Abstract
Hepadnavirus genome replication involves cytoplasmic and nuclear stages, requiring balanced targeting of cytoplasmic nucleocapsids to the nuclear compartment. In this study, we analyze the signals determining capsid compartmentalization in the duck hepatitis B virus (DHBV) animal model, as this system also allows us to study hepadnavirus infection of cultured primary hepatocytes. Using fusions to the green fluorescent protein as a functional assay, we have identified a nuclear localization signal (NLS) that mediates nuclear pore association of the DHBV nucleocapsid and nuclear import of DHBV core protein (DHBc)-derived polypeptides. The DHBc NLS mapped is unique. It bears homology to repetitive NLS elements previously identified near the carboxy terminus of the capsid protein of hepatitis B virus, the human prototype of the hepadnavirus family, but it maps to a more internal position. In further contrast to the hepatitis B virus core protein NLS, the DHBc NLS is not positioned near phosphorylation target sites that are generally assumed to modulate nucleocytoplasmic transport. In functional assays with a knockout mutant, the DHBc NLS was found to be essential for nuclear pore association of the nucleocapsid. The NLS was found to be also essential for virus production from the full-length DHBV genome in transfected cells and from hepatocytes infected with transcomplemented mutant virus. Finally, the DHBc additionally displayed activity indicative of a nuclear export signal, presumably counterbalancing NLS function in the productive state of the infected cell and thereby preventing nucleoplasmic accumulation of nucleocapsids.
Viruses that replicate in the nucleus have evolved means to transport their infecting genome through the cytoplasm towards and into the nucleus. As described previously for adenovirus and herpesviruses, viruses typically exploit the preexisting cellular nucleocytoplasmic transport machinery for targeting the nucleocapsid to the nuclear pore, where the genome is released and imported as a more or less complex nucleoprotein, as exemplified by lentiviruses or influenza viruses (12, 17, 21, 42). Nuclear targeting of the nucleocapsid is of particular importance in the case of the hepadnaviruses (hepatitis B viruses [HBVs]), small, enveloped animal viruses, which replicate their circular, partially double-stranded DNA genome only in part in the nucleoplasm. There, the incoming genome matures into an extrachromosomal, covalently closed, circular DNA molecule, the template for genomic RNA synthesis through the cellular transcription machinery (10, 29). The following steps, reverse transcription and plus-strand DNA synthesis, occur inside cytoplasmic nucleocapsids, giving rise to mature core particles. Of these, a fraction is diverted from the major pathway leading to the export of the enveloped virion, thereby serving to establish an elevated intranuclear pool of genome copies early on and to replenish this pool throughout persistent infection (41). For duck HBV (DHBV), it is well established that nuclear targeting of nucleocapsids is negatively influenced by the presence of the large envelope protein (L-protein), whose levels determine the fraction entering the export pathway and whose absence leads, by default, to much enhanced levels of intracellular nucleocapsids and of nuclear DNA genomes (40). However, comparable studies with HBV genomes lacking L-protein expression, using transfected or transduced cultured cells or HBV transgenic mice as experimental systems, failed to detect any evidence that such a mechanism might also be operating and of similar importance for the mammalian hepadnaviruses (M. Sprinzl, U. Protzer, C. Kuhn, and H. Schaller, unpublished data). Thus, an additional mechanism is expected to control import of progeny hepadnavirus genomes in general, possibly involving a (potentially regulatable) nuclear localization signal (NLS) on the nucleocapsid surface, i.e., in the amino acid sequence of the core protein, the only constituent of the outer shell of the nucleocapsid.
Early studies with the HBV prototype revealed the presence of one or several NLSs overlapping with a cluster of arginine repeats close to the C terminus of the capsid protein (7, 44). More-recent studies with recombinant capsids in digitonin-permeabilized cells further support the notion that these sequences participate in targeting HBV capsids to the nuclear pore in an importin α/β-mediated fashion (20). While these observations gave the first, although indirect, experimental evidence in support of the model that the NLS-containing carboxy terminus of the core protein subunit was exposed on the surface of the nucleocapsid, surface disposition of the core protein carboxy terminus had previously been demonstrated directly for DHBV by use of a sequence-specific antiserum (38). However, there are no comparable arginine-rich elements in this part of the avian viral protein. This discrepancy is paralleled by the fact that the core proteins of the avian and mammalian viruses differ in length (262 versus 183 residues, respectively) and display only little overall sequence homology (39). Thus, the question arises whether mammalian and avian HBV use different mechanisms for nuclear targeting of the virus genome to the cell nucleus.
Another more general, unresolved question is how the intracellular localization of the core protein (and of newly synthesized subunits) is regulated at different stages of the hepadnavirus life cycle. As outlined above, the incoming nucleocapsid, as well as a subfraction of progeny capsids, must attach to the nuclear pore, whereas this appears to be avoided for newly synthesized protein subunits. Reportedly, these remain predominantly (in the case of mammalian HBVs) or exclusively (in the case of avian HBVs) in the cytoplasm, 240 subunits each coassembling with genomic RNA and DNA polymerase-reverse transcriptase to cytosolic nucleocapsids. Intranuclear assembly of (empty) capsids may, nevertheless, occur under special experimental circumstances, such as in the HBV transgenic mouse (8, 13, 14), in chronically infected HBV patients (2), or in certain HBV core protein (HBc)-expressing cell lines after cell-cycle synchronization by serum starvation (45). These observations predict the existence of a yet unknown mechanism that normally prevents the core protein subunit (and probably also the immature RNA-containing capsid) from entering and accumulating in the nucleoplasm.
Results from various experimental systems suggest that phosphorylation may play a major role in regulating signal-mediated nucleocytoplasmic transport of the hepadnaviral core protein. While agreeing on the general importance of phosphorylation, these data are in conflict in detail: mutational analysis in transfected cells of phosphorylation target sites supports an inhibitory function (23), whereas NLS activation through phosphorylation is suggested by the results of capsid transport studies in a cell-free system (20).
We have started to address these questions with the DHBV animal model, as this system allows us to study hepadnavirus nucleocapsid targeting at the molecular level, including bona fide infection of cultured primary duck hepatocytes (PDHs). Following a previous report detecting a close correlation of capsid maturation with DHBV core protein (DHBc) dephosphorylation and envelope protein-independent membrane attachment as steps preceding cytosolic capsid envelopment (24), the present study concentrates on defining the signals that control nuclear targeting of nucleocapsids. Using fusions to the green fluorescent protein (GFP) as a functional assay, we have identified a single NLS in the DHBc sequence and show that this signal is of functional importance for the targeting of the DHBV nucleocapsid to the nuclear pore. Furthermore, we present evidence indicating that the DHBc also contains an activity directing its nuclear export, a property explaining its overall cytoplasmic localization despite the presence of the mapped NLS.
MATERIALS AND METHODS
Plasmids.
pCD16, previously also designated pCD0, a plasmid carrying the full-size DHBV genome (subtype 16 [see reference 25]), starting at position 2520, under the control of the cytomegalovirus (CMV) immediate-early promoter-enhancer (CMV promoter), and the helper construct pCD4 lacking part of the 5′-proximal RNA packaging signal ɛ have been described elsewhere (1, 31, 33). pCDcore, a plasmid expressing the DHBc as the only gene product, is a derivative of pCD4 lacking DHBV sequences between DHBV positions 863 and 2295. pCGFP was derived from pCD16-S-GFP (33), a derivative of pCD16 encoding the GFP open reading frame (ORF) instead of the S gene (from KpnI, position 1290, to BstEII, position 1847), by removing the DHBV sequence between the SalI and XhoI sites (1.7 kb) between positions 2522 and 1212, thereby placing the start site of CMV promoter-driven transcripts some 170 nucleotides upstream of the S gene AUG fused to the GFP gene. We furthermore removed the downstream sequence copy of the packaging signal ɛ (between AflII and XbaI, positions 2526 to 2662). To facilitate DHBc sequence fusions to the C terminus of GFP, we introduced a synthetic polylinker (multiple cloning site [MCS], GTCAAGCTTCATCGATTGCATGCGAATTCGCAGATCTCCCTCGCCTAGGAAATAAGGTTACC) at the 3′ end of the GFP ORF, replacing the GFP stop codon with a continuous coding sequence containing the unique recognition sequences (underlined) for restriction enzymes HindIII, ClaI, SphI, and EcoRI (in that order) followed by the last 21 nucleotides of the DHBc ORF, including the DHBc stop codon (containing unique BglII and AvrII restriction sites [also underlined]), and a BstEII restriction site. All cleavage sites in the MCS are placed so as to allow in-phase fusions to DHBc sequences using preexisting homologous sites in the DHBc gene. Deletion constructs pGFP/C67-262 and pGFP/C125-262 were obtained by inserting DHBc gene fragments from SphI to BglII (positions 2843 to 391), or from EcoRI to BglII (positions 3017 to 391), respectively. To create plasmid pGFP/C5-262, which contains a nearly full-length core protein sequence, we transferred a ClaI/BglII fragment from plasmid pCD10 (which differs from pCD16 by an A-to-G exchange at position 10 in the core gene, introducing a ClaI restriction site and resulting in an N-to-D exchange at codon 4 [see reference 1]). To generate core fusions terminating at amino acid position 226, we used as a donor for PCR amplification plasmid pC226 which carries a stop codon at core position 227 (38). pGFP/C184-226 was obtained after introducing an EcoRI restriction site upstream of codon 184 with primer CGGAATTCCCGACCATTGAAGCA and a reverse primer corresponding to DHBV 435 to 418. pGFP/C125-183 was generated using the reverse primer CGAGATCTTTAGGCATCTCTACC, ending at position 163, and introducing a stop at codon 184 (bold) followed by a BglII restriction site (underlined). The PCR products produced were digested with EcoRI and BglII and inserted into pCGFP-MCS.
For construction of a tandem repeat GFP dimer, we amplified by PCR the GFP gene from plasmid pCGFP using primers CTGTACAAGTCAATCGATGCC and CCGAAGCTTGATGTCTGGTACCATG, introducing HindIII and ClaI restriction sites (underlined) at the 5′ and 3′ termini of the GFP gene, respectively. The PCR product was then inserted into pGFP-MCS between ClaI and HindIII at the GFP-DHBc junction, generating the plasmid p2xGFP-MCS. To construct p2xGFP/C67-262, a DHBc sequence (from position 67 to 262) was introduced as described above for pGFP/C67-262. For the construction of the GGE mutation, inactivating the DHBc-NLS, we used two complementary mutation primers, CGAACCTAGAGGAGGAGAAGTTAAA and TTTAACTTCTCCTCCTCTAGGTTCG, and two external primers (positions 2546 to 61 and 435 to 418, respectively). The final PCR product produced was digested with SphI and AvrII and ligated into SphI/AvrII-digested pCGFP-MCS, creating pGFP/C67-262-GGE; the mutation was thereafter transferred into pCD16, creating pCD16-GGE.
GFP fluorescence and immunofluorescence analysis.
Cells from the human hepatoma cell line HuH7, cells from the embryonic kidney cell line 293, or HeLa cells were cultivated on coverglass chamber slides (Nunc) and transfected with the respective plasmids by using a standard calcium phosphate protocol (24). At 1 to 2 days after transfection, the GFP fluorescence was analyzed with a Leica TCS NT confocal laser scanning microscope (63×, 1.2 objective). For immunofluorescence analysis the cells were fixed 3 to 4 days after transfection with 4% formalin in phosphate-buffered saline (PBS) for 30 min, permeabilized with 0.25% Triton X-100 in PBS, and coimmunostained with the rabbit anti-DHBc (raised against E. coli-derived full-length DHBc [37]) and/or anti-nuclear pore complex (NPC) monoclonal antibody 414 (kindly provided by Dirk Görlich, ZMBH, Heidelberg, Germany) and, subsequently, with fluorescein- or tetramethylrhodamine-conjugated secondary antibodies. Fluorescence was analyzed with a Leica DM IRB inverted fluorescence microscope (20×, 0.40 objective) equipped with an automatic camera and the Leica TCS NT confocal laser scanning microscope as described above. Sequential excitation and scanning of the two fluorescent channels (separate excitations at 488 and 568 nm) were used to avoid cross-bleeding of the fluorochromes between channels.
Analysis of DHBc NLS knockout mutant (D16-GGE) in transfected cells.
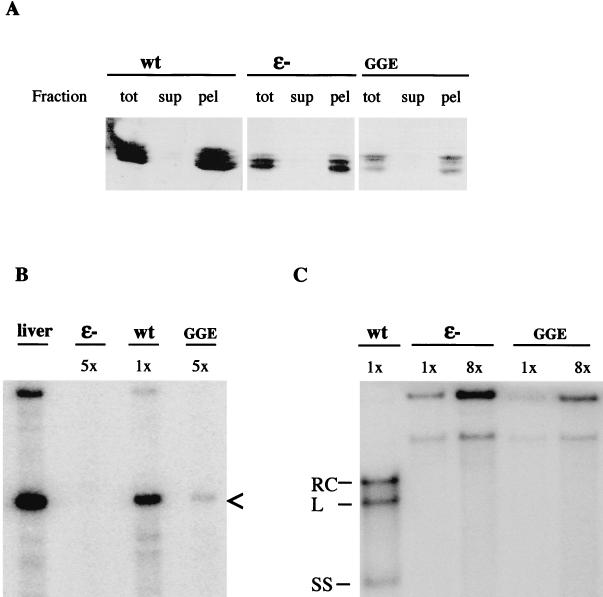
LMH cells, from a chicken hepatoma cell line (3), in a 10-cm-diameter cell culture dish (about 107 cells) were transfected with 20 μg of either plasmid pCD16, plasmid pCD4, or plasmid pCD16-GGE, and lysed 4 days posttransfection by suspension in 1 ml of 10 mM Tris-HCl (pH 7.5)–1 mM EDTA–1% NP-40. After removal of the cell nuclei by 10 min of centrifugation in a microcentrifuge, the cytosolic fraction was subdivided into several aliquots and capsids were pelleted by ultracentrifugation (TLA45 rotor, 44,000 rpm, 1 h). For Western blotting and primer extension analysis, this step included sedimentation through a step gradient consisting of 200 μl of 20% and 50 μl of 60% sucrose. To determine the relative fraction of DHBc in pelletable capsids, equal aliquots from the pellet and the supernatant, as well as of the cytosol prior to centrifugation, were analyzed with a Western blot probed with a polyclonal anti-core protein antiserum (D087) as described previously (24). Proteins were visualized by enhanced chemiluminescence (Amersham) according to the manufacturer's manual. For primer extension analysis detecting 5′ termini of genomic RNA (31), encapsidated RNA was liberated by prior proteinase K digestion, followed by extraction with phenol-chloroform and ethanol precipitation. The primer chosen (5′-CCCTGTGTAGTCTGCCAGAAGTCTTC-3′, nucleotide positions 2843 to 2818) yielded the expectedly sized cDNA extension product of 310 nucleotides (Fig. 4B). Before Southern blotting according to standard protocols (36), free DNA was largely removed by treating the pelleted capsids with pancreatic DNAse, before starting proteinase K digestion.
FIG. 4.
Effects on DHBV genome replication by the GGE NLS knockout mutation. Cytosolic fractions from about 107 LMH cells, transfected with plasmid pCD16 (wt), pCD4 (ɛ-), or pCD16-GGE (GGE), respectively, were divided into several aliquots. Capsids were pelleted by ultracentrifugation and analyzed separately for the presence of DHBV nucleocapsids and DHBV replication intermediates, as detailed in Materials and Methods. (A) Western blot detecting DHBc in aliquots (corresponding to 1.5% of the total cells) from the pellet (pel), the supernatant (sup), and before centrifugation (tot). (B) Primer extension analysis detecting 5′ termini of genomic RNA as the major (310-nucleotide-long) cDNA extension product (marked by the arrowhead) that was also produced abundantly from a positive control, RNA from DHBV-infected duck liver (liver) (see reference 30). The aliquots analyzed corresponded to 6 or 30% of the cells (1x and 5x, respectively). (C) Southern blot analysis identified encapsidated DNA replication products in aliquots corresponding to 0.9% or 7% of the cells (1x and 8x, respectively). Relaxed circular (RC), linear (L), and single-stranded (S) DHBV DNA were detectable only in cells transfected with pCD16; the more slowly migrating bands originate from traces of the transfecting plasmids not removed prior to analysis.
Virus production and PDH infection with DHBV-GGE.
For the production of recombinant DHBV, LMH cells in a 10-cm-diameter dish were cotransfected using a calcium phosphate protocol with 10 μg each of the pCD16-GGE DNA and helper pCD4. Cell culture medium containing recombinant virions collected from days 3 to 6 posttransfection was concentrated 10- to 50-fold by precipitation with 6.5% polyethylene glycol 20,000–0.9% NaCl at 0°C and stored in PBS–10% glycerol at −20°C until further use. Wild-type DHBV was produced by transfecting the pCD16 DNA, accordingly. Virus titers were determined (as DNA-containing enveloped viral particles) by density-gradient centrifugation and dot blot analysis relative to an DHBV DNA standard (31). PDHs were prepared and cultured essentially as previously described (15). For infection, wild-type or recombinant DHBV particles (to a multiplicity of infection of 10 or 100, respectively) were applied to a 12-well culture dish containing about 8 × 105 cells per well. After 14 h of incubation, the cells were washed and further cultivated. After 5 or 8 days, the cells were fixed and immunostained for the DHBc as described above.
Heterokaryon assay.
Nucleocytoplasmic shuttling was detected by using a heterokaryon assay (32, 35). HeLa cells were transfected with plasmid p2xGFP/C67-262, using the calcium phosphate method. Twenty-four h after transfection, the cells were trypsinized and 106 cells were seeded on a 6-well tissue culture dish together with 2 × 106 (murine) BALB/c 3T3 cells. After 18 h, the cells were treated for 30 min with culture medium containing 50 μg of cycloheximide (Sigma) per ml. To induce cell fusion, the cells were then covered with a solution of 50% (wt/vol) polyethylene glycol 8,000 (Sigma) in Dulbecco's modified Eagle's medium (DMEM) for 2 min at 37°C. The cells were washed extensively with PBS and further cultured in DMEM–10% PBS containing cycloheximide (50 μg/ml). Ninety minutes later, the cells were fixed with 4% paraformaldehyde in PBS for 15 min, followed by permeabilization with 0.2% Triton X-100 in PBS (20 min). Finally, cells were washed in PBS containing bis-benzimide (2 μg/ml) for staining of the nuclei. The nuclei staining pattern as well as the fluorescence was examined by a Leica DM IRB inverted fluorescence microscope (20×, 0.40 objective).
RESULTS
A fraction of intracellular core protein localizes to nuclear pores.
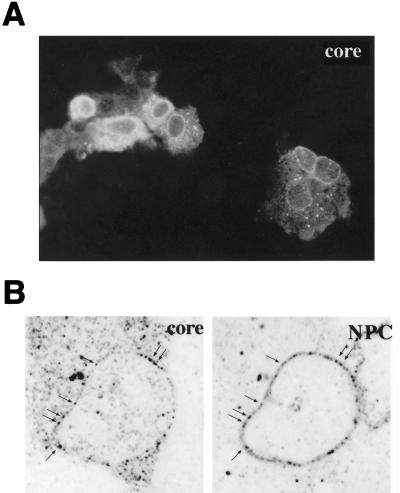
Indirect immunofluorescence experiments were performed to assess whether DHBV capsids bind to the NPC, as observed for their HBV counterparts in permeabilized cells (20). The hepatoma cell line HuH7 was transfected with plasmid pCD16, which produces a full-size, replication-competent DHBV RNA pregenome under the control of the CMV promoter (1, 31). Cells were fixed at day 3 to 4 posttransfection, immunostained for DHBc and analyzed by fluorescence microscopy. As shown in Fig. 1A, core protein-specific staining was predominantly localized in the cytoplasm of the transfected cells. While this was consistent with similar results from previous studies (e.g., see references 9 and 45), we additionally observed a distinct core-specific staining at the periphery of the cell nuclei. This perinuclear staining was particularly evident in cells with low cytoplasmic staining (Fig. 1A).
FIG. 1.
A fraction DHBV core antigen associates with nuclear pores. (A) HuH7 cells, transfected with pCD16, carrying an overlength DHBV genome, were fixed, immunostained for DHBc using a fluorescein-conjugated secondary antibody, and analyzed by conventional fluorescence microscopy. The perinuclear staining is seen best in cells with low cytoplasmic staining (cells at bottom right). (B) Analogously treated cells, costained for core protein using a fluorescein-conjugated secondary antibody (core [left] subpanel) and NPC using a tetramethylrhodamine-conjugated secondary antibody (NPC [right] subpanel) and analyzed with a confocal laser-scanning microscope. Arrows mark identical sites in the subpanels.
To examine whether the perinuclear core protein detected by conventional fluorescence microscopy was localized at the nuclear pores, DHBV-transfected cells were further analyzed at the higher resolution of the confocal laser scanning microscope after coimmunostaining with the NPC-specific monoclonal antibody 414. As shown in Fig. 1B, again most of the intracellular core protein was detected in the cytoplasm, with an additional punctuate staining at the nuclear membrane, the latter signal superimposable with the NPC staining (Fig. 1B). Nuclei, visualized by NPC staining, in neighboring, apparently untransfected cells (Fig. 1B, right panel) did not display any DHBc signal (Fig. 1B, left panel), demonstrating the selectivity of the antibody used. NPC binding of DHBc similar to that observed with the full-length DHBV genome, was also observed in analogous experiments with cells transfected with plasmid pCDcore which expresses the core protein as the only DHBV gene product (data not shown), indicating that nuclear pore association was an intrinsic property of the DHBc, not requiring the cooperation of any other viral gene product.
The DHBc sequence contains an NLS.
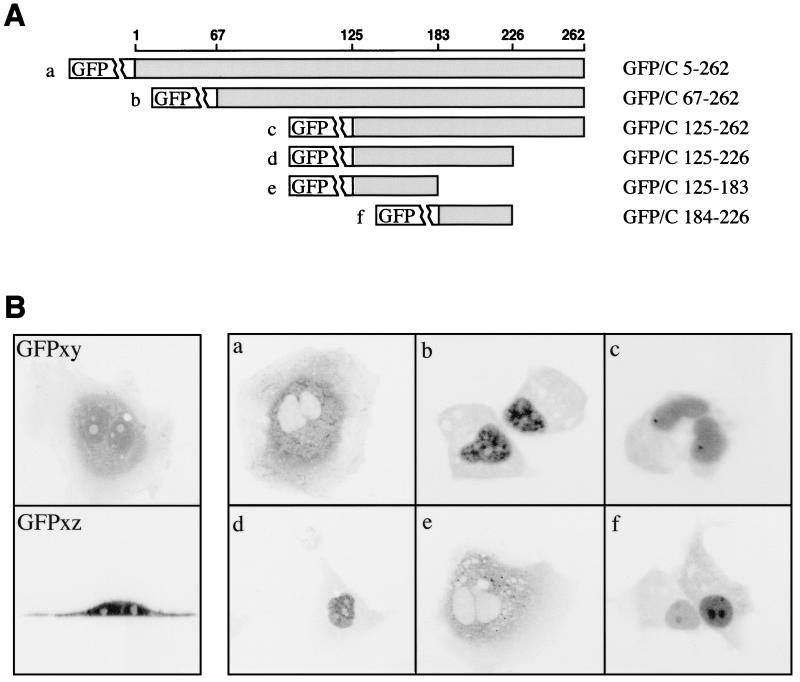
To confirm the presumed presence of an NLS in the DHBc, the full-length protein sequence, or different fragments thereof, were fused to the GFP. A series of such carboxy-terminal fusion constructs (schematically depicted in Fig. 2A) (for details, see Materials and Methods) was expressed in transfected HuH7 cells, and the cellular distribution of the respective fusion proteins was analyzed by confocal microscopy by detecting the GFP fluorescence in live cells. Proper expression and intactness of the fusion proteins was ascertained by Western blotting of cellular lysates with only proteins of the expected size, and no degradation products, being detected with both anti-core protein and anti-GFP antibodies (data not shown).
FIG. 2.
Deletion mapping of the DHBc NLS. (A) Schematic representation of the GFP-core fusion proteins used, with the numbers representing positions in the core protein amino acid sequence. The names, indicating the limits of the various fusions, are indicated on the right. The letters designating individual constructs (shown at left) correspond to images shown in panel B. (B) Cellular distribution after expression in transfected HuH7 cells of the proteins whose constructs are shown in panel A. GFP fluorescence in live cells was analyzed using a confocal microscope, at day 1 or 2 posttransfection. Two fluorescence micrographs showing the distribution of unfused GFP protein as a control are depicted on the left. The apparent decrease of the GFP signal at the periphery of the cell in the xy plane (GFP xy subpanel) reflects the shape of these cultured cells. Analysis of the signal in the xz plane (GFP xz subpanel) showed a homogeneous distribution of the protein.
Typical results of the fluorescence analysis are shown in Fig. 2B. First, in a control experiment testing expression of GFP unfused, the fluorescence was found to be distributed homogeneously throughout the cell (with somewhat weaker staining in the nucleoli), freely diffusing through the nuclear pores as expected for a protein of 30 kDa (Fig. 2B, subpanels GFPxy and GFPxz). Fusion of GFP to the essentially complete core sequence (GFP–C5-262) led to a cytoplasmic localization of the fluorescent GFP signal (Fig. 2Ba), probably reflecting the size of capsids assembled from GFP–C5-262, which precludes passage through the nuclear pore (20). In contrast, deletion of the first 67 core protein amino acids, expected to abrogate capsid assembly (43), yielded a predominantly nuclear fluorescence (Fig. 2Bb), thus confirming the presumed presence of an NLS in the DHBc. Deleting the core sequence further, N terminally up to amino acid 125 or C terminally to amino acid 226, did not affect the distinct nuclear localization of the respective fusion proteins (Fig. 2Bc and d), which locates the NLS in the DHBc sequence between amino acids 125 and 226. This sequence was further split into two fusions containing DHBc amino acids 125 to 183 or 184 to 226, respectively (Fig. 2A, constructs e and f). Of these, only the fusion protein GFP–C184-226 was found to locate to the nucleus, whereas the GFP fusion to the N-terminal part amino acids 125 to 183 remained exclusively cytoplasmic (Fig. 2Bf and e, respectively), a finding suggesting the presence of a nuclear export signal (NES). Interestingly, the various GFP fusions differed reproducibly in their intranuclear staining pattern, suggesting a specific interaction of the sequences flanking the NLS-containing core segment with cellular nucleoproteins in defined subnuclear domains; for example, fusion protein GFP–C67-262 always showed a granular nuclear pattern (Fig. 2Bb), whereas GFP–C184-226 appeared to be preferentially enriched in the nucleoli (Fig. 2Bf). Taken together, these data indicate the presence of an NLS between positions 184 and 226 of the DHBc amino acid sequence.
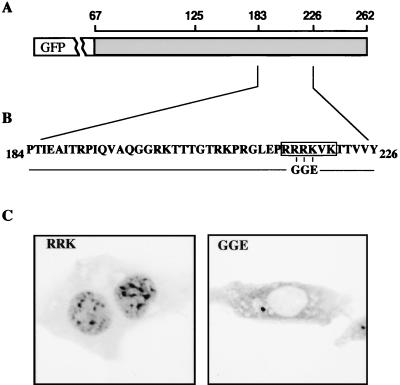
A basic stretch of amino acids within core protein amino acids 184 to 226 is essential for NLS function.
A classical NLS consists of either a single or bipartite stretch of basic amino acid residues (18, 19, 34). The DHBV core sequence from position 184 to 226 contains such a run of basic amino acids (214PRRRKVK220), with features resembling those of a classical signal (Fig. 3B), while a second basic element, 227RRRSKSRERR236, lies just outside of the DHBc segment showing NLS activity. To test whether the predicted signal was functionally important, we introduced, in the context of fusion protein GFP–C67-262, a triple mutation, changing amino acids 216 to 218 from RRK to GGE (Fig. 3B). The mutant and the corresponding wild-type plasmids were transfected in parallel into HuH7 cells, and the distribution of the fusion proteins as indicated by GFP fluorescence was analyzed by confocal microscopy (Fig. 3C). As already shown above (Fig. 2B), the GFP–C67-262 wild-type protein was located in the nuclei in a granular pattern (Fig. 3C, subpanel RRK). In contrast, introduction of the triple mutation led to an exclusively cytoplasmic localization, indicating the abrogation of nuclear import (Fig. 3C, subpanel GGE). We therefore conclude that amino acids 216RRK218 are an essential part of the DHBc NLS detected and that, furthermore, this signal probably encompasses the sequence 214PRRRKVK220, fitting the classical basic NLS type. This conclusion was further supported by the lack of nuclear import of a GFP fusion to DHBc 133-214, in which the signal proposed above had been deleted (pGFP/C133-214; M. Windisch, H. Mabit, and H. Schaller, unpublished data). Finally, our results also rule out that the presence of the neighboring basic element mentioned above, located just downstream of position 226, is essential for the function of the DHBc NLS identified. Thus, the DHBc appears to contain only a single, monopartite NLS sequence.
FIG. 3.
Mutations in DHBc NLS abrogate NLS function. (A and B) Schematic representation of the mutations introduced, with the numbers above the diagram indicating amino acid positions. The mutational exchanges in the DHBc sequence are shown below, with the cluster of basic residues boxed. (C) Cellular distribution in transfected HuH7 cells of GFP–C67-262 (RRK [left] subpanel) and GFP–C67-262RGGE (GGE [right] subpanel). GFP fluorescence was analyzed using a confocal microscope at day 1 posttransfection, without prior fixation.
The DHBV core NLS is essential for viral replication.
To examine the importance of the DHBc NLS defined above for the viral life cycle, we introduced the NLS knockout mutation (RRK to GGE) in the context of the complete replication-competent DHBV genome as present in plasmid pCD16 (care had been taken during construction of the GGE mutations in the DHBc ORF that these did not cause any amino acid changes in the overlapping polymerase frame). The resulting plasmid, pCD16-GGE, was transfected into LMH cells (this is a chicken hepatoma cell line known to produce high virus yields after pCD16 transfection [3]). Culture supernatants and cell lysates were investigated for the ability of the mutant DHBV genome (i) to form core particles, (ii) to encapsidate the pregenomic RNA, (iii) to undergo reverse transcription, and (iv) to produce viral particles. The results obtained are presented in Fig. 4 and summarized in Table 1. Firstly, Western blot analysis of the cytosolic fractions before and after pelleting through a 60% sucrose cushion showed that the amount of mutant capsids was about threefold reduced in comparison to the wild-type in mutant transfected cells and furthermore that the protein was in either case pelletable from the extract, indicative of the presence of core particles. As judged from primer extension analysis (Fig. 4B), the mutant capsids appeared to be additionally reduced about 10-fold in their capacity to encapsidate the pregenomic RNA. An even more severe defect was evident in Southern blots which failed to detect conversion of RNA pregenomes into DNA (Fig. 4C). Finally, and not surprising in view of the aforementioned results, analysis of cell culture supernatants for viral particles by sedimentation into a preformed CsCl density gradient followed by DNA dot blot analysis (31), as well as infection experiments, revealed that the NLS mutant genome was unable to produce detectable amounts of virus. Virus production was, however, restored by cotransfection with pCD4, a helper plasmid providing all viral proteins in trans, but lacking the RNA packaging signal ɛ and therefore by itself unable to produce virus particles (1, 33). The transcomplemented virus particles containing the defective D16-GGE genome were still able to infect PDHs, the number of cells infected per DNA-containing virus particle being comparable to that observed with the wild type. However, there was no production of infectious progeny virus from mutant-infected cells as visualized by the absence of spread from the initially infected single cells to neighboring hepatocytes, even after culturing the cells for up to 10 days postinfection (data not shown). Furthermore, and again in contrast to wild-type-virus-infected cells, accumulation of cytoplasmic DHBc to levels detectable by immunofluorescence was delayed to day 5 or 6 postinfection while wild-type-virus-infected cells required only 3 to 4 days to reach a comparable signal strength. This result is in keeping with the data described above, indicating a block in the production of cytoplasmic progeny DNA genomes caused by the GGE mutation, as well as with the lack of nuclear capsid targeting described below.
TABLE 1.
Characteristics of the DHBc NLS knockout genomea
| Plasmid(s) | Phenotype | Relative yield of intracellular capsids
|
Result of analysis of virus production
|
||||
|---|---|---|---|---|---|---|---|
| DHBcc | Particulatec | RNAd | DNAe | Relative yieldb | Infectivityf | ||
| pcD16 | wt | 1.0 | + | 1.0 | + | 1.0 | + |
| pCD16-GGE | NLS− | 0.15 | + | 0.02 | − | <0.05 | − |
| pCD4 | ɛ− | 0.25 | + | <0.02 | − | <0.05 | − |
| pCD16-GGE and pCD4 | ND | ND | ND | ND | 0.3 | + | |
Plasmids used for transfection of LMH cells are listed, as are phenotypes. For details, see Materials and Methods. wt, wild type; ND, not determined; +, positive signal; −, no signal.
Relative yields of virus produced day 5 posttransfection, as determined by dot blot analysis.
Relative yields of DHBc, as determined by Western blotting before (DHBc) or after (particulate) ultracentrifugation (Fig. 4A).
Relative yields of genomic RNA, as determined by primer extension (Fig. 4B).
Relative yields of DHBV DNA, as determined by Southern blotting (Fig. 4C).
Detected by DHBc immunostaining of infected hepatocytes.
The DHBV core NLS is essential for nuclear pore targeting.
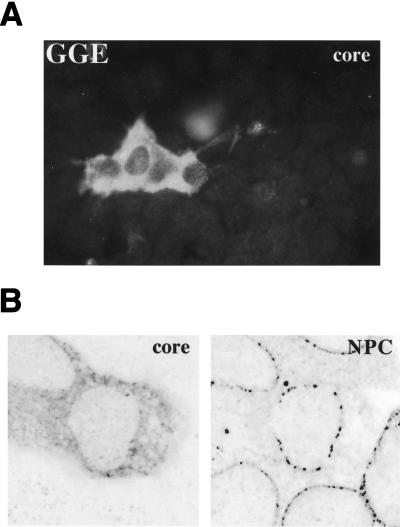
To further examine the function(s) of the DHBc NLS in the viral life cycle, the intracellular distribution of the mutant core protein from the NLS-defective genomic DHBV construct, pCD16-GGE, was also analyzed in transfected HuH7 cells. As shown by indirect immunofluorescence, cells transfected with the mutant genome did not show the characteristic perinuclear core protein staining (Fig. 5A), in contrast to the wild-type control (results shown in Fig. 1A [experiments performed in parallel]). Costaining with an anti-nuclear pore antibody and confocal microscopic analysis confirmed that mutant core protein was no longer associated with nuclear pores in the transfected cells (Fig. 5B). These data are consistent with the hypothesis that the NLS identified is important for proper nuclear pore targeting of the DHBV nucleocapsid also in the context of viral replication cycle. However, we cannot exclude the possibility that the NLS defect may have also contributed indirectly to the loss of nuclear targeting of the immature RNA-containing nucleocapsids (see also Discussion).
FIG. 5.
DHBc NLS function is required for nuclear pore targeting of capsids. (A) HuH7 cells transfected with pCD16-GGE carrying a mutated capsid NLS were fixed, immunostained for DHBc using a fluorescein-conjugated secondary antibody, and analyzed by conventional fluorescence microscopy. (B) Analogously treated cells, costained for core protein using a fluorescein-conjugated secondary antibody (core [left] subpanel) and NPC using a tetramethylrhodamine-conjugated secondary antibody (NPC [right] subpanel) were analyzed with a confocal laser-scanning microscope. The data shown should be compared to those obtained in parallel with the wild-type control (pCD16) shown in Fig. 1.
DHBc contains signals for nuclear export.
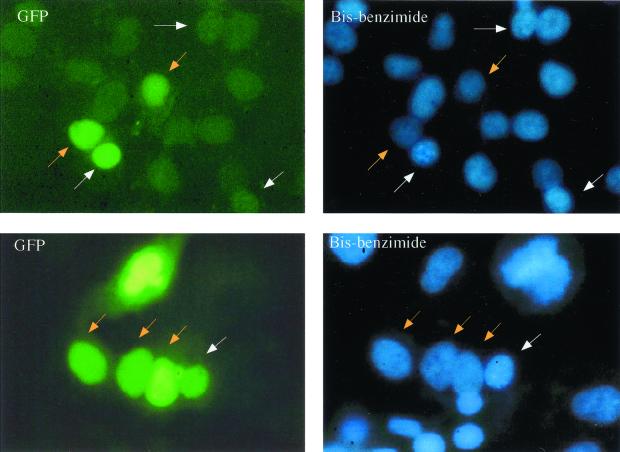
In the experiments testing for nuclear import of the GFP–DHBc fusion constructs, chimeric proteins lacking the DHBc-NLS had shown an exclusively cytoplasmic distribution (Fig. 2 and 5). This was particularly unexpected in the case of construct GFP–C125-183 which, according to its size of about 36 kDa, was expected to freely diffuse between the cytoplasm and nucleoplasm. An explanation for the exclusive cytoplasmic staining of such a small protein could be the presence of a nuclear export signal. To test for the presence of such a signal in the DHBc protein, we performed heterokaryon shuttle experiments, investigating protein translocation between heterologous nuclei in fused cells (5, 32). To preclude background by free nucleocytoplasmic diffusion, the largest DHBc sequence containing a functional NLS (C67-262) was chosen and fused to a tandemly repeated GFP dimer (2xGFP). When expressed in transfected HeLa cells, this protein (2xGFP–C67-262) localized exclusively to the nucleoplasm, whereas the control with unfused 2xGFP was, despite of its size of 60 kDa, exclusively cytoplasmic (not shown), both results supporting the validity of the assay system. In the subsequent shuttle experiments (results shown in Fig. 6), p2xGFP/C67-262-transfected HeLa cells were fused to NIH 3T3 mouse fibroblasts, and the GFP fusion protein was assayed for its potential to shuttle from HeLa cell nuclei to the mouse cell nuclei (and back). To this end, the cells were fixed and the nuclei stained with bis-benzimide, which marks the mouse nuclei with a distinct punctuate pattern (Fig. 6), thus allowing the nuclei from both cell types to be distinguished. At the time point of examination (1.5 h after cell fusion), the fluorescent fusion protein was found to be enriched equally well in mouse nuclei as in the adjacently located HeLa cell nuclei donating the fluorescent protein (Fig. 6), indicative of rapid nuclear export of the DHBc fusion protein.
FIG. 6.
The DHBc sequence contains a signal(s) functioning in nuclear export. Fluorescence micrographs of a heterokaryon examining a protein consisting of 2xGFP fused to DHBc amino acids 67 to 262. p2xGFP/C67-262-transfected HeLa cells were fused with mouse fibroblasts in the presence of cycloheximid. Cells were fixed 1.5 h postfusion, and cell nuclei were stained with bis-benzimide and examined with a fluorescence microscope. Mouse nuclei, identified by a typical bis-benzimide staining pattern, are marked with white arrows, and the human nuclei are marked with orange arrows.
DISCUSSION
The occurrence of regulated nuclear targeting of hepadnavirus nucleocapsids was initially predicted from the demonstration of nuclear import of progeny DNA genomes in DHBV-infected cultured duck hepatocytes (41). The present study now provides the first direct experimental evidence that the DHBc contains signals for bidirectional cyto-nucleoplasmic transport, thereby complementing and extending data obtained with HBV in transfected cells. A single, unipartite NLS was mapped within amino acids 184 to 226 of the DHBc sequence, most probably encompassing a stretch of basic amino acids (PRRRKVK) with similarity to the classical basic amino acid-type NLS of the simian virus 40 large T antigen (PKKKRKV [see references 18 and 19]). Furthermore, transfer of the NLS-containing sequence (in the context of amino acids 184 to 262) onto other GFP-fused polypeptides, which were by themselves incapable of nuclear import, demonstrated that the sequence was not only essential but also sufficient for NLS function (A. Knaust and H. Schaller, unpublished data). Interestingly, no comparable results were obtained in this assay with HBc segments encompassing the HBc NLS as characterized in other detection systems.
In the context of the complete DHBV genome, an NLS knockout mutation resulted in a complete loss of virus production from transfected cells or in infected hepatocytes, a result underlining the importance of the signal for the virus replication cycle. However, this mutation (RRK to GGE) also resulted in reduced RNA packaging and in a complete loss of reverse transcription of the viral RNA genome into DNA and, consequently, in a general block in capsid maturation. We therefore cannot exclude the possibility that these defects may have also contributed indirectly to the loss of nuclear nucleocapsids targeting. On the other hand, association of core protein with the nuclear membrane was also observed with a construct expressing the wild-type core protein (and empty core particles) in the absence of any other viral gene product. Hence, lack of core staining at the nuclear pore after transfection of the mutated genome has to be attributed to the lack of a functional NLS on the capsid.
Several studies have mapped and further investigated an NLS activity overlapping with four arginine clusters localized close to the carboxy terminus of the human HBV core protein (7, 20, 44), in a domain that lies outside of the domain required for assembly into the well-characterized HBV capsid structure (4, 28) but whose basic amino acid sequences play an essential role in genomic RNA encapsidation and reverse transcription in both HBV and DHBV (28, 38). Mutational analysis of this region in the HBc suggests that there is more than one NLS contained in this region in HBV and, furthermore, that several overlapping serine-proline motifs are subject to phosphorylation, resulting in a modulation of the several functions of the arginine repeats, including genomic RNA packaging, reverse transcription, and probably also NLS functionality (11, 22, 23). Extrapolating from these observations, and assuming that a glutamate residue mimics constitutive phosphorylation, we notice that the sequences from HBV and DHBV share a similar motif: SPRRRR in the HBc sequence and EPRRRK in the DHBc sequence. Thus, it is conceivable that both core proteins carry a basically similar arginine-rich NLS, and it seems likely that both viruses use a common cellular receptor for capsid import, most probably importin α/β, which has recently been shown to be essential for nuclear pore binding of HBV capsids (20). It should be emphasized, however, that the two signals are located at different positions in the respective polypeptide chains and that the phosphorylatable serine and threonine residues in the DHBc (positions 239, 245, 257, and 259 [see reference 46]) are located separate from the NLS. Thus, the two NLSs may respond quite differently to capsid phosphorylation, similar to the different influences observed on genomic RNA packaging and reverse transcription (11, 22).
Finally, we present experimental evidence suggesting that the core protein sequence might carry not only an NLS but also an NES function. Collectively, these data suggest an NES to be contained within the (leucine-rich) sequence between DHBc amino acids 125 and 183, which is sequentially separated from the NLS and which on its own appears to exclude GFP fusions from nuclear import (Fig. 2Be). They do not, however, rule out an alternative, more-complex nuclear shuttle signal unifying the NLS and NES functions in a single signal sequence interacting with both import and export factors which themselves shuttle (for a review, see reference 27).
Whatever type of signal may mediate nuclear export, the respective function may serve distinct roles in the hepadnaviral live cycle. Firstly, it may help to solve the obvious, but largely unnoticed, problem that the capsid subunit as synthesized in the cytoplasm is expected to be a substrate for the nuclear import machinery. As an alternative (although the two are not mutually exclusive) to NLS masking by protein folding and/or phosphorylation, we now propose that the NLS may be counterbalanced by an NES function, thereby preventing nuclear assembly of hepadnaviral capsids. Balancing nuclear localization through counteracting NLSs and NESs, as postulated here for hepadnaviruses, may well provide a more general mechanism used by viruses for avoiding nuclear accumulation of progeny capsids, while maintaining the essential nuclear targeting of the incoming nucleocapsid. In keeping with this hypothesis, a functional NES was recently found in the human immunodeficiency virus type 1 (HIV-1) matrix protein (6), an NLS-containing protein that is normally targeted to the plasma membrane as part of the HIV gag precursor. Secondly, we and others (W. Yang and J. Summers, personal communication) have observed a local accumulation of DHBc in distinct nuclear bodies. We furthermore found that these loci also accumulated pregenomic DHBV RNA (H. Mabit and H. Schaller, unpublished data). Thus, it is tempting to speculate that a shuttling capsid protein may also aid export of viral RNA genomes from the nucleus via core protein-RNA interaction. Such mechanisms have been described for other viruses that export an unspliced genomic RNA, a prominent example being the HIV Rev export system (5) and the viral ribonucleoprotein export complex of influenzavirus A using the NS2 and/or the M1 protein (26, 30, 42).
ACKNOWLEDGMENTS
We thank Bärbel Glass for preparation of PDHs, Christa Kuhn for antibodies, Marc Windisch for construction of plasmids, Matthias Dobbelstein for advice regarding the shuttle assay, Elizabeth Grgacic for comments on the manuscript, and Karin Coutinho for expert editorial assistance.
This work was supported by a ZMBH fellowship to H.M. and by the Fond der Chemischen Industrie.
REFERENCES
- 1.Bartenschlager R. Strukturelle und funktionelle Charakterisierung des P-Proteins der Hepatitis B Viren. Ph.D. thesis. Heidelberg, Germany: Ruprecht-Karls-Universität; 1990. [Google Scholar]
- 2.Chu C M, Liaw Y F. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology. 1987;92:220–225. doi: 10.1016/0016-5085(87)90863-8. [DOI] [PubMed] [Google Scholar]
- 3.Condreay L D, Aldrich C E, Coates L, Mason W S, Wu T T. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowther R A, Kiselev N A, Böttcher B, Berriman J A, Borisova G P, Ose V, Pumpens P. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 5.Cullen B R, Malim M H. The HIV-1 Rev protein: prototype of a novel class of eukaryotic post-transcriptional regulators. Trends Biochem Sci. 1991;16:346–350. doi: 10.1016/0968-0004(91)90141-h. [DOI] [PubMed] [Google Scholar]
- 6.Dupont S, Sharova N, DeHoratius C, Virbasius C M, Zhu X, Bukrinskaya A G, Stevenson M, Green M R. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature. 1999;402:681–685. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- 7.Eckhardt S G, Milich D R, McLachlan A. Hepatitis B core antigen has two nuclear localization sequences in the arginine-rich carboxy terminus. J Virol. 1991;65:575–582. doi: 10.1128/jvi.65.2.575-582.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farza H, Hadchouel M, Scotto J, Tiollais P, Babinet C, Pourcel C. Replication and gene expression of hepatitis B virus in a transgenic mouse that contains the complete viral genome. J Virol. 1988;62:4144–4152. doi: 10.1128/jvi.62.11.4144-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galle P R, Schlicht H J, Fischer M, Schaller H. Production of infectious duck hepatitis B virus in a human hepatoma cell line. J Virol. 1988;62:1736–1740. doi: 10.1128/jvi.62.5.1736-1740.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem D. Hepadnaviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2703–2737. [Google Scholar]
- 11.Gazina E V, Fielding J E, Lin B, Anderson D A. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J Virol. 2000;74:4721–4728. doi: 10.1128/jvi.74.10.4721-4728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 13.Guidotti L G, Martinez V, Loh Y-T, Rogler C E, Chisari F V. Hepatitis B virus nucleocapsid particles do not cross the hepatocyte nuclear membrane in transgenic mice. J Virol. 1994;68:5469–5475. doi: 10.1128/jvi.68.9.5469-5475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hild M, Weber O, Schaller H. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J Virol. 1998;72:2600–2606. doi: 10.1128/jvi.72.4.2600-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu H C, Su I J, Lai M Y, Chen D S, Chang M H, Chuang S M, Sung J L. Biologic and prognostic significance of hepatocyte hepatitis B core antigen expressions in the natural course of chronic hepatitis B virus infection. J Hepatol. 1987;5:45–50. doi: 10.1016/s0168-8278(87)80060-0. [DOI] [PubMed] [Google Scholar]
- 17.Izaurralde E, Kann M, Pante N, Sodeik B, Hohn T. Viruses, microorganisms and scientists meet the nuclear pore. EMBO Workshop Report. EMBO J. 1999;18:289–296. doi: 10.1093/emboj/18.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalderon D, Richardson W D, Markham A F, Smith A E. Sequence requirements for nuclear localisation of SV40 large T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 19.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 20.Kann M, Sodeik B, Vlachou A, Gerlich W H, Helenius A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 1999;145:45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasamatsu H, Nakanishi A. How do animal DNA viruses get to the nucleus? Annu Rev Microbiol. 1998;52:627–686. doi: 10.1146/annurev.micro.52.1.627. [DOI] [PubMed] [Google Scholar]
- 22.Lan Y T, Li J, Liao W, Ou J. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology. 1999;259:342–348. doi: 10.1006/viro.1999.9798. [DOI] [PubMed] [Google Scholar]
- 23.Liao W, Ou J H. Phosphorylation and nuclear localization of the hepatitis virus core protein: significance of serine in the three repeated SPRRR motifs. J Virol. 1995;69:1025–1029. doi: 10.1128/jvi.69.2.1025-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mabit H, Schaller H. Intracellular hepadnaviral nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J Virol. 2000;74:11472–11478. doi: 10.1128/jvi.74.24.11472-11478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 27.Michael W M. Nucleocytoplasmic shuttling signals: two for the price of one. Trends Cell Biol. 2000;10:46–50. doi: 10.1016/s0962-8924(99)01695-5. [DOI] [PubMed] [Google Scholar]
- 28.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepatitis. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 30.O'Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obert S, Zachmann-Brand B, Deindl E, Tucker W, Bartenschlager R, Schaller H. A spliced hepadnavirus RNA that is essential for virus replication. EMBO J. 1996;15:2565–2574. [PMC free article] [PubMed] [Google Scholar]
- 32.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 33.Protzer U, Nassal M, Chiang P W, Kirschfink M, Schaller H. Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection. Proc Natl Acad Sci USA. 1999;96:10818–10823. doi: 10.1073/pnas.96.19.10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 35.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficincy virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Salfeld J. Genprodukte des Hepatitis B-Virus. Struktur, Zuordnung zum Genom und Biosynthese. Ph.D. thesis. Heidelberg, Germany: Ruprecht-Karls-Universität; 1985. [Google Scholar]
- 38.Schlicht H J, Bartenschlager R, Schaller H. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J Virol. 1989;63:2995–3000. doi: 10.1128/jvi.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 40.Summers J, Smith P, Horwich A L. Hepadnaviral envelope proteins regulate amplification of covalently closed circular DNA. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuttleman J S, Pourcell C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 42.Wittaker G R, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 43.Yang W, Guo J, Ying Z, Hua S, Dong W, Chen H. Capsid assembly and involved function analysis of twelve core protein mutants of duck hepatitis B virus. J Virol. 1994;68:338–345. doi: 10.1128/jvi.68.1.338-345.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh C-T, Liaw Y-F, Ou J-H. The arginine-rich domain of hepatitis virus precore and core proteins contains a signal for nuclear transport. J Virol. 1990;64:6141–6147. doi: 10.1128/jvi.64.12.6141-6147.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeh C-T, Wong S W, Fung Y-K, Ou J-H. Cell cycle regulation of nuclear localization of hepatitis B virus core protein. Proc Natl Acad Sci USA. 1993;90:6459–6463. doi: 10.1073/pnas.90.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, Summers J. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J Virol. 1994;68:2965–2969. doi: 10.1128/jvi.68.5.2965-2969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]