Abstract
Detecting EGFR mutations in plasma using droplet digital PCR (ddPCR) assay offers a promising diagnostic tool for lung cancer patients. The performance of plasma-based ddPCR assay relative to traditional EGFR mutation testing in tissue biopsies among Asian patients with suspected lung cancer remains underexplored. Consecutive patients admitted for diagnostic workup for suspected lung cancer were recruited. Peripheral blood samples were collected on the same day of tissue biopsies. Tissue samples were subjected to EGFR mutation analysis via real-time PCR, whereas plasma samples were processed for ddPCR assay to evaluate for EGFR mutation status. The tissue re-biopsy rate was 43.8% while 0.7% of patients failed blood taking. Despite repeat biopsy, 15.2% of patients could not achieve histological diagnosis. Of the 202 patients newly diagnosed with lung cancer, EGFR mutations were detected in 13.4% of plasma samples, compared to 44.3% in tissue samples. Plasma ddPCR for EGFR mutations detection were barely detectable in stages I and II non-small cell lung cancer (NSCLC), but the sensitivity was 25.0%, 56.3%, and 75.0% in stages III, IVA, and IVB NSCLC, respectively. Plasma EGFR mutations were highly specific among all stages of lung cancer. Concordance rates of plasma ddPCR assay also rose with more advanced stages, recorded at 41.9% for stages I and II, 71.9% for stage III, 86.3% for stage IV. In stage IV lung cancer, the false negative rate for the plasma ddPCR assay was 34.4%, whereas that for the tissue testing was 19.2% due to insufficient tissue samples. Plasma-based EGFR genotyping using ddPCR is a non-invasive method that offers early diagnosis and serves as a valuable adjunct to tissue-based testing for patients with advanced-stage lung cancer. However, its usefulness is limited in the context of early-stage lung cancer, indicating a need for further research to improve its accuracy in these patients.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76890-0.
Keywords: EGFR mutations, NSCLC, Plasma, Lung cancer, ddPCR
Subject terms: Cancer, Biomarkers, Medical research
Background
Identification of EGFR mutations is critical for tailoring treatment plans for patients with advanced-stage non-small cell lung cancer (NSCLC). The National Comprehensive Cancer Network (NCCN) NSCLC Panel recommends testing for tumor tissue EGFR mutations in patients with metastatic NSCLC or stage IB-IIIA NSCLC after resection1,2. However, acquiring sufficient tumor tissue for molecular testing remains challenging. Many patients have high-risk factors for tissue biopsy due to comorbidities and the invasive nature of the procedures. In addition, the scheduling for pathological and molecular diagnoses can be time-consuming in many healthcare systems, possibly delaying the initiation of therapy. Blood-based testing, a prevalent form of liquid biopsy, is emerging as a non-invasive alternative for both diagnostic genotyping and treatment monitoring3. Unlike tissue biopsy, liquid biopsy allows for repeated sampling without increased risk and typically offers a much shorter turnaround time, usually measured in days4,5.
Circulating tumor DNA (ctDNA) is the DNA found in the bloodstream that originates from tumors. Reports have indicated a positive correlation between tumour burden and ctDNA mutant allele frequency6. The main challenges in early detection of EGFR mutations in NSCLC patients are from the low tumor burden and the difficulty in identifying the small quantity of ctDNA in the bloodstream7. In additional, plasma EGFR mutations may arise from non-tumor origin, such as clonal haematopoiesis of undetermined potential (CHIP) arising from haematopoietic progenitors6. Various platforms have been developed to identify circulating DNA in plasma, such as amplification-refractory mutation system PCR, next-generation sequencing, and droplet digital PCR (ddPCR)8–11. Studies utilizing CancerSEEK, an early cancer detection platform that incorporates NGS of cell-free DNA(cfDNA) plus protein biomarkers, or multiplex PCR (mPCR) assays, have shown that the largest proportion of patients with stage I NSCLC exhibit undetectable ctDNA6. Introduced in 1999, ddPCR is capable of quantifying absolute nucleic acids without using endogenous controls12. It has demonstrated high accuracy in detecting minute amounts of mutated DNA, enabling the identification of allele frequencies ranging from 0.001–0.4%12,13. This method is highly sensitive for detecting EGFR mutations14–17.
The latest molecular testing guideline from the College of American Pathologists, released in 2018, recommended that plasma DNA testing could be used to detect EGFR mutations in cases where tumor tissue samples are insufficient for molecular analysis. However, the effectiveness of ddPCR in determining plasma EGFR genotypes in patients with suspected lung cancer, spanning early to advanced stages, in comparison to tissue-based EGFR genotyping, remains uncertain This prospective study aims to evaluate the effectiveness of plasma ddPCR assay in detecting EGFR mutations among patients with suspected lung cancer and in those with confirmed lung cancer across various stages. Through a prospective design and pairing blood and tissue genotyping, the study seeks to address challenges related to temporal heterogeneity and provide insights into the clinical validity and practicality of using plasma ddPCR assay in the diagnosis and treatment of patients with lung cancer.
Methods
Subjects
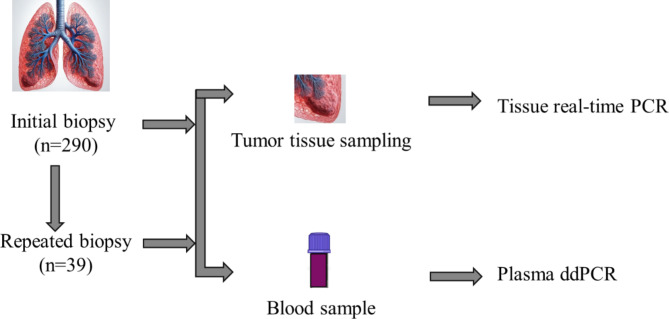
Between July 2019 and November 2020, a total of 290 consecutive adult patients undergoing evaluation for potential lung cancer were enrolled to assess the effectiveness of plasma ddPCR assay in detecting EGFR mutations in patients with suspected lung cancer and those with confirmed lung cancer across various stages. Follow-up for all participants continued until March 31, 2023, or until their decease. The study was approved by the Ethics Committee of Kowloon Central Cluster in Hong Kong (Approval number: KC/KE-19-0041/ER-3), all research was performed in accordance with relevant guidelines/regulations and written informed consent was obtained from all participants. The inclusion criteria were patients aged 18 years or older with clinical or radiological findings suggestive of lung cancer pending biopsy confirmation. The exclusion criteria were patients with known history of lung cancer. Various diagnostic procedures were utilized to obtain a definitive histopathological diagnosis, such as endobronchial biopsy, transbronchial needle aspiration, CT-guided needle aspiration, thoracocentesis and pleural biopsy, pericardiocentesis, ultrasound-guided lymph node biopsy, and surgical biopsy. Tissue and pleural fluid samples were analyzed using standard histopathological and molecular services according to molecular testing guidelines18–20. In cases where NSCLC was confirmed, tissue or cell block samples underwent EGFR mutation testing using Therascreen®EGFR RGQ PCR Kit at tertiary hospitals in Hong Kong. Demographic and clinical characteristics of all enrolled subjects were documented. Cases where NSCLC could not be further classified were recorded as NSCLC. The results were recorded specifically as NSCLC, adenocarcinoma, lymphoepithelioma-like carcinoma or squamous cell carcinoma (SCC). The flowchart of this study is shown in Fig. 1.
Fig. 1.
Flowchart outlining the study procedure.
Sample collection and DNA extraction
Patients who were recruited underwent blood collection on the same day as their tissue biopsy. A venous blood sample was collected using tripotassium ethylenediaminetetraacetic acid (K3EDTA) pre-filled polystyrene tubes (VACUETTE®, Greiner Bio-One, Kremsmünster, Austria) and was centrifuged within 2 h to prepare the plasma. The supernatant plasma was then separated and stored at -80 °C until analysis. Plasma DNA was extracted using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The extracted DNA was stored at -20 °C until genotyping was performed. All subsequent laboratory procedures were conducted at the respiratory research laboratory of the Department of Medicine at the University of Hong Kong.
Droplet digital PCR assay for plasma EGFR mutation detections
Plasma cfDNA was genotyped for EGFR mutations using ddPCR with the QX200 Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA) following the manufacturer’s instructions15. Molecular probes (Bio-Rad, Hercules, CA, USA) were used to detect EGFR mutations for 19Del, L858R, T790M, G719A/C/S, and L861Q. ddPCR was applied with modification21. The 20ul PCR-mix was heated to 95 °C for 30 s to denature the dsDNA templates, then cooled down to 65 °C for 1 min to allow primer-template binding, and then further cooled down and held at 12 °C. After denaturation, droplets were generated using a QX100 droplet generator (Bio-Rad, Hercules, CA, USA), and PCR amplification was performed using a thermal cycler (Bio-Rad, Hercules, CA, USA). The cycling conditions for PCR reactions included an initial incubation at 95 °C for 9.5 min, 45 cycles of 94 °C for 30 s and 55 °C for 60 s, prolonged incubation at 55 °C for 5 min, enzyme inactivation at 98 °C for 10 min, and hold at 12 °C overnight. After thermal cycling, the plates were transferred to a QX200 Droplet reader (Bio-Rad, Hercules, CA, USA) for counting fluorescence-positive and fluorescence-negative droplets.
Data were processed using QuantaSoft software (Bio-Rad, Hercules, CA, USA). The thresholds for the ddPCR results were determined using QuantaSoft and manually inspected for further validation. All technical staff performing plasma ddPCR were blinded to tissue EGFR mutation results. In this study, positive plasma EGFR mutations referred to tyrosine kinase inhibitor (TKI)-sensitive mutations (i.e., 19 Del, L858R, G719X, and L861Q). Given the limitation of validated assays available from the manufacturer at the onset of our study, only 19 Del multiplex assay reported wildtype results along with the mutant allele frequency (MAF). The endogenous control gene RPP3022 was used as an internal control within the assay panel, which enabled the quantification of the relative mutant amounts. The MAF of 19 Del was calculated as MAF (%) = mutant copies per µL / total copies per uL × 100, with the total copies representing the sum of mutant and wildtype copies. The relative mutant amount of EGFR mutations was calculated as Relative Mutant Amount (per k copies ref) = mutant copies per µL / RPP30 copies per µL × 1000, with the “per k copies ref” means per 1000 copies of detected reference gene RPP30.
Statistical analyses
Continuous variables were reported as mean and standard deviation or median, and frequencies were reported as number and proportion. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and concordance of EGFR mutation between blood and tissue genotyping results were calculated using SPSS version 23.0 (SPSS, Inc., Chicago, IL, USA).
Results
Demographic characteristics
A total of 290 consecutive Asian patients with suspected lung cancer were recruited. Among them, 39 patients had repeated blood tests when they underwent repeated procedures. All patients were treatment naïve for TKIs. The characteristics of the enrolled cases are shown in Table 1. The mean age was 69.8 ± 11.8 years, with 118 (40.7%) of patients being female. Out of the 290 patients, 157 (54.1%) patients were former or current smoker, 83 (28.6%) had COPD, and 21 (7.2%) had concurrent cancers including thyroid carcinoma, prostate cancer, stomach cancer, nasal pharyngeal carcinoma, colonic cancer, corpus cancer, renal cell carcinoma, breast cancer, oral squamous cell carcinoma, and pancreatic neuroendocrine tumors. Initial tissue biopsy pathology confirmed primary lung cancer in 138 individuals out of the 290 patients, as detailed in Table 2. Final tissue pathology following repeat biopsies identified primary lung cancer in 202 (69.7%) patients, with 193 (95.5%) of these cases being NSCLC, as shown in Table 2. Benign lung diseases were diagnosed in 32 (11.0%) patients.
Table 1.
Baseline characteristics of the study population.
| Characteristics | Total | Tissue EGFR mutations | Plasma EGFR mutations* | ||
|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | ||
| Number | 290 | 88 | 70 | 259 | 29 |
| Gender (F %) | 118 (40.7) | 22 (25) | 46 (65.7) | 95 (36.7) | 22 (75.9) |
| Age(years) | 69.8 ± 11.8 | 70.6 ± 10.8 | 69.0 ± 12.3 | 70.0 ± 11.6 | 69.2 ± 14.0 |
| BMI, kg/m2 | 22.5 ± 3.9 | 22.5 ± 4.1 | 22.9 ± 3.6 | 22.4 ± 4.0 | 23.6 ± 3.8 |
| ECOG | |||||
| 0–2 | 273 (94.1) | 84 (95.4) | 68 (97.2) | 248 (95.8) | 24 (82.8) |
| 3–4 | 17 (5.9) | 4 (4.6) | 2 (2.8) | 11 (4.2) | 5 (17.2) |
| Smoking | |||||
| Never smoker | 104 (35.9) | 18 (20.5) | 42 (60.0) | 86 (33.2) | 18 (62.1) |
| 2nd hand smoke exposure | 29 (10) | 5 (5.7) | 10 (14.3) | 23 (8.9) | 5 (17.2) |
| Former smoker | 81 (27.9) | 27 (30.7) | 11 (15.7) | 77 (29.7) | 4 (13.8) |
| Current smoker | 76 (26.2) | 38 (43.2) | 7 (10) | 73 (28.2) | 2 (6.9) |
| Hx of COPD | 83 (28.6) | 42 (47.7) | 7 (10) | 81 (31.1) | 1 (3.4) |
| Hx of TB | 29 (10) | 8 (9.1) | 6 (8.6) | 26 (10.0) | 3 (10.3) |
| Current Cancera | 21 (7.2) | 7 (8) | 3 (4.3) | 20 (7.7) | 0 (0) |
| Serum CEA level (ng/ml) | 55.7 ± 336.6 | 48.7 ± 144.4 | 78.1 ± 198.3 | 26.6 ± 99.0 | 317.0 ± 996.5 |
| Tumor volume (cm3) | 62.4 ± 146.3 | 79.0 ± 172.1 | 27.2 ± 58.7 | 62.1 ± 149.2 | 69.4 ± 127.0 |
| SUVs | 8.2 ± 5.4 | 10.6 ± 6.2 | 6.8 ± 3.4 | 8.2 ± 5.6 | 9.2 ± 3.2 |
| Stages | |||||
| I/II/III | 144 (49.7) | 38 (43.8) | 38 (54.3) | 140 (54.1) | 3 (10.3) |
| IV | 146 (50.3) | 49 (56.3) | 32 (45.7) | 119 (45.9) | 26(89.7) |
| Final Tis EGFR | |||||
| Wild | 88 (55.7) | 88(100) | 0 (0) | 87 (64.4) | 0 (0) |
| L858R | 39 (24.7) | 0 (0) | 39 (55.7) | 25 (18.5) | 13 (54.2) |
| 19Del | 26 (16.5) | 0 (0) | 26 (37.1) | 16 (11.9) | 10 (41.7) |
| Uncommonb | 5 (3.2) | 0 (0) | 5 (7.1) | 3 (2.9) | 1 (4.2) |
| Final Tis Patho | |||||
| No malignancy | 32 (13.0) | 0 (0) | 0 (0) | 32 (14.7) | 0 (0.0) |
| Adenocarcinoma/NSCLCc | 174 (70.7) | 83 (95.4) | 70 (100) | 146(67.3) | 26 (96.3) |
| Other malignancyd | 40 (16.3) | 4 (4.6) | 0 (0) | 39 (18.0) | 1 (3.7) |
*2 patients declined to provide blood samples.
a active cancer originating from a primary tumor other than lung cancer.
b including G719x, L861x, Ins20.
c adenocarcinoma and NSCLC were mutually exclusive.
d including SCC, SCLC, metastatic tumors, thymoma, mesothelioma and lymphoma.
RG, reference group; Mets, metastases; Blank, not applicable.
Table 2.
Pathological diagnoses according to paired tissues and final tissues.
| Pathological diagnoses | Paired tissue (No.) | Paired tissue (%) | Final tissue (No.) | Final tissue (%) |
|---|---|---|---|---|
| No malignancy | 88 | 30.3 | 16 | 5.5 |
| NSCLC | 26 | 9.0 | 32 | 11.0 |
| Adenocarcinoma | 89 | 30.7 | 138 | 47.6 |
| SCC | 15 | 5.2 | 19 | 6.6 |
| SCLC | 6 | 2.1 | 9 | 3.1 |
| LELC | 2 | 0.7 | 4 | 1.4 |
| Mesothelioma | 2 | 0.7 | 2 | 0.7 |
| Lymphoma | 1 | 0.3 | 1 | 0.3 |
| Thymoma | 0 | 0 | 1 | 0.3 |
| 2nd carcinoma | 2 | 0.7 | 8 | 2.8 |
| IgG4 related disease | 0 | 0 | 1 | 0.3 |
| Chronic inflammation | 0 | 0 | 2 | 0.7 |
| Abscess/necrosis | 3 | 1.0 | 3 | 1.0 |
| Granulation tissue | 5 | 1.7 | 8 | 2.8 |
| Organized pneumonia | 1 | 0.3 | 2 | 0.7 |
| Unknown/atypical cells | 7/43 | 2.4/14.8 | 44 | 15.1 |
| Total | 290 | 100 | 290 | 100.0 |
LELC, lymphoepithelioma-like carcinoma.
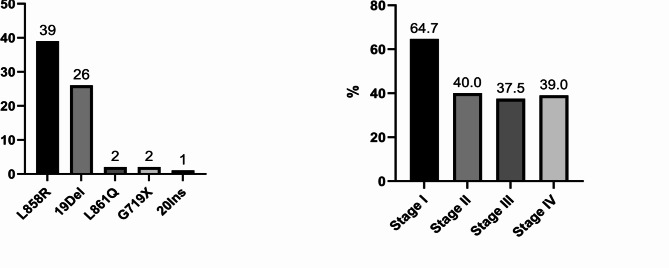
Of the 202 patients with a definitive diagnosis of lung cancer, 45 (22.3%) were at stage I, 13 (6.4%) at stage II, 38 (18.8%) at stage III, and 106 (52.5%) at stage IV of the disease. In addition, 12 patients (5.9%) were found to have other types of lung malignancies including lung metastases from other primary cancers, lymphoma, thymoma, and mesothelioma. Among the 202 patients with a diagnosis of lung cancer, 158 cases had tissue EGFR mutation testing results available. Among these, 88 (55.7%) were EGFR wild-type, while 70 (44.3%) had EGFR mutations. The most frequently identified EGFR mutations in tissue samples were L858R substitution (39 cases), and exon 19 deletions (19Del, 26 cases). Rarer mutations such as L861Q and G719X were found in 2 out of 70 patients each, and there was one case of an exon 20 insertion mutation (Fig. 2A). T790M mutation was not detected in any of treatment-naïve NSCLC tissue samples, and no cases of double mutations were observed.
Fig. 2.
Tissue EGFR genotyping and mutation rates by lung cancer stage. The prevalent EGFR mutations identified in tissue samples were L858R and Exon 19 deletions. EGFR mutation rate varied across different stages of lung cancer; the rate was highest at 64.7% in stage I NSCLC and decreased to 40.0% in stage II, 37.5% in stage III, and 39.0% in stage IV NSCLC.
The presence of tissue EGFR mutations varied by sex, with a higher rate of 67.6% in females compared to 26.7% in males. Variation in EGFR mutation rates was also observed across different stages of lung cancer, with the highest rate at 64.7% (22/34) in stage I NSCLC, decreasing to 40.0% (4/10) in stage II, 37.5% (12/32) in stage III, and 39.0% (32/82) in stage IV NSCLC, as shown in Fig. 2B. Variations in EGFR mutation rates were further observed when comparing patients with a history of smoking to non-smokers or those exposed to secondhand smoke. Non-smokers or second-hand smokers had a tissue EGFR mutation rate of 59.8%, whereas this rate dropped to 17% among former or current smokers. Among patients with tissue EGFR mutations, 25.7% were former or current smokers, while in those with wild-type EGFR, a higher proportion of 73.6% were former or current smokers.
Re-biopsy rate and time to final diagnosis
Out of 290 patients, 163 (56.2%) had a pathological diagnosis after the first tissue biopsy, while the remaining 127 (43.8%) required a repeat biopsy. Despite extensive investigations, 44 individuals (15.2%) remained without a histological diagnosis. The likelihood of requiring a repeat biopsy was lower in those with radiological stage IV lung cancer (34.2%) compared to those with stages I/II (52.3%). The median time from initial enrollment to the final diagnosis was 55.5 days. When stratified by cancer stages, the median time to diagnosis was at 131 days for stage I/II lung cancer patients, shorter for stage III (52 days) and stage IV (34 days) lung cancer patients, and 50 days for patients diagnosed with other thoracic malignancies.
Plasma EGFR mutation status in patients with suspected lung cancer
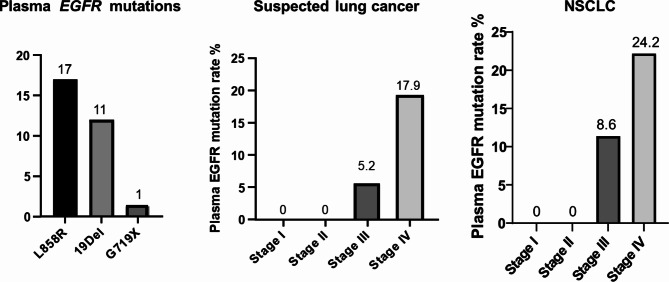
Plasma genotyping using ddPCR assay was successfully conducted for 288 of the 290 patients, with 2 patients declining to provide blood samples. MAF and relative mutant amount detected by ddPCR in plasma of the EGFR mutant cases are displayed in supplementary data. Of the analyzed cases, 259 (89.9%) exhibited no EGFR mutations in plasma, while 29 (10.1%) showed EGFR mutations. Specifically, 17 patients had the L858R substitution, 11 had 19Del, and 1 had the G719X mutation, as illustrated in Fig. 3. Out of 202 lung cancer patients, 27 (13.5%) demonstrated EGFR mutations in plasma, including 15 cases of the L858R substitution, 11 of 19Del and 1 of the G719X mutation. In contrast, none of the 32 patients with benign lung conditions had detectable plasma EGFR mutations. In the 44 patients with radiological lung cancer but lacking a definitive histological diagnosis, 2 cases (4.5%) had EGFR mutations in their plasma, both being the L858R substitution. One patient with SCC, representing 5.3% of the SCC group, had L858R substitution detected in plasma.
Fig. 3.
Plasma EGFR genotyping and mutation rates by lung cancer stage. The prevalent EGFR mutations identified in plasma samples are L858R and Exon 19 deletions. EGFR mutations were not detected in the plasma of patients with stage I and II lung cancer, but detection rates were notably higher in stage IV lung cancer, similar findings were found in patients with suspected lung cancer.
Plasma EGFR mutations varied across lung cancer stages (Fig. 3), with no detectable plasma EGFR mutations in stages I (n = 67) and stage II (n = 18) suspected lung cancer, a rate of 5.2% in patients with stage III suspected lung cancer (3 out of 58), and 17.9% in those suspected of stage IV lung cancer (26 out of 145). This trend was consistent in patients with a confirmed NSCLC, where stages I (n = 44) and stage II (n = 13) patients exhibited no detectable plasma EGFR mutations, while stage III patients had a plasma EGFR mutation rate of 8.6% (3 out of 35), and stage IV patients had a rate of 24.2% (24 out of 99). Differences in plasma EGFR mutation rates were observed between patients with a history of smoking and nonsmokers or those exposed to secondhand smoke. Non-smokers or second-hand smokers had a plasma EGFR mutation rate of 15.9%, whereas this rate dropped to 7.1% among former or current smokers. Among patients with plasma EGFR mutations, 55.6% were non-smokers or second-hand smokers, while in those with wild-type plasma EGFR, a higher proportion of 56.6% were former or current smokers.
In a subset of 39 patients whose initial tissue biopsy did not yield a cancer diagnosis, matched blood and tissue samples were collected both at initial recruitment and during the repeat biopsy, with plasma ddPCR assays performed on both plasma samples. Among these patients, 5 showed EGFR mutation on first plasma ddPCR assessment, which were consistently present upon repeat testing. These mutations were subsequently confirmed through analysis of tissue samples with which the diagnosis of lung cancer was confirmed. Among the 21 patients with concurrent cancer, none of them showed EGFR mutations in plasma.
Diagnostic performance of ddPCR in plasma samples
Tissue-based EGFR genotyping in NSCLC patients was conducted in accordance with the 2018 guidelines for molecular testing in lung cancer20. The sensitivity, specificity, PPV, NPV, and kappa statistics of plasma ddPCR assay in NSCLC patients were shown in Table 3. The sensitivity of the plasma ddPCR assay increase with the stage of lung cancer, being 0% for stages I and II, 25.0% for stage III, 65.6% for stage IV, 56.3% for stage IVA, and 75.0% for stage IVB NSCLC. The PPV remained consistently high at 100% across all lung cancer stages. Concordance rates of plasma ddPCR assay also rose with advanced stages, with rates recorded at 41.9% for stages I and II, 71.9% for stage III, 86.3% for stage IV, 82.9% for stage IVA, and 89.7% for stage IVB. Kappa statistics, which assessed agreement beyond chance, improved from 0 in stages I and II (no agreement) to 0.29 in stage III (fair agreement), 0.70 in stage IV (substantial agreement), 0.61 in stage IVA, and 0.78 in stage IVB. These findings were consistent with results obtained when paired tissue-based genotyping was used as the reference for NSCLC patients (Table 3).
Table 3.
Sensitivity, specificity, PPV, NPV and kappa statistics of plasma ddPCR assay in patients with stages I/II, III, IV, IVA and IVB lung cancer, respectively.
| EGFR mutation status (Count) | Plasma | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Concordance (%) | Kappa statistics | |
|---|---|---|---|---|---|---|---|---|
| WT | Mutated | |||||||
| Stage I/II | ||||||||
| Paired Tissuea | ||||||||
| WT | 10 | 0 | 0 | 100 | UD | 37.0 | 37.0 | 0.00 |
| Mutated | 17 | 0 | ||||||
| Final tissueb | ||||||||
| WT | 18 | 0 | 0 | 100 | UD | 41.9 | 41.9 | 0.00 |
| Mutated | 25 | 0 | ||||||
| Stage III | ||||||||
| Paired Tissuea | ||||||||
| WT | 14 | 0 | 27.3 | 100 | 100 | 63.6 | 68.0 | 0.30 |
| Mutated | 8 | 3 | ||||||
| Final tissueb | ||||||||
| WT | 20 | 0 | 25.0 | 100 | 100 | 69.0 | 71.9 | 0.29 |
| Mutated | 9 | 3 | ||||||
| Stage IV | ||||||||
| Paired Tissuea | ||||||||
| WT | 43 | 0 | 68.0 | 100 | 100 | 84.3 | 88.2 | 0.73 |
| Mutated | 8 | 17 | ||||||
| Final tissueb | ||||||||
| WT | 48 | 0 | 65.6 | 100 | 100 | 81.4 | 86.3 | 0.70 |
| Mutated | 11 | 21 | ||||||
| Stage IVA | ||||||||
| Paired Tissuea | ||||||||
| WT | 23 | 0 | 58.3 | 100 | 100 | 82.1 | 85.7 | 0.65 |
| Mutated | 5 | 7 | ||||||
| Final tissueb | ||||||||
| WT | 25 | 0 | 56.3 | 100 | 100 | 78.1 | 82.9 | 0.61 |
| Mutated | 7 | 9 | ||||||
| Stage IVB | ||||||||
| Paired Tissuea | ||||||||
| WT | 19 | 0 | 76.9 | 100 | 100 | 86.4 | 90.6 | 0.80 |
| Mutated | 3 | 10 | ||||||
| Final tissueb | ||||||||
| WT | 23 | 0 | 75.0 | 100 | 100 | 85.2 | 89.7 | 0.78 |
| Mutated | 4 | 12 | ||||||
aCollected on enrollment with paired blood samples.
bTissue with final diagnosis achieved either at initial biopsy tissue with pathological diagnosis achieved or at repeat biopsy tissue with pathological diagnosis achieved.
WT, wild-type; PPV, positive predictive value; NPV, negative predictive value; UD, undefined.
Discordance between plasma and tissue EGFR mutation genotyping in patients with suspected lung cancer
Discordance was observed when comparing genotyping results for EGFR mutations in plasma and tissue samples. Specifically, 24 patients had EGFR mutations detected in both tissue and plasma, 45 had EGFR mutations solely detected in tissue samples, while no patient had EGFR mutations detected exclusively in plasma. Using tissue EGFR mutation results as the standard, the overall false negative rate of plasma ddPCR assay was 65.2% (45/69). Among patients with stage IV lung cancer, the false negative rate was lower, at 34.4% (11/32). While using plasma EGFR results as the standard, tissue-based testing resulted in false negatives, primarily attributed to inadequate tissue sampling for molecular testing. The overall false negative rate of initial paired tissue-based testing was 31.0% (9 out of 29 patients) and the false negative rate of final tissue-based testing was 17.2% (5 out of 29 patients). Among patients with stage IV lung cancer, the false negative rate of tissue-based testing was 19.2% (5/26).
Discussion
In this prospective study, we assessed the effectiveness of ddPCR assay, a highly sensitive method for detecting EGFR mutations in plasma, in patients with suspected lung cancer. Our findings indicated that plasma ddPCR assay could facilitate earlier detection of EGFR mutations in patients with suspected advanced lung cancer compared to traditional tissue-based testing. Given the high PPV of plasma-based EGFR genotyping, this approach could serve as an alternative to tissue biopsy when sufficient tissue sampling is unavailable in advanced lung cancer cases. However, for early-stage lung cancer, plasma-based EGFR genotyping using ddPCR did not detect any EGFR mutation even when EGFR mutations were identified in tissue samples. Our study underscores the limited sensitivity of plasma ddPCR analysis in the detection of EGFR mutations compared to tumor tissue analysis in early-stage lung cancer.
Tissue biopsy has traditionally served as the standard method for detecting targetable mutations. However, the accuracy of tissue-based EGFR genotyping may be compromised by insufficient tissue samples, potentially leading to missed detections of EGFR mutations. A prospective study in 2016 showed that ddPCR-based plasma genotyping assay could rapidly and accurately detect EGFR mutations in a real-world clinical setting, supported the potential use of this assay to guide clinical decisions23. Our research confirmed these findings by employing a prospective approach that incorporated the simultaneous collection of paired blood and tissue specimens, precise documentation of clinical data, and blinding of laboratory personnel involved in both tissue and plasma genotyping assessments. In our study, plasma genotyping was successfully performed in 99.3% of patients with presumed lung cancer. Among the 44 patients with radiological lung cancer lacking histological confirmation, 2 cases exhibited EGFR mutations in their plasma, providing valuable information for making treatment decisions regarding the use of TKI treatment options. In contrast, only 56.2% of patients received a histological and molecular diagnosis following the initial tissue biopsy. Furthermore, plasma genotyping facilitated an earlier diagnosis, with a median duration of 55.5 days, compared to tissue-based genotyping.
The prevalence of EGFR mutations is known to differ among various ethnic groups24. Liang et al. analysed data from 1134 advanced NSCLC patients in China and found a tissue EGFR mutation rate of 44.1%25. Similarly, Zhou et al. examined 261 NSCLC patients in Western China and reported an EGFR mutation rate of 48.7%26. In our study, the overall tissue EGFR mutation rate among NSCLC patients was 44.3%, with the highest rate observed in stage I and II patients at 59.1%. The rate decreased to 39% in stage IV patients. This finding aligns with a previous report that the EGFR mutation rate was 51.5% (16/33) in early-stage NSCLC patients in Taiwan27. Lung cancer in never smokers ranks the fifth most common cause of cancer-related deaths worldwide in 2023, preferentially affecting in women and Asian populations28. Previous studies suggested lung cancer in never smokers may be driven by distinct driver mutations which were different from the genetic pathways in smokers29. Our study found formal or current smokers had higher rate of tissue EGFR mutation and plasma EGFR mutation, consisting with previous study in Chinese patients30.
A highly sensitive method for EGFR mutation detection is essential to increase the detection rate of patients who would benefit from EGFR-TKIs. The ddPCR assay used in this study has demonstrated to be more sensitive than Sanger sequencing and amplification-refractory mutation system PCR technology, making it a valuable tool for detecting EGFR mutations in liquid biopsies11,31,14–17. However, there are limitations of digital PCR. Due to its probe-based PCR design, the performance of digital PCR can be affected by factors such as primer/probe design and thermal cycling conditions, similar to traditional PCR methods32. For instance, the melting temperature of the manufacturer designed assays is at a relatively low 55 ℃, which can yield higher signal-to-background ratio but result in a more scattered cloud of droplets. To address these issues, minor optimizations of protocols were applied to partially mitigate these limitations in this study21. Furthermore, the determination of a valid positive signal is a common concern in digital PCR. The sensitivity of most assays could be as low as 0.1% when conducting absolute quantification on diluted positive control specimen. However, samples with very low DNA input or those with few positive droplets in the results can pose challenges in accurately determining the genotype33,34. Despite the use of ddPCR assay, our study did not detect significant EGFR mutations in plasma samples from early-stage lung cancer patients. This contrasts with a retrospective study where ddPCR identified EGFR mutations in the serum of 12.0% (12/100) of early-stage lung cancer patients who had tissue-confirmed EGFR mutations35. Our findings suggest that plasma ddPCR plays a very limited role in the diagnosis of early-stage lung cancer. These results support the latest NCCN guidelines, which advise against the routine use of ctDNA for clinical decision-making outside of advanced or metastatic disease scenarios2. The application of plasma ddPCR assay for EGFR mutation detection in early-stage lung cancer remains a challenge, with the scarcity of ctDNA in the blood creating a bottleneck that limits the ddPCR sensitivity. The amounts of cfDNA and ctDNA in the circulation are regulated by factors such as cell turnover, degradation, and clearance mechanisms like nuclease digestion, renal excretion, and uptake by macrophages and the phagocyte system in the liver. The rapid clearance of cfDNA means that a blood draw of 10 ml yields a limited amount of cfDNA. Efforts to enhance sensitivity have primarily focused on ex vivo strategies, such as sampling, analytical processes and bioinformatics36. Martin-Alonso et al. reported an alternative strategy of transiently attenuating cfDNA clearance in vivo to increase its concentration in blood samples37. Two priming agents, liposomal nanoparticles and DNA-binding antibody, given 1–2 h before blood collection, improved the sensitivity and robustness of ctDNA testing in tumor bearing mice37. This approach led to in a more than 10-fold increase of ctDNA recovery and improved the sensitivity for detecting small tumors from less than 10% to over 75%37.
As NGS becomes more prevalent, the significance of ddPCR for single gene evaluations has diminished, given that high-throughput NGS-based multigene liquid biopsy tests can detect a range of genomic alterations. However, NGS typically requires a higher allele frequency compared to ddPCR38. Blood-based NGS might not detect driver alterations due to insufficient shedding of tumor DNA28. Whether the use of multigene panels offers improved clinical outcomes compared to single-gene assays remains to be determined36.
Our study revealed an ascending sensitivity in ddPCR plasma assay from early-stage (I/II) to advanced-stage (IVB) lung cancer. The sensitivity increased from 25% for stage III, 56.3% for stage IVA, and to 75% for stage IVB lung cancer. The assay demonstrated a high PPV of 100% in stage III and IV lung cancer patients, consisting with earlier trials involving Asian populations39,40. In the Asia-centric LUX-Lung 6 trial, the plasma EGFR mutation detection rate using the Therascreen real-time PCR assay was 60.5% for stages III and IV lung cancer patients39,40. Studies from Japan and India have reported sensitivities ranging from 75.8 to 81.8% and a specificity from 87.5 to 100% in advanced lung cancer patients using plasma ddPCR assay14–17. Furthermore, our study revealed that the ddPCR plasma assay exhibited a high PPV of 100%. However, both plasma and tissue testing demonstrated considerable false negative rates in stage IV lung cancer − 34.4% for plasma ddPCR assay and 19.2% for tissue testing approach, the latter primarily due to insufficient tissue samples. These findings suggest that integrating plasma and tissue testing in advanced stage lung cancer could potentially expedite the detection of actionable mutations, thereby facilitating the prompt initiation of targeted therapies.
This study represents the largest cohort of data evaluating plasma ddPCR assay for EGFR genotyping in patients with suspected lung cancer. However, there were several limitations. Firstly, the cohort included a relatively small number of patients with early-stage lung cancer compared to advanced stage lung cancer. Further research, incorporating larger-scale studies and exploring novel diagnostic methods, or an integration of multiple approaches, are essential to enhance our understanding of early-stage lung cancer detection. Secondly, while paired blood and tissue samples were collected on the same day, 45.7% of the blood samples were obtained immediately following tissue sampling, potentially introducing interference in the blood test results due to the invasive nature of the procedure. In future studies, ensuring that blood is collected prior to any invasive procedure would filter out potential interference. Lastly, EGFR mutations are not exclusive to lung cancer. EGFR was upregulated in glioblastoma, head and neck squamous cell carcinoma, kidney renal cell carcinoma, and NSCLC, while it was downregulated in breast invasive carcinoma, colon adenocarcinoma, pheochromocytoma and paraganglioma, prostate adenocarcinoma, rectum adenocarcinoma, and uterine corpus endometrial carcinoma41,42. In our study, EGFR mutation testing from concurrent tumor tissues in patients with multiple cancers were beyond the scope of our research. However, none of the 21 patients with concurrent cancers exhibited EGFR mutations in plasma. Moreover, EGFR mutation detected in plasma may not always indicate a tumor origin. In cases of CHIP, where haematopoietic progenitor lead to clonal expansion without haematological neoplasia, genes such as DNMT3A, TET2, PPM1D and TP53 were commonly mutated6. Screening white blood cell DNA and cfDNA can filter out CHIP-related mutations. As far as our knowledge goes, no EGFR mutations linked to CHIP have been reported. In our study, all the EGFR mutations detected in plasma were consistent with positive findings in lung cancer tumor tissues.
Conclusions
This prospective study underscores the clinical utility of a plasma-based ddPCR assay for identifying EGFR mutations in patients with suspected advanced lung cancer, particularly beneficial for those unable to undergo tissue biopsy. This non-invasive approach has the potential to influence treatment decisions, ultimately improving the quality of life for patients with advanced lung cancer. However, its usefulness is limited in the context of early-stage lung cancer, indicating a need for further research on novel priming agents or combined diagnostic strategies to enhance detection accuracy43.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to express our gratitude to Prof Mary Sau-Man Ip for her invaluable advice, thorough review and revisions of the manuscript, to Mr Steven Yuk-Fai Lau and Prof Benny Chung-Ying Zee for their advice on data analysis. We thank Dr Wilma Shum, Dr Derek Lee, Dr Chun-Lee Kwok, Dr Yee-Yan Sze, Dr Cee-Kin Tseng, Dr Nicolas Yiu-Hung Tang, and Dr Emmanuel Le for their generous support in patient screening. We also extend our appreciation to Ms Wing-Han Tsui, Mr Wing-Hei Yip and Ms Ruby Chan for their assistance with data input.
Author contributions
L.S. conceptualized the study, acquired funding, analyzed and interpreted the patient data, administered the project, and contributed to writing the original draft . J.D. performed ddPCR assay, contributed to data collection and draft review and editing. H.K. and N.L. were responsible for blood sample processing. S.T. contributed to patient screening, data collection. L.N. and W.Y. were involved in conceptualization, project administration, supervision. D.L. was responsible for conceptualization, project administration, supervision, and manuscript review & editing, also contributed to funding acquisition. All authors reviewed the manuscript.
Funding
This research was supported by the Hong Kong Kowloon Central Cluster Research Grant (Ref: KCC/RC/1920-A04) and the Lee and the Ho Families Respiratory Health Research Fund at the University of Hong Kong. The funding body has no role in the design of the study, the collection, analysis, and interpretation of data, or the writing of the manuscript.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Kowloon Central Cluster in Hong Kong (Approval number: KC/KE-19-0041/ER-3), and written informed consent was obtained from all participants.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: LynnYim-Wah Shong and Jun-Yang Deng.
References
- 1.Ettinger, D. S. et al. NCCN Guidelines Insights: Non-small cell lung cancer, Version 2.2021. J. Natl. Comp. Cancer Netw.19(3), 254–266. 10.6004/jnccn.2021.0013 (2021). [DOI] [PubMed] [Google Scholar]
- 2.National Comprehensive Cancer Network. Clinical Practice Guidelines in Oncology (NCCN Guidelines): Non–Small Cell Lung Cancer, Version 10.2024. (https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1450).
- 3.Corcoran, R. B. & Chabner, B. A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med.379(18), 1754–1765. 10.1056/NEJMra1706174 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Overman, M. J. et al. Use of research biopsies in clinical trials: Are risks and benefits adequately discussed?. J. Clin. Oncol.31(1), 17–22. 10.1200/JCO.2012.43.1718 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanderlaan, P. A. et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer84(1), 39–44. 10.1016/j.lungcan.2014.01.013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbosh, C., Birkbak, N. J. & Swanton, C. Early stage NSCLC—Challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol.15(9), 577–586. 10.1038/s41571-018-0058-3 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Schwarzenbach, H., Hoon, D. S. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer11(6), 426–437. 10.1038/nrc3066 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Liu, J., Zhao, R., Zhang, J. & Zhang, J. ARMS for EGFR mutation analysis of cytologic and corresponding lung adenocarcinoma histologic specimens. J. Cancer Res. Clin. Oncol.141(2), 221–227. 10.1007/s00432-014-1807-z (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, X. et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: Comparison of methodologies. J. Clin. Pathol.66(12), 1065–1069. 10.1136/jclinpath-2013-201728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vendrell, J. A. et al. Circulating cell free tumor DNA detection as a routine tool for lung cancer patient management. Int. J. Mol. Sci.10.3390/ijms18020264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sacher, A. G., Komatsubara, K. M. & Oxnard, G. R. Application of plasma genotyping technologies in non-small cell lung cancer: A practical review. J. Thorac. Oncol.12(9), 1344–1356. 10.1016/j.jtho.2017.05.022 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem.83(22), 8604–10. 10.1021/ac202028g (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein, B. & Kinzler, K. W. Digital PCR. Proc. Natl. Acad. Sci. USA96(16), 9236–9241 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suryavanshi, M. et al. The detection of primary and secondary EGFR mutations using droplet digital PCR in patients with nonsmall cell lung cancer. Lung India35(5), 384–389. 10.4103/lungindia.lungindia_472_17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahama, T. et al. Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study). Oncotarget7(36), 58492–58499. 10.18632/oncotarget.11303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellert, H. et al. Development and clinical utility of a blood-based test service for the rapid identification of actionable mutations in non-small cell lung carcinoma. J. Mol. Diagn.19(3), 404–416. 10.1016/j.jmoldx.2016.11.004 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Zhu, G. et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J. Mol. Diagn.17(3), 265–272. 10.1016/j.jmoldx.2015.01.004 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Planchard, D. et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol.29(Suppl 4), iv192–iv237. 10.1093/annonc/mdy275 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Lindeman, N. I. et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med.142(3), 321–346. 10.5858/arpa.2017-0388-CP (2018). [DOI] [PubMed] [Google Scholar]
- 20.Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol.36(16), 1631–1641. 10.1200/jco.2017.76.8671 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Fitarelli-Kiehl, M. et al. Denaturation-enhanced droplet digital PCR for liquid biopsies. Clin. Chem.64(12), 1762–1771. 10.1373/clinchem.2018.293845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowlands, V. et al. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci. Rep.9(1), 12620. 10.1038/s41598-019-49043-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sacher, A. G. et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol.2(8), 1014–22. 10.1001/jamaoncol.2016.0173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Midha, A., Dearden, S. & McCormack, R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: A systematic review and global map by ethnicity (mutMapII). Am. J. Cancer Res.5(9), 2892–2911 (2015). [PMC free article] [PubMed] [Google Scholar]
- 25.Liang, H. et al. Real-world data on EGFR/ALK gene status and first-line targeted therapy rate in newly diagnosed advanced non-small cell lung cancer patients in Northern China: A prospective observational study. Thorac. Cancer10(7), 1521–1532. 10.1111/1759-7714.13090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, J. et al. Prevalence and clinical profile of EGFR mutation in non- small-cell lung carcinoma patients in Southwest China. Asian Pac. J. Cancer Prev.17(3), 965–71. 10.7314/apjcp.2016.17.3.965 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Wu, Y. H. et al. Old age and EGFR mutation status in inoperable early-stage non-small cell lung cancer patients receiving stereotactic ablative radiotherapy: A single institute experience of 71 patients in Taiwan. Thorac. Cancer14(7), 654–661. 10.1111/1759-7714.14786 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LoPiccolo, J., Gusev, A., Christiani, D. C. & Jänne, P. A. Lung cancer in patients who have never smoked—An emerging disease. Nat. Rev. Clin. Oncol.21(2), 121–146. 10.1038/s41571-023-00844-0 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman, A. M., Sun, K. Y., Ruestow, P., Cowan, D. M. & Madl, A. K. Lung cancer mutation profile of EGFR, ALK, and KRAS: Meta-analysis and comparison of never and ever smokers. Lung Cancer102, 122–134. 10.1016/j.lungcan.2016.10.010 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Bai, H. et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J. Clin. Oncol.27(16), 2653–9. 10.1200/jco.2008.17.3930 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Akhoundova, D. et al. The role of the liquid biopsy in decision-making for patients with non-small cell lung cancer. J. Clin. Med.10.3390/jcm9113674 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, S. C., Carbonneau, J., Shelton, D. N. & Boivin, G. Optimization of droplet digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: Clinical implications for quantification of Oseltamivir-resistant subpopulations. J. Virol. Methods224, 58–66. 10.1016/j.jviromet.2015.08.014 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Léonce, C. et al. Comparison and validation of rapid molecular testing methods for theranostic epidermal growth factor receptor alterations in lung cancer: Idylla versus digital droplet PCR. Int. J. Mol. Sci.10.3390/ijms242115684 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lettig, L. et al. EGFR T790M detection rate in lung adenocarcinomas at baseline using droplet digital PCR and validation by ultra-deep next generation sequencing. Transl. Lung Cancer Res.8(5), 584–592. 10.21037/tlcr.2019.09.18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito, M. et al. Sensitivity and optimal clinicopathological features for mutation-targeted liquid biopsy in pN0M0 EGFR-mutant lung adenocarcinoma. J. Cancer Res. Clin. Oncol.148(6), 1419–1428. 10.1007/s00432-021-03721-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ignatiadis, M., Sledge, G. W. & Jeffrey, S. S. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat. Rev. Clin. Oncol.18(5), 297–312. 10.1038/s41571-020-00457-x (2021). [DOI] [PubMed] [Google Scholar]
- 37.Martin-Alonso, C. et al. Priming agents transiently reduce the clearance of cell-free DNA to improve liquid biopsies. Science383(6680), eadf2341. 10.1126/science.adf2341 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen, M. & Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genomics13(1), 34. 10.1186/s40246-019-0220-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, Y. L. et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol.15(2), 213–22. 10.1016/s1470-2045(13)70604-1 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Wu, Y. L. et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: Analysis of LUX-Lung 3 and 6. Br. J. Cancer116(2), 175–185. 10.1038/bjc.2016.420 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, B. et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol.17(1), 174. 10.1186/s13059-016-1028-7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makawi, A., Khalafallah, S. A., Faris, I. M. & Alfaki, M. Comprehensive analysis reveals epithelial growth factor receptor as a potential diagnostic biomarker in glioblastoma multiforme. Cureus16(7), e64506. 10.7759/cureus.64506 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moser, T. & Heitzer, E. Surpassing sensitivity limits in liquid biopsy. Science383(6680), 260–261. 10.1126/science.adn1886 (2024). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.