Abstract
Blood oxygen level dependent (BOLD) MRI time series with maternal hyperoxia can assess placental oxygenation and function. Measuring precise BOLD changes in the placenta requires accurate temporal placental segmentation and is confounded by fetal and maternal motion, contractions, and hyperoxia-induced intensity changes. Current BOLD placenta segmentation methods warp a manually annotated subject-specific template to the entire time series. However, as the placenta is a thin, elongated, and highly non-rigid organ subject to large deformations and obfuscated edges, existing work cannot accurately segment the placental shape, especially near boundaries. In this work, we propose a machine learning segmentation framework for placental BOLD MRI and apply it to segmenting each volume in a time series. We use a placental-boundary weighted loss formulation and perform a comprehensive evaluation across several popular segmentation objectives. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. Biomedically, our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. We further find that boundary-weighting increases placental segmentation performance by 8.3% and 6.0% Dice coefficient for the cross-entropy and signed distance transform objectives, respectively.
Keywords: Placenta, Fetus, Segmentation, BOLD MRI, Shape
1. Introduction
Biomedical motivation.
The placenta delivers oxygen and nutrients to support fetal growth. Placental dysfunction causes pregnancy complications that affect fetal development, leading to a critical need to assess placental function in vivo. Blood oxygen level dependent (BOLD) MRI images oxygen transport within the placenta (Sørensen et al., 2013; Abaci Turk et al., 2019) and has emerged as a promising tool to non-invasively study placental function. Temporal analysis of BOLD MRI with maternal oxygen administration can identify contractions (Abaci Turk et al., 2020; Sinding et al., 2016), biomarkers of fetal growth restriction (Luo et al., 2017; Sørensen et al., 2015), predict placental age (Pietsch et al., 2021), and can study congenital heart disease (You et al., 2020; Steinweg et al., 2021).
Challenges and current approaches.
Despite its importance for many downstream clinical research tasks, placental segmentation is often performed manually and can take a significant amount of time, even for a trained expert. For temporal BOLD MRI studies, manual segmentation is rendered more challenging due to the sheer number of MRI scans acquired and rapid signal changes due to common experimental designs. For example, maternal oxygenation experiments acquire several hundred whole-uterus MRI scans to observe signal changes in three stages: normoxia (baseline), hyperoxia, and return to normoxia. During the hyperoxic stage, BOLD signals increase rapidly, giving the placenta a hyperintense appearance. Further, placental shape undergoes large deformation caused by maternal breathing, contractions, and fetal movements which are typically stronger during hyperoxia (You et al., 2015), as illustrated in Figure 1.
Figure 1:
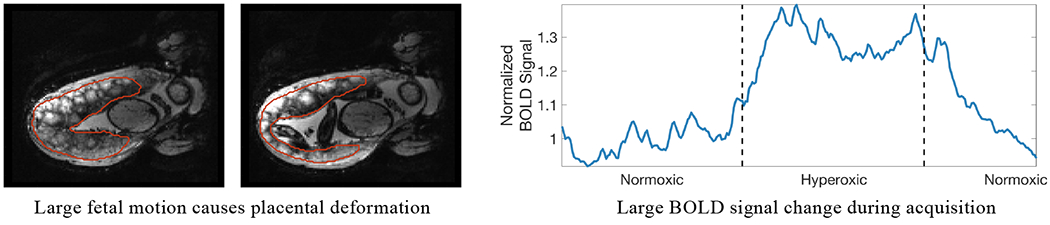
During maternal oxygen administration, the placenta undergoes large shape deformation caused by fetal motion (left) and strong BOLD signal changes caused by maternal hyperoxia (right). Placental boundaries are marked in red. Signal normalization is based on averaging all MRI intensities in the first normoxic period.
Current practice analyzes BOLD signals with respect to a template volume. Deformable registration of all volumes in the time series to the template is performed to enable spatiotemporal analysis (Abaci Turk et al., 2017; You et al., 2015; Chi et al., 2023). However, template-to-volume registration within the uterus can lead to large errors, necessitating outlier detection and possibly rejecting a significant number of volumes (Abaci Turk et al., 2017; You et al., 2015). These registration difficulties arise from the placenta, fetus, and mother being subject to highly disparate deformation models along the temporal sequence. For example, the fetus undergoes piecewise-rigid motion whereas the placenta deforms highly non-rigidly, thus precluding the direct use of standard registration frameworks such as ANTs (Tustison et al., 2020) and VoxelMorph (Balakrishnan et al., 2019) on temporal whole-uterus images.
Contributions.
To address these challenges, we propose a deep network framework to automatically segment the placenta in BOLD MRI time series. Our model is trained on volumes obtained during the normoxic and hyperoxic phases from each patient so as to capture placental shape and appearance variability during maternal oxygenation. As the placenta is a thin and elongated organ, we use a boundary-weighted formulation (parameterized by thresholded signed distance function approximations) of several popular region and/or shape-based segmentation objectives which yield significant gains in both placental segmentation accuracy and surface overlap over their non-boundary-weighted equivalents. Our model demonstrates consistency in the predicted segmentation label maps on a large dataset of unseen BOLD MRI and generalizes to a broad range of gestational ages and pregnancy conditions, thus enabling improved non-invasive pregnancy studies via maternal oxygenation. Finally, to demonstrate the feasibility of our method for clinical research, we illustrate an application based on relative BOLD signal increases.
This paper extends our preliminary analysis of placental segmentation (Abulnaga et al., 2022b) first presented at the International Workshop on Perinatal, Preterm and Paediatric Image Analysis held in conjunction with the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) in 2022. We expand on it by providing more motivation for the work in Section 1 and relevant context for boundary-weighted and shape-aware segmentation losses in Section 2. In our methods in Section 3, we provide additional details, illustrations, and justification for the chosen additive boundary-weighted loss formulation. We then experiment with several additional shape-aware and boundary-weighted loss functions in our experiments (Section 4) alongside introducing qualitative measures of temporal segmentation performance and providing failure cases. Lastly, we provide a substantially expanded discussion of our work in Section 5 which overviews its place in the shape-aware segmentation literature and its utility for clinical research.
2. Related Work
Placenta segmentation in structural MRI.
Machine learning segmentation models for the placenta have been previously proposed and include both semi-automatic (Wang et al., 2015) and automatic (Alansary et al., 2016; Torrents-Barrena et al., 2019a; Pietsch et al., 2021; Specktor-Fadida et al., 2021) approaches. While semi-automatic methods have achieved success in predicting segmentation label maps with high accuracy, these approaches are infeasible for segmenting BOLD MRI time series due to the large number of volumes that would require manual annotation. The majority of automatic methods focus on segmentation in structural images as opposed to BOLD MRI as in this work. For example, Alansary et al. (2016) proposed a model for segmenting T2-weighted (T2w) images based on a 3D CNN followed by a dense CRF for segmentation refinement and validated it on a singleton cohort that included patients with fetal growth restriction (FGR). Torrents-Barrena et al. (2019a) developed a segmentation framework based on super-resolution and a support vector machine and validated it using a singleton and twin cohort of T2w MRI. Specktor-Fadida et al. (2021) focused on transferring segmentation networks across MRI sequences using a self-training model yielding successful segmentation of steady-state precession MRI sequences. For a detailed treatment of fetal MRI segmentation, we refer the reader to the survey by Torrents-Barrena et al. (2019b).
Placenta segmentation in BOLD MRI.
BOLD MR images of the placenta differ greatly from anatomical images, as BOLD images have lower in-plane resolution and the contrast between the placental boundary and surrounding anatomy is less pronounced. Anatomical images may also benefit from super-resolution approaches to increase SNR in the acquired image (Uus et al., 2020). Pietsch et al. (2021) are the first to consider placental segmentation in functional MRI. They proposed a 2D patch-based U-Net model for functional image segmentation and demonstrated a successful application of age prediction using the estimated T2* values. They focused on a cohort of singleton subjects and demonstrated successful applications to abnormal pregnancy conditions including preeclampsia. In contrast to their approach that segments derived T2* maps, we evaluate our segmentation model on BOLD MRI time series. Furthermore, our 3D model operates on the entire volume rather than patches, thereby helping to better resolve the boundaries of the placenta.
Boundary-weighted segmentation objectives.
Popular supervised segmentation losses such as cross-entropy underperform in the presence of highly imbalanced classes common in radiology volumes. As the interface between organs can be obfuscated by motion-related artifacts and/or isointense appearance, strongly penalizing incorrect predictions near the boundary has been demonstrated to improve segmentation performance across several biomedical applications (Ma et al., 2021). For example, Kervadec et al. (2021) and Zhang et al. (2020) implemented distance metrics on organ contour predictions in addition to region-based losses such as soft Dice to improve performance. In parallel, several works regress ground truth distance transforms which are strongly weighted near the organ boundary (Hoopes et al., 2022). As illustrative examples, Huang et al. (2021) regressed ground truth distance maps that are normalized using the Heaviside function to penalize near-boundary misclassifications, while Karimi and Salcudean (2019) proposed a soft-Hausdorff distance loss parameterized by distance transforms. Lastly, the boundary weighting formulation we use is most similar to that of Caliva et al. (2019), wherein a thresholded signed distance transform is used to upweigh boundary neighborhoods in region-based losses.
3. Methods
We train a model parameterized by that takes volumes from a BOLD MRI time series and independently predicts a placental segmentation label map for each time point , where is the total number of time points at which MRI scans were acquired. For a given BOLD time series, we have a small number of frames with ground truth labels (), where is an MRI scan and is the ground truth placenta label map. The model predicts segmentation label maps on each 3D volume in the time series independently.
3.1. Architecture and data considerations
We use a standard 3D U-Net (Ronneberger et al., 2015) with 4 blocks in the contracting and expanding paths each. Each block consists of two consecutive Conv-BatchNorm-ReLU blocks using filters of size . Each block is followed by max pooling (contraction path) or transpose convolution (expansion path). We employ batch normalization before ReLU activation. We augment the images using random affine transforms, flips, whole-image brightness shifts, contrast changes, random noise, and elastic deformations, all using TorchIO (Pérez-García et al., 2021). Specific to segmentation with maternal oxygenation, we simulate the effects of maternal normoxia and hyperoxia with a constant intensity shift in the placenta. All augmentation decisions were made based on cross-validation performance.
To capture the MRI signal and placental shape changes resulting from maternal hyperoxia and fetal motion, we specifically train on several manually segmented volumes in the normoxic or hyperoxic phase. This allows the model to learn from the realistic variations that arise during maternal oxygenation.
3.2. Additive Boundary Loss
To emphasize placental boundary details during training, we construct an additive boundary-weighting which is compatible with any per-voxel segmentation loss function . Given a ground truth placental label map , we denote its boundary as . We use a signed distance function that measures the signed distance, , of voxel to the boundary, where when inside of the placenta and when outside. The boundary weighting is additive for voxels within -distance of ,
| (1) |
The weighted loss is then,
| (2) |
where is a per-voxel class weighting. In practice, we set , to account for class weighting and to penalize outside voxels more heavily and learn to distinguish the placenta from its surrounding anatomy.
We note that several forms of boundary-weighted losses exist in the literature (Ma et al., 2021) with ours being most similar to Caliva et al. (2019). Many of these use a decaying weight for voxels further from the boundary, while we use a constant weighting. For computational efficiency, rather than computing for all voxels as in Caliva et al. (2019), we approximate the region of distance from using convolutional kernels. To find voxels with , we estimate a -wide boundary by an average pooling filter on with kernel size and take the smoothed outputs to lie in the boundary. A larger produces a wider boundary, penalizing more misclassified voxels. See Figure 2 for an illustration. Computing the boundary using a convolution operator is advantageous as it does not require computing the signed distance transform directly, which is computationally expensive and bottlenecks deep network training.
Figure 2:
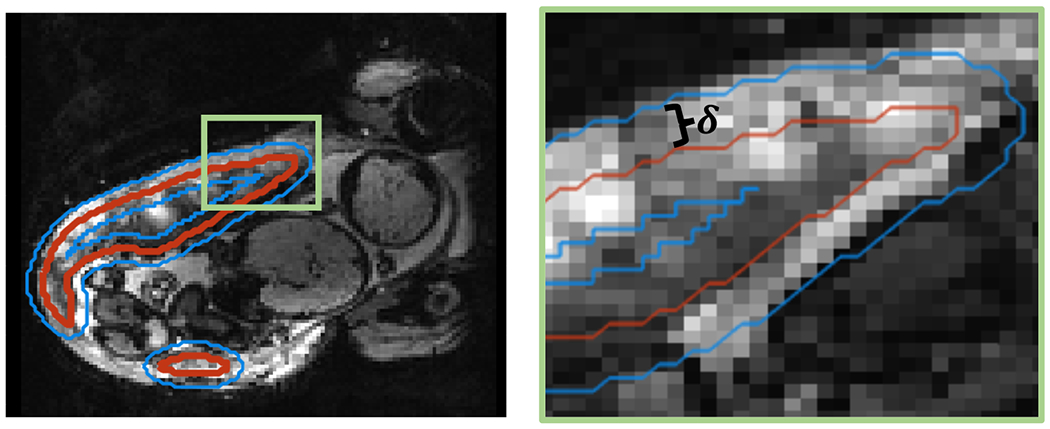
Illustration of inner and outer boundaries used in the additive boundary loss. Right image is zoomed in from the top left corner of the placenta (green box) to closely illustrate boundaries. Placenta segmentation outline is shown in red and inner and outer boundaries δ-distance away are shown in blue.
3.3. Implementation Details
We train using a learning rate with linear decay for 5500 epochs and select the model with the best Dice score on the validation set. For the additive boundary loss, we set , and . For training, we use a simple preprocessing pipeline. All images are normalized by scaling the percentile intensity value to 1 without thresholding or clipping any values. We crop or pad all volumes in the dataset to have dimension and train on the entire 3D volume. We use a batch size of 8 MRI volumes in training. We augment our data with random translations of up to 10 voxels, rotations up to , Gaussian noise sampled with , elastic deformations with 5 control points and a maximum displacement of 10 voxels, whole volume intensity shifts up to , and whole-placenta intensity shifts of normalized intensity values. These values were determined by cross-validation on the training set. When evaluating the model on our test set, we post-processed produced label maps by taking the largest connected component to eliminate islands. Our segmentation code and trained model are available at https://github.com/mabulnaga/automatic-placenta-segmentation.
4. Experiments
4.1. Data
Our dataset consists of BOLD MRI scans taken from two clinical research studies. Data were collected from 91 subjects of which 78 were singleton pregnancies (gestational age (GA) at MRI scan of 23 weeks (wk), 5 days (d) to 37wk6d), and 13 were monochorionic-diamniotic (Mo-Di) twins (GA at MRI scan of 27wk5d – 34wk5d). Of these, 63 pregnancies were controls, 16 had fetal growth restriction (FGR), and 12 had high maternal body mass index (BMI, BMI > 30). Obstetrical ultrasound was used to classify subjects with FGR. For singleton subjects, FGR classification was done based on having fetuses with estimated weight less than the 10th percentile. For twin subjects, FGR classification was determined by monochorionicity and discordance in the estimated fetal weight by growth restriction ( percentile) in one or both fetuses; and/or ii) growth discordance () between fetuses. Table 1 shows patient demographics and GA ranges per group.
Table 1:
Demographic information for subjects in our analysis. GA is gestational age.
| Group | Criteria | Control | FGR | High BMI |
|---|---|---|---|---|
| Singleton | # subjects | 60 | 6 | 12 |
| GA at MRI | 23wk5d – 37wk6d | 26wk6d – 34wk5d | 26wk4d – 36wk6d | |
|
| ||||
| Twin | # subjects | 3 | 10 | 0 |
| GA at MRI | 31wk2d – 34wk5d | 27wk5d – 34wk5d | N/A | |
MRI BOLD scans were acquired on a 3T Siemens Skyra scanner (GRE-EPI, interleaved with 3mm isotropic voxels, TR = 5.8–8s, TE = 32 – 47 ms, FA = 90°). To eliminate intravolume motion artifacts, we split the acquired interleaved volumes into two separate volumes with spacing , then linearly interpolate to . In our analysis, we only consider one of two split volumes, as the signals are redundant between pairs. Maternal oxygen supply was alternated during the BOLD acquisition via a nonrebreathing facial mask to have three 10-minute or 5-minute consecutive episodes: 1. normoxia (), 2. hyperoxia (), and 3. a return to normoxia .
To generate training data, the placenta was manually segmented by a trained observer. Each BOLD MRI time series had 1 to 6 manual segmentations, yielding a total of 176 ground truth labels. The data was then split into training, validation, and test sets: (: subjects). Data was stratified to have proportional distributions of subjects with singleton and twin pregnancies, then to proportionally distribute healthy controls, subjects with FGR, and subjects with high BMI. Our test set had 15 singleton pregnancies and 2 twins. Of the singleton pregnancies, 12 were healthy controls, 1 had FGR, and 2 had high BMI. Both twin subjects had FGR. Our test set had a total of 31 labeled images, none of which were used before final evaluation.
Each subject in the training set had up to ground truth segmentations in the BOLD time series. To prevent bias in sampling images, we train by randomly sampling 1 of ground truth segmentations for each subject. The length of one epoch is the number of subjects rather than the number of images. Subject-wise random sampling was used to reduce bias from subjects with more ground truth labels.
4.2. Evaluation
Performance measures.
We first compare the predicted segmentation label maps to ground truth segmentations. We measure similarity using the Dice score (Dice), the 95th-percentile Hausdorff distance (HD95), and the Average Symmetric Surface Distance (ASSD). To evaluate the feasibility of the produced segmentations for clinical research studying whole-organ signal changes, we evaluate the percentage error in the mean BOLD values between our prediction and the ground truth (BOLD error), defined as , where and denote the mean BOLD signal in the ground truth and in the predicted segmentation, respectively. As estimating whole-organ BOLD signal changes is an important clinical research task, this metric quantifies the appropriate error caused by using automatic placental segmentations.
Benchmarked segmentation loss functions.
We benchmark several popular loss functions and their boundary-weighted extensions to assess their performance on placental segmentation. We quantify improved performance using the boundary-weighting approach (Eq. 1) by comparing performance of the cross-entropy loss and the signed distance transform (SDT) loss () with their boundary-weighted counterparts (). The SDT loss poses segmentation as a regression problem and predicts the signed distance transformation to the placenta boundary. The loss computes the mean-squared error of the predicted SDT from ground truth (Hoopes et al., 2022). We also benchmark performance on the widely used soft Dice loss () (Milletari et al., 2016) and a boundary-weighted Focal loss () with (Lin et al., 2017). As segmentation often benefits from hybrid loss functions (Ma et al., 2021), we also evaluate various combinations of losses. Lastly, we evaluate two boundary-focused signed distance-based loss functions from Huang et al. (2021) () and from Karimi and Salcudean (2019) .
Consistency with hyperoxia.
We evaluate our model’s sensitivity to oxygenation by comparing the accuracy of predictions in normoxia and hyperoxia for subjects with multiple ground truth annotations. We compute segmentation performance using metric in normoxia ), and in hyperoxia (, where denotes the similarity using metric of our predicted segmentation to the ground truth for subject in normoxia. For the evaluation metric , we use the Dice score, HD95, ASSD, and percentage BOLD error. To evaluate consistency with oxygenation, we compute the mean absolute error between segmentation performance in both oxygenation phases . A low error indicates predicted segmentations are consistent with oxygenation changes in the placenta.
Temporal consistency.
We assess the consistency of our predictions by applying our model to all volumes in the BOLD time series of the test set. Since our volumes are acquired interleaved and split into two separate volumes, we apply our model to every second volume in the time series, yielding a mean of volumes per subject. We measure consistency by comparing the Dice score between consecutive volumes. We qualitatively evaluate segmentation performance across the time series and visualize robustness to fetal motion and oxygenation change.
4.3. Results
Table 2 reports the performance of several segmentation losses on the test set. Figure 3 presents box-and-whisker plots of each model’s performance. Both our best performing models trained using and achieve a mean Dice score of 82.80, though produces slightly lower variance. This model also achieves low relative BOLD error (), indicating that our model’s segmentations are suitable for clinical research studies assessing whole-organ signal changes. Similar performance is achieved for the other loss functions. Our model also outperforms the two shape-based baselines and on all metrics, though performance improvements are not statistically significant. These shape-based baselines demonstrate less consistent performance as they have higher variance with outliers (Figure 3).
Table 2:
Test results produced by our 3D U-Net model trained using different loss functions. Numbers in bold indicate the best result in each column. We evaluate commonly used loss functions and the shape-based loss () of (Huang et al., 2021) and the soft-Hausdorff distance loss of Karimi and Salcudean (2019).
| Loss | Dice | HD95 (mm) | ASSD (mm) | BOLD error (%) |
|---|---|---|---|---|
| 82.80 ± 3.25 | 13.31 ± 6.4 | 4.01 ± 1.0 | 411 ± 3.0 | |
| 81.98 ± 5.3 | 12.61 ± 4.66 | 4.08 ± 1.02 | 4.30 ± 4.52 | |
| 76.47 ± 7.43 | 17.92 ± 11.02 | 5.96 ± 2.1 | 5.22 ± 2.53 | |
| 77.13 ± 9.74 | 22.51 ± 18.02 | 6.14 ± 3.95 | 8.95 ± 11.37 | |
| 79.82 ± 6.57 | 15.91 ± 7.93 | 4.44 ± 1.3 | 4.61 ± 2.59 | |
| 81.44 ± 6.42 | 12.87 ± 8.9 | 4.14 ± 1.42 | 5.84 ± 8.28 | |
| 81.82 ± 4.71 | 12.88 ± 5.04 | 4.10 ± 0.97 | 4.44 ± 3.4 | |
| 82.80 ± 3.91 | 12.75 ± 5.58 | 4.02 ± 0.92 | 4.06 ± 1.75 | |
| 81.96 ± 6.19 | 14.02 ± 8.99 | 4.27 ± 1.62 | 6.32 ± 7.25 | |
| 80.63 ± 5.96 | 16.63 ± 12.4 | 4.68 ± 1.92 | 5.64 ± 6.35 | |
| 76.06 ± 8.67 | 20.14 ± 14.25 | 6.01 ± 2.51 | 7.09 ± 8.86 | |
| 80.39 ± 7.16 | 16.16 ± 12.17 | 4.62 ± 1.98 | 6.35 ± 9.52 | |
| 81.54 ± 6.30 | 16.06 ± 13.28 | 4.65 ± 2.26 | 5.25 ± 4.59 |
Figure 3:
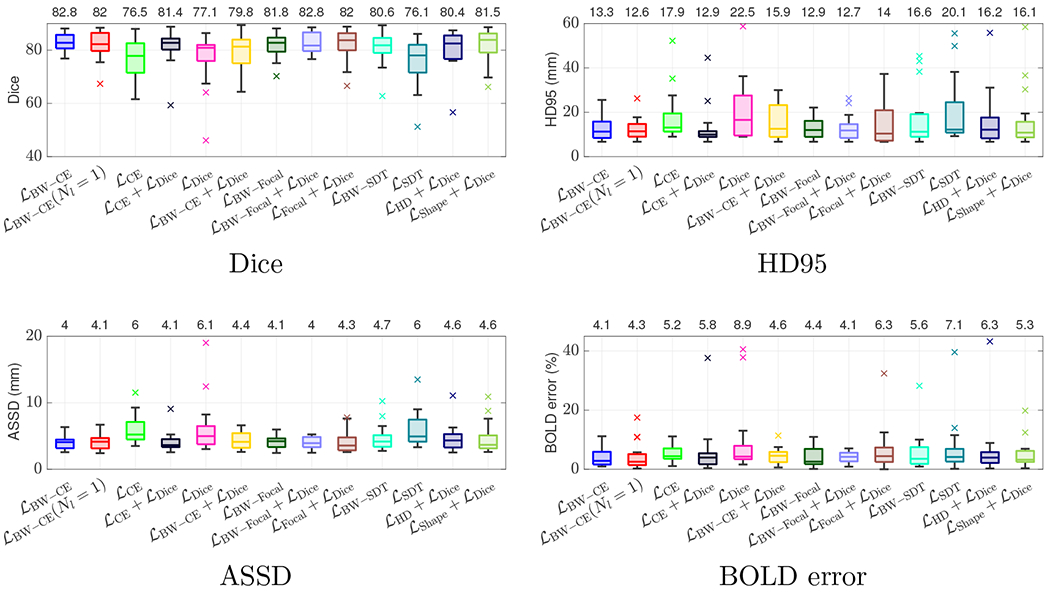
Box-and-whisker plots of Dice (top left), HD95 (top right), ASSD (bottom left), and BOLD error (bottom right) on the test set across all models. We evaluate several commonly used loss functions with and without our boundary weighting and evaluate the baseline shape-based loss () of Huang et al. (2021) and the soft-Hausdorff distance loss () of Karimi and Salcudean (2019). Black horizontal lines inside the box indicate the median. The boxes extend to the and percentiles and the whiskers reach to the most extreme values not considered outliers (black bars). The outliers, shown as crosses, are points farther than 1.5 times the interquartile range. Mean values are indicated on the top horizontal axis. Boundary weighting results in higher mean values with more narrow distributions of distortion compared to their non-boundary weighted counterparts. We further observe improved performance compared with baseline losses in terms of mean, variance, and number of outliers.
The boundary weighting improves the performance of several loss functions compared to their non-boundary weighted counterparts. Training the model with our boundary weighting results in a statistically significant increase in performance. When trained using , we achieve a mean Dice of 82.5 with boundary weighting in the loss compared to 76.5 without. Similarly, for we achieve a mean Dice of 80.6 with boundary weighting compared to 76.1 without. However, training with an additive Dice loss improves this performance gap: achieves a Dice of 81.4 compared to 82.8 for , and achieves a Dice of 82.0 compared to 82.8 for . The boundary-weighting also improves mean performance of ASSD and BOLD error and demonstrates more narrow distributions of distortion with fewer outliers (see Figure 3.) Using only the first segmented volume of the BOLD MRI series in normoxia also results in a small drop in performance . Adding labeled examples from the hyperoxic phase helps generalization, as the placental shape and intensity patterns can change greatly.
Our performance is consistent across pregnancy conditions, as we achieve Dice scores of on the two subjects with twin pregnancies, on the singletons , on the FGR cohort on the controls and on the two BMI cases.
Our model performs consistently well in the normoxic and hyperoxic phases. For the 5 subjects with ground truth segmentations in both the normoxia and hyperoxia, we achieve a mean absolute difference between predictions in normoxia and hyperoxia of Dice, HD95, ASSD, and relative BOLD error. These results suggest that our model is robust to contrast changes in the placenta resulting from maternal hyperoxia, and can be used in studies quantifying oxygen transport in the organ. A larger number of subjects are needed to assess statistical significance.
Figure 4 compares the predicted label maps with ground truth on 5 subjects with increasing Dice scores using the BW-CE model. The model accurately identifies the location of the placenta, but in the worst cases misses boundary details.
Figure 4:
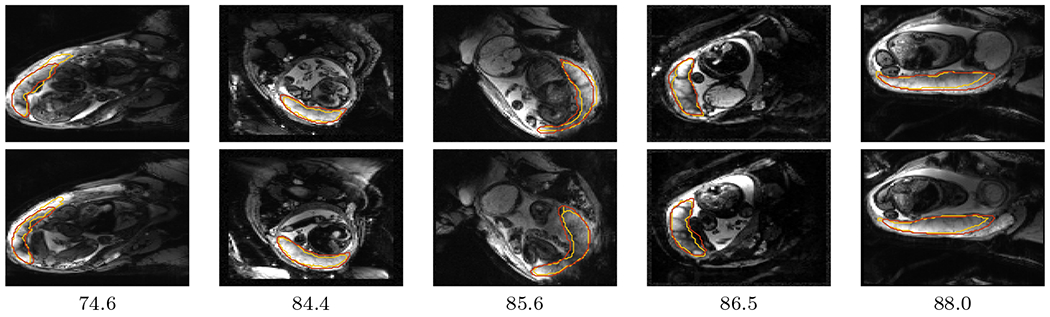
Example predictions on 5 subjects from the test set. Ground truth segmentations are shown in yellow and predictions in red. Dice scores are indicated below each column. Two slices are shown for each subject, spaced 18mm apart.
BOLD Time Series Evaluation
Figure 5 presents example predicted segmentations at multiple points in the BOLD MRI time series for 3 subjects. The predicted segmentations are robust to large fetal deformations and placental signal changes. Figure 6 (top) presents distributions of Dice score between predicted label maps of consecutive frames in the BOLD time series for all subjects in the test set. Distributions have high medians (Dice > 90) for all but one case, with high density at high Dice scores (Dice ). Dice differences are highly affected by fetal and maternal motion that cause placental deformation. We visually verified that modest drops in Dice () were mainly due to fetal motion, but 3 subjects had a small number of frames with large drops (Dice ) that were caused by errors in the produced label maps. Figure 6 (bottom) shows 3D models of failed segmentations from two subjects from frames with Dice . Our model omitted parts of the placenta for Subject 9 and added a large region for Subject 15. In practice, these failures ocurred in a small number of frames, of frames for Subject 9 and of frames for Subject 15. Overall, predicted label maps are consistent between consecutive volumes of the MRI time series, achieving a Dice of and a BOLD difference of . The small differences between the relative mean-BOLD values suggest these produced segmentations may be suitable for research studies assessing placental function.
Figure 5:
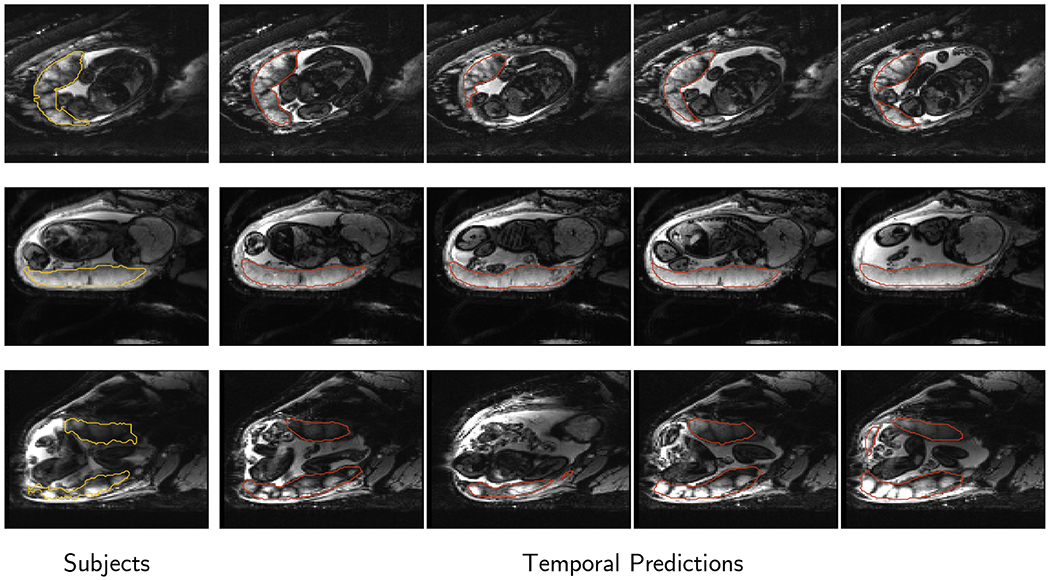
Consistency of predictions throughout the time series on 3 subjects from the test set. Each row corresponds to a subject. The leftmost column shows the ground truth segmentation outlined in yellow, and temporal-predicted segmentations in columns are shown in red. The same axial slice is shown for each subject, and images span the time series. Segmentations are consistent across time series, achieving consecutive BOLD differences of , and and Dice scores of , and (top, middle, and bottom). Variations in these metrics are expected due to fetal motion and changes in oxygenation due to the BOLD protocol.
Figure 6:
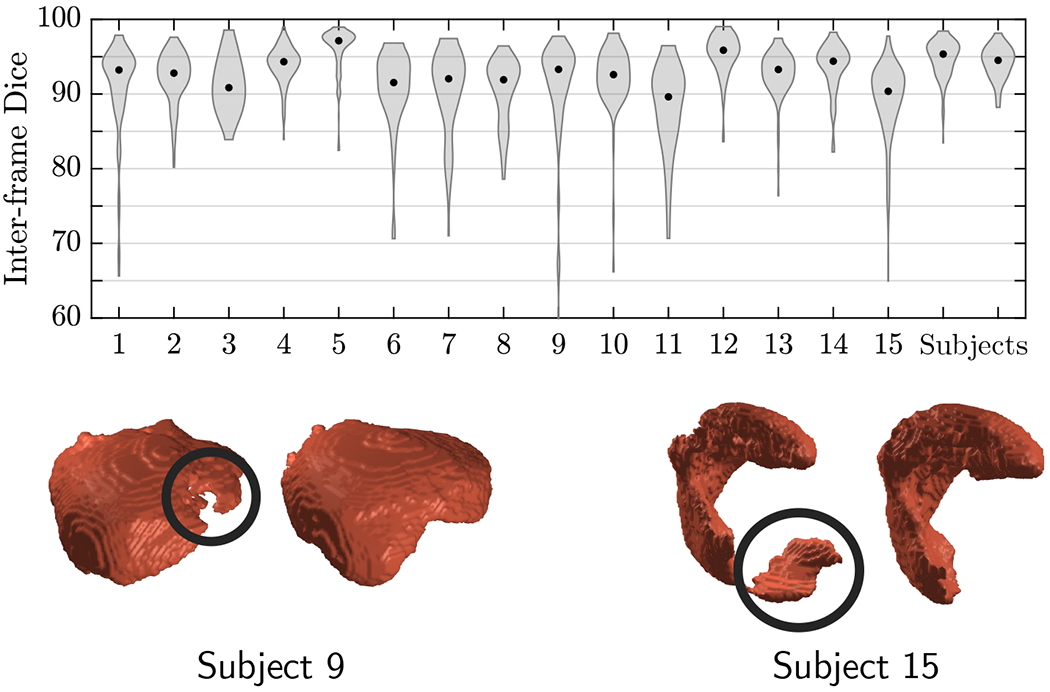
Top: Per-subject density distributions of Dice scores between consecutive predictions in BOLD MRI time series. Dots inside distributions indicate the median. Bottom: Example failure cases at frames with Dice . 3D models of placentae predicted at two consecutive time points are shown for two subjects. Left: The region in black is distorted in the predicted segmentation of subject 9. Right: A large portion of the placenta (encircled) is wrongfully predicted in the placenta of subject 15. Failure cases are likely to occur with large drops in consecutive Dice (Dice ).
Automatic segmentation of each volume in BOLD MRI time series is advantageous as it can enable whole-organ spatiotemporal analysis without requiring inter-volume registration, which may fail under the presence of large motion. We illustrate a possible application of automatic placenta segmentation by investigating the percentage increase in BOLD signal in response to maternal hyperoxia. We calculate the percentage increase over the baseline period: , where denotes the mean BOLD signal over the baseline period, and denotes the mean of the signal in the last 10 frames of the hyperoxic period. Figure 7 shows a scatter plot of the hyperoxia response for all subjects in the test set and two examples of the BOLD signal time course in the produced placenta segmentation label maps. In the control subjects (), we observe an increase of . The observed increase for the healthy controls is consistent with previous studies that demonstrated an increase of (Sørensen et al., 2015) and from to throughout gestation () (Sinding et al., 2018).
Figure 7:
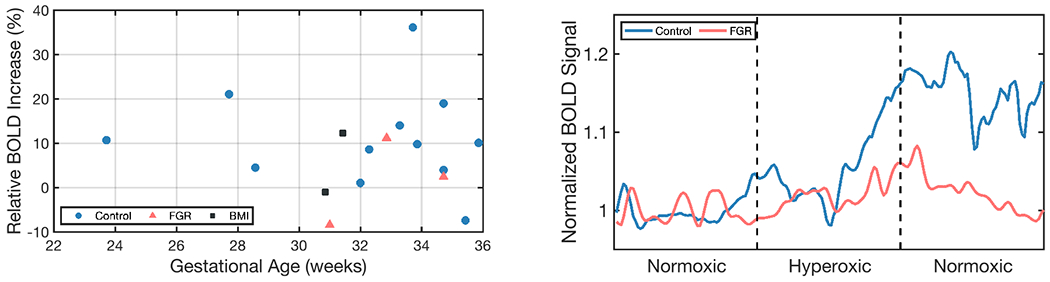
Example application using our model’s produced placenta segmentations in BOLD time series to characterize oxygenation response from maternal hyperoxia. Left: Observed increase relative to normoxic baseline for the test set. Right: Example time series for one singleton control (GA=33wk2d, Dice=85.5, ) and one singleton FGR subject (GA=34wk5d, Dice=83.5, .
5. Discussion
We proposed a model to automatically segment the placenta in BOLD MRI time series and achieved close matching to ground truth labels with consistent performance in predicting segmentations in both the normoxic and hyperoxic phases. Our solution was developed to be resilient to the variability caused by large signal changes in the BOLD experiment protocol.
Shape-aware segmentation.
Identification of the placental boundary is challenging as the organ is a thin and elongated structure with limited contrast with surrounding anatomy. In this work, we emphasized these challenging aspects of placental shape during network training by using a simple additive boundary-weighted loss function. As hypothesized, boundary weighting significantly improved placenta segmentation performance when integrated with the popular cross-entropy and signed distance transform (SDT) losses as compared with their non-boundary weighted counterparts. We then performed an extensive evaluation over shape-based losses including our chosen formulation and the losses of Karimi and Salcudean (2019), and Huang et al. (2021) and found that our adopted loss outperforms others in several key aspects of placenta segmentation. Lastly, we broadly find that shape-based losses outperform losses without shape information (cross-entropy, Dice, and SDT losses), demonstrating that including shape information can aid in the identification of the placenta.
Utility for clinical research.
The main objective of this work was to develop a segmentation model to assess whole-organ signal changes in BOLD MRI time series. We achieve low BOLD error (4%) compared to ground truth with performance that is consistent across oxygenation period. Segmenting each volume in the BOLD MRI time series can be advantageous for clinical research assessing whole-organ temporal changes. We illustrated one possible study in assessing placental response during hyperoxia and observed an increase in signal intensity consistent with prior work. However, our cohort is limited, and several factors, including maternal position, gestational age, and contractions are covariates not considered. Producing segmentations that are resilient to placental oxygenation can also enable essential post-processing such as motion correction (Abaci Turk et al., 2017), reconstruction (Uus et al., 2020), and mapping to a standardized representation (Miao et al., 2017; Abulnaga et al., 2019, 2022a; Chi et al., 2023). These tasks are often essential in clinical research studies assessing placental function.
We provide access to our code and trained model for use in future clinical research studies. Our model is robust to gestational age, pregnancy condition, and oxygenation. Further, the model requires simple preprocessing and can be used with a variety of EPI scans. In this work however, we only trained and tested on isotropic MRI with TE=32 – 47 ms. It is unclear how well the model would work on scans with earlier GA, nonistropic images, or different imaging protocols.
A limitation of relying only on segmentations is that they can only be used to quantify whole-organ signal changes, such as mean or mean BOLD increase. One is often interested in assessing functional differences within subregions of the placenta, for example across twins in Mo-Di pregnancies (Luo et al., 2017; Shnitzer et al., 2022), within vessels (Torrents-Barrena et al., 2019a), or across cotyledons (Dey et al., 2023). Localized analysis requires deformable registration (Abaci Turk et al., 2017) to track changes within these regions. Having reliable segmentations for each point in the BOLD MRI time series can be used to improve registration, for example by treating segmentations as spatial priors. One may also consider methods that jointly learn registration and segmentation such as that of Xu and Niethammer (2019).
Comparison to prior work.
The closest work to ours is that of Pietsch et al. (2021) that proposes a 2D U-Net based model to automatically segment the placenta in functional MRI (BOLD, T2*). They achieve a Dice of 58 on a cohort of 108 subjects of low- and high-risk singleton subjects of a wide GA range. Their performance was comparable to the inter-rater variability of two radiologists (Dice=68), which represents an upper limit. In contrast, we achieve a Dice of 82.8 on a cohort of singleton and twin subjects from healthy pregnancies, subjects with FGR, and subjects with high BMI. While comparing our work with that of Pietsch et al. (2021) provides context for our performance, direct comparison with this work is not feasible due to differences in data set size and patient demographics, imaging protocols, and MRI study design. An interesting direction of future work is to quantify the improvement in performance due to model design versus dataset composition.
Limitations and Future Work.
The main limitation of this work is the inability to quantify segmentation performance across the entire BOLD MRI study. While we demonstrated low absolute differences in predictions between normoxia and hyperoxia, we only had 5 subjects with ground truth images in multiple time points. We measured the consistency of consecutive predictions via Dice overlap and percentage BOLD differences. However, without correction for inter-frame placental deformation, the reported scores are subject to noise caused by motion. We performed visual quality control and found that for many subjects, modest drops in Dice , were often due to fetal motion displacing the placenta. In a small number of cases, we observed large drops (Dice ) that were caused by segmentation error (Figure 6 bottom). Since we apply the model to each volume in the time series independently, imaging artifacts, such as intensity and geometric artifacts, can affect the predicted segmentations.
We performed a comprehensive evaluation of several commonly used loss functions with and without our boundary weighting approach, and compared with shape-based baselines of two previous works: of Huang et al. (2021) and of Karimi and Salcudean (2019). While our boundary-weighting outperformed these loss functions, we observed that any shape-based loss improved performance over conventional loss functions (e.g. , demonstrating the benefit in capturing the placenta boundary accurately. Additional loss functions exist that we did not compare with, such as the distance transform-based boundary loss of Kervadec et al. (2021), and the boundary contour-based loss functions of Specktor-Fadida et al. (2021) and Jurdi et al. (2021). Similar to our baselines, these loss functions are additive and aim to improve boundary capture and reduce the Hausdorff distance. Consequently, we do not expect significant differences over our proposed model, though future work should compare with additional baselines. Since our proposed loss is a boundary-based weighting rather than a separate loss function, it is versatile to be used with any existing loss.
Future work can investigate semi-supervised learning approaches to incorporate all unlabeled volumes in the BOLD MRI time series, increasing the variety of available data to potentially improve temporally consistent segmentation. As there are often a few hundred unlabeled volumes in each BOLD time series, these approaches can more accurately capture the rapid signal changes resulting from fetal motion and maternal oxygenation. The unlabeled data can be incorporated using non-rigid registration as in (Xu and Niethammer, 2019; Zhao et al., 2019; Chartsias et al., 2020) or by using unsupervised shape-regularization losses (Mirikharaji and Hamarneh, 2018; Young et al., 2023).
6. Conclusion
We developed a model to automatically segment the placenta in BOLD MRI time series. Our model performed consistently well at different oxygenation phases of the BOLD protocol, and across a variety of pregnancy conditions and gestational ages. We demonstrated one potential clinical research application of this work in quantifying BOLD increase due to hyperoxia that matched reported values from the literature. Automatic segmentation in BOLD MRI time series can be used to investigate oxygenation dynamics in the placenta. For example, temporal segmentations can be used to derive T2* maps to perform whole-organ signal comparisons across population groups, enabling quantitative analysis of placental function with the ultimate goal of developing biomarkers of placental and fetal health.
7. Acknowledgments
This work was supported in part by NIH NIBIB NAC P41EB015902, NIH NICHD R01HD100009, R01EB032708, R21HD106553, MIT-IBM Watson AI Lab, NSERC PGS D, NSF GRFP, and a MathWorks Fellowship.
Footnotes
Ethical Standards
Written informed consent was obtained from all subjects. The data used in this work came from two clinical resarch studies. The study protocols were reviewed and approved by Boston Children’s Hospital institutional review board (IRB), from IRB protocol #P00012416 and #P00012586. All methods were carried out in accordance with institutional guidelines and regulations.
Conflicts of Interest
The authors declare no conflicts of interest.
Contributor Information
S. Mazdak Abulnaga, CSAIL/EECS, Massachusetts Institute of Technology, Cambridge, MA, USA; MGH/HST Martinos Center for Biomedical Imaging, Harvard Medical School, Boston, MA, USA.
Neel Dey, CSAIL, Massachusetts Institute of Technology, Cambridge, MA, USA.
Sean I. Young, MGH/HST Martinos Center for Biomedical Imaging, Harvard Medical School, Boston, MA, USA; CSAIL, Massachusetts Institute of Technology, Cambridge, MA, USA
Eileen Pan, CSAIL/EECS, Massachusetts Institute of Technology, Cambridge, MA, USA.
Katherine I. Hobgood, CSAIL, Massachusetts Institute of Technology, Cambridge, MA, USA
Clinton J. Wang, CSAIL/EECS, Massachusetts Institute of Technology, Cambridge, MA, USA
P. Ellen Grant, Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Harvard Medical School, Boston, MA, USA.
Esra Abaci Turk, Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Harvard Medical School, Boston, MA, USA.
Polina Golland, CSAIL/EECS, Massachusetts Institute of Technology, Cambridge, MA, USA.
Data availability
The data is currently not approved for public release. The data is being de-anonymized with plans for a future public release. The code and model weights are available at https://github.com/mabulnaga/automatic-placenta-segmentation.
References
- Turk Esra Abaci, Luo Jie, Gagoski Borjan, Pascau Javier, Bibbo Carolina, Robinson Julian N., Grant P. Ellen, Adalsteinsson Elfar, Golland Polina, and Malpica Norberto. Spatiotemporal alignment of in utero BOLD-MRI series. Journal of Magnetic Resonance Imaging, 46(2):403–412, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk Esra Abaci, Stout Jeffrey N., Ha Christopher, Luo Jie, Gagoski Borjan, Yetisir Filiz, Golland Polina, Wald Lawrence L., Adalsteinsson Elfar, Robinson Julian N., Roberts Drucilla J., Barth William H. Jr., and Grant P. Ellen. Placental MRI: Developing accurate quantitative measures of oxygenation. Topics in Magnetic Resonance Imaging, 28(5):285–297, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk Esra Abaci, Abulnaga S. Mazdak, Luo Jie, Stout Jeffrey N., Feldman Henry A., Turk Ata, Gagoski Borjan, Wald Lawrence L., Adalsteinsson Elfar, Roberts Drucilla J., Bibbo Carolina, Robinson Julian N., Golland Polina, Grant P. Ellen, and Barth William H. Jr. Placental MRI: Effect of maternal position and uterine contractions on placental BOLD MRI measurements. Placenta, 95:69–77, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abulnaga S. Mazdak, Turk Esra Abaci, Bessmeltsev Mikhail, Grant P. Ellen, Solomon Justin, and Golland Polina. Placental flattening via volumetric parameterization. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019, pages 39–47, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abulnaga S. Mazdak, Turk Esra Abaci, Bessmeltsev Mikhail, Grant P. Ellen, Solomon Justin, and Golland Polina. Volumetric parameterization of the placenta to a flattened template. IEEE Transactions on Medical Imaging, 41(4):925–936, 2022a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abulnaga S. Mazdak, Young Sean I., Hobgood Katherine, Pan Eileen, Wang Clinton J., Grant P. Ellen, Turk Esra Abaci, and Golland Polina. Automatic segmentation of the placenta in BOLD MRI time series. In MICCAI International Workshop on Preterm, Perinatal and Paediatric Image Analysis, pages 1–12. Springer Nature Switzerland, 2022b. [Google Scholar]
- Alansary Amir, Kamnitsas Konstantinos, Davidson Alice, Khlebnikov Rostislav, Rajchl Martin, Malamateniou Christina, Rutherford Mary, Hajnal Joseph V., Glocker Ben, Rueckert Daniel, and Kainz Bernhard. Fast fully automatic segmentation of the human placenta from motion corrupted MRI. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 589–597. Springer, 2016. [Google Scholar]
- Balakrishnan Guha, Zhao Amy, Sabuncu Mert R., Guttag John V., and Dalca Adrian V.. Voxelmorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging, 38(8):1788–1800, 2019. [DOI] [PubMed] [Google Scholar]
- Caliva Francesco, Iriondo Claudia, Martinez Alejandro Morales, Majumdar Sharmila, and Pedoia Valentina. Distance map loss penalty term for semantic segmentation. arXiv preprint arXiv:1908.03679, 2019.
- Chartsias Agisilaos, Papanastasiou Giorgos, Wang Chengjia, Semple Scott, Newby David E., Dharmakumar Rohan, and Tsaftaris Sotirios A.. Disentangle, align and fuse for multimodal and semi-supervised image segmentation. IEEE transactions on medical imaging, 40(3):781–792, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Zeen, Cong Zhongxiao, Wang Clinton J, Liu Yingcheng, Turk Esra Abaci, Grant P Ellen, Abulnaga S Mazdak, Golland Polina, and Dey Neel. Dynamic neural fields for learning atlases of 4d fetal mri time-series. arXiv preprint arXiv:2311.02874, 2023.
- Dey Neel, Abulnaga S Mazdak, Billot Benjamin, Turk Esra Abaci, Grant P Ellen, Dalca Adrian V, and Golland Polina. Anystar: Domain randomized universal star-convex 3d instance segmentation. arXiv preprint arXiv:2307.07044, 2023.
- Hoopes Andrew, Mora Jocelyn S., Dalca Adrian V., Fischl Bruce, and Hoffmann Malte. Synthstrip: skull-stripping for any brain image. NeuroImage, 260:119474, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Quanwei, Zhou Yuezhi, and Tao Linmi. Dual-term loss function for shape-aware medical image segmentation. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), pages 1798–1802, 2021. [Google Scholar]
- Jurdi Rosana E. L., Petitjean Caroline, Honeine Paul, Cheplygina Veronika, and Abdallah Fahed. A surprisingly effective perimeter-based loss for medical image segmentation. In Medical Imaging with Deep Learning, pages 158–167. PMLR, 2021. [Google Scholar]
- Karimi Davood and Salcudean Septimiu E.. Reducing the hausdorff distance in medical image segmentation with convolutional neural networks. IEEE Transactions on medical imaging, 39(2):499–513, 2019. [DOI] [PubMed] [Google Scholar]
- Kervadec Hoel, Bouchtiba Jihene, Desrosiers Christian, Granger Eric, Dolz Jose, and Ayed Ismail Ben. Boundary loss for highly unbalanced segmentation. Medical image analysis, 67:101851, 2021. [DOI] [PubMed] [Google Scholar]
- Lin Tsung-Yi, Goyal Priya, Girshick Ross, He Kaiming, and Dollár Piotr. Focal loss for dense object detection. In 2017 IEEE International Conference on Computer Vision (ICCV), pages 2999–3007, 2017. [Google Scholar]
- Luo Jie, Turk Esra Abaci, Bibbo Carolina, Gagoski Borjan, Roberts Drucilla J., Vangel Mark, Tempany-Afdhal Clare M., Barnewolt Carol, Estroff Judy, Palanisamy Arvind, Barth William H. Jr., Zera Chloe, Malpica Norberto, Golland Polina, Adalsteinsson Elfar, Robinson Julian N., and Grant P. Ellen. In vivo quantification of placental insufficiency by BOLD MRI: a human study. Scientific Reports, 7(1):3713, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Jun, Chen Jianan, Ng Matthew, Huang Rui, Li Yu, Li Chen, Yang Xiaoping, and Martel Anne L.. Loss odyssey in medical image segmentation. Medical Image Analysis, 71:102035, 2021. [DOI] [PubMed] [Google Scholar]
- Miao Haichao, Mistelbauer Gabriel, Karimov Alexey, Alansary Amir, Davidson Alice, Lloyd David F. A., Damodaram Mellisa, Story Lisa, Hutter Jana, Hajnal Joseph V., Rutherford Mary, Preim Bernhard, Kainz Bernhard, and Gröller Eduard M.. Placenta maps: in utero placental health assessment of the human fetus. IEEE Transactions on Visualization and Computer Graphics, 23(6):1612–1623, 2017. [DOI] [PubMed] [Google Scholar]
- Milletari Fausto, Navab Nassir, and Ahmadi Seyed-Ahmad. V-net: Fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth International Conference on 3D Vision (3DV), pages 565–571, 2016. [Google Scholar]
- Mirikharaji Zahra and Hamarneh Ghassan. Star shape prior in fully convolutional networks for skin lesion segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 737–745. Springer, 2018. [Google Scholar]
- Pietsch Maximilian, Ho Alison, Bardanzellu Alessia, Zeidan Aya Mutaz Ahmad, Chappell Lucy C., Hajnal Joseph V., Rutherford Mary, and Hutter Jana. APPLAUSE: Automatic prediction of placental health via U-net segmentation and statistical evaluation. Medical Image Analysis, 72:102145, 2021. ISSN 1361-8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-García Fernando, Sparks Rachel, and Ourselin Sébastien. Torchio: A python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. Computer Methods and Programs in Biomedicine, 208:106236,2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger Olaf, Fischer Philipp, and Brox Thomas. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, pages 234–241, 2015. [Google Scholar]
- Shnitzer Tal, Abulnaga S. Mazdak, Bibbo Carolina, Grant P. Ellen, Golland Polina, Solomon Justin, and Turk Esra Abaci. Automatic segmentation of twin regions in Mo-Di placentae based on geometric analysis of spatiotemporal BOLD MRI signals. In Proceedings of the International Society for Magnetic Resonance in Medicine, 2022. [Google Scholar]
- Sinding Marianne, Peters David A, Frøkjær Jens B, Christiansen Ole B, Uldbjerg Niels, and Sørensen Anne. Reduced placental oxygenation during subclinical uterine contractions as assessed by BOLD MRI. Placenta, 39:16–20, 2016. [DOI] [PubMed] [Google Scholar]
- Sinding Marianne, Peters David A., Poulsen Sofie S., Frøkjær Jens B., Christiansen Ole B., Petersen Astrid, Uldbjerg Niels, and Sørensen Anne. Placental baseline conditions modulate the hyperoxic BOLD-MRI response. Placenta, 61:17–23, 2018. [DOI] [PubMed] [Google Scholar]
- Sørensen Anne, Peters David, Simonsen Carsten, Pedersen Michael, Stausbøl-Grøn Brian, Christiansen Ole B., Lingman Göran, and Uldbjerg Niels. Changes in human fetal oxygenation during maternal hyperoxia as estimated by BOLD MRI. Prenatal Diagnosis, 33(2):141–145, 2013. [DOI] [PubMed] [Google Scholar]
- Sørensen Anne, Sinding Marianne, Peters David A., Petersen Astrid, Frøkjær Jens B., Christiansen Ole B., and Uldbjerg Niels. Placental oxygen transport estimated by the hyperoxic placental BOLD MRI response. Physiological reports, 3(10):e12582, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specktor-Fadida Bella, Link-Sourani Daphna, Ferster-Kveller Shai, Ben-Sira Liat, Miller Elka, Ben-Bashat Dafna, and Joskowicz Leo. A bootstrap self-training method for sequence transfer: State-of-the-art placenta segmentation in fetal MRI. In Uncertainty for Safe Utilization of Machine Learning in Medical Imaging, and Perinatal Imaging, Placental and Preterm Image Analysis, pages 189–199. Springer, 2021. [Google Scholar]
- Steinweg Johannes K., Hui Grace Tin Yan, Pietsch Maximilian, Ho Alison, van Poppel Milou P. M., Lloyd David, Colford Kathleen, Simpson John M., Razavi Reza, Pushparajah Kuberan, Rutherford Mary, and Hutter Jana. T2* placental MRI in pregnancies complicated with fetal congenital heart disease. Placenta, 108:23–31, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents-Barrena Jordina, Piella Gemma, Masoller Narcís, Gratacós Eduard, Eixarch Elisenda, Ceresa Mario, and Ballester Miguel Ángel González. Fully automatic 3D reconstruction of the placenta and its peripheral vasculature in intrauterine fetal MRI. Medical image analysis, 54:263–279, 2019a. [DOI] [PubMed] [Google Scholar]
- Torrents-Barrena Jordina, Piella Gemma, Masoller Narcís, Gratacós Eduard, Eixarch Elisenda, Ceresa Mario, and Ballester Miguel Ángel González. Segmentation and classification in mri and us fetal imaging: Recent trends and future prospects. Medical Image Analysis, 51:61–88, 2019b. [DOI] [PubMed] [Google Scholar]
- Tustison Nicholas J., Cook Philip A., Holbrook Andrew J., Johnson Hans J., Muschelli John, Devenyi Gabriel A., Duda Jeffrey T., Das Sandhitsu R., Cullen Nicholas C., Gillen Daniel L., et al. Antsx: A dynamic ecosystem for quantitative biological and medical imaging. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uus Alena, Zhang T, Jackson LH, Roberts TA, Rutherford MA, Hajnal JV, and Deprez M. Deformable slice-to-volume registration for motion correction of fetal body and placenta MRI. IEEE transactions on medical imaging, 39(9):2750–2759, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Guotai, Zuluaga Maria A, Pratt Rosalind, Aertsen Michael, David Anna L, Deprest Jan, Vercauteren Tom, and Ourselin Sebastien. Slic-seg: slice-by-slice segmentation propagation of the placenta in fetal mri using one-plane scribbles and online learning. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 29–37. Springer, 2015. [Google Scholar]
- Xu Zhenlin and Niethammer Marc. Deepatlas: Joint semi-supervised learning of image registration and segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part II 22, pages 420–429. Springer, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Wonsang, Serag Ahmed, Evangelou Iordanis E., Andescavage Nickie, and Limperopoulos Catherine. Robust motion correction and outlier rejection of in vivo functional MR images of the fetal brain and placenta during maternal hyperoxia. In SPIE Medical Imaging, volume 9417, pages 177–189. SPIE, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Wonsang, Andescavage Nickie N., Kapse Kushal, Donofrio Mary T., Jacobs Marni, and Limperopoulos Catherine. Hemodynamic responses of the placenta and brain to maternal hyperoxia in fetuses with congenital heart disease by using blood oxygen–level dependent MRI. Radiology, 294(1):141–148, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young Sean I., Dalca Adrian V., Ferrante Enzo, Golland Polina, Metzler Christopher A., Fischl Bruce, and Iglesias Juan Eugenio. Supervision by denoising. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2023. [DOI] [PubMed] [Google Scholar]
- Zhang Mo, Dong Bin, and Li Quanzheng. Deep active contour network for medical image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part IV 23, pages 321–331. Springer, 2020. [Google Scholar]
- Zhao Amy, Balakrishnan Guha, Durand Fredo, Guttag John V., and Dalca Adrian V.. Data augmentation using learned transformations for one-shot medical image segmentation. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pages 8543–8553, 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data is currently not approved for public release. The data is being de-anonymized with plans for a future public release. The code and model weights are available at https://github.com/mabulnaga/automatic-placenta-segmentation.


