SUMMARY
Rotavirus vaccines (RVV) protect against childhood gastroenteritis caused by rotavirus (RV) but have decreased effectiveness in low- and middle-income settings. This proof-of-concept, randomized-controlled, open-label trial tested if microbiome modulation can improve RVV immunogenicity. Healthy adults were randomized and administered broad-spectrum (oral vancomycin, ciprofloxacin, metronidazole), narrow-spectrum (vancomycin), or no antibiotics and then vaccinated with RVV, 21 per group per protocol. Baseline anti-RV IgA was high in all subjects. Although antibiotics did not alter absolute anti-RV IgA titers, RVV immunogenicity was boosted at 7 days in the narrow-spectrum group. Further, antibiotics increased fecal shedding of RV while also rapidly altering gut bacterial beta diversity. Beta diversity associated with RVV immunogenicity boosting at day 7 and specific bacterial taxa that distinguish RVV boosters and RV shedders were identified. Despite the negative primary endpoint, this study demonstrates that microbiota modification alters the immune response to RVV and supports further exploration of microbiome manipulation to improve RVV immunogenicity.
In Brief
Rotavirus vaccines (RVV) are less effective in poor-resourced settings. This randomized-controlled trial in adults tested the effect of microbiome modulation via broad-spectrum, narrow-spectrum, or no antibiotics on RVV performance. Absolute anti-RV IgA titer did not change. However, antibiotics resulted in higher day-7 boosting and increased RV-antigen shedding.
Graphical Abstract

INTRODUCTION
Rotavirus (RV) is one of the leading causes of severe gastroenteritis in children under 5 years of age in low-income countries (Tate et al., 2016). Rotavirus vaccines (RVV) have demonstrated high effectiveness in high-income countries; however, their efficacy and effectiveness is markedly lower in low-income countries in sub-Saharan Africa and Asia, where the majority of rotavirus-related deaths continue to occur (Jonesteller et al., 2017). Addressing this gap in vaccine effectiveness has become a public health priority, as even small improvements in rotavirus vaccine effectiveness could prevent tens of thousands of rotavirus-related deaths per year (PATH, 2006).
Numerous hypotheses have been postulated to explain the gap in RVV effectiveness between high- and low-income country settings, including histo-blood group antigen expression, maternal serum and breast milk antibody levels, concurrent oral polio virus vaccination, and enterovirus carriage (Velasquez et al., 2018). However, mounting evidence also supports a role for the intestinal microbiome in determining RVV immunogenicity. The intestinal bacterial microbiome is implicated in the pathogenesis of numerous enteric viruses, including poliovirus, norovirus, and murine rotavirus (Pfeiffer and Virgin, 2016). Gnotobiotic mice and mice treated with broad-spectrum antibiotics have reduced RV replication and increased late anti-RV antibody responses (Uchiyama et al., 2014). The bacterial microbiome also regulates intestinal immune responses. Early germfree mice research demonstrated that bacterial colonization is required for normal gut-associated lymphoid tissue (GALT) development, and specific microbiota can increase the secretion of IgA, known to be important in RV immunity (Benveniste et al., 1971). Murine studies also suggest that intestinal bacteria can serve as adjuvants in vaccination, as demonstrated by the capacity of flagellated bacteria to restore influenza antibody response following antibiotic treatment and influenza vaccination (Oh et al., 2014).
Human data also support a link between microbiome composition and RVV immunogenicity. These correlations are geography specific. We have demonstrated through two nested case-control studies that microbiome composition correlates significantly with RVV immunogenicity in human infants in low-income country settings: Gammaproteobacteria correlate with increased anti-RV IgA responses in an urban infant population in Asia, while Bacteroidetes species correlate with low anti-RV IgA responses in a rural population in sub-Saharan Africa (Harris et al., 2017a, 2017b). A recent retrospective case-control study in India showed no differences in microbiome composition between RVV responders and non-responders; however, RVV responders had significantly higher numbers of bacteria with pathogenic potential, primarily from the Proteobacteria phylum, prior to vaccination (Parker et al., 2018).
Extrapolating these findings to designing microbiome-based interventions to improve RVV immunogenicity for human infants is the logical and necessary extension of these studies. To date, investigators have chosen to employ empiric probiotic therapies as a way to alter the microbiome in human infants (Isolauri et al., 1995; Lazarus et al., 2018), with modest to no significant effect on RVV immunogenicity. However, these empiric probiotic approaches do not incorporate existing epidemiologic data correlating microbiome composition with vaccine response into their approaches. They also do not answer the critical question of whether the microbiome is causally linked to RVV immunogenicity and, if it is linked, how. Studies addressing these questions in humans are sorely lacking. This current trial was therefore designed as a proof-of-concept study, hypothesizing that if the bacterial microbiome is causal in determining RVV immunogenicity, then prospective alteration of the microbiome, specifically amplification of Gammaproteobacteria and reduction of Bacteroidetes (as observed in infant studies in low-income countries), should boost anti-RV immune responses in humans. Previous human studies have shown that, when given to adults, oral vancomycin significantly depletes the phylum Bacteroidetes and increases the phylum Proteobacteria (Isaac et al., 2017). While vancomycin also effects other changes in the microbiota, such as reduction in Firmicutes abundance, this antibiotic most closely achieves the microbiome changes we sought to elicit. Though the ideal study population would be low-income-country infants, we considered a proof-of-concept antibiotic intervention study to be unethical in children. Additionally, if alterations in RVV immunogenicity could be elicited in an adult population (known to have a higher threshold for RV illness and immunity; Ward et al., 1989) while simulating some of the bacterial microbiome phenotypes correlating with RVV immunogenicity in infants in low-income countries, they would likely remain relevant to an infant population. We therefore selected to alter the microbiota in adults prior to rotavirus vaccination: with broad-spectrum antibiotics—a positive control expected to deplete most bacterial taxa in the intestine; with vancomycin—expected to decrease Bacteroidetes and increase Proteobacteria; or with no antibiotics— a negative control. Pneumococcal polysaccharide (a T cell-independent) vaccine and tetanus toxoid (a T cell-dependent) vaccine were administered intramuscularly as immunologic controls and given their predictable and well-described antibody responses following adult immunization (Orange et al., 2012). Should RVV immunogenicity differ between arms, we could both establish a causative role for the microbiome in RV vaccination and identify promising microbial candidates for augmentation of RVV protection in infant populations.
RESULTS
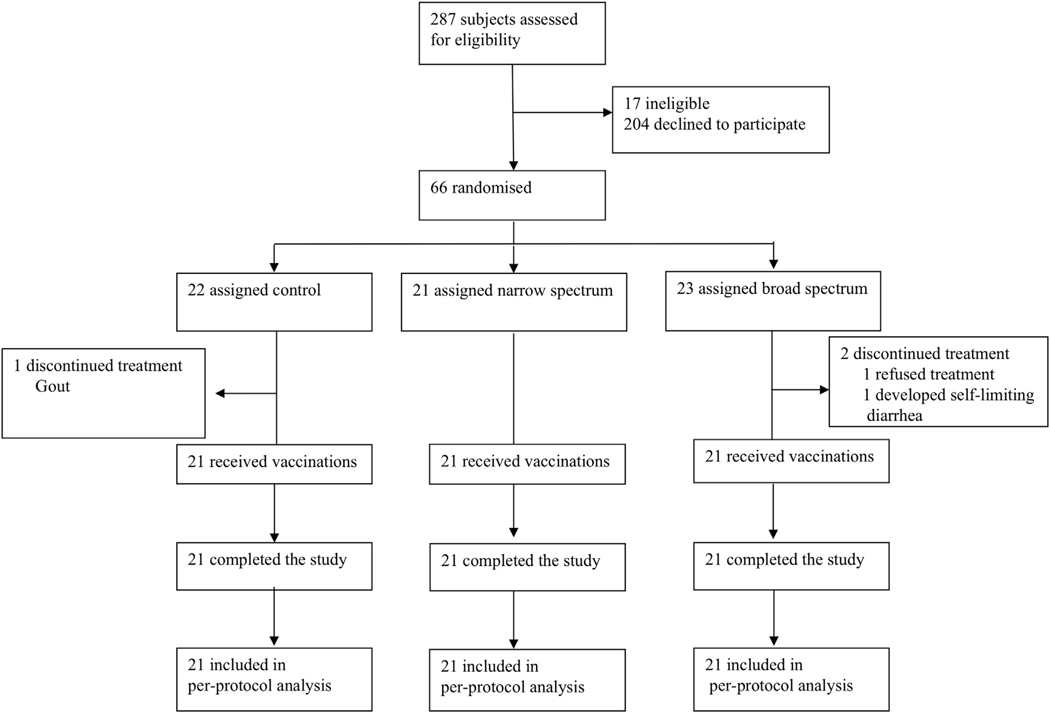
Between September 1, 2015, and January 6, 2017, 66 healthy human subjects were randomly assigned: 23 to receive open-label broad-spectrum antibiotics (7 days of oral vancomycin, ciprofloxacin, and metronidazole), 21 to receive open-label narrow-spectrum antibiotics (7 days of oral vancomycin), and 22 to receive no antibiotics (Figure 1). Thirty-six hours after antibiotic therapy (wash-out period to eliminate antibiotics), subjects were vaccinated with oral Rotarix, Pneumo23 (polysaccharide pneumococcal), and Tetanus toxoid vaccines. Stool was collected prior to antibiotics and vaccination and 7 days post-vaccination for microbiome analysis and RV shedding. Blood was collected prior to antibiotics; vaccination; and 7, 14, and 28 days following vaccination for vaccine serology. Three participants did not complete the study per study protocol as described in Figure 1 and were excluded from the per-protocol study analysis. Study was complete when 21 subjects per arm completed the study per-protocol. Therefore, 21 of 23 (91%) of those in the broad-spectrum group, 21 (100%) of 21 in the narrow-spectrum group, and 21 (95%) of 22 in the control group completed the study per protocol. No participants developed a Clostridium difficile infection. Baseline characteristics for all participants are listed in Table 1.
Figure 1.
Trial Profile
Table 1.
Baseline Characteristics of Study Participants
| Control (n = 21) | Broad spectrum (n = 21) | Narrow spectrum (n = 21) | |
|---|---|---|---|
|
| |||
| Sex | |||
| Male | 21/21 | 21/21 | 21/21 |
| (100%) | (100%) | (100%) | |
| Age (years) | 23.5 (4.0) | 24.2 (4.5) | 23.8 (4.3) |
| Ethnicity | |||
| Caucasian | 20 (95%) | 16 (76%) | 17 (81%) |
| Surinam | 0 (0%) | 2 (10%) | 1 (5%) |
| North-African | 1 (5%) | 0 (0%) | 1 (5%) |
| Dutch Antilles | 0 (0%) | 1 (5%) | 1 (5%) |
| Persian | 0 (0%) | 2 (10%) | 1 (5%) |
| BMI (kg/m2) | 23.1 (2.2) | 22.9 (2.7) | 22.5(1.6) |
| Asthma | 4(19%) | 3 (14%) | 2 (10%) |
| Eczema | 3(14%) | 3 (14%) | 2 (10%) |
| Allergies | 3(14%) | 4 (19%) | 4 (19%) |
| Alcohol units per week (SD) | 8.3 (7.5) | 5.8 (5.6) | 6.5 (4.4) |
Data are n (%) or mean (SD). Asthma, eczema and allergies based on self-report.
Antibiotics Do Not Alter Absolute Anti-RV IgA Titers
Anti-RV IgA geometric mean titers did not change significantly by time or by treatment allocation (Table 2; Figure 2A, time series ANOVA, p = 0.268, time; p = 0.171, treatment allocation). Pre-vaccination anti-RV IgA GMT, indicative of prior rotavirus exposure, was high in all groups prior to vaccination and did not differ significantly from control (baseline anti-RV IgA GMTs for control 201.6 [95% CI 95.3–426.3], broad 160.0 [83.8–305.4], and narrow 390.1 [187.2–812.8]; one-way ANOVA, p = 0.848 for broad and p = 0.296 for narrow; Table 2; Figure 2A). There were no differences in the primary endpoint: anti-RV IgA GMT at 28 days (day 28 anti-RV IgA GMT for control 185.1 [91.9–373.0], broad 140.2 [87.2–225.4], and narrow 245.7 [128.2–470.8]; one-way ANOVA, p = 0.730 for broad and p = 0.722 for narrow; Figure 2A; Table 2). There were also no differences in the absolute anti-RV IgA titer at any other time points (Figure 2A; Table 2) or 4-fold titer change at 28 days (control 0/21, broad 1/21, narrow 1/21 subjects, Fisher’s p > 0.99 both comparisons to control).
Table 2.
Primary and Secondary Endpoints
| Endpoint | Control | Broad spectrum | Narrow spectrum |
|---|---|---|---|
|
| |||
| Anti-RV IgA boosting (≥2-fold increase by 7 days) | |||
| Exploratory | 1/21 (4.7%) | 1/21 (4.7%) | 8/21 (38%) |
| RR = 1.0, 95% CI 0.11–9.22, p > 0.99 | RR = 0.125, 95% CI 0.02–0.67, p = 0.021* | ||
| RV shedding (proportion with any shedding) | |||
| Secondary | 1/21 (4.7%) | 8/21 (38%) | 8/21 (38%) |
| RR = 0.125, 95% CI 0.02–0.67, p = 0.021* | RR = 0.125, 95% CI 0.02–0.67, p = 0.021* | ||
| Anti-RV IgA titer (GMT, 95% CI) | |||
| Primary (d28), Secondary (d7,14) | |||
| Pre-vaccination | 201.6 (95.3–426.3) | 160.0 (83.8–305.4), p = 0.848 | 390.1 (187.2–812.8), p = 0.296 |
| Day 7 | 237.8 (125.0–452.4) | 140.2 (73.7–266.9), p = 0.407 | 485.0 (229.8–1023.9), p = 0.219 |
| Day 14 | 245.7 (136.8–441.4) | 188.7 (131.9–270.1), p = 0.686 | 377.4 (204.2–697.6), p = 0.390 |
| Day 28 | 185.1 (91.9–373.0) | 140.2 (87.2–225.4), p = 0.730 | 245.7 (128.2–470.8), p = 0.722 |
| Anti-RV IgG titer (GMT, 95% CI) | |||
| Secondary | |||
| Pre-vaccination | 1050 (576.4–1913.0) | 920.2 (516.0–1641.0), p = 0.927 | 1121.7 (598.6–2101.9), p = 0.981 |
| Day 7 | 1121.7 (598.6–2101.9) | 1085.3 (659.6–1785.6), p = 0.995 | 1085.3 (617.7–1906.7), p = 0.995 |
| Day 14 | 1238.4 (672.4–2281.1) | 1085.3 (680.3–1731.2), p = 0.906 | 1085.3 (659.6–1785.6), p = 0.906 |
| Day 28 | 1159.3 (630.4–2132.1) | 1280 (801.6–2043.9), p = 0.949 | 1238.4 (720.5–2128.8), p = 0.977 |
| Anti-Pneumococcal IgG (GMT, 95% CI) | |||
| Secondary | |||
| Pre-vaccination | 42.4 (31.8–56.4) | 48.6 (30.2–78.3), p = 0.809 | 39.5 (28.2–55.4), p = 0.948 |
| Day 7 | 118.7 (93.0–151.5) | 128.9 (88.8–187.0), p = 0.908 | 107.7 (74.5–155.8), p = 0.876 |
| Day 14 | 280.7 (225.1–349.9) | 375.8 (277.4–509.3), p = 0.269 | 285.7 (198.6–411.1), p = 0.994 |
| Day 28 | 305.5 (245.2–380.6) | 386.7 (288.3–518.7), p = 0.410 | 300.4 (207.4–435.0), p = 0.995 |
| Tetanus IgG (GMT, 95% CI) | |||
| Secondary | |||
| Pre-vaccination | 0.8 (0.5–1.4) | 1.1 (0.6–1.8), p = 0.631 | 1.5 (0.9–2.6), p = 0.153 |
| Day 7 | 4.1 (3.0–5.6) | 4.2 (2.9–6.1), p = 0.991 | 4.3 (3.1–6.0), p = 0.963 |
| Day 14 | 9.8 (7.0–13.7) | 10.5 (7.9–13.9), p = 0.908 | 8.7 (6.8–11.1), p = 0.767 |
| Day 28 | 8.6 (6.3–11.7) | 9.5 (7.3–12.4), p = 0.807 | 7.9 (6.2–10.1), p = 0.889 |
Proportions of RVV boosting and RV shedding according to treatment group with significance evaluated using Fisher’s exact test. Antibody responses over time for anti-RV IgA, IgG, anti-pneumococcal total IgG, and anti-tetanus toxoid IgG according to study group. Geometric mean titers with 95% confidence intervals for each antibody response per treatment group and time point. Significance evaluated comparing treatment group to control per time point using one-way ANOVA with Dunnet’s correction.
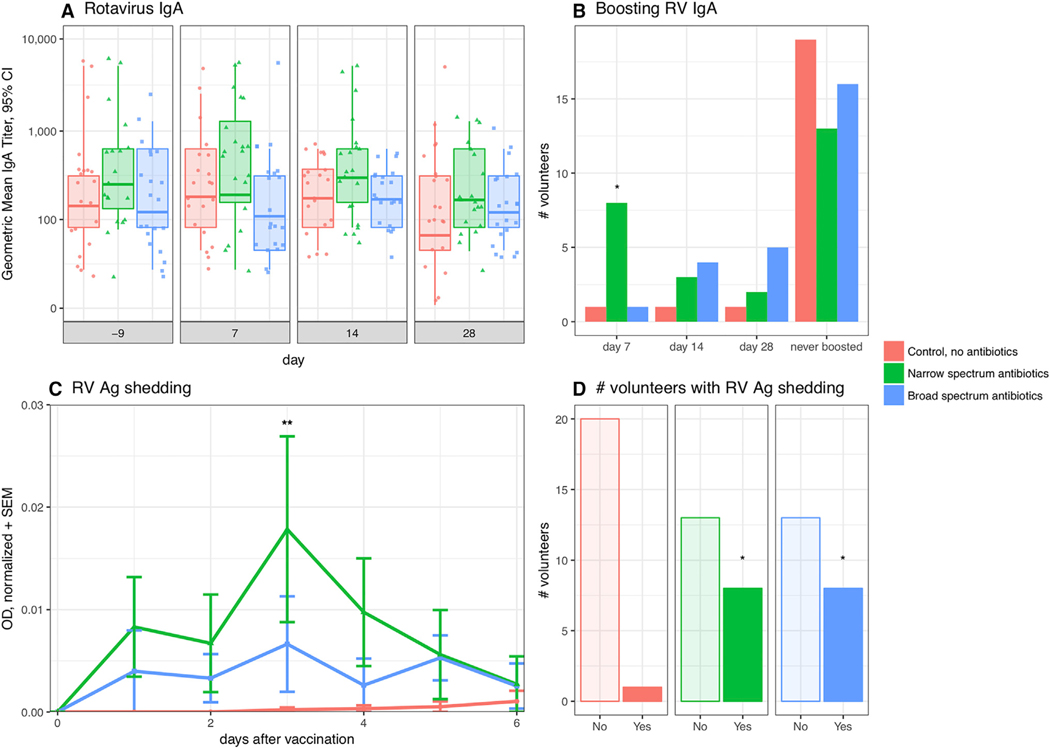
Figure 2. Vancomycin Minimally Boosts 7-Day Anti-RV IgA and Increases RV Ag Shedding.
(A) RV-specific IgA geometric mean titer (GMT) per treatment arm and over time with 95% CI. One-way ANOVA with Dunnet’s.
(B) Proportion of subjects with RV-specific IgA boosting per treatment arm over time. Fisher’s exact test.
(C) Mean normalized OD + SEM of RV antigen shedding per treatment group over time. Friedman’s with Dunn’s multiple comparison test.
(D) Proportion of subjects per treatment arm that ever shed. Fisher’s exact test. See also Figures S1 and S2.
Vancomycin Boosts RVV Immunogenicity at 7 Days
Given the high baseline anti-RV IgA titers and RV sero-positivity in our subjects, we performed an exploratory analysis evaluating differences in the proportion of volunteers who had secondary immune responses or IgA boosting to vaccination, defined as a ≥2-fold anti-RV titer increase by 7 days post-vaccination between treatment groups and control. Anti-RV IgA boosting was significantly higher in the narrow-spectrum antibiotic group (Figure 2B). Eight of 21 (38%) volunteers in the narrow-spectrum antibiotic group compared to 1 of 21 (4.7%) in the control group had IgA boosting by 7 days post-vaccination (RR = 0.125, 95% CI 0.02–0.67, Fisher’s p = 0.021). There was no significant difference between broad-spectrum boosting (1/21, 4.7%) and control group boosting (1/21, 4.7%) (RR = 1.0, 95% CI 0.11–9.22, Fisher’s p > 0.99). There were no differences in 2-fold titer change at any other time point (Figure 2B).
Antibiotics Increase RV Ag Fecal Shedding
There was both a higher amount and proportion of RV shedding in the narrow and broad-spectrum groups following RV vaccination compared to control. Subjects in the narrow-spectrum group had higher overall mean RV shedding levels post-vaccination than control (Friedman’s w/Dunn’s p adj = 0.0027; Figures 2C and S1A). Rotavirus antigen was detected in the stool in 8/21 volunteers in both the narrow-spectrum and broad-spectrum antibiotic groups as compared to 1/21 volunteers in the control group (RR 8, 95% CI 1.5–47.4, Fisher’s p = 0.021; Figure 2D). Subjects with anti-RV IgA boosting had higher overall mean RV shedding post-vaccination than non-boosters (mean difference 0.009, 95% CI of difference 0.0019–0.0152, Wilcoxon matched-pairs signed rank test, p = 0.031; Figure S2A). Subjects who had IgA boosting had no significant differences in proportion of RV shedding (5/10, 50%) compared to subjects that did not boost (12/53, 22.6%) (RR 2.2, 95% CI 0.92–4.5, Fisher’s p = 0.116; Figure S2B).
There were no differences in anti-RV IgG titers over time (two-way ANOVA p = 0.197) and between study groups (two-way ANOVA, p = 0.989; Table 2; Figure S1B).
Finally, both anti-pneumococcal and anti-tetanus IgG increased significantly over time post-vaccination (time since vaccination accounting for 57.4% of the variance, two-way ANOVA p < 0.0001 and 53.1% of the variance p < 0.0001, respectively; Figures S1C and S1D). Variance was not explained by treatment allocation for either total pneumococcal IgG (two-way ANOVA, p = 0.449) or anti-tetanus IgG (two-way ANOVA, p = 0.718).
Antibiotics Rapidly Alter Bacterial Diversity
Fecal microbiome analysis first evaluated bacterial diversity. There were no significant differences in mean alpha diversity (data not shown) between treatment arms and control prior to antibiotic treatment. The control arm had no changes in mean alpha diversity over time (Figures S3A and S3B). Seven-day treatment with either broad- or narrow-spectrum antibiotics significantly decreased both richness and Shannon diversity by vaccination at day 0 (narrow p = 2.0e-6/p = 3.3e-5 and broad p = 1.8e-4/p = 4.8e-7, respectively) with a partial recovery (yet still significantly different) by 7 days post-vaccination as compared to pre-treatment (narrow p = 1.5e-2/p = 2.5e-2 and broad p = 2.0e-3/p = 4.1e-6, respectively; Figures S3A and S3B). Mean Shannon diversity was the same between all treatments by day 7. Mean sample richness remained slightly lower in broad spectrum-treated samples when compared to narrow spectrum (p = 0.043), but was similar to day 7 control samples (p = 0.120) (data not shown).
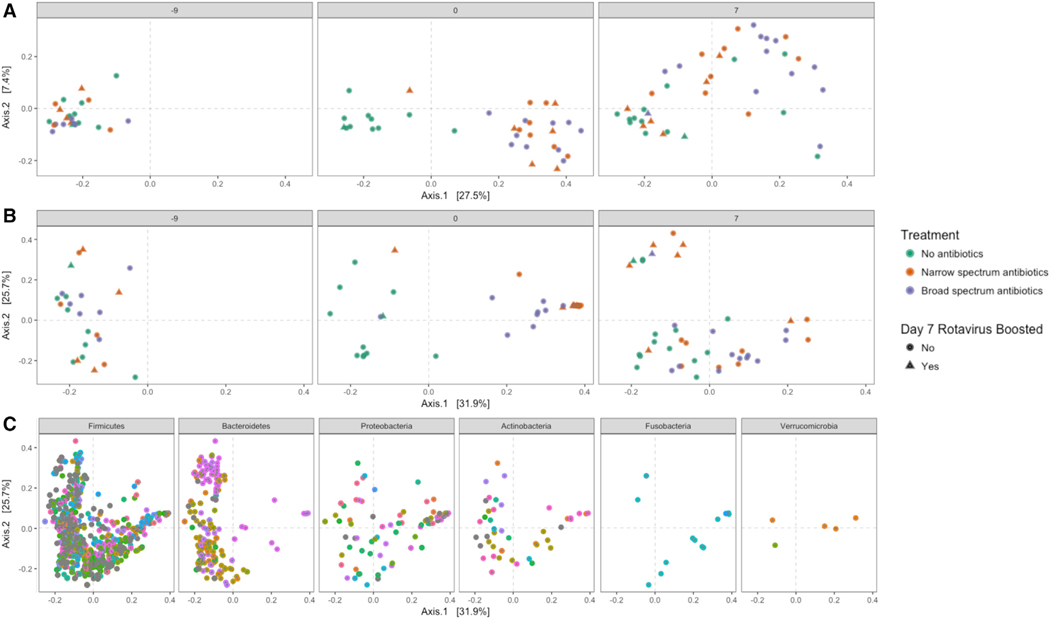
Similar patterns were observed in beta diversity. There were no significant differences in community composition as determined by treatment arm prior to antibiotic treatment (Figures 3A and 3B; ADONIS p = 0.895, R2 = 0.044 control versus narrow; p = 0.415, R2 = 0.068 control versus broad; p = 0.858, R2 = 0.056 narrow versus broad). Following antibiotic treatment at day 0, variance in community composition was significantly explained by treatment arms (ADONIS p = 0.001, R2 = 0.195 control versus narrow; p = 0.001, R2 = 0.154 control versus broad), and both antibiotic treatments yielded distinct communities (p = 0.001, R2 = 0.129 narrow versus broad) with diminished, but still some significant differences by day 7 (ADONIS p = 0.023, R2 = 0.055 control versus narrow; p = 0.008, R2 = 0.064, control versus broad; p = 0.064, R2 = 0.047, narrow versus broad) (Figures 3A and 3B).
Figure 3. Beta Diversity Associations with RVV Boost.
(A and B) (A) Principle Coordinates Analysis (PCoA) of unweighted UniFrac (UniFrac) and (B) weighted UniFrac (wUniFrac) distances of samples at each sampling time. Samples are colored by treatment, and shape is defined by whether or not the patient had significant RV boosting by day 7. Note that two green control points are overlapping at day 7 in the upper-left quadrant.
(C) PCoA of wUniFrac distances for bacterial taxa from the six phyla found to be altered by antibiotic treatment at one or more sampling time point. Each color indicates a unique bacterial genus within each phylum (legend excluded for space). See also Figure S3.
Beta Diversity Associates with RVV Boosting at Day 7
PCoA analysis of unweighted UniFrac (Figure 3A) and weighted UniFrac (Figure 3B) at each time point revealed distinct findings. In the weighted UniFrac analysis there was a distinct group of nine samples at day 7 post-vaccination (Figure 3B, day 7, upper-left-hand quadrant). Six of these nine samples (67%) were from RVV-boosted subjects (five from narrow-, one from broad-spectrum treatment groups). Analysis of what types of bacterial taxa associated with this group revealed an overlap with a genera of Bacteroidetes (Figure 3C, upper left-hand quadrant), but not genera from other phyla. The observation that unweighted UniFrac was unable to identify group separation at day 7 suggests that it is not the presence or absence of a particular bacteria (ASV) that leads to the group distinctions. Rather, the wUniFrac analysis grouping (which considers not only the presence or absence of a specific ASV, but also each ASV relative abundance) observed at day 7 must be due to the property of ASV abundance.
Narrow and Broad-Spectrum Antibiotics Differentially Impact Microbiome
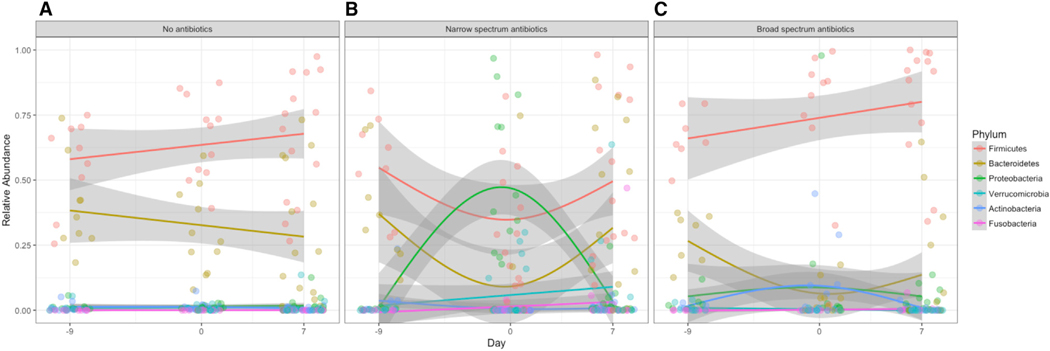
The fecal microbiome analysis next evaluated bacterial community composition over time and treatment arm. ASV from 11 different phyla were detected in the study, but the mean abundances of only six phyla were significantly altered by treatment (Figure S4), while mean ASV abundance from the Elusimicrobia, Lentisphaerae, Spirochaetes, Synergistetes, and Tenericutes were similar across all treatment groups at each sampling time. At the taxonomic level of phylum, both broad- and narrow-spectrum antibiotic treatments led to notable reduction of Bacteroidetes at day 0 (Figures 4 and S4; Kruskal-Wallis with Dunn’s, narrow p = 9.6e-4, broad p = 4.8e-4). Only narrow-spectrum antibiotics decreased the mean abundance of Firmicutes by day 0 (Figures 4 and S4; Kruskal-Wallis with Dunn’s, narrow p = 5.8e-3, broad p = 0.1101), and only narrow-spectrum antibiotics led to a marked increase in mean Proteobacteria at day 0 (Figures 4 and S4; Kruskal-Wallis with Dunn’s, narrow p = 3.3e-4, broad p = 0.6200). While the mean abundances of both Bacteroidetes and Firmicutes returned to control levels at day 7, the Bacteriodetes remained lower (p = 0.024) and Firmicutes increased (p = 7.5e-3) in broad-spectrum-treated samples. The three other phyla had relatively low abundance in all samples yet varied significantly between treatment arm and control: Fusobacteria were significantly more abundant at day 0 (p = 0.036) and day 7 (p = 0.042) in narrow-spectrum antibiotics samples compared to control samples. Mean levels of Actinobacteria increased in broad-spectrum antibiotics-treated samples at day 0 (p = 1.6e-3) but returned to control levels by day 7. Mean levels of Verrucomicrobia were reduced in broad-spectrum-treated samples in day 7 samples when compared to control samples (p = 0.011) but were not significantly different at day 0 (p = 0.210) (Figure S4).
Figure 4. Temporal Trajectories of Treatment Altered Bacterial Phyla.
Changes in the relative abundance of the six bacterial phyla found to be altered by treatment over time in (A) no antibiotics, control, (B) narrow-spectrum antibiotic, and (C) broad-spectrum antibiotic treatment arms. Lines indicate the loess smoothed fit across the 3 days, and the shaded error is the confidence interval. See also Figure S4.
In order to identify discriminating taxa between treatments at each sampling time, normalized (rlog) values were assessed for differential abundance between groups at each independent sampling time (nine unique pairwise comparisons: control versus narrow, control versus broad, and narrow versus broad at days −9, 0, and 7). The rlog function transforms the ASV counts to the log scale while minimizing differences for ASV with small counts and normalizing with respect to library size (Love et al., 2014). Twenty-eight unique taxa (derived from 41 unique ASV) were identified with significant differential abundance (threshold: base mean > 100 and adjusted p ≤ 0.05; Table S1; Figure S5). The only bacteria that distinguished narrow-spectrum antibiotics from both the broad-spectrum and the control arms at day 0 were taxa from the Proteobacteria phylum (Figure S5; Table S1).
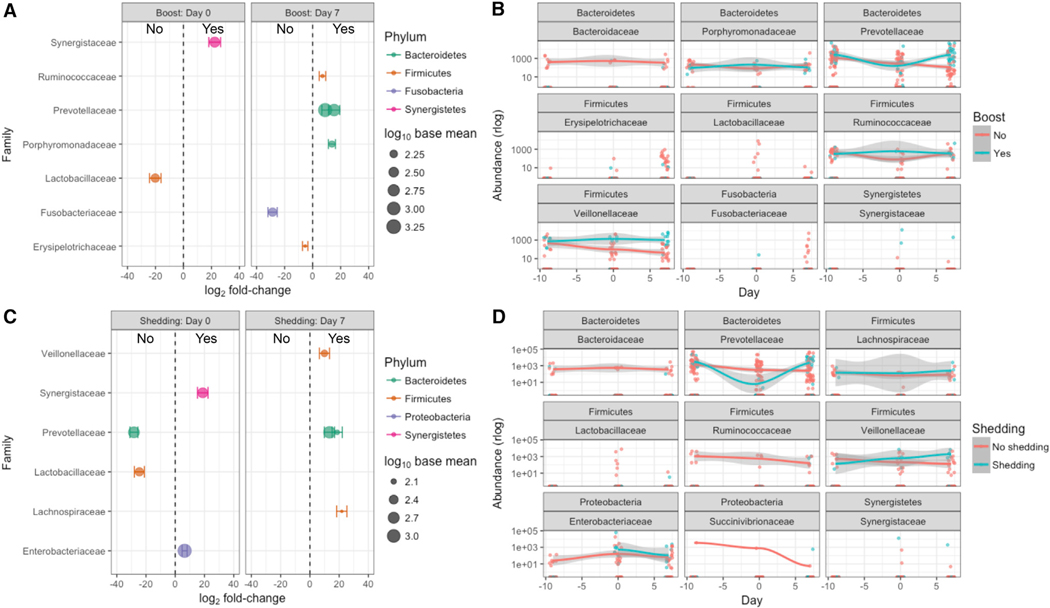
Bacterial Taxa Distinguish RVV Boosters and RV Shedders
Next, we assessed which microbiota differences distinguished RVV boosters and shedders, independent of treatment arm allocation. Each sequence variant and its rlog normalized abundance was assessed for differential abundance between groups at days 0 and 7 post-vaccination: RVV boosters versus nonboosters, RV shedders versus non-shedders. Several differentially abundant taxa distinguished RVV boosters and RV shedders (Figure 5; Table S1). Abundance of Cloacibacillus everynsis (Synergistaceae family) as well as Proteobacteria from the Escherichia/Shigella genus (Enterobacteriaceae family) correlated significantly with RV shedding at day 0 (Figures 5C and 5D; Table S1). A diminished abundance of Bacteroidetes, particularly members of the family Prevotellaceae, correlated significantly with no RV shedding at day 0 (Figures 5C and 5D; Table S1). Reflective of the association of Bacteroidetes in RVV boosters’ samples at day 7 in the wUniFrac analysis and shown in Figure 3, members of the family Prevotellaceaea were also found to be more abundant in RVV boosters at day 7 (Figures 5A and 5B; Table S1). Similarly, members of the Prevotellaceae were also associated with RV shedding at day 7 (Figures 5C and 5D; Table S1). Members of the Firmicutes phylum were represented in various groups, including being associated with the lack of both RVV boosting and RV shedding at day 0 as well as with RVV boosting and RV shedding by day 7 (Figure 5; Table S1).
Figure 5. Differentially Abundant Bacterial Taxa Associated with RVV Boosting and RV Shedding.
(A) Bacterial taxa differentially abundant (p < 0.05, Wald test as implemented in DESeq2) between samples from RVV-boosted (Yes) or non-boosted (No) patient samples.
(B) Temporal trajectories of the normalized (rlog) abundances of differentially abundant taxa from (A) selected at level of Phylum (top header) and Family (bottom header).
(C and D) As in (A) and (B), but for RV shedder (Yes) or non-shedder (No) subject samples.
Error bars in (A) and (C) indicate the SE of the log2 fold-change. Lines in (B) and (D) indicate the loess smoothed fit across the 3 days, and the shaded error is the confidence interval. See also Table S1 and Figure S5.
DISCUSSION
In this randomized, controlled proof-of-concept study, there were no differences in absolute anti-RV IgA titers at any time point between treatment arms. However, prospective modulation of the intestinal microbiome with vancomycin, but not broad-spectrum antibiotics, resulted in slightly increased anti-RV IgA boosting at 7 days post-vaccination. Both antibiotic arms, but particularly vancomycin, increased RV shedding when compared to no antibiotic treatment in adult volunteers. Vancomycin therapy and broad-spectrum antibiotics differentially affected the intestinal microbiome. Both antibiotic arms caused major alterations to overall bacterial community structure and depleted Bacteroidetes, but only vancomycin led to a reduction of Firmicutes and an expansion of Proteobacteria directly following antibiotic treatment. Independent of treatment arm, several bacterial taxa correlated with RV shedding and boosting. Among them were a higher abundance of Enterobacteriaceae (Proteobacteria) and a lower abundance of Prevotellaceaea (Bacteroidetes) at the time of vaccination, and an outgrowth of Prevotellaceaea by day 7 post-vaccination. Antibiotic therapy did not alter immune responses to either pneumococcal 23-valent polysaccharide vaccination or the tetanus toxoid vaccination. This study suggests that targeted modulation of the intestinal microbiota alters both RVV boosting and shedding in healthy adults.
Given that specific bacterial taxa correlate with RVV boosting and RV shedding and that vancomycin, not broad-spectrum antibiotics, resulted in a more robust RVV immune phenotype (with both higher RVV shedding and slight boosting), it is likely that defined bacterial taxa and not microbiota depletion alone alter RVV immunogenicity. These taxa may lead to increased RVV-strain replication and therefore enhanced anti-RV IgA responses. There are numerous illustrations of direct interactions between enteric viruses and bacteria leading to enhanced viral replication and transmission (Pfeiffer and Virgin, 2016). For example, poliovirus binding to bacterial cell wall components such as lipopolysaccharide (LPS) and peptidoglycan leads to stabilization of the virion, limiting premature RNA release (Robinson et al., 2014). Potentially, differentially abundant expression of bacterial surface polysaccharides (for example, Gram-negative Enterobacteriaceae in the Proteobacteria phylum) may have facilitated RV replication or host cell binding and subsequent boosting.
Antibiotic-induced changes in the microbiota can also induce profound changes in bacterial metabolism. For example, vancomycin is known to alter bile-acid metabolism, decreasing fecal secondary bile acids, likely through reduction of Firmicutes (Vrieze et al., 2014). Recent advances in norovirus culturing in enteroid systems have demonstrated that (species-specific) bile is needed for some norovirus strain replication and that bile’s effect is mediated not through the virus but with the cell potentially as a binding co-factor (Ettayebi et al., 2016). Such mechanisms may also be at play with rotavirus and vancomyin-induced changes to the microbiota may therefore have altered bacterial metabolism and secondarily host susceptibility to the rotavirus vaccine strain.
The commensal microbiota is known to regulate intestinal immunity and antibiotic treatment may alter this regulation—either by reducing bacteria that induce immune tolerance or by increasing bacteria that enhance mucosal immunity. It is known, for example, that LPS expressed by Proteobacteria and Bacteroidetes have different structures: LPS derived from Proteobacteria are hexa-acylated and can lead to strong inflammatory cytokine responses, whereas LPS derived from many Bacteroidetes species are penta- or tetra-aceylated and can downregulate cytokine expression (Vatanen et al., 2016). Differential expression of these cell membrane components with increased Proteobacteria and decreased Bacteroidetes species may therefore have augmented (adjuvanted) innate and adaptive immune responses to the RVV in the vancomycin arm.
Finally, an intriguing study finding was the appearance of a distinct subset of subjects with increased regrowth of Bacteroidetes abundance, by day 7 post-vaccination. This re-growth, seen in the beta-diversity analyses and differential abundance testing, correlated with both boosting and shedding. Recent work suggests that some Bacteroidetes (Bacteroides fragilis) utilize IgA for mucosal colonization (Donaldson et al., 2018). Perhaps subjects with increased shedding and boosting had higher mucosal IgA induction following vaccination, secondarily leading to enhanced mucosal colonization by those bacteria that utilize IgA for colonization.
Antibiotics had no effect on pneumococcal or tetanus vaccine immunogenicity, implying that alteration of the microbiome may not affect immunity to systemic vaccines. Other studies have evaluated the effect of antibiotics on vaccine responses. Oral polio vaccine immunogenicity was evaluated in infants in India who were randomized to either placebo or treatment with azithromycin. Azithromycin treatment did not improve polio immunogenicity (Grassly et al., 2016). Azithromycin was chosen in that study not to achieve specific shifts in the microbiota but for its immune-modulatory effects and for targeting of enteric pathogens. An ongoing study is evaluating the effect of antibiotics on influenza vaccine immunogenicity on adult volunteers (clinicaltrials.gov NCT02154061). This current study used a different approach by employing vancomycin to try and simulate some of the microbiota compositions that already correlated with RVV response in human infants in Ghana and in Pakistan (Harris et al., 2017a, 2017b). Interactions between the microbiome and vaccines are therefore likely to be microbiota and vaccine specific. The caveat to our approach is that vancomycin has broader effects than Proteobacteria and Bacteroidetes alteration alone, and bacterial taxa outside of these phyla also correlated with RVV shedding and boosting.
A major limitation in this study is that the study population consisted of adult men. We considered it to be unethical to perform this non-therapeutic antibiotic intervention in an at-risk infant population and therefore conducted a proof-of-concept study in an adult volunteer population, which, despite its limitations, was considered to be the closest alternative experimental population. Adults have significantly different intestinal microbiomes than infants (Yatsunenko et al., 2012); however, we used antibiotics to alter the adult microbiome to imperfectly recapitulate some of the microbiome phenotype correlating with RVV immunogenicity in low-income-country infants. We also expect that an adult volunteer model still permits evaluation of viral-bacterial interactions, changes in bacterial metabolism, and adjuvant or immune-tolerant effects of the microbiome.
Perhaps an even more significant limitation to using adult volunteers is that RV is a childhood disease, and while adults can be infected, they have significantly fewer clinical symptoms due to RV infection. Adults have had repeated RV exposure. RVV are targeted toward seronegative infants in the first 2 months of life, and the definitions of seroconversion by 28 days (either an anti-RV IgA > 20 IU/mL or a ≥4-fold rise in titer from baseline) are imperfect correlates of vaccine protection (Velasquez et al., 2018). There are no accepted correlates of protection for evaluating RVV in seropositive individuals or adults, particularly given that nearly all adults are previously exposed. Because adequate recruitment of adults with low seropositivity to this study was unsuccessful, the 28-day anti-RV IgA time point that was proposed as the primary study endpoint may be invalid. Our study actually evaluated memory antibody responses or boosting to RVV. Secondary antibody responses are more brisk than primary responses. Additionally, in contrast to the 20-day half-life of most IgGs, IgA has a very short, 6-day half-life. Therefore, we speculate that anti-RV IgA titer rises in seropositive individuals might best be captured at early (7 day) time points (Murphy and Weavery, 2017). A 2-fold or greater antibody rise between convalescent and acute-phase sera, as has been used in this study, has been used in RV adult volunteer challenge studies, and early-phase Rotarix vaccination studies in adults used any antibody rise by days 14 and 21 (Ward et al., 1989). Despite earlier precedent, it is quite possible that a 2-fold rise falls within a margin of error of the IgA assay, and this finding is due to chance, particularly as there were no significant differences in absolute anti-RV IgA titers at day 7. RV shedding may be a more stringent outcome than boosting, as it reflects the efficiency of RVV strain replication and correlates significantly with subsequent antibody response in infants (Anderson, 2008). In RV-naive infants in low-income countries, ~5%–13% of infants shed Rotarix following the first vaccination when co-administered with OPV (Armah et al., 2016). Adults generally do not shed Rotarix—only 1 out of 50 of all the adults given Rotarix in pre-licensure studies shed rotavirus, and 0 of the 21 control subjects in this study’s control arm shed (Anderson, 2008). The significantly increased shedding which we now report in the vancomycin group with a biologic shedding peak detected 3 days post-vaccination is therefore highly suggestive that vancomycin therapy increases the efficiency of vaccine-strain replication and take.
Another potential limitation in this study is that antibiotics themselves could be altering immunity through off-target effects. For example, total serum and fecal IgA was not measured, but antibiotics could have altered their levels in relation to control. However, should antibiotics be impacting RVV immunity through off-target effects, we would have expected the broad-spectrum rather than the narrow-spectrum antibiotic group to show the greatest effects. The higher boosting and shedding in the vancomycin arm indirectly suggest microbiota alteration, not antibiotic effects, are mediating immunity.
This study provides a human demonstration that alteration of the bacterial intestinal microbiome can alter a vaccine’s immunogenicity, as evidenced by the slight RVV boosting and significant shedding following oral vancomycin treatment as compared to control. Our findings may chart a path to improving rotavirus efficacy in low-income country settings through microbiome alteration. While vancomycin was used in adults in this study, its use in very young infants at a public health level is both impractical and not without risk. Rather, this study provides an evidence base and potential candidates for testing of future interventions to improve rotavirus vaccine performance in these settings. Options include employing bacteria that correlated with RVV shedding and boosting for use as probiotics prior to rotavirus vaccination, testing differentially expressed metabolites that could enhance vaccine-strain stability or susceptibility, and assessing differentially abundant bacteria’s cell wall components for their potential as novel mucosal adjuvants. An understanding and application of these options may help improve the performance of rotavirus and other oral vaccines in settings where they are needed most.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vanessa Harris (v.c.harris@amc.uva.nl).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study Design
This was a single center, randomized (1:1:1), open-label, controlled study conducted in Amsterdam, the Netherlands. The study took place in the Academic Medical Center’s internal medicine outpatient clinic. The Academic Medical Center’s ethical committee approved the study (METC 2015_045, NL 52510.018.15). We undertook the study in accordance with the 2013 Declaration of Helsinki and all relevant national, and local rules and legislation. All participants provided written informed consent.
Participants
Volunteers were recruited via posters in hospitals and university campuses in Amsterdam as well as with print and electronic advertisements. In a screening visit, volunteers were questioned about their past medical history, had a physical examination, and if they met the preliminary eligibility requirements, they underwent a blood draw for routine chemistry and hematology as well as baseline anti-RV, pneumococcal and tetanus antibodies. Volunteers were eligible if they were healthy men (determined by history, physical and blood draw) between the ages of 18–35 with a normal defecation pattern and no medication or antibiotic use in the preceding 12 months. Initial eligibility criteria included a baseline anti-RV IgA titer of < 40, however only 1 in 10 initial volunteers were able to meet this threshold, therefore we subsequently accepted any baseline IgA titer. Exclusion criteria included any major illness in the prior 3 months, any chronic illness, participation in another study with use of an investigational product in the three months prior to study commencement, or subjects with a history of any immune deficiency, malignancy, thrombocytopenia or bleeding disorder, gastrointestinal disorder, liver disease, or alcoholism.
Randomization
Following inclusion, subjects were randomly assigned to one of three treatment groups: broad-spectrum antibiotics, narrow-spectrum antibiotics or no antibiotic control. Randomization sequence was generated using statistical software (“Sealed Envelope Ltd,” https://www.sealedenvelope.com/simple-randomiser/v1/lists) with a 1:1:1 allocation using random block sizes of 3 and 6 by an investigator who did not enroll participants. Allocation designation was only opened after enrolled participants completed all the baseline assessments, had signed informed consent, and it was time to allocate the intervention.
Study Procedures
Following randomization, subjects received antibiotic therapy according to their randomization arm: 7 days of broad-spectrum antibiotics (oral vancomycin 500 mg three times a day, oral ciprofloxacin 500mg twice a day and metronidazole 500 mg three times a day), narrow-spectrum antibiotics (oral vancomycin 500 mg three times a day) or control (no antibiotics) according to standardized protocols. After a 36-hour period without antibiotic administration (“wash-out period”), all subjects were simultaneously vaccinated with 1 dose of the oral RVV (Rotarix), and intramuscular injections with the polysaccharide pneumococcal vaccine (Pneumo 23) and tetanus toxoid vaccine delivered in contralateral arms. Subjects returned to the outpatient clinic 7, 14, and 28 days following vaccination for blood draws to measure vaccine antibody response. Subjects additionally provided fecal samples prior to antibiotic administration, following antibiotic administration on the day of vaccination, and daily 1–7 days following vaccination. If fecal samples were collected on the same day as a hospital visit, they were delivered at room temperature and subsequently stored at −80°C. If there was more than 12 hours between fecal sample production and a hospital visit, volunteers stored the feces at home at −20°C and transported the samples in cooling units to the hospital. These samples were subsequently stored at −80°C. Visual study overview is provided in the Figure S6.
METHOD DETAILS
Laboratory Procedures
All RV serology was performed at the Centers for Disease Control, Atlanta, GA. RV-specific IgA and IgG in serum samples were determined using enzyme-linked immunosorbent assay, as described previously (Moon et al., 2016). Briefly, microplate wells were coated with rabbit hyperimmune serum to rhesus rotavirus (RRV) and incubated with diluted RV1 strain or blotto (5% skim milk in phosphate-buffered saline [PBS]). After washing, serum samples (1:10–1:10 240) that were serially diluted in diluent buffer (1% skim milk and 0$5% [v/v] of 10% polyoxyethylene ether W1 in PBS) were added to the wells, followed by biotin-conjugated goat anti-human IgA antibodies (KPL, Gaithersburg, Maryland). After incubation and washing, extravidin (Sigma, St. Louis, Missouri) was added to the wells and incubated, and then the reactions were developed with 3,3′,5,5′-tetramethylbenzidine (TMB; Sigma) and stopped with 1N hydrogen chloride. Optical density (OD) was determined at 450 nm with an enzyme immunoassay reader (MRX Revelation; Dynex Technologies, Chantilly, Virginia). RV-specific IgG in serum samples was tested and analyzed in the same manner as IgA except that 0$5% normal rabbit serum was added to the biotin-conjugate solution. IgA and IgG titers in serum were calculated as the reciprocal of the highest dilution that gave a mean OD greater than the cutoff value (3 standard deviations above the mean OD of the negative-control serum wells). Rotavirus IgA seropositivity at baseline was defined as an IgA titer ≥ 40 before vaccination. Rotavirus IgA boosting was defined as a ≥ 2-fold increase in IgA titer 7 days after vaccination, as has been used in other adult RVV studies (Ward et al., 1989).
Tetanus and pneumococcal serology was performed at the Academic Medical Center in Amsterdam, the Netherlands using anti-Tetanus Toxoid IgG Enzyme Immunoassay Kit and anti-PCP IgG Enzyme Immunoassay Kit (The Binding Site, Birmingham, England). Total IgG and IgG to pneumococcal subtypes were determined using a pneumococcal multiplex assay at the Netherlands National Institute for Public Health and Environment (RIVM) as described elsewhere (Elberse et al., 2010).
Fecal RV shedding (Rotavirus Ag) was determined using a commercially available enzyme-linked immunosorbent assay (ProspecT Rotavirus microassay, Oxoid, UK) using polyclonal capture and detector antibodies against RV structural proteins. An in-house quantitative negative control was established by testing all pre-vaccination (presumed RV negative) fecal samples. A mean OD + SD was calculated for all the pre-vaccination samples and determined to be maximally 2.7 times higher than the kit negative control. All day 1–7 fecal samples were subsequently tested in duplicate using the kit. Fecal sample ODs were determined by taking an average of the duplicate and subtracting 2.7 x (kit negative control). All samples ≤ 0 were considered negative.
Fecal microbiome analysis and bacterial 16S rRNA gene amplicon sequencing was performed by extracting stool total nucleic acid from aliquots of pulverized human stool as described previously (Reyes et al., 2013). Primer selection and polymerase chain reaction were performed following previously described methods (Caporaso et al., 2011). Briefly, DNA was phenol/chloroform-extracted and amplified in triplicate with Golay-barcoded primers specific for the V4 region of the 16S rRNA gene. Amplicons were pooled and purified with 0.6 × Agencourt Ampure XP beads (Beckman-Coulter) prior to sequencing at the Center for Genome Sciences, Washington University School of Medicine using the 2 × 250-bp protocol on the Illumina MiSeq platform. All samples were processed on the same day with one batch of reagents by the same technician and sequenced on a single MiSeq run to minimize batch effects.
16S rRNA Gene Amplicon Analysis
An average of 24,463 sequences per sample were obtained (sd ± 23,974). Several samples in each treatment group had fewer than 100 sequences and were removed from subsequent analysis. Data analysis was performed on unrarified data. Amplicon sequence variants (ASV) were selected using dada2 (Callahan et al., 2016). ASVs were filtered when they were not assigned to the kingdom Bacteria or were assigned to the Family Mitochondria, the Class of Chloroplast or the Phylum of Cyanobacteria/Chloroplast. Following filtering, 1,588 ASV remained. Taxonomy was assigned using the Ribosomal Database Project (RDP) 16S rRNA gene sequence database (Cole et al., 2014). All subsequent ecological analysis was completed using PhyloSeq and other R packages (McMurdie and Holmes, 2013). A fully reproducible workflow for both the dada2 sequence preprocessing as well as all of the analysis presented in this manuscript can be found at: https://github.com/shandley/human_volunteer_rotabiome along with all required analysis data files.
Differences in alpha diversity (richness and Shannon diversity) were evaluated between groups and over time using the Kruskal-Wallis one-way analysis of variance with Dunn’s multiple comparison test. Differences in beta-diversity groups was determined using Permutational Multivariate Analysis of Variance (ADONIS). Statistical tests and parameters are indicated in-line in results, and table and figure legends indicate which statistical test and parameters were used.
ASV differentially abundant between 1) treatment groups, 2) RVV boosters and non-boosters and 3) RV shedders and non-shedders, were identified by comparing their rlog normalized abundances using DESeq2 (Love et al., 2014). Comparisons were completed independently at each sampling time (Days −9, 0 and 7). Differential abundance was determined using the Wald test with significance at p < 0.05. Only taxa with a base mean > 100 were selected for further subsequent analysis. The base mean is calculated as the average of the normalized counts divided by size factors (which control for differences in per sample sequencing depth) taken over all samples. The Kruskal-Wallis test with Dunn’s test for multiple comparisons was used for comparing the mean abundances between groups of ASV selected at specific taxonomic levels (e.g., Phylum).
QUANTIFICATION AND STATISTICAL ANALYSIS
Study Outcomes
The primary outcome for the study was the difference in the absolute height of the anti-RV IgA serum titer 28-days post-vaccination between the randomization groups. Secondary endpoints were differences in day 7 and 14 post-vaccination anti-RV IgA titers, differences in absolute and proportion of RV antigen fecal shedding days 1–7 post-vaccination, differences in anti-pneumococcal and anti-tetanus IgG 7, 14, and 28 days post-vaccination between randomization groups, and bacterial microbiome composition pre and post-antibiotics.
The primary study outcome assumed that recruited adults would be seronegative and able to increase IgA levels by 28-days post-vaccination. However, as we were unable to recruit RV seronegative adults and given that serum IgA has a typical 6 day half-life, (Murphy and Weavery, 2017) we considered an exploratory outcome of 7 day anti-RV boosting to be a physiologically relevant study endpoint.
Subjects were questioned about any adverse events (including diarrhea possibly related to Clostridium difficile) during every follow-up clinic visit following antibiotic therapy.
Statistical Analysis
This was a proof-of-concept study where effects of microbiome modulation could not safely be estimated. To gain an indication for sample size we used a previous published study evaluating the effect of rotavirus (Rotateq) vaccination in elderly adults to estimate a mean 28 day post-vaccination IgA geometric mean titers (GMT) response of 1026 dilution units in controls and estimate a projected standard deviation of approximately 400 dilution units (Lawrence et al., 2014). We calculated an enrollment of 21 volunteers per treatment group would provide 80% power with α = 0.05 to detect a difference in mean anti-RV IgA titer of 35% between control and treatment groups.
Statistical analyses were performed with Prism7 software (Graphpad software, San Diego, CA). The geometric mean titers (GMT) were defined as the exponential of mean logarithmic transformation of the anti-RV IgA titers, anti-RV IgG titers, anti-pneumococcal IgG titers, and anti-tetanus IgG titers. The log transformed antibody titers (in order to attain a Gaussian distribution) were compared between the treatment groups and control at individual time points using one-way ANOVA and for time courses using two-way ANOVA, both with Dunnett’s multiple comparison correction. Time and treatment group were considered independent variables. Antibody fold changes were calculated between baseline and post-vaccination day 7 (≥2-fold) and 28 (≥4-fold) with fold change defined as (post titer – pre titer)/(pre titer). RV antigen shedding was evaluated days 0–7 post-vaccination and expressed as mean OD and the proportion of subjects shedding per treatment group. Mean OD was considered non-parametric and the mean rank was compared between treatment groups and control over time using a Friedmann’s test with Dunn’s multiple comparison test (3 matched groups) and between boosters and non-boosters over time using a Wilcoxon matched-pairs signed rank test (2 paired groups). All proportions were compared between groups using Fisher’s exact test and 95% confidence intervals. A 2-sided p and significance level of 0$05 were used for all statistical tests.
DATA AND SOFTWARE AVAILABILITY
The accession number for the data reported in this paper is ENA: PRJEB25634. Full analysis workflows for microbiome dada2 ASV resolution, statistical analysis and plotting are available at: https://github.com/shandley/human_volunteer_rotabiome. Unprocessed sequence data are available at EMBL accession PRJEB25634
ADDITIONAL RESOURCES
This study is registered with Clinicaltrials.gov identifier, number NCT02538211. The full study protocol and Consort Checklist for clinical trials can also be accessed at: https://github.com/shandley/human_volunteer_rotabiome.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Pneumococcal serotype multiplex in-house assay (PCV13-MIA) | RIVM, the Netherlands, Elberse et al., 2010 | IIV-IMS-MO22-MIA Pneumo |
| Biotinylated Anti-Human IgG (gamma) Antibody | KPL, Gaithersberg Maryland | Cat# 16-10-02; RRID:AB_2732814 |
| Biotinylated Anti-Human IgA (alpha chain) Antibody | KPL, Gaithersberg Maryland | Cat# 16–10-01; RRID:AB_2732815 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| Rotavirus antigen, supernatant from clarified Rotarix extract (~106 FFU/ml), Rotarix-infected MA104 cell cultures are clarified by centrifugation. | CDC, Atlanta, GA | N/A |
|
| ||
| Biological Samples | ||
|
| ||
| Rabbit hyperimmune serum to Rhesus rotavirus | CDC, Atlanta, GA | CD 08, 10–5-01 |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| Extravidin | Sigma Aldrich, St Louis, MO (Sigma-Aldrich, RRID:SCR_008988 | Cat# E2886, ExtrAvidin FITC, Sigma-Aldrich; RRID:SCR_013728 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| ProspecT Rotavirus microassay | Thermo Fischer Scientific, Oxoid, UK | Cat# R240396; RRID:AB_2732820 |
| anti-Tetanus Toxoid IgG Enzyme Immunoassay Kit | The Binding Site, Birmingham, England (Binding Site, RRID:SCR_004051) | Cat# MK010; RRID:AB_2732816 |
| anti-PCP IgG Enzyme Immunoassay Kit | The Binding Site, Birmingham, England (Binding Site, RRID:SCR_004051) | Cat# MK012; RRID:AB_2732817 |
|
| ||
| Deposited Data | ||
|
| ||
| Sequence data | This paper | ENA Accession Number PRJEB25634 |
| Full analysis workflows for microbiome dada2 ASV resolution, statistical analysis and plotting | This paper | https://github.com/shandley/human_volunteer_rotabiome |
|
| ||
| Software and Algorithms | ||
|
| ||
| Prism7 software | Graphpad, San Diego, CA | Version 7.0c; RRID:SCR_002798 |
| RStudio | RStudio, Inc | Version 1.0.143; RRID:SCR_000432 |
| PhyloSeq | McMurdie and Holmes, 2013 | https://joey711.github.io/phyloseq/; RRID:SCR_013080 |
| Dada2 | Callahan et al., 2016 |
https://github.com/benjjneb/dada2; RRID:SCR_008205 |
Highlights.
Rotavirus vaccine immunogenicity tested following antibiotic administration in adults
Antibiotics did not alter absolute anti-RV IgA titers
Narrow-spectrum antibiotics increased anti-RV IgA boosting and RV shedding by day 7
Microbiome composition correlates with RVV boosting and shedding
ACKNOWLEDGMENTS
This research was funded by a research grant from the Emma Children’s Hospital Foundation, Academic Medical Center, Amsterdam (Stichting Emma Foundation) (project number WAR004-2015-03-001 to M.B.H. and V.CH.). The study authors would like to thank the participants in this study for their willingness to enroll in this study, their commitment, their time, and their enthusiasm. We would also like to thank Rosan vd Lee at the Center for Molecular Medicine in the AMC for their laboratory time and excellent technical assistance. We would like to thank Sung-Sil Moon, Yuhuan Wang, and Dr. Umesh Parashar at the Division of Viral Diseases Centers for Disease Control and Prevention and Professor Carlo Giaquinto at the Department of Pediatrics at the University of Padova for their support, advice, and insight. We would also like to thank Herbert “Skip” W. Virgin for his support of the microbiome library preparation within his laboratory facility. Finally, this study would not have been possible without the enthusiasm and commitment of Professor Joseph (Joep) Lange, who died prior to its publication.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interest.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and one table and can be found with this article at https://doi.org/10.1016/j.chom.2018.07.005.
REFERENCES
- Anderson EJ (2008). Rotavirus vaccines: viral shedding and risk of transmission. Lancet Infect. Dis 8, 642–649. [DOI] [PubMed] [Google Scholar]
- Armah G, Lewis KDC, Cortese MM, Parashar UD, Ansah A, Gazley L, Victor JC, McNeal MM, Binka F, and Steele AD (2016). A randomized, controlled trial of the impact of alternative dosing schedules on the immune response to human rotavirus vaccine in rural Ghanaian infants. J. Infect. Dis 213, 1678–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste J, Lespinats G, and Salomon J. (1971). Serum and secretory IgA in axenic and holoxenic mice. J. Immunol 107, 1656–1662. [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, and Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, and Knight R. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108 (Suppl 1 ), 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, and Tiedje JM (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, et al. (2018). Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elberse KEM, Tcherniaeva I, Berbers GAM, and Schouls LM (2010). Optimization and application of a multiplex bead-based assay to quantify serotype-specific IgG against Streptococcus pneumoniae polysaccharides: response to the booster vaccine after immunization with the pneumococcal 7-valent conjugate vaccine. Clin. Vaccine Immunol 17, 674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, et al. (2016). Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassly NC, Praharaj I, Babji S, Kaliappan SP, Giri S, Venugopal S, Parker EPK, Abraham A, Muliyil J, Doss S, et al. (2016). The effect of azithromycin on the immunogenicity of oral poliovirus vaccine: a double-blind randomised placebo-controlled trial in seronegative Indian infants. Lancet Infect. Dis 16, 905–914. [DOI] [PubMed] [Google Scholar]
- Harris VC, Ali A, Fuentes S, Korpela K, Kazi M, Tate J, Parashar U, Wiersinga WJ, Giaquinto C, de Weerth C, et al. (2017a). Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes 3, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, Tate J, de Weerth C, Giaquinto C, Wiersinga WJ, et al. (2017b). Significant correlation getween the infant gut microbiome and rotavirus vaccine response in rural Ghana. J. Infect. Dis 215, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac S, Scher JU, Djukovic A, Jimé nez N, Littman DR, Abramson SB, Pamer EG, and Ubeda C. (2017). Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J. Antimicrob. Chemother 72, 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isolauri E, Joensuu J, Suomalainen H, Luomala M, and Vesikari T. (1995). Improved immunogenicity of oral D x RRV reassortant rotavirus vaccine by Lactobacillus casei GG. Vaccine 13, 310–312. [DOI] [PubMed] [Google Scholar]
- Jonesteller CL, Burnett E, Yen C, Tate JE, and Parashar UD (2017). Effectiveness of rotavirus vaccination: a systematic review of the first decade of global postlicensure data, 2006–2016. Clin. Infect. Dis 65, 840–850. [DOI] [PubMed] [Google Scholar]
- Lawrence J, He S, Martin J, Schödel F, Ciarlet M, and Murray AV (2014). Safety and immunogenicity of pentavalent rotavirus vaccine in a randomized, double-blind, placebo-controlled study in healthy elderly subjects. Hum. Vaccin. Immunother 10, 2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus RP, John J, Shanmugasundaram E, Rajan AK, Thiagarajan S, Giri S, Babji S, Sarkar R, Kaliappan PS, Venugopal S, et al. (2018). The effect of probiotics and zinc supplementation on the immune response to oral rotavirus vaccine: A randomized, factorial design, placebo-controlled study among Indian infants. Vaccine 36, 273–279. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, and Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S-S, Groome MJ, Velasquez DE, Parashar UD, Jones S, Koen A, van Niekerk N, Jiang B, and Madhi SA (2016). Prevaccination rotavirus serum IgG and IgA are associated with lower immunogenicity of live, oral human rotavirus vaccine in South African infants. Clin. Infect. Dis 62, 157–165. [DOI] [PubMed] [Google Scholar]
- Murphy K, and Weavery C. (2017). Janeway’s Immunobiology, Ninth Edition (Garland Science, Taylor & Francis Group; ), p. 193. [Google Scholar]
- Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, et al. (2014). TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange JS, Ballow M, Stiehm ER, Ballas ZK, Chinen J, De La Morena M, Kumararatne D, Harville TO, Hesterberg P, Koleilat M, et al. (2012). Use and interpretation of diagnostic vaccination in primary immunodeficiency: a working group report of the Basic and Clinical Immunology Interest Section of the American Academy of Allergy, Asthma & Immunology. J. Allergy Clin. Immunol 130 (3, Suppl), S1–S24. [DOI] [PubMed] [Google Scholar]
- Parker EPK, Praharaj I, Zekavati A, Lazarus RP, Giri S, Operario DJ, Liu J, Houpt E, Iturriza-Gómara M, Kampmann B, et al. (2018). Influence of the intestinal microbiota on the immunogenicity of oral rotavirus vaccine given to infants in south India. Vaccine 36, 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PATH Centers for Disease Control and Prevention, World Health Organization (2006). Accelerating the introduction of rotavirus vaccines into GAVI-Eligible Countries. Investment case for GAVI secretariaat. http://www.nitag-resource.org/uploads/media/default/0001/01/ea8101072b609db7271b4a6fe0b1886e338a889e.pdf (Accessed May 17, 2018). [Google Scholar]
- Pfeiffer JK, and Virgin HW (2016). Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351, aad5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Wu M, McNulty NP, Rohwer FL, and Gordon JI (2013). Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. USA 110, 20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CM, Jesudhasan PR, and Pfeiffer JK (2014). Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 15, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, and Parashar UD; World Health Organization–Coordinated Global Rotavirus Surveillance Network (2016). Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin. Infect. Dis 62 (Suppl 2 ), S96–S105. [DOI] [PubMed] [Google Scholar]
- Uchiyama R, Chassaing B, Zhang B, and Gewirtz AT (2014). Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis 210, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hämäläinen A-M, et al. ; DIABIMMUNE Study Group (2016). Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez DE, Parashar U, and Jiang B. (2018). Decreased performance of live attenuated, oral rotavirus vaccines in low-income settings: causes and contributing factors. Expert Rev. Vaccines 17, 145–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, van Nood E, Holleman F, Knaapen M, Romijn JA, et al. (2014). Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol 60, 824–831. [DOI] [PubMed] [Google Scholar]
- Ward RL, Bernstein DI, Shukla R, Young EC, Sherwood JR, McNeal MM, Walker MC, and Schiff GM (1989). Effects of antibody to rotavirus on protection of adults challenged with a human rotavirus. J. Infect. Dis 159, 79–88. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the data reported in this paper is ENA: PRJEB25634. Full analysis workflows for microbiome dada2 ASV resolution, statistical analysis and plotting are available at: https://github.com/shandley/human_volunteer_rotabiome. Unprocessed sequence data are available at EMBL accession PRJEB25634