Abstract
Human parechovirus 1 (HPEV-1) is a prototype member of parechoviruses, a recently established picornavirus genus. Although there is preliminary evidence that HPEV-1 recognizes αV integrins as cellular receptors, our understanding of early events during HPEV-1 infection is still very limited. The aim of this study was to clarify the entry mechanisms of HPEV-1, including the attachment of the virus onto the host cell surface and subsequent internalization. In blocking experiments with monoclonal antibodies against different receptor candidates, antibodies against αV and β3 integrin subunits, in particular in combination, appeared to be the most efficient ones in preventing the HPEV-1 infection. To find out whether HPEV-1 uses clathrin-coated vesicles or other routes for the entry into the host cell, we carried out double-labeling experiments of virus-infected cells with anti-HPEV-1 antibodies and antibodies against known markers of the clathrin and the caveolin routes. At the early phase of infection (5 min postinfection [p.i.]) HPEV-1 colocalized with EEA1 (early endosomes), and later, after 30 min p.i., it colocalized with mannose-6-phosphate receptor (late endosomes), whereas no colocalization with caveolin-1 was observed. The data indicate that HPEV-1 utilizes the clathrin-dependent endocytic pathway for entry into the host cells. Interestingly, endocytosed HPEV-1 capsid proteins were observed in the endoplasmic reticulum and cis-Golgi network 30 to 60 min p.i. Depolymerization of microtubules with nocodazole inhibited translocation of the virus to the late endosomes but did not block HPEV-1 replication, suggesting that the RNA genome may be released early during the entry process.
Early events in viral infection include specific attachment of the virion onto the cell surface receptor(s) followed by entry into the cell and subsequent release of the genome. Successful completion of this process is a prerequisite for the initiation of the infection cycle, and these events play an important role in tissue tropism and pathogenesis. Recently, numerous cell surface molecules, with wide variation in their structures and normal physiological functions, have been identified as virus receptors.
The routes by which extracellular ligands, including viruses, are internalized into the cell include clathrin-mediated endocytosis, uptake via caveolae, macropinocytosis, phagocytosis, and other pathways that presently are poorly characterized. For some virus systems, the entry events have been described in detail. For instance, both adenovirus type 2, a nonenveloped DNA virus, and Semliki Forest virus, an enveloped RNA virus, enter the host cells through a clathrin-mediated pathway (7, 8, 19, 39, 42). Different members of the polyomavirus family are known to enter the host cell by distinct mechanisms; simian virus 40 uses the caveola-dependent endocytic route, while the human polyomavirus JC virus enters the cells through clathrin-mediated endocytosis (24, 32).
Picornaviruses include several important human pathogens which belong to the Enterovirus, Hepatovirus, Parechovirus, and Rhinovirus genera. The virion consists of an icosahedral protein capsid surrounding the single-stranded RNA genome directly acting as mRNA when released into the cytoplasm. Several picornavirus receptors have been identified, and they include, for instance, members of the immunoglobulin superfamily and integrins. Some of the cell surface receptors are preferably accessory molecules, while others bring about the essential conformational changes to the virion needed for the eventual release of the infectious genome from the virion. The exact entry routes of picornaviruses are still relatively poorly understood. However, it has been shown that human rhinovirus 14 uses the clathrin-dependent endocytotic pathway, whereas polioviruses, members of the Enterovirus genus, may release their genome through the plasma membrane directly after attachment to their specific receptor (2, 5, 23, 35). More recently, caveola-mediated endocytosis has been demonstrated for another enterovirus, echovirus 1 (V. Marjomäki, V. Pietiäinen, H. Matilainen, J. Ivaska, L. Nissinen, H. Reunanen, P. Huttunen, T. Hyypiä, and J. Heino, unpublished data).
Human parechovirus 1 (HPEV-1) and HPEV-2 were recently reclassified in the new Parechovirus genus on the basis of their exceptional molecular and biological properties among picornaviruses (34). In addition to the overall genetic distance from members of the other genera (12), these properties include the lack of the maturation cleavage of the capsid protein precursor VP0 to VP2 and VP4 polypeptides, in contrast to the case for virtually all other picornaviruses, and a distinctive form of the 2A protein (10, 33). Moreover, there is an N-terminal extension to the VP3 capsid protein, which is not seen in other picornaviruses. An arginine-glycine-aspartic acid (RGD) motif was found at the C terminus of the VP1 capsid polypeptide, and various approaches indicated that it may play a role in cell surface interactions of the virion by interacting with αV integrins (25, 28, 33). The aim of the present study was to further illuminate the early events of HPEV-1 interactions, including specific receptor recognition by the virus and subsequent entry events leading to the initiation of a productive infection cycle.
MATERIALS AND METHODS
Viruses and cells.
Coxsackievirus A9 (CAV9) (Griggs strain) and coxsackievirus B3 (CBV3) (Nancy strain) were originally obtained from the American Type Culture Collection (ATCC). The HPEV-1 (Harris strain) cDNA clone (12) was kindly provided by Glyn Stanway, Department of Biological Sciences, University of Essex, Colchester, United Kingdom. In vitro-transcribed full-length viral RNA was used for transfection of cells to produce HPEV-1 stock virus. The A549 (human lung carcinoma) cell line (ATCC) was used in all the experiments.
Abs.
HPEV-1 antisera were obtained by immunizing rabbits and mice with sucrose gradient-purified viruses as described previously (14). Rabbits were immunized with three sequential 20- to 100-μg doses injected at 2- to 4-week intervals using the popliteal lymph node method (17), with Freund's complete adjuvant included in the first dose. The sera were collected 2 weeks after the last injection. The mice were immunized subcutaneously with three sequential doses of purified virus at 2-week intervals, and Freund's complete adjuvant was used in the first dose. Monoclonal antibody (MAb) 90BB10 against the β3 integrin subunit (45) was a gift from Ismo Virtanen, Department of Anatomy, University of Helsinki. MAbs against coxsackievirus-adenovirus receptor (CAR) and decay-accelerating factor (DAF) were kindly provided by Jeffrey Bergelson, Children's Hospital, University of Pennsylvania. In addition, MAbs against the following cell surface molecules were used: integrin αV subunit (1.2 mg/ml) (L230; ATCC) (16), integrin α2 subunit (1 mg/ml) (Chemicon), and integrin β1 subunit (1 mg/ml) (Chemicon). Moreover, polyclonal rabbit antibodies (Abs) against β2 microglobulin (Chemicon) were used in the blocking experiments.
To study virus entry, the following collection of polyclonal rabbit Abs against different cellular structures was used: Ab against the cation-independent mannose-6-phosphate receptor (CI-MPR) was used for the detection of late endosomes (18); EEA1 Ab, a kind gift from Harald Stenmark, Norwegian Radium Hospital, Oslo, Norway, was used to stain early endosomes (20); Ab against the trans-Golgi network (GB2) (1) was obtained from George Banting, School of Medical Sciences, Bristol, England, and Ab against the cis-Golgi network (p23) (29) was from Jean Gruenberg, University of Geneva, Geneva, Switzerland. Furthermore, MAb 1D3, kindly provided by Steve Fuller, European Molecular Biology Laboratory, Heidelberg, Germany, was utilized for the detection of endoplasmic reticulum (ER) (11), and MAb against caveolin-1 (Zymed) was used to identify caveolae. The microtubulus network was stained with a MAb recognizing tubulin (Chemicon).
Infectivity titration.
The cell monolayers were grown to full confluency in 24-well cell culture plates (Costar). The cells were washed once with Hanks' balanced salt solution, and 150 μl of HPEV-1 or CBV3 (multiplicity of infection, 5), diluted in Hanks' solution supplemented with 0.6% fetal calf serum (FCS), was added to duplicate wells. After incubation for 1 h at room temperature (RT), the cells were washed three times with Hanks' solution, and then 1 ml of Ham's F-12 culture medium (Gibco BRL) supplemented with 1% FCS (Gibco BRL) was added and the plates were incubated in CO2 atmosphere at 37°C. Samples were collected after different time intervals, and the cells and supernatant were separated (4,000 rpm in an Eppendorf centrifuge for 2 min at 4°C) and stored at −70°C until analyzed. For titration, samples were diluted from 10−1 to 10−6, added to the A549 cells, and incubated for 30 min at 37°C. The virus dilutions were then replaced by 0.5% carboxymethyl cellulose in the culture medium (minimal essential medium [Gibco BRL] supplemented with 0.2% bovine serum albumin, 1% FCS, and 20 mM HEPES [pH 7.4]), and the incubation was continued for 48 h at 37°C. The cells were stained using crystal violet solution prior to counting the number of viral plaques.
Blocking of infection with cell surface Abs.
A549 cells were grown as monolayer cultures on six-well plates (Costar). The cells were washed once with Hanks' solution, and 60 μl of Ab dilutions in Hanks' solution containing 0.6% FCS was added and incubated at RT for 45 min. Forty microliters (approximately 100 PFU) of purified HPEV-1, CAV9, or CBV3, diluted in Hanks' solution containing 0.6% FCS, was added to the wells and incubated at RT for 15 min. The virus solution was then replaced by 0.5% carboxymethyl cellulose in culture medium, and the number of plaques was determined as described above.
Immunofluorescence labeling and confocal microscopy.
Subconfluent A549 cells grown on glass coverslips were infected with HPEV-1 (multiplicity of infection, 2) and fixed with methanol for 6 min at −20°C after different times postinfection (p.i.). The following chemicals were also used to study the viral entry procedures: nocodazole (20 μM) (Sigma) for depolymerization of microtubules, chlorpromazine (25 and 50 μM) (Sigma) to inhibit the formation of clathrin-coated pits, and cycloheximide (100 μg/ml) (Sigma) to prevent de novo protein synthesis in the cells. The chemicals were added in the cell culture medium prior to attachment of the virus (nocodazole, 90 min; chlorpromazine and cycloheximide, 30 min), and they were also present during the course of infection. The efficiency of viral replication was analyzed by counting the number of immunofluorescence-positive cells by confocal microscopy after 6 h (chlorpromazine) or 8 h (nocodazole) p.i. For immunofluorescence labeling, the Abs were diluted in phosphate-buffered saline (PBS) containing 3% bovine serum albumin, and the cells were incubated with the primary and secondary Abs at RT for 1 h and 30 min, respectively. Goat Abs against rabbit (Alexa Red 546 nm; Molecular Probes, Inc.) and mouse (Alexa Green 488 nm) immunoglobulins were used as secondary Abs. Following washing four times with PBS, the cells were mounted in Mowiol and viewed with a confocal microscope (Zeiss LSM510) equipped with a 458/488/514-nm argon-krypton and a 543-nm helium-neon laser. In order to quantitate HPEV-1 and CBV3, the detector gain and offset values were first set to similar levels for each labeling experiment and cell type using anti-HPEV-1 or anti-CBV3-labeled noninfected cells to give the negative background level.
Measurement of transferrin-HRP uptake.
To determine the effect of chlorpromazine on the clathrin-mediated endocytosis, transferrin uptake by the cells was measured (22). A549 cells were first washed two times with PBS and then incubated in serum-free medium for 1 h without or with chlorpromazine (25 or 50 μM). After this, a 5-min pulse of 25-μg/ml transferrin-horseradish peroxidase (transferrin-HRP) (Biotrend, Cologne, Germany) was added to the cells. The cells were then washed two times with PBS on ice and scraped from the dish. After pelleting (1,000 × g, 5 min) and permeabilization with 0.2% Triton X-100 (30 min on ice), the lysate was centrifuged at 15,000 × g for 5 min and the HRP activity and protein content in the supernatant were measured.
Percoll fractionation of infected cell homogenates and immunoblotting.
Subconfluent cultures of A549 cells were infected with HPEV-1 (1 h on ice), washed with PBS, and treated with HRP (Sigma, type II; 2 mg/ml in Dulbecco minimal essential medium). For the detection of early endosomes, the cells were allowed to internalize HRP for 5 min at 37°C. After the treatment, the cells were immediately transferred onto ice, washed twice with cold PBS, and detached from the dishes by gentle scraping. Following centrifugation for 5 min at 150 × g, the cell pellet was suspended in homogenization buffer (250 mM sucrose, 1 mM EDTA, and 3 mM imidazole [pH 7.4]). The suspension was pelleted for 5 min at 650 × g, diluted in 1 ml of homogenization buffer, and further homogenized through a syringe. This homogenate was centrifuged for 10 min at 2,000 rpm, and the postnuclear supernatant was collected.
Percoll gradients (20%) were prepared as previously described (27). The postnuclear supernatant samples were centrifuged at 35,000 × g for 2 h in a Sorvall SS34 rotor Fractions of 500 μl were collected, and HRP activities were determined as previously described (27). The following samples were pooled from the gradient: fractions 5 to 7 (pool I), fractions 10 to 12 (pool II), and fractions 18 to 20 (pool III). The pooled samples were treated with 1% Triton-X 100 (KEBOLab) for 30 min on ice and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The polypeptides were blotted onto nitrocellulose membranes (Schleicher & Schuell) which were subsequently incubated in PBS containing 5% milk powder (Valio) (PBS-milk) for 15 min at RT to block nonspecific binding. HPEV-1 rabbit antiserum (diluted 1:1,000 in PBS-milk) or Abs against the integrin αV (1:100) or β3 (1:5,000) subunit were added onto the membrane, and after 2 h of incubation at RT with the antiserum, the membranes were washed two times with PBS containing 0.05% Tween 20 and rinsed with PBS. The blot was then treated with HRP-conjugated goat anti-rabbit or anti-mouse Ab (Bio-Rad Laboratories, Inc.) and visualized with Super Signal substrate (Pierce).
RESULTS
Viral replication cycle.
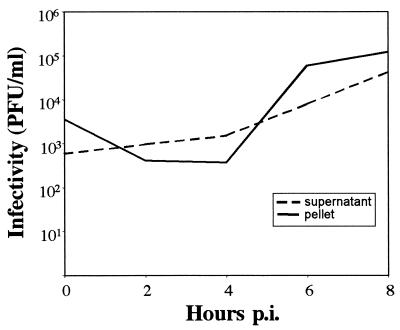
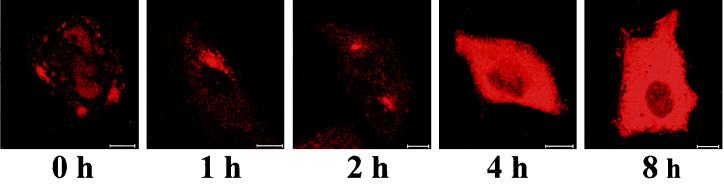
The infection cycle of HPEV-1 was studied in A549 cells; the cells and supernatants were collected separately after different time intervals, and the infectivity of the samples was tested by plaque assay (Fig. 1). After the attachment of the virus (0 h), the cells contained more virus than the supernatant. Subsequently, some virus, which did not become internalized, was released into the supernatant. The replication cycle of HPEV-1 was approximately 6 to 8 h. We also used immunofluorescence to study localization of viral capsid proteins during the replication cycle (Fig. 2). After the attachment, HPEV-1 was clearly located on the cell surface, and 1 h later the viral proteins were found near the perinuclear area. Newly synthesized capsid polypeptides became detectable at 4 h p.i. in the cytoplasm of the infected cells.
FIG. 1.
Production of infectious virus during HPEV-1 infection in A549 cells. The amount of virus was determined from the cells and the culture supernatants using plaque assay.
FIG. 2.
Localization of HPEV-1 capsid proteins in A549 cells during the infection cycle, visualized by indirect immunofluorescence. Rabbit antiserum against purified HPEV-1 was used as the primary antibody. Bars, 10 μm.
Cell surface interactions of HPEV-1.
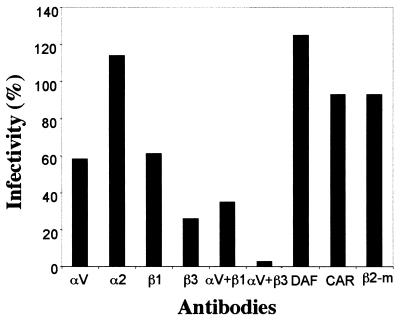
We first tested the ability of Abs against selected picornavirus receptor candidates to prevent HPEV-1 infection in A549 cells (Fig. 3). MAbs against αV and β3 integrin subunits blocked HPEV-1 infection most efficiently (42 and 74% inhibition, respectively), and when these two Abs were used in combination, the infection was prevented nearly completely (97%). MAb against β1 integrin showed inhibitory effects of 39% alone and 65% when used in combination with αV MAb, while other cell surface antibodies tested (α2 integrin, β2 microglobulin, CAR, and DAF) had no significant blocking effect (less than 7%). Abs against β2 microglobulin inhibited the CAV9 infection almost completely (95%), and CBV3 infection was inhibited efficiently by anti-CAR (92%) and partially by anti-DAF (44%) Abs (data not shown).
FIG. 3.
Blocking of HPEV-1 infection by Abs against different cell surface proteins. αV, α2, β1, β2 microglobulin (β2-m), and DAF Abs were used at a dilution of 1:100 while anti-CAR and -β3 supernatants were used undiluted. After incubation with the Abs, the cells were infected with the virus (100 PFU), and the number of plaques was counted after 48 h of incubation. The results are expressed as proportional numbers of plaques compared to those appearing in the cells infected in the absence of Abs.
Localization of integrin subunits in infected cells.
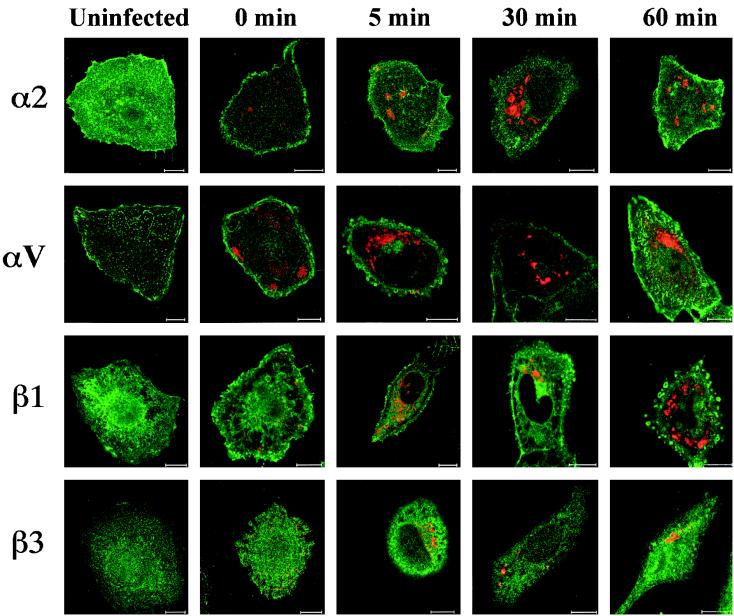
Because Abs against αV and β3 integrin chains were the most efficient ones in inhibiting HPEV-1 infection in the blocking experiments, we used confocal microscopy to study further their colocalization with the viral capsid proteins and possible redistribution of the subunits during the HPEV-1 internalization process. In uninfected cells, the αV integrin subunit was located in nail-like structures and β3 integrin was located in punctate, batch-like structures on the cell surface (Fig. 4). For comparison, the distributions of α2 and β1 integrin subunits were also studied, in uninfected cells both α2 and β1 were diffusely located on the plasma membrane. After virus attachment (1 h, 0°C), the distribution of these integrin subunits did not change significantly, but some colocalization of HPEV1 with β3 integrin was observed. At 5 min p.i., HPEV-1 was internalized into early endosomes (Fig. 5), but no colocalization or significant redistribution was observed with any of the integrin subunits studied. When the virus was translocated to the late endosomes near the perinuclear area at 30 to 60 min p.i., again no colocalization with the integrin subunits was seen.
FIG. 4.
Localization of HPEV-1 capsid proteins and integrin subunits (α2, αV, β1, and β3) during the early steps of infection. Red, virus; green, integrin subunits; yellow, colocalization. Bars, 10 μm.
FIG. 5.
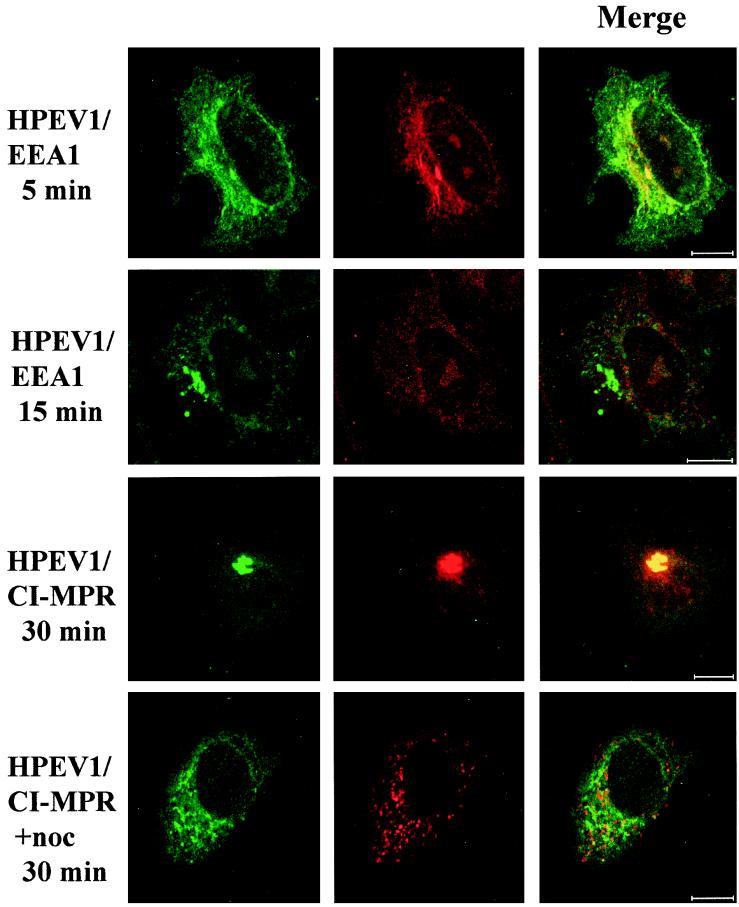
Colocalization of HPEV-1 with early and late endocytic markers. EEA1 was used to stain the early endosomes at 5 and 15 min p.i. CI-MPR was used for the detection of late endosomes at 30 min p.i. in A549 cells with or without nocodazole (noc) treatment. Green, virus; red, EEA1 or CI-MPR; yellow, colocalization. Bars, 10 μm.
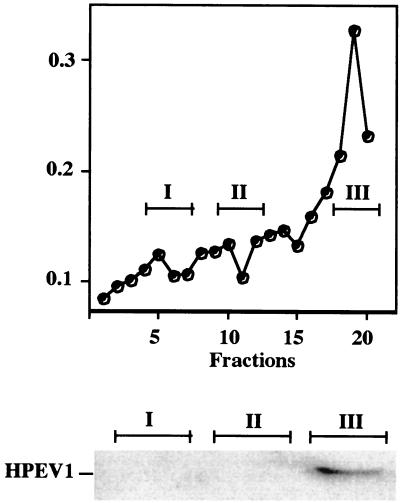
To further investigate possible colocalization of HPEV-1 with the integrin subunits, we used fractionation of homogenized cellular samples in 20% Percoll gradients to discriminate light early endosomes from heavier late endosomes and lysosomes. Infected cells were also allowed to internalize HRP during the last 5 min of the experiment in order to label early endosomes. Peroxidase activity was, typically, highest in the top fractions (Fig. 6). From the gradient, three fractions were pooled together from the bottom part (pool I), from the middle part (pool II), and from the top (pool III) of the gradient. Immunoblotting with HPEV-1 Abs revealed the presence of HPEV-1 capsid proteins in the top fractions at only 5 min p.i. When immunoblotting with Abs against αV and β3 was performed with the early endosomal fraction, the fraction was not shown to contain either of these integrin subunits (data not shown). Thus, it seems that αVβ3 integrin is involved mainly in the early interactions of the virus on the cell surface, whereas subsequent internalization steps may need other cellular macromolecules.
FIG. 6.
Subcellular fractionation of cellular homogenates from HPEV-1-infected cells in 20% Percoll gradients. A549 cells were allowed to internalize HRP for 5 min at 37°C in order to label early endosomes. Three adjoining fractions were pooled from the bottom (fractions 5 to 7; pool I), from the middle (fractions 10 to 12; pool II), and from the top (fractions 18 to 20; pool III) of the gradient. The pooled fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted using HPEV-1 rabbit Abs.
Entry route.
To investigate whether HPEV-1 enters the host cell via the clathrin or caveolin endocytic pathway, we used immunofluorescence labeling with Abs against the virus capsid and different markers of the entry pathways. Double labeling with anti-HPEV-1 and anti-EEA1 antibodies at 5 min p.i. showed colocalization, suggesting that the virus enters early endosomes (Fig. 5), also supporting the results obtained by immunoblotting of the early endosomal fraction. No colocalization was seen at 15 min p.i., indicating that the virus has already been translocated from the early endosomes (Fig. 5). HPEV-1 colocalized with CI-MPR, a marker for late endosomes, at 30 min p.i. (Fig. 5) and remained in these structures until 60 min p.i. (data not shown). Many of the infected cells showed accumulation of viral capsid proteins together with CI-MPR. While in uninfected cells, CI-MPR is localized in membranous late endosomes in the perinuclear region, HPEV-1–CI-MPR colocalization was relatively frequently seen also in peripheral structures. Thus, it seems that HPEV-1 infection and accumulation of viral capsid proteins in the clathrin pathway may somehow perturbe the normal recycling of CI-MPR.
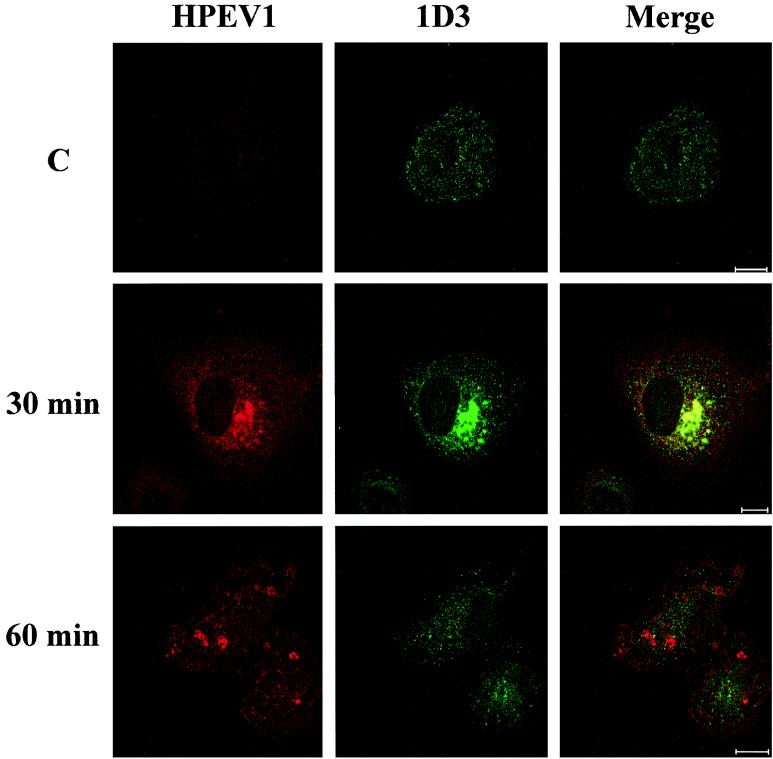
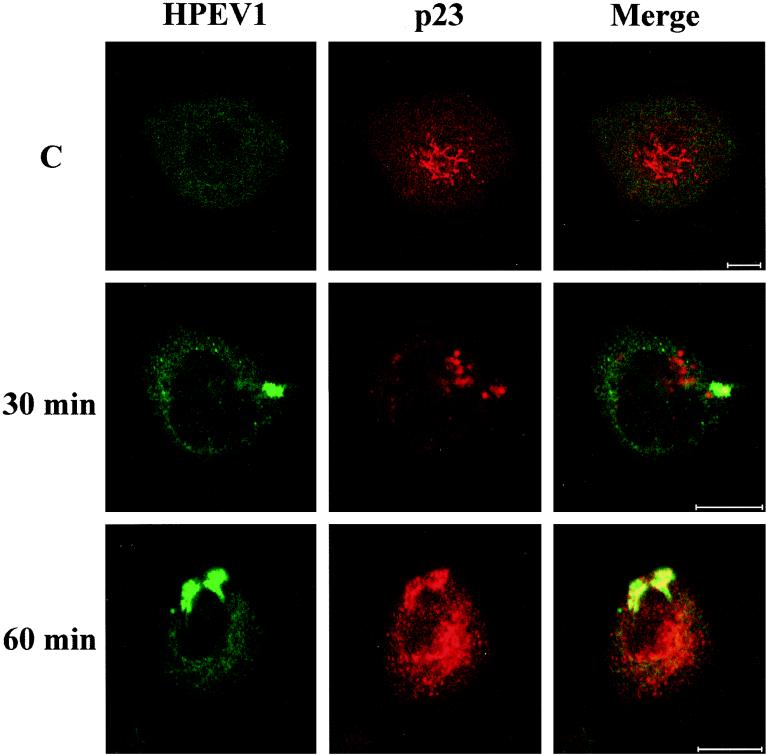
Interestingly, HPEV-1 capsid proteins were also found to enter the ER at 30 min p.i., based on colocalization with the 1D3 marker, but at 1 h p.i. colocalization was no longer detectable (Fig. 7). Cycloheximide treatment during the infection did not prevent localization of HPEV-1 in the ER, suggesting that the observed proteins were not newly synthesized viral polypeptides (data not shown). Double-labeling studies with known markers for Golgi (TGN-46 for the trans-Golgi network and p23 for the cis-Golgi) showed viral proteins to be translocated to the cis-Golgi network after 30 to 60 min p.i. (Fig. 8), whereas no colocalization with the trans-Golgi was observed (data not shown). Double labeling with HPEV-1 and caveolin-1 Abs showed no colocalization (data not shown).
FIG. 7.
Colocalization of HPEV-1 capsid proteins with ER (1D3 Ab) at 30 and 60 min p.i. 1D3 detects protein disulfide isomerase in the endoplasmic reticulum. C, uninfected control. Red, virus; green, 1D3; yellow, colocalization. Bars, 10 μm.
FIG. 8.
Colocalization of HPEV-1 capsid proteins with cis-Golgi network (p23 Ab) at 30 and 60 min p.i. C, uninfected control. Green, virus; red, p23; yellow, colocalization. Bars, 10 μm.
Effects of clathrin route inhibitors on viral replication.
Clathrin-dependent entry can be inhibited at different stages with various chemicals: nocodazole blocks endosomal traffic between peripheral early and late endosomes by depolymerizing the microtubules (6), whereas chlorpromazine has been shown to cause disappearance of clathrin-coated pits from the plasma membrane, hence inhibiting initiation of the clathrin-dependent endocytic pathway (40). We studied, by immunofluorescence, whether treatment by these chemicals could inhibit HPEV-1 infection in A549 cells. Although nocodazole prevented the colocalization of HPEV-1 capsid proteins with late endosomes (Fig. 5), it had no effect on virus replication: at 8 h p.i. 66% of the nocodazole-treated cells were infected, while the corresponding value with nontreated cells was 61%. Tubulin labeling showed that microtubules were totally depolymerized after nocodazole pretreatment (data not shown). In contrast, chlorpromazine at nontoxic concentrations (25 μM) prevented approximately 55% of HPEV-1 infection (Table 1). In order to monitor the inhibitory effect of chlorpromazine on clathrin-dependent endocytosis, internalization of HRP-transferrin in A549 cells was measured at different chlorpromazine concentrations; it was shown that 25 μM chlorpromazine prevented 43% of transferrin internalization (Table 1.).
TABLE 1.
Percentage of HPEV1-infected A549 cells (6 h p.i.) and amount of internalized HRP-transferrin (5 min p.i.) in cells treated with different chlorpromazine concentrations
| Chlorpromazine (μM) | % Infectivity (no. of cells examined) | Transferrin-HRP (μg of protein, 103) | % Toxicitya |
|---|---|---|---|
| 0 | 47.4 (542) | 1.92 | 0.2 |
| 25 | 26.5 (599) | 1.09 | 6.9 |
| 50 | 9.9 (392) | 0.885 | 45.9 |
Chlorpromazine toxicity (6 h after treatment) was determined by trypan blue staining.
DISCUSSION
HPEV-1 and -2 were originally isolated during studies of summer diarrhea by Wigand and Sabin (44) and classified as echoviruses 22 and 23, respectively, in the Enterovirus genus according to the criteria used at the time of their first characterization. However, comparison of the growth properties of these two viruses with those of previously identified enteroviruses suggested that HPEVs exhibit exceptional characteristics in their intracellular replication (13, 31, 36, 44).
Sequence analysis revealed that HPEVs are molecularly distant from enteroviruses and represent an independent picornavirus genus (12, 33). An RGD motif was seen in the predicted sequence of the C terminus of the VP1 capsid polypeptide, and because such elements had been shown to recognize integrins during cell-matrix and cell-cell interactions (30) and also function in receptor recognition of microbial pathogens, studies were performed to illuminate the role of the HPEV-1 RGD motif in the early events of infection. Indeed, it was shown that the infection can be blocked with RGD-containing peptides (33), and HPEV-1 competes for cell surface binding with CAV9, an enterovirus with a similar RGD sequence (28). We have recently shown that the RGD-containing region of VP1 is antigenic in both HPEV-1 and CAV9 and that significant antigenic cross-reactivity occurs between these viruses (14, 26).
Four human picornaviruses are known to interact with integrins. In addition to HPEV-1 and CAV9, which have been shown to attach to αV integrins, one echovirus 9 strain (Barty) also contains a functional RGD motif (46, 47), while echovirus 1 binds to α2β1 integrin (a collagen receptor) (3). Moreover, echovirus 1 apparently interacts with a complex containing β2 microglobulin, since the initiation of infection can be blocked by Abs against this molecule (41). Interestingly, CAV9 infection can also be blocked by β2 microglobulin Abs (37), suggesting similarities in the entry processes of these enteroviruses. However, no blocking of HPEV-1 infection with β2 microglobulin Abs was observed in the preset study, indicating significant differences in the entry pathways of integrin-recognizing picornaviruses.
Previous investigations have suggested that HPEV-1 interacts with αVβ1 and/or αVβ3 integrins (25, 28), and a more recent study on HPEV-1 receptor interactions supports these findings (38). Our present results strongly support the idea that HPEV-1 preferably recognizes αVβ3 integrin during the attachment onto the cell surface, while the subsequent entry process may be brought about by another molecule(s), as indicated, for instance, by the lack of colocalization of the virus and the integrin subunits during the entry process.
Viruses can enter the cells through different internalization pathways, and this variation may largely correlate with the interactions with specific cell surface receptor molecules. After attachment to the receptor, the virus penetrates into the host cell, and this process is followed by uncoating and release of the viral genome. Our results show that HPEV-1 capsids migrate from diffusely scattered locations on the cell surface to early endosomes, reactive with EEA1 Abs, within 5 min after the shift of the temperature from 0 to 37°C. Ten minutes later, the capsids are no longer found in these structures, and after a 30-min incubation, period the HPEV-1 capsids are localized in late endosomes (detected by CI-MPR Abs). Due to the high particle/PFU ratio in HPEV-1 preparations, it is not possible to conclude how many of the particles are actually capable of initiating a productive infection cycle. However, when chlorpromazine led to 55% inhibition of the clathrin-dependent endocytic pathway, HPEV-1 infection also was inhibited in the same range, supporting the idea that HPEV-1 entry takes place through the clathrin route.
To get an idea of where the release of the RNA genome could take place, we used chlorpromazine and nocodazole, drugs which inhibit the clathrin route at different stages. While chlorpromazine inhibits the route by preventing the formation of clathrin-coated pits, nocodazole depolymerizes microtubules, which leads to accumulation of endocytosed material into early endosomes and carrier vesicles and prevents translocation to the late endosomes via microtubules. Chlorpromazine, at a nontoxic concentration, inhibited 55% of HPEV-1 infection, whereas nocodazole prevented the colocalization of the virus with late endosomes but had no significant effect on virus replication. These data suggest that internalization of HPEV-1 capsids in early endosomes is sufficient for the initiation of a productive replication cycle and that the viral genome may be released at the very early stages of the entry process.
Interestingly, HPEV-1 was found in the ER at 30 min p.i. and subsequently in the cis-Golgi network (60 min p.i.). The overall appearance of viruses within ER is not well understood. However, simian virus 40, a DNA virus, which is known to enter the host cell via caveolae, has been shown to localize in various cellular compartments, including the ER (15). In HPEV-1 infection the ER could serve as an acceptor compartment for internalized HPEV-1 capsid proteins, in particular because treatment with cycloheximide did not inhibit this colocalization.
For picornaviruses, different entry mechanisms have been proposed. Recently, internalization of human rhinovirus 14 and poliovirus 1 was studied using dynamin mutant cells (5). Dynamin is a protein that facilitates budding of clathrin-coated pits, leading to formation of coated vesicles. Human rhinovirus 14 was incapable of infecting dynamin mutant cells, suggesting that it uses the clathrin route for its entry, while the infection of mutant cells by poliovirus 1 was not affected. However, recent data have shown that dynamin not only is specific for the clathrin route but is also needed for the formation of caveolae (9, 21). For poliovirus entry, different mechanisms have been suggested; in a recent model, direct pore formation on the cell membrane followed by passage of viral RNA through the pore into the cytoplasm is a generally supported mechanism (2, 35).
It has recently been shown that in echovirus 1 infection the entry process makes use of caveolae instead of clathrin-coated pits (V. Marjomäki, V. Pietiäinen, H. Matilainen, J. Ivaska, L. Nissinen, H. Reunanen, P. Huttunen, T. Hyypiä, and J. Heino, unpublished data). Echovirus 1 capsid proteins were found to be internalized in a complex with α2β1 integrin, a receptor for echovirus 1, and caveolin-1. The importance of integrins in internalization of viruses using the clathrin pathway has also been shown in other virus systems. Adenovirus 2 uses integrins αVβ3 and αVβ5 for entry and it has been shown that blocking of these integrins by antibodies inhibits virus internalization without affecting the attachment (43), while another cell surface protein (CAR) is responsible for virus binding (4).
In conclusion, our results confirm that HPEV-1 interacts with αVβ3 integrin on the cell surface and subsequently enters the host cell through the clathrin-dependent endocytic pathway. However, no significant internalization of the integrin subunits or their colocalization with the virus during the early stages of HPEV-1 infection was observed. Further studies are needed to determine the potential cellular components playing a role in the early events of HPEV-1 entry.
ACKNOWLEDGMENTS
We thank George Banting, Jeffrey Bergelson, Steve Fuller, Jean Gruenberg, Harald Stenmark, and Ismo Virtanen for providing us with antibodies used in the assays. Tapani Hovi, Marikki Laiho, Vilja Pietiäinen, Tuija Pöyry, and Glyn Stanway are acknowledged for helpful discussions, and Maaria Vainio is acknowledged for technical assistance.
The study was financially supported by the Academy of Finland and Helsinki Biomedical Graduate School.
REFERENCES
- 1.Banting G, Maile R, Roquemore E P. The steady state distribution of humTGN46 is not significantly altered in cells defective in clathrin-mediated endocytosis. J Cell Sci. 1998;111:3451–3458. doi: 10.1242/jcs.111.23.3451. [DOI] [PubMed] [Google Scholar]
- 2.Belnap D M, Filman D J, Trus B L, Cheng N, Booy F P, Conway J F, Curry S, Hiremath C N, Tsang S K, Steven A C, Hogle J M. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson J M, Shepley M P, Chan B M, Hemler M E, Finberg R W. Identification of the integrin VLA-2 as a receptor for echovirus 1. Science. 1992;27:1718–1720. doi: 10.1126/science.1553561. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson J M, Cunnigham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.DeTulleo L, Kirchhausen T. The clathrin endocytic pathway in viral infection. EMBO J, 1998;17:4585–4593. doi: 10.1093/emboj/17.16.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruenberg J, Griffiths G, Howell K E. Characterization of the early endosome and putative endocytic carrier vesicles in vivo and with an assay of vesicle fusion in vitro. J Cell Biol. 1989;108:1301–1316. doi: 10.1083/jcb.108.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helenius A, Kartenbeck J, Simons K, Fries E. On the entry of Semliki Forest virus into BHK-21 cells. J Cell Biol. 1980;84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helenius A, Kielian M, Wellsteed J, Mellman I, Rudnick G. Effects of monovalent cations on Semliki Forest virus entry into BHK-21 cells. J Biol Chem. 1985;260:5691–5697. [PubMed] [Google Scholar]
- 9.Henley J R, Krueger E W, Oswald B J, NcNiven M A. Dynamin-mediated internalization of caveolae. J Cell Biol. 1998;141:85–99. doi: 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes P J, Stanway G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J Gen Virol. 2000;81:201–207. doi: 10.1099/0022-1317-81-1-201. [DOI] [PubMed] [Google Scholar]
- 11.Huovila A-P J, Eder A, Fuller S D. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J Cell Biol. 1992;147:401–416. doi: 10.1083/jcb.118.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyypiä T, Horsnell C, Maaronen M, Khan M, Kalkkinen N, Auvinen P, Kinnunen L, Stanway G. A distinct picornavirus group identified by sequence analysis. Proc Natl Acad Sci USA. 1992;89:8847–8851. doi: 10.1073/pnas.89.18.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamison R M. An electron microscopic study of the intracellular development of echovirus 22. Arch Gesamte Virusforsch. 1974;44:184–194. doi: 10.1007/BF01240606. [DOI] [PubMed] [Google Scholar]
- 14.Joki-Korpela P, Roivainen M, Lankinen H, Pöyry T, Hyypiä T. Antigenic properties of human parechovirus 1. J Gen Virol. 2000;81:1709–1718. doi: 10.1099/0022-1317-81-7-1709. [DOI] [PubMed] [Google Scholar]
- 15.Kartenbeck J, Stukenbrock H, Helenius A. Endocytosis of simian virus 40 into the endoplasmic reticulum. J Cell Biol. 1989;109:2721–2729. doi: 10.1083/jcb.109.6.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koistinen P, Pulli T, Uitto V J, Nissinen L, Hyypiä T, Heino J. Depletion of αV integrins from osteosarcoma cells by intracellular antibody expression induces bone differentiation marker genes and suppresses gelatinase (MMP-2) synthesis. Matrix Biol. 1999;18:239–251. doi: 10.1016/s0945-053x(99)00022-0. [DOI] [PubMed] [Google Scholar]
- 17.Leinonen M. Serological methods for the study of bacterial surface antigens. In: Korhonen T K, Dawes E A, Mäkelä P H, editors. Enterobacterial surface antigens: methods for molecular characterization. Amsterdam, The Netherland: Elsevier; 1985. pp. 179–206. [Google Scholar]
- 18.Marjomäki V S, Huovila A P, Surkka M A, Jokinen I, Salminen A. Lysosomal trafficking in rat cardiac myocytes. J Histochem Cytochem. 1990;38:1155–1164. doi: 10.1177/38.8.2164059. [DOI] [PubMed] [Google Scholar]
- 19.Marsh M, Kielian M C, Helenius A. Semliki Forest virus entry and the endocytic pathway. Biochem Soc Trans. 1984;12:981–983. doi: 10.1042/bst0120981. [DOI] [PubMed] [Google Scholar]
- 20.Mu F T, Callaghan J M, Steele-Mortimer O, Stenmark H, Parton R G, Campbell P L, McCluskey J, Yeo J P, Tock E P, Toh B H. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 21.Oh P, McIntosh D P, Schnizer J E. Dynamin at the neck of caveolae mediates their budding to form transport vesicles by GTP-driven fission from the plasma membrane of endothelium. J Cell Biol. 1998;141:101–114. doi: 10.1083/jcb.141.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parton R G, Joggerst B, Simons K. Regulated internalization of caveolae. J Cell Biol, 1994;127:1199–1215. doi: 10.1083/jcb.127.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez L, Carrasco L. Entry of poliovirus into cells does not require a low-pH step. J Virol. 1993;67:4543–4548. doi: 10.1128/jvi.67.8.4543-4548.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pho M T, Ashok A, Atwood W J. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J Virol, 2000;74:2288–2292. doi: 10.1128/jvi.74.5.2288-2292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulli T, Koivunen E, Hyypiä T. Cell-surface interactions of echovirus 22. J Biol Chem. 1997;272:21176–21180. doi: 10.1074/jbc.272.34.21176. [DOI] [PubMed] [Google Scholar]
- 26.Pulli T, Lankinen H, Roivainen M, Hyypiä T. Antigenic sites of coxsackievirus A9. Virology. 1998;240:202–212. doi: 10.1006/viro.1997.8908. [DOI] [PubMed] [Google Scholar]
- 27.Punnonen E L, Marjomäki V S, Reunanen H. 3-Methyladenine inhibits transport from late endosomes to lysosomes in rat and mouse fibroblasts. Eur J Cell Biol. 1994;65:14–25. [PubMed] [Google Scholar]
- 28.Roivainen M, Piirainen L, Hovi T, Virtanen I, Riikonen T, Heino J, Hyypiä T. Entry of coxsackievirus A9 into host cells: specific interactions with αVβ3 integrin, the vitronectin receptor. Virology. 1994;203:357–365. doi: 10.1006/viro.1994.1494. [DOI] [PubMed] [Google Scholar]
- 29.Rojo M, Pepperkok R, Emery G, Kellner R, Stang E, Parton R G, Gruenberg J. Involvement of the transmembrane protein p23 in biosynthetic protein transport. J Cell Biol. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruoslahti E, Pierschbacher M D. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–493. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 31.Shaver D N, Barron A L, Karzon D T. Distinctive cytopathology of ECHO viruses types 22 and 23. Proc Soc Exp Biol Med. 1961;106:648–652. [Google Scholar]
- 32.Stang E, Kartenbeck J, Parton R G. Major histocompatibility complex class I molecules mediate association of SV40 with caveolae. Biol Cell. 1997;8:47–57. doi: 10.1091/mbc.8.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanway G, Kalkkinen N, Roivainen M, Ghazi F, Khan M, Smyth M, Meurman O, Hyypiä T. Molecular and biological characteristics of echovirus 22: a representative of a new picornavirus group. J Virol. 1994;68:8232–8238. doi: 10.1128/jvi.68.12.8232-8238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanway G, Hyypiä T. Parechoviruses. J Virol. 1999;73:5249–5254. doi: 10.1128/jvi.73.7.5249-5254.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tosteson M T, Chow M. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J Virol. 1997;71:507–511. doi: 10.1128/jvi.71.1.507-511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trant C M, Jamison R M. Electron microscopic study of the morphogenesis of echovirus 23. Exp Mol Pathol. 1975;22:55–64. doi: 10.1016/0014-4800(75)90051-9. [DOI] [PubMed] [Google Scholar]
- 37.Triantafilou M, Triantafilou K, Wilson K M, Takada Y, Frenandez N, Stanway G. Involvement of β2-microglobulin and integrin αVβ3 molecules in the coxsackievirus A9 infectious cycle. J Gen Virol. 1999;80:2591–2600. doi: 10.1099/0022-1317-80-10-2591. [DOI] [PubMed] [Google Scholar]
- 38.Triantafilou K, Triantafilou M, Takada Y, Fernandez N. Human parechovirus 1 utilizes integrins αVβ3 and αVβ1 as receptors. J Virol. 2000;74:5856–5862. doi: 10.1128/jvi.74.13.5856-5862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, Huang S, Kapoor-Munshi A, Nemerow G. Adenovirus internalization and infection require dynamin. J Virol. 1998;72:3455–3458. doi: 10.1128/jvi.72.4.3455-3458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L-H, Rothberg K G, Anderson R G W. Mis-assemly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward T, Powell R M, Pipkin P A, Evans D J, Minor P D, Almond J W. Role for β2-microglobulin in echovirus infection of rhabdomyosarcoma cells. J Virol. 1998;72:5360–5365. doi: 10.1128/jvi.72.7.5360-5365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White J, Kartenbeck J, Helenius A. Fusion of Semliki forest virus with the plasma membrane can be induced by low pH. J Cell Biol. 1980;87:264–272. doi: 10.1083/jcb.87.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins αVβ3 and αVβ5 promote adenovirus but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 44.Wigand R, Sabin A B. Properties of ECHO types 22, 23, and 24 viruses. Arch Gesamte Virusforsch. 1961;11:224–247. doi: 10.1007/BF01241688. [DOI] [PubMed] [Google Scholar]
- 45.Ylänne J, Hormia M, Järvinen M, Vartio T, Virtanen I. Platelet glycoprotein IIb/IIIa complex in cultured cells. Localization in focal adhesion sites in spreading HEL cells. Blood. 1988;72:1478–1486. [PubMed] [Google Scholar]
- 46.Zimmermann H, Eggers H J, Nelsen-Salz B. Molecular cloning and sequence determination of the complete genome of the virulent echovirus 9 strain barty. Virus Genes. 1996;12:149–154. doi: 10.1007/BF00572953. [DOI] [PubMed] [Google Scholar]
- 47.Zimmermann H, Eggers H J, Nelsen-Salz B. Cell attachment and mouse virulence of echovirus 9 correlate with an RGD motif in the capsid protein. Virology. 1997;233:149–156. doi: 10.1006/viro.1997.8601. [DOI] [PubMed] [Google Scholar]