi, J. C. et al. Lifespan extension induced by caffeine in Caenorhabditis elegans is partially dependent on adenosine signaling. Front. Aging Neurosci.7, 220. 10.3389/fnagi.2015.00220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arantes, L. P. et al. Mechanisms involved in anti-aging effects of guarana (Paullinia cupana) in Caenorhabditis elegans. Braz. J. Med. Biol. Res.51(9), e7552. 10.1590/1414-431X20187552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Machado, M. L. et al. Ilex paraguariensis modulates fat metabolism in Caenorhabditis elegans through purinergic system (ADOR-1) and nuclear hormone receptor (NHR-49) pathways. PLoS ONE. 13(9), e0204023. 10.1371/journal.pone.0204023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Di Rocco, M. et al. Phenotypic assessment of pathogenic variants in GNAO1 and response to caffeine in C. Elegans models of the disease. Genes. 14(2), 319. 10.3390/genes14020319 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1010386.