Abstract
The rubella virus (RV) structural proteins capsid, E2, and E1 are synthesized as a polyprotein precursor. The signal peptide that initiates translocation of E2 into the lumen of the endoplasmic reticulum remains attached to the carboxy terminus of the capsid protein after cleavage by signal peptidase. Among togaviruses, this feature is unique to RV. The E2 signal peptide has previously been shown to function as a membrane anchor for the capsid protein. In the present study, we demonstrate that this domain is required for RV glycoprotein-dependent localization of the capsid protein to the juxtanuclear region and subsequent virus assembly at the Golgi complex.
Rubella virus (RV) is the sole member of the genus Rubivirus within the family Togaviridae. The virus is a human pathogen and causes a mild self-limiting disease that is characterized by low-grade fever and a rash in adults. However, RV infection during the first trimester of pregnancy often results in a characteristic pattern of severe birth defects in the fetus collectively known as congenital rubella syndrome.
The structure and replication of RV have been studied extensively (for reviews, see references 5 and 20). Virions consist of a host-derived membrane, three virus-encoded structural proteins, and a single molecule of 40S genomic RNA. The virus genome serves as an mRNA for translation of the nonstructural proteins, whereas the structural proteins are translated as a polyprotein precursor from a 24S subgenomic RNA (16, 17). The structural proteins are the RNA-binding protein, capsid protein, and membrane glycoproteins E2 and E1. The structural polyprotein precursor contains two signal peptides (SPs) that are necessary for directing translocation of the glycoproteins into the lumen of the endoplasmic reticulum (ER); two signal peptidase-mediated cleavages within the polyprotein produce the three structural proteins capsid, E2, and E1 (7, 10). The E2 SP remains attached to the carboxy terminus of the capsid protein after cleavage by the signal peptidase. Retention of the hydrophobic SP at the carboxy terminus of capsid proteins is unique to RV among the members of the Togaviridae and may have functional consequences. Indeed, SPs are known to have a variety of other functions in addition to initiating translocation of their cognate proteins into the ER (15). In this particular case, the E2 SP can function as a membrane anchor for the capsid protein and it has been suggested that this is important for the membrane-dependent assembly of nucleocapsids (19). However, this has yet to be proved experimentally. In the present study, we have investigated whether the E2 SP has functions in RV assembly apart from initiating translocation of E2 into the ER.
Cells expressing the RV structural proteins have been shown to assemble and secrete RV-like particles (RLPs) which are virtually indistinguishable from RV virions in terms of morphology and antigenicity (9). Accordingly, RLPs have proved to be a useful model system with which to study RV assembly (6). In order to assay the importance of E2 SP in virus assembly and secretion, COS cells were transiently cotransfected with plasmids encoding capsid proteins with or without E2 SP, CapE2SP and CapΔSP, respectively (Fig. 1), and glycoproteins E2 and E1 (8). RLP secretion was detected by using an immunoblot-based assay (6). CapE2SP and CapΔSP were constructed by PCR amplification with Pwo polymerase (Roche Molecular Biochemicals, Laval, Quebec, Canada), using primers containing EcoRI or BglII sites (underlined). The forward primer capsidF (5′ CGCGAATTCATGGCTTCCACTACCC 3′) was used in combination with capsidR (5′ GGTCAGATCTCTAGGCGCGCGCGGTGC 3′) or CapΔSPR (5′ ACTGAGATCTAGCGGATGCGCCAAGGATG 3′) to produce cDNA products encoding CapE2SP and CapΔSP, respectively. PCR products were digested with EcoRI and BglII and then ligated into the mammalian expression vector pCMV5 (1). The authenticity of each construct was confirmed by DNA sequencing. COS cells (1.5 × 105) in 35-mm-diameter culture dishes were transfected with 1 μg of each plasmid combined with 5 μl of Fugene 6 transfection reagent (Roche Molecular Biochemicals). Forty-eight hours posttransfection, media from the transfected cells were precleared by centrifugation at 10,000 × g to remove cell-associated material, followed by a second centrifugation at 100,000 × g for 1 h to pellet RLPs (6). In parallel, lysates were prepared from the transfected cells in order to demonstrate capsid protein expression at the cellular level. Cell lysates and 100,000 × g pellets were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with anticapsid antibody (2). When cells were cotransfected with plasmids encoding CapE2SP and E2E1, capsid protein was detected in the cell lysates and 100,000 × g medium pellets (Fig. 2, lanes 1 and 2). The presence of capsid protein in the 100,000 × g medium pellets indicated that RLPs were assembled and secreted from the cells (6). Cells expressing E2E1 and capsid protein lacking the E2 SP (CapΔSP) produced high levels of capsid protein which were detectable in cell lysates but not in the 100,000 × g medium pellets (Fig. 2, lanes 7 and 8). The E2E1 construct also contains the E2 SP (Fig. 1) (8), and we confirmed that processing of E2 and E1 occurred normally in the doubly transfected cells (data not shown). These results indicate that the presence of an SP on the capsid protein is required for RLP secretion.
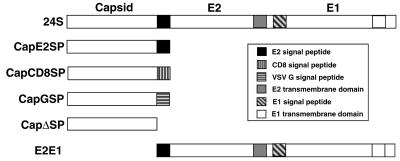
FIG. 1.
Schematic of RV protein constructs. The 24S cDNA encodes all three RV structural proteins in the order capsid-E2-E1. The rest of the constructs are named according to the proteins and heterologous domains that they encode. For example, CapCD8SP encodes capsid protein with the CD8 SP attached to its carboxy terminus. The E2E1 construct encodes RV E2 and E1 and contains the SP from E2. SPs and transmembrane domains are indicated by differently shaded boxes.
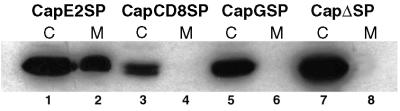
FIG. 2.
The E2 SP is required for RLP secretion. Capsid protein constructs with an E2, VSV G, or CD8 SP were cotransfected with the E2E1 expression plasmid into COS cells. Forty-eight hours posttransfection, media from the transfected cells were precleared of cell-associated material and then the medium samples (M) were subjected to centrifugation at 100,000 × g to pellet RLPs. Cell lysates (C) were also prepared from the transfected cells. Lysates (C) and membrane pellets (M) were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membranes, and immunoblotted with anticapsid antibody, followed by enhanced chemiluminescence detection. Capsid proteins which are incorporated into secreted RLPs are detected in the membrane pellet fractions.
The experiments shown in Fig. 2 demonstrated that deletion of the E2 SP from the capsid protein abrogates secretion of RLPs; however, they did not address whether this domain functions simply as a membrane anchor or if it has an additional role in virus assembly and/or secretion. If the former were true, SPs from non-RV glycoproteins should be able to functionally replace the E2 SP. To determine if capsid proteins containing heterologous SPs could function in virus assembly, the SPs from two other type I membrane glycoproteins, CD8 and vesicular stomatitis virus (VSV) G, were fused onto the carboxy terminus of the capsid protein in place of the E2 SP to create CapCD8SP and CapGSP, respectively (Fig. 1). The construction of CapCD8SP is described elsewhere (4), and CapGSP was generated by using the megaprimer and PCR overlap methods as previously described (13, 18). The primers 5′ CCATCCTTGCGCATCCGCATGAAGTGCCTTTTGTACTTAG 3′ and 5′ ATATCAGCGCGGGGCTGGAGCCCGCAATTCACCCCAATGAATAA 3′ were used in a PCR to create a cDNA that encodes the carboxy terminus of the capsid protein fused to the VSV G SP sequence. This PCR product was then used as a megaprimer in combination with the primers 5′ CGCGAATTCATGGCTTCCACTACCC 3′ or 5′ CCGACGCGCAAGGTGC 3′ in two separate PCRs by using the RV 24S cDNA as template. The products of these two PCRs were combined, and PCR overlap extension was used to produce the final product, which was subcloned into the 24S cDNA using EcoRI and BstEII. Like the CapE2SP construct, CapGSP and CapCD8SP contain stop codons immediately following the SP sequences.
The CD8 and VSV G SPs are 21 and 16 amino acids in length, respectively, whereas the E2 SP is 23 amino acids long. However, all three of these hydrophobic peptides should be long enough to span the ER and Golgi membranes to function as transmembrane domains (3). Plasmids encoding the capsid constructs were transiently cotransfected with the E2E1 plasmid into COS cells, and RLP secretion was assayed as described above. RLP secretion was observed only in cells expressing CapE2SP and E2E1, despite the fact that relatively high levels of capsid proteins were detected in all cell lysates (Fig. 2).
There are two obvious possible reasons to account for the failure of cells expressing CapGSP and CapCD8SP to secrete RLPs. The first possibility is that these capsid chimeras do not stably associate with membranes and are therefore unable to support RLP assembly. An alternative explanation is that the E2 SP interacts with other virus components in a sequence-specific manner during assembly and/or secretion. To address the first hypothesis, we used a previously described membrane copelleting assay (19) to determine whether the chimeric capsid proteins could stably associate with membranes. Briefly, 35S-labeled capsid proteins were synthesized in vitro by a coupled transcription/translation system (TnT; Promega, Madison, Wis.) either in the presence or absence of dog pancreatic microsomes. Ten percent of the reaction mixtures (5 μl) were extracted on ice with 50 mM sodium carbonate (50 μl) for 30 min, followed by centrifugation through a sucrose cushion (50 μl) containing 0.2 M sucrose, 30 mM HEPES (pH 11.5), 150 mM potassium acetate, 2.5 mM magnesium acetate, and 1 mM dithiothreitol in a Beckman Airfuge set at 25 lb/in2. The supernatants and the pelleted membrane fractions were subjected to SDS-PAGE and fluorography. Under these conditions, only capsids that are stably associated with microsomes are recovered in the pellet fractions.
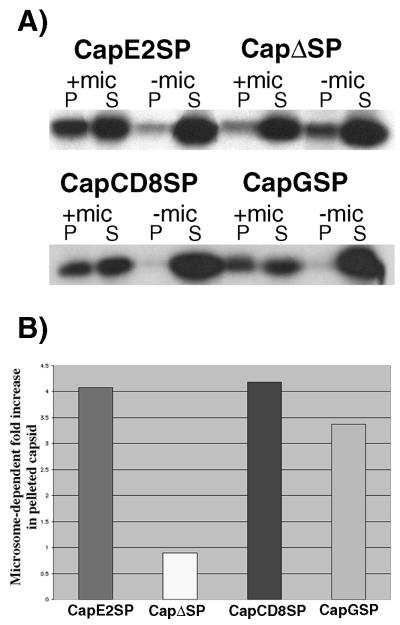
Figure 3A illustrates the results from a representative membrane-copelleting assay. Quantitation of the capsid protein bands by densitometry indicated that translation in the presence of microsomes results in a 3.4- to 4.1-fold increase in the amounts of membrane-associated capsid proteins for all three constructs that contain SPs but not for those containing CapΔSP (Fig. 3B). Our results are similar to those of Suomalainen et al. (19), who showed that 38 and 11%, respectively, of capsid proteins with and without the E2 SP stably associate with microsomes under these conditions. Moreover, these experiments show that replacement of E2 SP with CD8 or VSV G SPs did not decrease the ability of the capsid protein to stably associate with membranes.
FIG. 3.
Capsids with heterologous SPs stably associate with membranes in vitro. Capsid protein constructs with an E2, VSV G, or CD8 SP were transcribed and translated in vitro either in the presence or in the absence of microsomes (mic). Samples were extracted with 50 mM sodium carbonate (pH 11.5), and the membranes were pelleted through a sucrose cushion by using a Beckman airfuge set at 25 lb/in2. The supernatant (S) and the pelleted membranes (P) were subjected to SDS-PAGE and fluorography. (A) Fluorographs from a representative experiment are shown. (B) Graphic representation of membrane-associated capsid proteins (average of two independent experiments). The y axis represents the fold increase in the proportion of pelleted capsid proteins translated in the presence of microsomes relative to those when translation was performed in the absence of microsomes.
In light of our recent work which demonstrated that alteration of the E1 membrane-spanning or cytoplasmic domains blocks secretion but not RLP assembly (6), we thought it would be important to determine whether the E2 SP is required at a pre- or post-virus assembly step. Transient expression of CapE2SP in CHO cells stably expressing E2 and E1 (CHO-E2E1) (11) resulted in the formation of RLPs that can be visualized by electron microscopy (Fig. 4A, arrowheads). RLPs are readily visible in the Golgi complex of cells expressing RV structural proteins by this method, whether they are secreted or not (6). This method was used to determine if coexpression of mutant capsid proteins in CHO-E2E1 cells resulted in formation of RLPs that are not secreted.
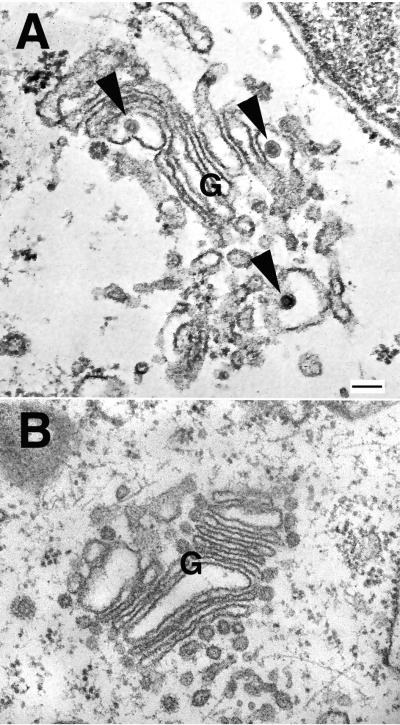
FIG. 4.
E2 SP is required for RLP assembly. CHO-E2E1 cells were transfected with expression vectors encoding different capsid protein constructs, and at 48 h posttransfection, cells were prepared for routine morphologic examination by embedding them in Epon. (A) Electron micrograph of CHO-E2E1 cells transfected with CapE2SP. RLPs (arrowheads) can be seen in the Golgi complex (G) of these cells. No RLPs were observed in the Golgi complex of CapCD8SP-transfected cells (B). Bar, 100 μm.
CHO-E2E1 cells were transfected with the four capsid constructs, and at 48 h posttransfection, cells were fixed with 2.5% glutaldehyde in 0.1 M cacodylate buffer (pH 7.4) and then pelleted at 2,400 × g for 4 min. Cell pellets were embedded in Epon and processed for routine morphologic changes as described previously (6). Transfection efficiencies were monitored by indirect immunofluorescence, using rabbit anticapsid antibodies. Routinely, >10% transfection efficiency was achieved for each capsid construct (data not shown). At least 100 cells from each sample were examined for the presence of RLPs in the Golgi cisternae or associated vesicles. In CHO-E2E1 cells transfected with CapE2SP (Fig. 4A), RLPs were seen in the Golgi complex of approximately 10% of the cells examined, thus confirming that the proportion of cells containing RLPs paralleled the transfection efficiency. No RLPs were detected in CHO-E2E1 cells transfected with CapCD8SP (Fig. 4B) or CapΔSP (data not shown). Among more than 100 cells analyzed, a single RLP was detected in a CHO-E2E1 cell transfected with CapGSP (data not shown). Together, these data argue that the E2 SP does not simply function as a membrane anchor but is also required for efficient virus assembly.
A recent study from this laboratory demonstrated that transport of RV structural proteins to the perinuclear region was required for efficient assembly of RLPs (6). When transport of one or more of the structural proteins was impaired, RLP assembly did not occur. Therefore, we decided to examine the subcellular localization of the different capsid constructs to determine if the E2 SP was required for transport of the capsid protein to the same perinuclear region in which glycoproteins E2 and E1 accumulate. COS cells grown on coverslips were transiently transfected with plasmids encoding the capsid protein, E2, and E1 (24S) or E2E1 plus different capsid constructs. Samples were processed for double-label indirect immunofluorescence at 24 h posttransfection by using rabbit anticapsid and mouse anti-E1 antibodies, as previously described (12). Primary antibodies were detected with fluorescein isothiocyanate-labeled donkey anti-rabbit immunoglobulin G (IgG) and Texas Red-labeled goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, Pa.), and cells were examined using a Zeiss 510 confocal microscope. A total of 20 optical sections (0.5 μm) was collected from each sample. Captured images were examined in the X-Y, X-Z, and Y-Z planes, and final images were processed using Adobe Photoshop 5.0.
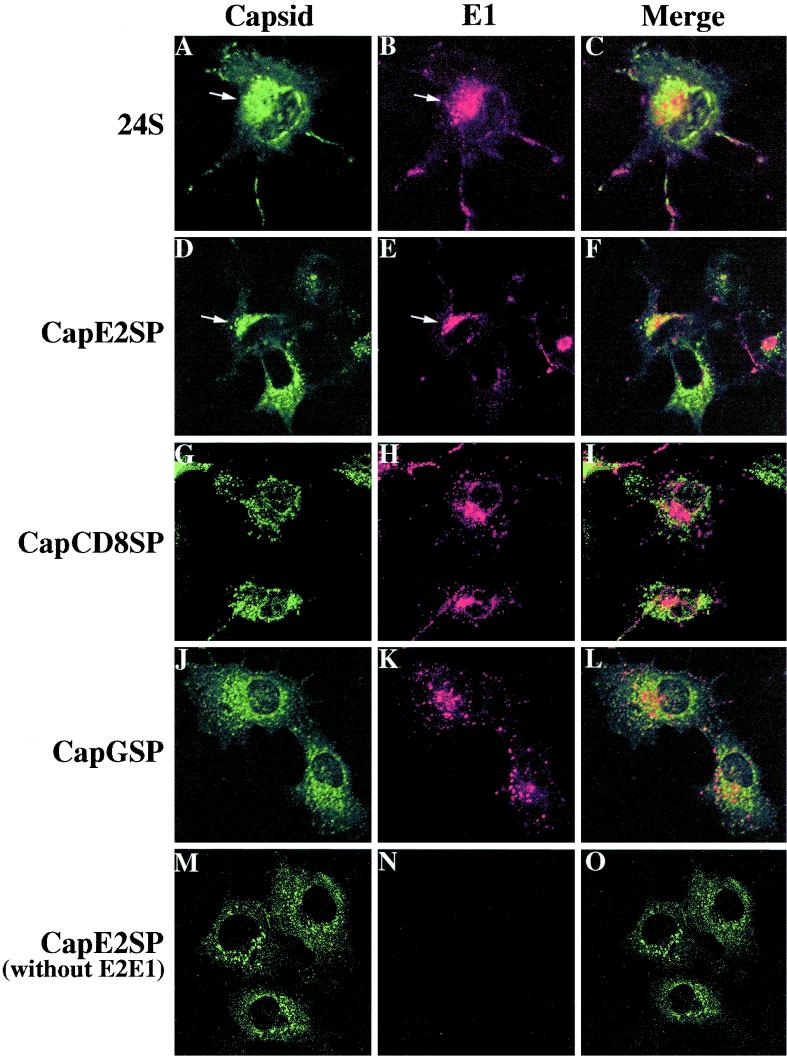
In cells expressing 24S cDNA (Fig. 5A through C, arrows) and CapE2SP plus E2E1 (Fig. 5D through F, arrows), the capsid protein was often localized in compact juxtanuclear structures. Significant but not complete colocalization with E1 was evident in these cells, suggesting that a pool of E1 and capsid proteins was accumulating at the same intracellular site (8, 9). The localization of CapE2SP to these compact juxtanuclear structures was dependent on coexpression of RV glycoproteins (Fig. 5M to O). The panels in Fig. 5 represent single optical sections from each sample shown in the X-Y orientation. Colocalization between capsid protein and E1 was also observed when X-Z and Y-Z sections were examined (data not shown). The lack of complete colocalization between capsid protein and E1 was not unexpected, since a significant proportion of capsid protein but not E1 or E2 associates with mitochondria (2, 14). In contrast to the situation with CapE2SP, CapCD8SP and CapGSP exhibited punctate or reticular staining throughout the cytoplasm and did not colocalize with E1 (Fig. 5G to L). Double staining of these cells with a mouse monoclonal antibody to the ER protein calnexin and rabbit anticapsid antibody revealed similar staining patterns, indicating that large portions of CapCD8SP and CapGSP were localized to membranes of the ER (reference 4 and data not shown). CapΔSP formed punctate structures in the cytoplasm that did not overlap with E1 or ER membranes (data not shown). Collectively, our data indicate that the E2 SP is required for E2- and E1-dependent recruitment of capsid protein to the perinuclear region.
FIG. 5.
Capsid proteins with heterologous SPs do not colocalize with E1 in the juxtanuclear region. COS cells were transfected with expression vectors that encode CapE2SP, CapGSP, or CapCD8SP and E2E1 (A through L). Panels M through O show cells transfected with CapE2SP only. Cells were fixed, permeabilized with methanol, and double labeled with rabbit anti-capsid (A, D, G, J, and M) and mouse anti-E1 (B, E, H, K, and N) antibodies and then examined by confocal microscopy. Optical sections (0.5 μm) from the X-Y plane are shown. Primary antibodies were detected with fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. The FITC channel is shown on the left (A, D, G, J, and M), and the Texas Red channel is shown in the middle (B, E, H, K, and N). The merged images are shown on the right (C, F, I, L, and O).
The well-studied alphaviruses have served as useful paradigms for understanding many aspects of RV biology, but the more that is learned about these two genera of viruses, the more it appears that they are quite different. In the present study we have investigated one of these differences, processing of the capsid protein and its role in virus assembly. Retention of an SP at the carboxy end of the RV capsid protein is unique within the Togaviridae. Experimental evidence has shown that this domain, in addition to initiating translocation of E2 into the ER, can also function as a membrane anchor (reference 19 and this study). Suomalainen et al. (19) hypothesized that this may provide the mechanism to account for the membrane-dependent assembly of RV nucleocapsids. Results from this study are certainly consistent with this notion and, in addition, clearly show that the E2 SP has an additional function at an early step in the RV assembly pathway. Specifically, this domain is necessary for E2- and E1-dependent targeting of capsid to the Golgi region where virus budding occurs. Previous studies by members of our laboratory demonstrated that the folding and transport of E2E1 heterodimers from the ER to the Golgi is a highly coordinated process whose rate-limiting step is the maturation of E1 in the ER (12). Presumably, E2 and E1 are transported from the ER to the Golgi in a COPII/COPI-dependent manner similar to that for other viral and cellular membrane proteins; however, nothing is known about how the capsid protein reaches the Golgi complex, which is the site of virus assembly. The indirect immunofluorescence data presented in this paper clearly show that the E2 SP is required for transport of the capsid protein to the Golgi region (Fig. 5), and it is tempting to speculate that an interaction between the E2 SP and one or more of the three other membrane-spanning domains on E2 or E1 directs capsid protein to the same ER-derived transport vesicles as the glycoproteins. This process would serve to coordinate transport of capsid protein to the Golgi with that of E2 and E1. Moreover, since E2 has a Golgi retention signal that functions to retain the glycoprotein heterodimer at the Golgi (13), capsid protein by virtue of its stable association with E2 and/or E1 would be prevented from traveling beyond the virus assembly site. Furthermore, interactions between the E2 SP and the transmembrane domains of E2 and/or E1 in the Golgi membranes may augment assembly of the virus. It will now be of interest to determine which of the RV glycoprotein transmembrane domains interacts with the E2 SP. Preliminary studies have so far failed to elucidate whether the E2 SP interacts preferentially with E2 or E1.
Acknowledgments
We are grateful to Chris Nicchitta for providing canine pancreatic microsomes.
This work was supported by a grant from the Canadian Institutes of Health Research. T.C.H. is the recipient of a Senior Medical Scholar award from the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 2.Beatch D M, Hobman T C. Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J Virol. 2000;74:5569–5576. doi: 10.1128/jvi.74.12.5569-5576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bretscher S M, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 4.Duncan R, Esmaili A, Law L J, Bertholet S, Hough C, Hobman T C, Nakhasi H L. Rubella virus capsid protein induces apoptosis in transfected RK13 cells. Virology. 2000;275:20–29. doi: 10.1006/viro.2000.0467. [DOI] [PubMed] [Google Scholar]
- 5.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbutt M, Law L J, Chan H, Hobman T C. Role of rubella virus glycoprotein domains in assembly of virus-like particles. J Virol. 1999;73:3524–3533. doi: 10.1128/jvi.73.5.3524-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hobman T C, Gillam S. In vitro and in vivo expression of rubella virus E2 glycoprotein: the signal peptide is located in the C-terminal region of capsid protein. Virology. 1989;173:241–250. doi: 10.1016/0042-6822(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 8.Hobman T C, Lundstrom M L, Gillam S. Processing and transport of rubella virus structural proteins in COS cells. Virology. 1990;178:122–133. doi: 10.1016/0042-6822(90)90385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobman T C, Lundstrom M L, Mauracher C A, Woodward L, Gillam S, Farquhar M G. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology. 1994;202:574–585. doi: 10.1006/viro.1994.1379. [DOI] [PubMed] [Google Scholar]
- 10.Hobman T C, Shukin R, Gillam S. Translocation of rubella virus glycoprotein E1 into the endoplasmic reticulum. J Virol. 1988;62:4259–4264. doi: 10.1128/jvi.62.11.4259-4264.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobman T C, Woodward L, Farquhar M G. The rubella virus E1 glycoprotein is arrested in a novel post-ER, pre-Golgi compartment. J Cell Biol. 1992;118:795–811. doi: 10.1083/jcb.118.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobman T C, Woodward L, Farquhar M G. The rubella virus E2 and E1 spike glycoproteins are targeted to the Golgi complex. J Cell Biol. 1993;121:269–281. doi: 10.1083/jcb.121.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobman T C, Woodward L, Farquhar M G. Targeting of a heterodimeric membrane protein complex to the Golgi: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol Biol Cell. 1995;6:7–20. doi: 10.1091/mbc.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J Y, Marshall J A, Bowden D S. Localization of rubella virus core particles in Vero cells. Virology. 1999;265:110–119. doi: 10.1006/viro.1999.0016. [DOI] [PubMed] [Google Scholar]
- 15.Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 16.Oker-Blom C. The gene order for rubella virus structural proteins is NH2-C-E2–E1-COOH. J Virol. 1984;51:354–358. doi: 10.1128/jvi.51.2.354-358.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oker-Blom C, Kalkkinen N, Kaariainen L, Pettersson R F. Rubella virus contains one capsid protein and three envelope glycoproteins, E1, E2a, and E2b. J Virol. 1983;46:964–973. doi: 10.1128/jvi.46.3.964-973.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 19.Suomalainen M, Garoff H, Baron M D. The E2 signal sequence of rubella virus remains part of the capsid protein and confers membrane association in vitro. J Virol. 1990;64:5500–5509. doi: 10.1128/jvi.64.11.5500-5509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolinsky J. Rubella. In: Fields B N, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 899–929. [Google Scholar]