Summary
DataLad is a Python-based tool for the joint management of code, data, and their relationship, built on top of a versatile system for data logistics (git-annex) and the most popular distributed version control system (Git). It adapts principles of open-source software development and distribution to address the technical challenges of data management, data sharing, and digital provenance collection across the life cycle of digital objects. DataLad aims to make data management as easy as managing code. It streamlines procedures to consume, publish, and update data, for data of any size or type, and to link them as precisely versioned, lightweight dependencies. DataLad helps to make science more reproducible and FAIR (Wilkinson et al., 2016). It can capture complete and actionable process provenance of data transformations to enable automatic re-computation. The DataLad project (datalad.org) delivers a completely open, pioneering platform for flexible decentralized research data management (RDM) (Hanke, Pestilli, et al., 2021). It features a Python and a command-line interface, an extensible architecture, and does not depend on any centralized services but facilitates interoperability with a plurality of existing tools and services. In order to maximize its utility and target audience, DataLad is available for all major operating systems, and can be integrated into established workflows and environments with minimal friction.
Statement of Need
Code, data and computing environments are core components of scientific projects. While the collaborative development and use of research software and code is streamlined with established procedures and infrastructures, such as software distributions, distributed version control systems, and social coding portals like GitHub, other components of scientific projects are not as transparently managed or accessible. Data consumption is complicated by disconnected data portals that require a large variety of different data access and authentication methods. Compared with code in software development, data tend not to be as precisely identified because data versioning is rarely or only coarsely practiced. Scientific computation is not reproducible enough, because data provenance, the information of how a digital file came to be, is often incomplete and rarely automatically captured. Last but not least, in the absence of standardized data packages, there is no uniform way to declare actionable data dependencies and derivative relationships between inputs and outputs of a computation. DataLad aims to solve these issues by providing streamlined, transparent management of code, data, computing environments, and their relationship. It provides targeted interfaces and interoperability adapters to established scientific and commercial tools and services to set up unobstructed, unified access to all elements of scientific projects. This unique set of features enables workflows that are particularly suited for reproducible science, such as actionable process provenance capture for arbitrary command execution that affords automatic re-execution. To this end, it builds on and extends two established tools for version control and transport logistics, Git and git-annex.
Why Git and git-annex?
Git is the most popular version control system for software development1. It is a distributed content management system, specifically tuned towards managing and collaborating on text files, and excels at making all committed content reliably and efficiently available to all clones of a repository. At the same time, Git is not designed to efficiently handle large (e.g., over a gigabyte) or binary files (see, e.g., Kenlon, 2016). This makes it hard or impossible to use Git directly for distributed data storage with tailored access to individual files. Git-annex takes advantage of Git’s ability to efficiently manage textual information to overcome this limitation. File content handled by git-annex is placed into a managed repository annex, which avoids committing the file content directly to Git. Instead, git-annex commits a compact reference, typically derived from the checksum of a file’s content, that enables identification and association of a file name with the content. Using these identifiers, git-annex tracks content availability across all repository clones and external resources such as URLs pointing to individual files on the web. Upon user request, git-annex automatically manages data transport to and from a local repository annex at a granularity of individual files. With this simple approach, git-annex enables separate and optimized implementations for identification and transport of arbitrarily large files, using an extensible set of protocols, while retaining the distributed nature and compatibility with versatile workflows for versioning and collaboration provided by Git.
What does DataLad add to Git and git-annex?
Easy to use modularization.
Research workflows impose additional demands for an efficient research data management platform besides version control and data transport. Many research datasets contain millions of files, but a large number of files precludes managing such a dataset in a single Git repository, even if the total storage demand is not huge. Partitioning such datasets into smaller, linked components (e.g., one subdataset per sample in a dataset comprising thousands) allows for scalable management. Research datasets and projects can also be heterogeneous, comprising different data sources or evolving data across different processing stages, and with different pace. Beyond scalability, modularization into homogeneous components also enables efficient reuse of a selected subset of datasets and for recording a derivative relationship between datasets. Git’s submodule mechanism provides a way to nest individual repositories via unambiguously versioned linkage, but Git operations must still be performed within each individual repository. To achieve modularity without impeding usability, DataLad simplifies working with the resulting hierarchies of Git repositories via recursive operations across dataset boundaries. With this, DataLad provides a “monorepo”-like user experience in datasets with arbitrarily deep nesting, and makes it trivial to operate on individual files deep in the hierarchy or entire trees of datasets. A testament of this is datasets.datalad.org, created as the project’s initial goal to provide a data distribution with unified access to already available public data archives in neuroscience, such as crcns.org and openfmri.org. It is curated by the DataLad team and provides, at the time of publication, streamlined access to over 260 TBs of data across over 5,000 subdatasets from a wide range of projects and dozens of archives in a fully modularized way.
Re-executable annotation of changes.
Digital provenance is crucial for the trustworthiness and reproducibility of a research result, and contributes to the reusability aspect of the FAIR principles (Wilkinson et al., 2016). Knowing which code and data were used is essential, but, for changes that are programmatically introduced, how a command or script was invoked is another key piece of information to capture. One approach is to include this information in the Git commit message that accompanies a change, but doing so manually is tedious and error prone. To solve this, DataLad supports executing a command and automatically generating a commit message that includes a structured record with comprehensive details on the invocation. In addition to providing reliable information about past command-line invocations, these machine-readable records make it possible to easily re-execute commands (e.g., to verify if a result is computationally reproducible or to apply an analog change to a different dataset state).
Targeted interfaces and interoperability adapters.
Interoperability with scientific or commercial computing and storage services allows researchers to integrate data management routines into their established workflows with minimal friction. Git can already interact with other local or remote repositories via standard or custom network transport protocols. DataLad implements support for additional services that require custom protocols, such as the Open Science Framework (OSF) (Hanke, Poldrack, et al., 2021). Git-annex readily provides access to a wide range of external data storage resources via a large set of protocols. DataLad builds on this support and adds, for example, more fine-grained access (e.g. direct access to individual components contained in an archive hosted on cloud storage) or specialized services, such as XNAT (http://www.xnat.org/). Efficient and seamless access to scientific data is implemented using the special remote protocol provided by git-annex (Hess, 2013), through which external tools, like DataLad, can provide custom transport functionality transparently to a user. With this approach, DataLad and other projects can jointly facilitate access to an ever-growing collection of resources (Hess, 2011) and overcome technological limitations of storage solutions, like file size or inode limits.
Metadata management.
Metadata are essential for scientific discovery, as they are routinely used to complete all data analyses. Metadata is the core concept behind Git and git-annex functioning: Git records and uses metadata about each change (author, date, description, original state, etc) for each commit. Git-annex manages metadata about content availability and allows to associate additional arbitrary key-value pairs to any annexed content. Files managed by git and git-annex can in turn be of standardized file formats comprised of data with rich metadata records. Moreover, entire repositories might conform to a standard (e.g., BIDS (Gorgolewski et al., 2016)) or provide a standardized dataset descriptor (e.g., Frictionless data package). To facilitate metadata availability and utility, DataLad provides an extensible framework for metadata extraction and aggregation. Metadata for each file (contained in the file or recorded by git and git-annex) or associated with the entire dataset can be extracted into a collection of machine-readable (JSON) records and aggregated across all contained sub-datasets. Such simple mechanism makes it possible to provide immediate access to metadata about all contained data within a larger super-dataset (such as datasets.datalad.org).
Overview of DataLad and its ecosystem
Design principles
Besides the free software nature and open processes of the DataLad project, the development of DataLad is guided by four principles to ensure its open and domain agnostic nature, to maximize the long-term utility of its datasets and to minimize users’ technical debt:
Datasets and the files they comprise are the only two recognized entities
A dataset is a Git repository with an optional annex
Minimization of custom procedures and data structures
Complete decentralization, with no required central server or service, but maximum interoperability with existing 3rd-party resources and infrastructure
In conjunction, these principles aim to reduce the risk of adoption for DataLad users. They foster the resilience of an ecosystem using DataLad datasets as a standard package format for any digital objects by avoiding any critical dependency on service deployments governed by central entities, and even on DataLad itself, for access to any resources managed with DataLad.
DataLad core
The datalad Python package provides both a Python library and a command line tool which expose core DataLad functionality to fulfill a wide range of decentralized RDM use cases for any domain. All DataLad commands operate on DataLad datasets. On a technical level, these datasets are Git repositories with additional metadata. On a conceptual level, they constitute an overlay structure that allows to version control files of any size, track and publish files in a distributed fashion, and record, publish, and execute actionable provenance of files and file transformations. Figure 1 summarizes key commands and concepts for local or distributed data and provenance management.
Figure 1:
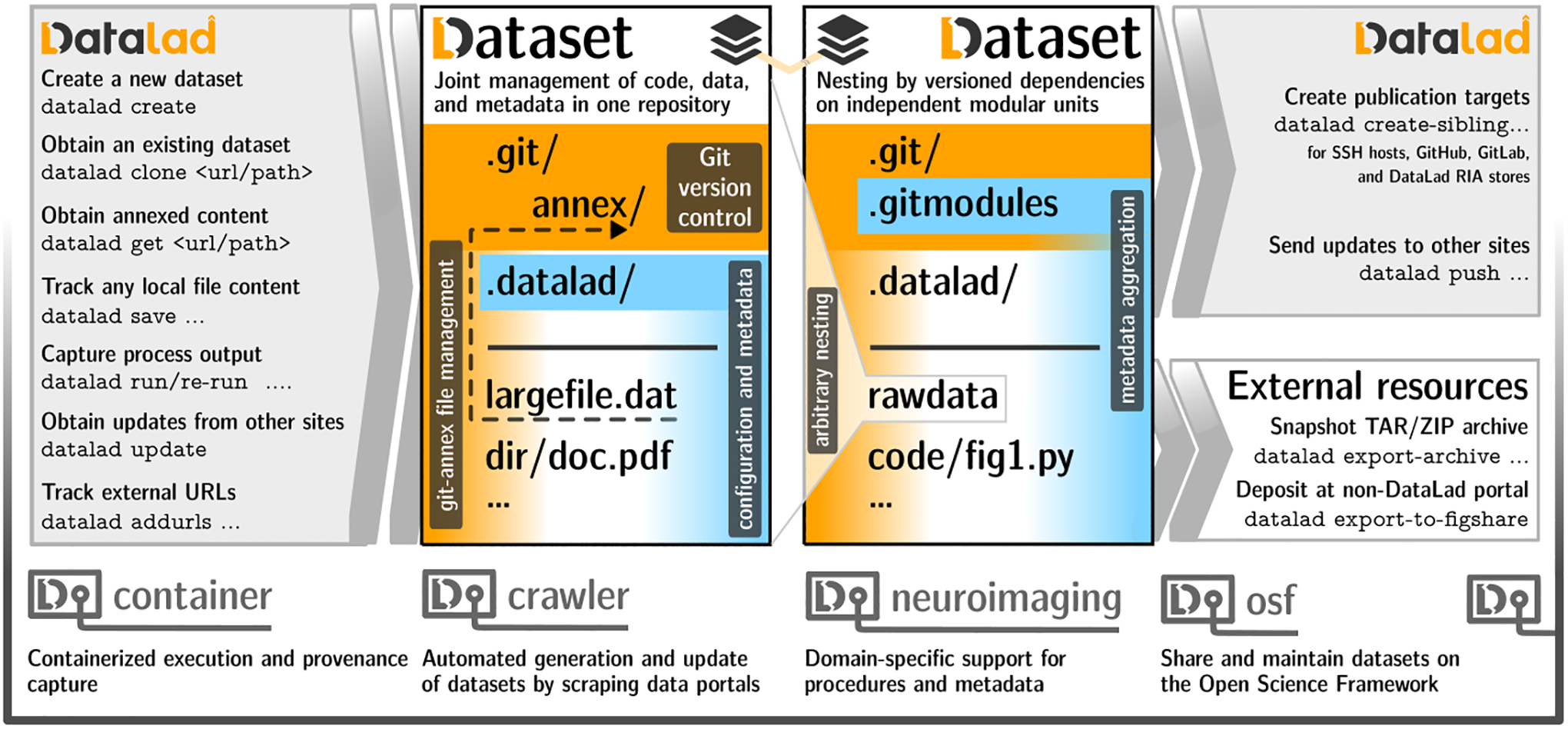
Schematic overview of a dataset, datasets nesting, and selected commands for content and dataset management. A more comprehensive cheatsheet is provided in the DataLad handbook (Wagner, 2020).
DataLad’s features can be flexibly integrated into standard scientific workflows. For example, by using the concept of dataset nesting to modularize the evolution of a research project, DataLad can fulfill the YODA principles for reproducible science (YODA Team, 2021), and, with this simple paradigm, facilitate efficient access, composition, scalability, reuse, sharing, and reproducibility of results (see Figure 2). With core commands that aim to simplify operation of the underlying tools, DataLad makes RDM workflows more accessible to novices and experts alike. Importantly, compatibility with all Git/git-annex functionality is retained.
Figure 2:
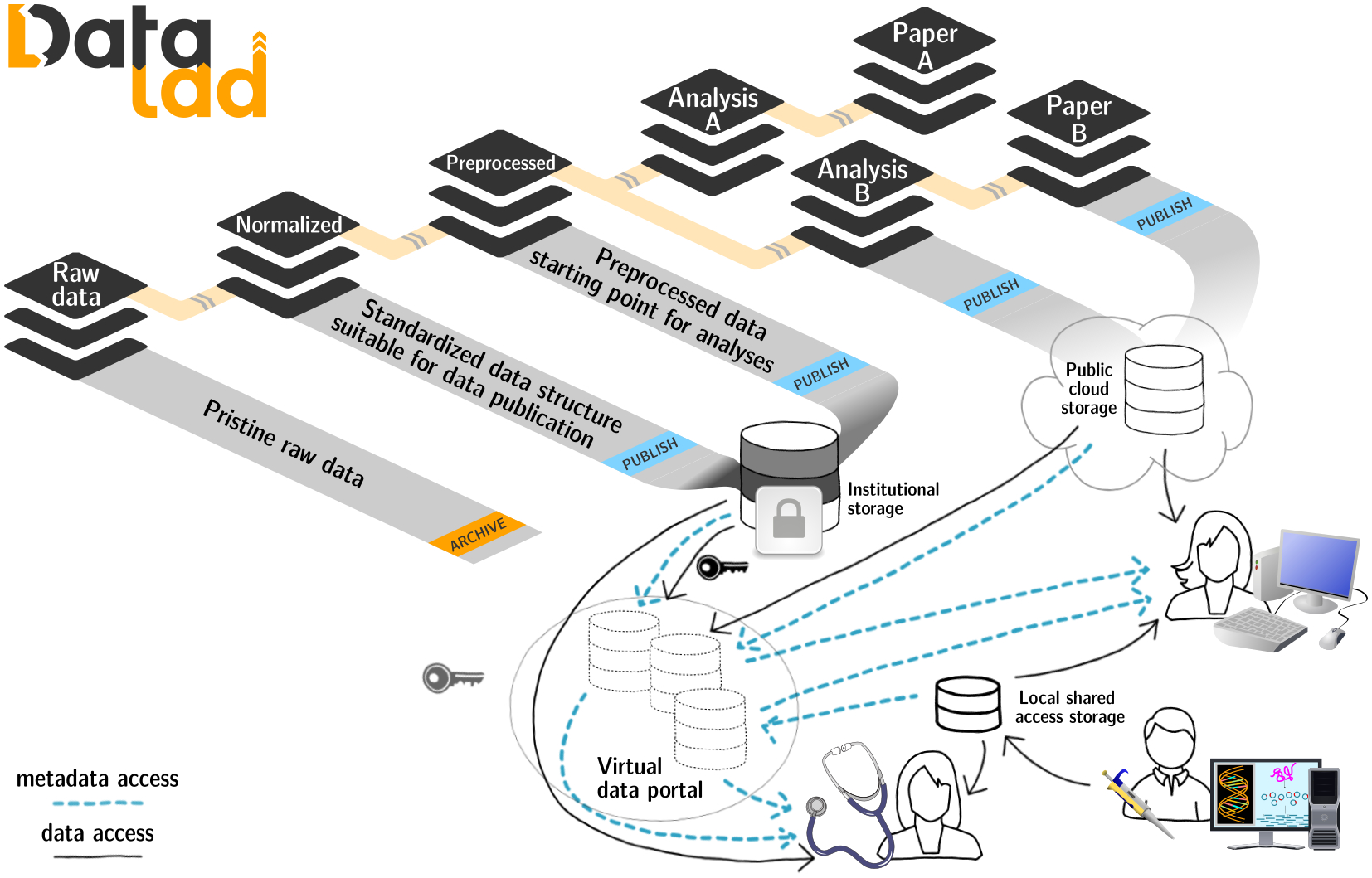
DataLad datasets are reusable modular components, which can be nested to establish a complete provenance trail all the way from a publication to the original data. Various access schemes to datasets and data are provided, and further extensibility is a key architectural property.
Extensions
Like Git and git-annex, DataLad core is a generic tool that is not specifically tuned to particular data types or use cases. It offers a robust foundation to build more specialized solutions on top of. DataLad extensions, stand-alone Python packages with additional DataLad functionality, extend DataLad with domain-focused or technology-specific features. A dedicated datalad-extension-template repository provides a starting point for creating new DataLad extensions. Some established extensions include:
datalad-container (Meyer et al., 2021) to simplify management and use of Docker and Singularity containers typically containing complete computational environments
datalad-crawler (Halchenko et al., 2021) to automate creation and updates of DataLad datasets from external resources
datalad-neuroimaging (Hanke et al., 2020) to provide neuroimaging-specific procedures and metadata extractors
datalad-osf (Hanke, Poldrack, et al., 2021) to collaborate using DataLad through the Open Science Framework (OSF)
datalad-ukbiobank (Hanke, Waite, et al., 2021) obtain and BIDS-normalize imaging data releases of the UKBiobank
The same mechanism of extensions is used for rapid development of new functionality to later be moved into the core tool (e.g., datalad-metalad). The datalad-extensions repository provides a list of extensions and continuous integration testing of their released versions against released and development versions of the DataLad core.
External uses and integrations
DataLad can be used as an independent tool to access and manage data (see e.g. Wittkuhn & Schuck (2021), Gautheron et al. (2021), Gautheron (2021)) or as a core technology behind another tool or a larger platform (e.g. Far et al. (2021)). TemplateFlow (Ciric et al., 2021) uses DataLad for the management of neuroimaging templates. OpenNeuro uses DataLad for data logistics with data deposition to a public S3 bucket. CONP-PCNO adopts aforementioned features for modular composition and nesting to deliver a rich collection of datasets with public or restricted access to data. ReproMan integrates with DataLad to provide version control and data logistics. www.datalad.org/integrations.html provides a more complete list of DataLad usage and integration with other projects, and Hanke, Pestilli, et al. (2021) provides a systematic depiction of DataLad as a system for decentralized RDM used by a number of projects.
Documentation
Developer-focused technical documentation at docs.datalad.org, with detailed descriptions of the command line and Python interfaces, is automatically generated from the DataLad core repository. A comprehensive handbook (Wagner et al., 2021a) provides user-oriented documentation with an introduction to research data management, and numerous use case descriptions for novice and advanced users of all backgrounds (Wagner et al., 2021b).
Installation
The handbook provides installation instructions for all major operating systems. DataLad releases are distributed through PyPI, Debian, NeuroDebian, brew, and conda-forge. The datalad-installer (also available from PyPI) streamlines the installation of DataLad and its dependencies, in particular git-annex, across a range of deployment scenarios, such as continuous integration systems, or high-performance computing (HPC) environments.
Development
DataLad has been developed openly in a public repository (github.com/datalad/datalad) since its inception in 2013. At the time of this publication, the repository amassed over 13.5k commits, 2.5k merged PRs, and 2.3k closed (+700 open) issues from over 30 contributors. Issue tracker, labels, milestones, and pull requests are used to coordinate development. The development process of DataLad is not isolated from its foundational building blocks. For every new feature or bug fix the most appropriate software layer is determined to maximize the size of the benefitting user base and, importantly, also the associated developer audience. This strategy aims to achieve a robust integration with the larger open source software ecosystem, and simultaneously minimize the total technical debt carried solely by the DataLad development team. Consequently, DataLad development is tightly connected to and involves frequent communication with the git-annex project and its main developer Joey Hess (Hess & DataLad Team, 2016). To guarantee robust operation across various deployments, DataLad heavily utilizes continuous integration platforms (Appveyor, GitHub actions, and Travis CI) for testing DataLad core, building and testing git-annex (in a dedicated github.com/datalad/git-annex), and integration testing with DataLad extensions (datalad-extensions).
Contributions
DataLad is free and open source software and encourages unconstrained use and reuse in any context. Therefore, DataLad is released under DFSG- and OSI-compliant MIT/Expat license. License terms for reused components in the code-base are provided in the COPYING file. The project aims to promote contributions rather than detached developments in forks and anyone is highly welcome to contribute to DataLad in any form under these terms. Technical and procedural guidelines for such contributions can be found in the CONTRIBUTING.md file shipped within DataLad’s source repository. Contributors are acknowledged on the project website, and also credited in the form of co-authorship in the Zenodo-based archival of software releases. All co-authors of this paper as well as the contributors acknowledged below have added to the project with code- or non-code-based contributions, and we thank past, present, and future contributors of this community for their involvement and work.
Supplementary Material
Acknowledgements
We express our gratitude to Joey Hess for the development and maintenance of git-annex, and for years of productive collaboration with the DataLad team. We would like to extend our gratitude to Joey Zhou, Matteo Visconti di Oleggio Castello, John T. Wodder II, Satya Ortiz-Gagné, Jörg Stadler, Andrew Connolly, John Lee, Nolan Nichols, Elizabeth DuPre, Cécile Madjar, Gergana Alteva, Timo Dickscheid, Alex Waite for notable contributions to the codebase, bug reports, recommendations, and promotion of DataLad.
DataLad development was made possible thanks to support by NSF 1429999, 1912266 (PI: Halchenko) and BMBF 01GQ1411, 01GQ1905 (PI: Hanke) through the CRCNS program. It received significant contributions from ReproNim 1P41EB019936-01A1 (PI: Kennedy) and DANDI 5R24MH117295-02 (PIs: Ghosh, Halchenko) NIH projects. It also received contributions from the Canadian Open Neuroscience Platform and the NeuroHub (Co-PI: Poline) projects thanks in part to funding from a Brain Canada Platform Support Grant Competition Award in addition to funds and in-kind support from sponsor organizations, and from the Canada First Research Excellence Fund, awarded through the Healthy Brains, Healthy Lives initiative at McGill University, and the Brain Canada Foundation with support from Health Canada. This development was supported by the European Regional Development Fund (Project: Center for Behavioral Brain Sciences Magdeburg, Imaging Platform, PI: Hanke), the European Union’s Horizon 2020 research and innovation programme under grant agreements Human Brain Project (SGA3, H2020-EU.3.1.5.3, grant no. 945539; Co-Investigators: Eickhoff, Hanke), and Virtual Brain Cloud (H2020-EU.3.1.5.3, grant no. 826421; PI: Eickhoff), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under grants SFB 1451 (431549029; Co-PIs: Eickhoff, Hanke) and IRTG 2150 (269953372; Co-PIs: Eickhoff, Hanke).
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- Ciric R, Lorenz R, Thompson W, Goncalves M, MacNicol E, Markiewicz C, Halchenko Y, Ghosh S, Gorgolewski K, Poldrack R, & Esteban O (2021). TemplateFlow: A community archive of imaging templates and atlases for improved consistency in neuroimaging. 10.21203/rs.3.rs-264855/v1 [DOI] [Google Scholar]
- Far MS, Stolz M, Fischer JM, Eickhoff SB, & Dukart J (2021). JTrack: A digital biomarker platform for remote monitoring in neuropsychiatric and psychiatric diseases. CoRR, abs/2101.10091. https://arxiv.org/abs/2101.10091 [Google Scholar]
- Gautheron L (2021). The LAAC superdataset: Datasets of infant day-long recordings. https://github.com/LAAC-LSCP/datasets
- Gautheron L, Rochat N, & Cristia A (2021). Managing, storing, and sharing long-form recordings and their annotations. 10.31234/osf.io/w8trm [DOI] [Google Scholar]
- Gorgolewski KJ, Auer T, Calhoun VD, Craddock RC, Das S, Duff EP, Flandin G, Ghosh SS, Glatard T, Halchenko YO, Handwerker DA, Hanke M, Keator D, Li X, Michael Z, Maumet C, Nichols BN, Nichols TE, Pellman J, … Poldrack RA (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data, 3(1). 10.1038/sdata.2016.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halchenko Y, Hanke M, Meyer K, Olson T, Chaselgrove, & Poldrack B (2021). Datalad-crawler: DataLad extension for crawling external resources. Zenodo. 10.5281/ZENODO.2558512 [DOI] [Google Scholar]
- Hanke M, Halchenko Y, Poldrack B, & Meyer K (2020). datalad-neuroimaging: DataLad extension for neuroimaging. Zenodo. 10.5281/ZENODO.3874225 [DOI] [Google Scholar]
- Hanke M, Pestilli F, Wagner AS, Markiewicz CJ, Poline J-B, & Halchenko YO (2021). In defense of decentralized research data management. Neuroforum, 27(1). 10.1515/nf-2020-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke M, Poldrack B, Wagner AS, Huijser D, Sahoo AK, Boos M, Steinkamp SR, Guenther N, & Appelhoff S (2021). datalad-osf: DataLad extension for integration with OSF.io. Zenodo. 10.5281/ZENODO.3900277 [DOI] [Google Scholar]
- Hanke M, Waite LK, Poline J-B, & Hutton A (2021). DataLad extension for working with the UKbiobank (Version 0.3.3) [Computer software]. Zenodo. 10.5281/zenodo.4773629 [DOI] [Google Scholar]
- Hess J (2011). Git-annex: Special remotes. https://git-annex.branchable.com/special_remotes/
- Hess J (2013). Git-annex: External special remote protocol. https://git-annex.branchable.com/design/external_special_remote_protocol/
- Hess J, & DataLad Team. (2016). Git-annex: DataLad project - bug and todo report. https://git-annex.branchable.com/projects/datalad
- Kenlon S (2016). How to manage binary blobs with git. https://opensource.com/life/16/8/how-manage-binary-blobs-git-part-7
- Meyer K, Hanke M, Halchenko Y, Poldrack B, & Wagner A (2021). datalad-container: DataLad extension for working with computational containers. Zenodo. 10.5281/ZENODO.2431914 [DOI] [Google Scholar]
- Wagner AS (2020). The DataLad Handbook: Cheatsheet. http://handbook.datalad.org/r.html?cheatsheet [Google Scholar]
- Wagner AS, Waite LK, Meyer K, Heckner MK, Kadelka T, Reuter N, Waite AQ, Poldrack B, Markiewicz CJ, Halchenko YO, Vavra P, Chormai P, Poline J-B, Paas LK, Herholz P, Mochalski LN, Kraljevic N, Wiersch L, Hutton A, … Hanke M (2021a). The DataLad Handbook. Zenodo. 10.5281/ZENODO.4495560 [DOI] [Google Scholar]
- Wagner AS, Waite LK, Meyer K, Heckner MK, Kadelka T, Reuter N, Waite AQ, Poldrack B, Markiewicz CJ, Halchenko YO, Vavra P, Chormai P, Poline J-B, Paas LK, Herholz P, Mochalski LN, Kraljevic N, Wiersch L, Hutton A, … Hanke M (2021b). The DataLad Handbook: Use Cases. http://handbook.datalad.org/usecases/intro.html
- Wilkinson MD, Dumontier M, Aalbersberg Ij. J., Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, … Mons B (2016). The FAIR guiding principles for scientific data management and stewardship. Scientific Data, 3(1). 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkuhn L, & Schuck NW (2021). Dynamics of fMRI patterns reflect sub-second activation sequences and reveal replay in human visual cortex. Nature Communications, 12(1). 10.1038/s41467-021-21970-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- YODA Team. (2021). YODA: README. https://github.com/myyoda/myyoda [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


