Abstract
Argonaute (Ago) proteins are programmable nucleases found in all domains of life, playing a crucial role in biological processes like DNA/RNA interference and gene regulation. Mesophilic prokaryotic Agos (pAgos) have gained increasing research interest due to their broad range of potential applications, yet their molecular mechanisms remain poorly understood. Here, we present seven cryo-electron microscopy structures of Kurthia massiliensis Ago (KmAgo) in various states. These structures encompass the steps of apo-form, guide binding, target recognition, cleavage, and release, revealing that KmAgo employs a unique DDD catalytic triad, instead of a DEDD tetrad, for DNA target cleavage under 5′P-DNA guide conditions. Notably, the last catalytic residue, D713, is positioned outside the catalytic pocket in the absence of guide. After guide binding, D713 enters the catalytic pocket. In contrast, the corresponding catalytic residue in other Agos has been consistently located in the catalytic pocket. Moreover, we identified several sites exhibiting enhanced catalytic activity through alanine mutagenesis. These sites have the potential to serve as engineering targets for augmenting the catalytic efficiency of KmAgo. This structural analysis of KmAgo advances the understanding of the diversity of molecular mechanisms by Agos, offering insights for developing and optimizing mesophilic pAgos-based programmable DNA and RNA manipulation tools.
Graphical Abstract
Graphical Abstract.
Introduction
Argonaute (Ago) proteins are nucleic acid-guided programmable nucleases that play a crucial role in the DNA/RNA interference pathways across all three domains of life (1,2). Eukaryotic Ago (eAgo) proteins mediate RNA interference by utilizing short RNAs as guides to target complementary RNA sequences (3,4). In contrast to eAgos, which exclusively target RNAs (5), prokaryotic Ago (pAgo) proteins employ guide DNAs or guide RNAs to target both DNAs and/or RNAs (6–16). Consequently, pAgos demonstrate a broader range of functions, such as targeting bacteriophages and plasmids through DNA interference (6,7,17–19), unlinking replicating chromosomes (20), and enhancing homologous recombination (21,22).
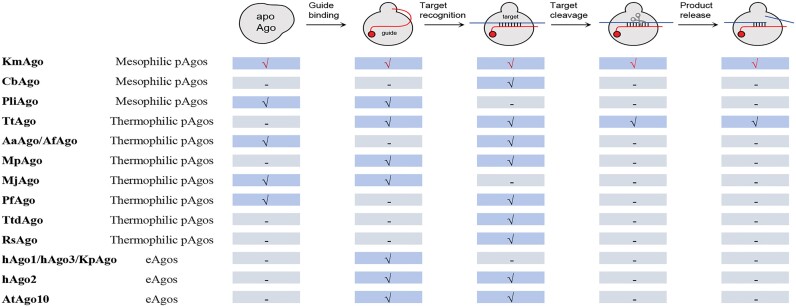
pAgos are prevalent in archaea, accounting for approximately 25% of all sequenced genomes, and in bacteria, accounting for around 17% of genomes (23). pAgos can be divided into long pAgos and short pAgos. Structures of apo-form Agos and their guide/target strands bound forms have been reported (10,24–38). eAgos and the majority of long pAgos consist of six main domains: N (N-terminal), L1 (Linker 1), PAZ (Piwi-Argonaute-Zwille), L2 (Linker 2), MID (Middle) and PIWI (P-element induced wimpy testis) (2,24). The catalytically active Agos PIWI contains a DEDX (X represents D, H, N or K) catalytic tetrad coordinates two divalent metal cations crucial for target cleavage (10,30,33). The MID creates a base pocket that accommodates the 5′-end of the guide strand through stacking and polar interactions. This process involves a divalent metal cation bound in certain eAgos and pAgos (10,25,28,39). The PAZ binds to the 3′-end of the guide strand, which is subsequently released during target recognition. The initial few nucleotides of the guide strand from the ‘seed’ region are pre-arranged in a helical conformation and play a role in the initial target recognition. The precise positioning of the guide within the Ago molecule determines the site of target cleavage, typically taking place between the 10th and 11th guide nucleotides in most studied pAgos and eAgos (2,24). Short pAgos retain the MID and PIWI important for guide-mediated target binding but lack cleavage activity. Emerging insights reveal that various short pAgos interact with different accessory ‘effector’ enzymes (17,23,40).
Due to their relatively small protein size and the absence of strong requirements for specific motifs during guide binding or target recognition, pAgos have the potential to be utilized as tools for nucleic acid manipulation and genomic technologies (41,42). A few pAgos have been well characterized and structurally studied (24). Notably, most of these enzymes are derived from thermophilic bacteria and archaea (Supplementary Table S1) (6,9,10,25,26,32,33). Nevertheless, their high optimal activity temperature, which ranges from 55 to 99.9°C, hinders their usefulness in vivo gene editing of species of interest and restricts their applications in detection-based diagnostic devices (6,8,34). Consequently, there has been an accelerated discovery of mesophilic pAgos that can carry out cleavage at moderate temperatures, enabling their translation to in vivo genome editing and other applications (11,13–16,43). However, structures of mesophilic pAgos in the active state that have both metal ions and target bound within the PIWI catalytic pocket have not yet been determined, although several structures of mesophilic pAgos have been determined (14,16,27). To understand how the mesophilic pAgos cleave targets, it is critical to elucidate the structures of mesophilic pAgo complexes in their catalytically competent states, with both metal ions and substrate bound in the PIWI catalytic pocket.
KmAgo from the mesophilic bacterium Kurthia massiliensis exhibits an unexpectedly broad specificity for guide and target molecules (11,43). KmAgo is capable of cleaving both DNA and RNA targets using both DNA and RNA guides, making it potentially applicable in various nucleic acid biotechnology applications. In this study, we determined seven cryo-electron microscopy (cryo-EM) structures of KmAgo in diverse states. Together with mutagenesis analysis and molecular dynamics simulations, we offer snapshots of distinct key steps within the catalytic cycle, providing fundamental insights into a diverse and yet enigmatic part of the Ago superfamily.
Materials and methods
Protein expression and purification
The KmAgo gene and its double mutant (KmAgo-DM, D527A, D596A) have been cloned into pET28a expression vectors with the N-terminal His6 tag in the previous study and stored in our laboratory. Point mutants used for in vitro activity assays were constructed by polymerase chain reaction (PCR) mediated site-directed mutagenesis (44). All cloned constructs were verified by DNA sequencing. The purification protocol for KmAgo and KmAgo-DM was adapted from our previous study (11). Briefly, the plasmids were transformed into Escherichia coli strain BL21 (DE3) (Novagen). The cultures were incubated at 37°C in an LB medium containing 50 μg/mL kanamycin until reaching an optical density (OD600= 0.6–0.8). Protein expression was then induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM. After induction, the cultures were incubated for 20 h at 18°C. Then, cells were harvested by centrifugation at 6000 rpm for 10 min, flash-frozen and stored at −80°C for subsequent purification.
KmAgo and KmAgo-DM proteins used for structures were prepared through a three-step purification. The cells were suspended in buffer A (50 mM Tris–HCl, pH 7.5, 500 mM NaCl) and then lysed using a JN-02C cell disrupter (JNBIO, Inc.). After centrifugation at 14 000 rpm for 30 min, the lysate was clarified, and the resulting soluble fraction was applied to a 4 ml packed Ni-NTA resin (Qiagen). The resin was washed with buffer A containing 60 mM imidazole, and then the target protein was eluted using buffer A containing 250 mM imidazole. The eluted protein was diluted in 50 mM Tris–HCl, pH 7.5, to reduce the final salt concentration to 150 mM NaCl. The diluted protein was loaded onto a Heparin column (HiTrap Heparin HP, GE Healthcare) that had been washed with buffer B (50 mM Tris–HCl, pH 7.5, 125 mM NaCl) and then eluted using a linear gradient of 0.125–2 M NaCl. Fractions containing target protein were concentrated using an Amicon 50K filter unit (Millipore) through ultrafiltration. The final purification step involved size-exclusion chromatography using a Superdex 200 10/300 increase column (GE Healthcare) equilibrated with buffer C (20 mM HEPES, pH 7.5, 150 mM NaCl and 5 mM DTT). The purified proteins were concentrated in buffer C to a final concentration of 2 mg/ml, then aliquoted and stored at −80°C. Only Ni-NTA resin purification was performed for KmAgo and its mutants used in vitro activity analysis.
Activity assays
Activity assays were conducted using synthetic guides and 5′FAM labeled targets. The oligonucleotide sequences can be found in Supplementary Table S2. Unless otherwise specified, 800 nM KmAgo or its mutant was mixed with 400 nM guides and incubated for 10 min at 37°C in a reaction buffer containing 10 mM HEPES-NaOH (pH 7.5), 5 mM MnCl2, 100 mM NaCl and 5% glycerol for guide loading. Then, target nucleic acids were added to the final concentration of 200 nM. The cleavage reactions were carried out in PCR tubes at 37°C and stopped after 15 min by mixing the samples with equal volumes of 2 × RNA loading dye, which consisted of 95% formamide, 18 mM EDTA, 0.025% sodium dodecyl sulfate (SDS), and 0.025% bromophenol blue. The samples were heated for 5 min at 95°C. The cleavage products were separated by 20% denaturing polyacrylamide gel electrophoresis (PAGE), visualized using the GelDoc Go Gel Imaging System (Bio-Rad), and analyzed using the NIH program ImageJ and Prism 8 (GraphPad).
Cryo-EM grids preparation and data acquisition
KmAgo and KmAgo-DM were purified as described above (Supplementary Figure S2). For the formation of KmAgogDNA binary complex grids, purified KmAgo was mixed with 18 nt 5′-phosphorylated guide DNA (5′P-gDNA) in a 1:1 molar ratio and incubated at 37°C for 30 min. A 4 μl drop of 1.0 mg/ml was added to glow-discharged 300 mesh holey gold grids (R1.2/1.3, Quantifoil), blotted for 4–5 s in 100% humidity at 4°C, and then plunged into liquid ethane using an FEI Vitrobot Mark IV (Thermo Fisher Scientific). To prepare ternary complex grids, the protein–nucleic acid complex was incubated at 37°C for 30 min or 60 min with a final molar ratio of 1:1:1 (protein: 5′P-gDNA: target DNA). The grid preparation procedure was identical to that of the KmAgogDNA binary complex. The final concentration of manganese in all reactions is 5mM.
The cryo-EM grids were loaded onto a 300 kV Titan Krios microscope (Thermo Fisher Scientific) equipped with a Gatan K3 direct electron detector and a BioQuantum energy filter, which operated at a slit width of 20 eV. Micrographs were collected automatically by EPU using standard procedures. Images were captured using a calibrated magnification of 105 000×, resulting in a pixel size of 0.851 Å. The defocus range was set between −1.1 μm and −1.5 μm. Each micrograph comprised 40 frames with a total exposure time of 2.5 s, resulting in an accumulated dose of about 54 e−/Å2.
Image processing
For KmAgoapo, KmAgogDNA, pre2-KmAgogDtD, pre1-KmAgo-DMgDtD, KmAgo-DMgDtR (cleaved, released)-KmAgogDtD, a total of 2739, 4026, 2931, 4715, 12 939 and 2581 image stacks, respectively, were summed and corrected for drift and beam-induced motion using MotionCor2 (45). The following data processing steps were performed with cryoSPARCv4.2.1 (46). The contrast transfer function parameters for each micrograph were estimated using the CryoSPARC patch CTF algorithm. For KmAgoapo, 5219822 raw particles were blob picked with a particle diameter range of 6–12 nm and subjected to ab-initio reconstruction and heterogeneous refinement. Then, each class was subjected to 2D classification individually to remove bad particles and contaminants. Good classes showed clear features were combined and subjected to another four rounds of ab-initio, heterogeneous refinement, and 2D classification, yielding 637006 good particles for non-uniform refinement. The refined particles were split into five classes for 3D classification. Three classes containing 386173 particles showed detailed features and were subjected to non-uniform refinement, resulting in a map at 2.85 Å resolution. For the rest of the datasets, the map of KmAgoapo was used to generate 2D templates with a particle diameter of 6 nm for template-picking. The final dataset of KmAgogDNA, pre2-KmAgogDtD, pre1-KmAgo-DMgDtD, KmAgo-DMgDtR contained 439341, 199254, 556763, and 439557 particles, which generated maps with resolutions at 2.66, 2.96, 2.68 and 2.84 Å, respectively. For (cleaved, released)-KmAgogDtD samples, two states were clearly defined after the final 3D classification. One with cleaved target DNA containing 308521 particles was further refined to 2.76 Å resolution, and the other with released target DNA containing 288 696 particles yielded a 2.76 Å density map. Local resolution estimation was performed with the CryoSPARC local resolution estimation algorithms. All parameters for image processing were summarized in Table 1A,B and processing procedures of all datasets were shown in Supplementary Figure S3, S4, S5, S6. All 3D density maps were displayed using UCSF Chimera (47).
Table 1A.
Cryo-EM data collection, refinement and validation statistics
| KmAgoapo (EMD-38354) (PDB: 8XHS) | KmAgogDNA (EMD-38355) (PDB: 8XHV) | pre1-KmAgo-DMgDtD (EMD-38409) (PDB: 8XJX) | KmAgo-DMgDtR (EMD-38408) (PDB: 8XJW) | |
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 105 000 | 105 000 | 105 000 | 105 000 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 54 | 54 | 54 | 54 |
| Defocus range (μm) | −1.0 ∼ −1.5 | −1.0 ∼ −1.5 | −1.0 ∼ −1.5 | −1.0 ∼ −1.5 |
| Pixel size (Å) | 0.851 | 0.851 | 0.851 | 0.851 |
| Symmetry imposed | C1 | C1 | C1 | C1 |
| Initial particle images (no.) | 2 644 446 | 10 959 937 | 13 369 141 | 11 662 653 |
| Final particle images (no.) | 386 173 | 439 341 | 556 763 | 439 557 |
| Map resolution (Å) | 2.85 | 2.66 | 2.68 | 2.84 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.44–42.73 | 2.30–39.22 | 2.48–41.04 | 2.43–44.28 |
| Refinement | ||||
| Initial model used | AlphaFold2 | KmAgoapo | KmAgoapo | KmAgoapo |
| Model resolution (Å) | 2.82 | 2.63 | 2.64 | 2.82 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | n/a | n/a | n/a | n/a |
| Map sharpening B factor (Å2) | −132.9 | −112.1 | −129.4 | −130.2 |
| Model composition | ||||
| Non-hydrogen atoms | 5076 | 6205 | 6310 | 6247 |
| Protein residues | 618 | 721 | 693 | 690 |
| Ligand | 0 | 1 | 1 | 1 |
| B-factors (Å2) | ||||
| Protein | 11.35/100.10/43.16 | 35.94/120.37/55.79 | 29.99/111.22/57.56 | 19.94/51.65/33.65 |
| Nucleotide | — | 27.27/122.98/60.44 | 29.83/71.22/43.37 | 21.23/56.87/34.33 |
| Ligand | — | 29.16/29.16/29.16 | 66.88/66.88/66.88 | 28.24/28.24/28.24 |
| Water | 9.05/33.39/18.20 | 15.27/33.84/24.58 | 19.99/77.57/32.76 | 15.05/28.27/23.57 |
| R.m.s. deviations | ||||
| Bond lengths(Å) | 0.003 | 0.003 | 0.003 | 0.002 |
| Bond angles (o) | 0.548 | 0.583 | 0.567 | 0.579 |
| Validation | ||||
| MolProbity score | 1.63 | 1.66 | 1.76 | 1.79 |
| Clashscore | 4.66 | 5.96 | 7.80 | 9.16 |
| Poor rotamer (%) | 0.00 | 0.00 | 0.00 | 0.00 |
| Ramachandran plot | ||||
| Favored (%) | 95.72 | 95.24 | 95.20 | 95.76 |
| Allowed (%) | 4.28 | 4.76 | 4.80 | 4.24 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 | 0.00 |
Table 1B.
(Cont.). Cryo-EM data collection, refinement and validation statistics
| Pre2-KmAgogDtD (EMD-38412) (PDB: 8XK0) | Cleaved-KmAgogDtD (EMD-38414) (PDB: 8XK3) | Released-KmAgogDtD (EMD-38415) (PDB: 8XK4) | |
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 105 000 | 105 000 | 105 000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 54 | 54 | 54 |
| Defocus range (μm) | −1.0 ∼ −1.5 | −1.0 ∼ −1.5 | −1.0 ∼ −1.5 |
| Pixel size (Å) | 0.851 | 0.851 | 0.851 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 8 545 993 | 7 443 662 | 7 443 662 |
| Final particle images (no.) | 199 254 | 308 521 | 288 696 |
| Map resolution (Å) | 2.96 | 2.76 | 2.76 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.48–41.04 | 2.42–39.41 | 2.41–42.82 |
| Refinement | |||
| Initial model used | KmAgoapo | KmAgoapo | KmAgoapo |
| Model resolution (Å) | 2.91 | 2.72 | 2.75 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Model resolution range(Å) | n/a. | n/a. | n/a. |
| Map sharpening B factor (Å2) | −129.4 | −120.4 | −119.9 |
| Model composition | |||
| Non-hydrogen atoms | 6301 | 6309 | 6049 |
| Protein residues | 692 | 692 | 689 |
| Ligand | 2 | 3 | 3 |
| B-factors (Å2) | |||
| Protein | 28.95/102.36/48.40 | 24.12/71.89/41.39 | 40.02/118.37/60.99 |
| Nucleotide | 30.69/101.34/50.77 | 13.22/68.90/39.99 | 44.27/112.92/58.20 |
| Ligand | 40.00/53.16/46.58 | 36.44/49.50/42.26 | 51.88/73.39/65.68 |
| Water | 13.62/41.51/24.93 | 12.73/36.00/25.53 | 25.17/27.24/26.21 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | 0.003 | 0.003 |
| Bond angles (o) | 0.670 | 0.545 | 0.573 |
| Validation | |||
| MolProbity score | 1.85 | 1.63 | 1.67 |
| Clashscore | 5.94 | 5.29 | 7.56 |
| Poor rotamer (%) | 0.00 | 0.00 | 0.00 |
| Ramachandran plot | |||
| Favored (%) | 92.57 | 95.04 | 96.18 |
| Allowed (%) | 7.43 | 4.96 | 3.82 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 |
Model building and refinement
The initial template of KmAgoapo was generated using AlphaFold2 (48), and then the rigid body was fitted into a cryo-EM density map with Chimera, followed by manual adjustment and rebuilding in COOT (49). The model was further refined by several rounds of real-space refinement in Phenix (50). For the rest of the six structures, the well-refined KmAgoapo model was used as a starting template, followed by iterative manual adjustment in COOT and real-space refinement in PHENIX. The final model statistics were validated and provided by MolProbity (51) and summarized in Table 1. Chimera was used for preparing the structural figures.
Molecular dynamics simulations
Based on the cryo-EM structure of cleaved-KmAgogDtD, we constructed a complex model for a system with reconstructed intact target DNA, which mimics the intermediate state. First, the target DNA in the cleaved-KmAgogDtD structure was reconstructed using COOT to repair the scissile phosphate (49). Then, the missing residues in the protein were modeled using MODELLER (52). The water molecules in the original structure were removed except for four waters close to Mn2+ A and Mn2+ B that were expected to be important for enzymatic activity. Hence, the initial model of the intermediate state was obtained.
CHARMM-GUI was used to build the initial structure of MD simulations (53). MD simulations were conducted in GROMACS 2020 using the all-atom CHARMM36m force field and the modified TIP3P water model, with the addition of 150 mM Na+ and Cl− ions to neutralize the system (54,55). Energy minimization was performed using the steepest descent algorithm for 50 000 steps. System equilibration was carried out for 1 ns with constraints on the hydrogen bonds in the isothermal-isochoric (NVT) and isothermal-isobaric (NPT) ensembles, respectively (56). The temperature was maintained at 298 K using the v-rescale method, and the pressure was kept constant at 1 bar by semi-isotropic coupling to a Parrinello-Rahman barostat with τp = 2.0 ps and a compressibility of 4.5 × 10−5 bar (57,58). Subsequently, three independent 500 ns production runs were performed for each simulation. H bonds were constrained using the LINCS algorithm, with a time step of 2 fs (59). The cutoff values for electrostatic interactions and van der Waals interactions were both set to 1.2 nm. Long-range electrostatic interactions were computed using the particle mesh Ewald (PME) method (60). The trajectories were analyzed using tools implemented in GROMACS and in-house scripts (61).
Results
Overall structure of the apo-form KmAgo with D713 and E562 positioned away from the catalytic center
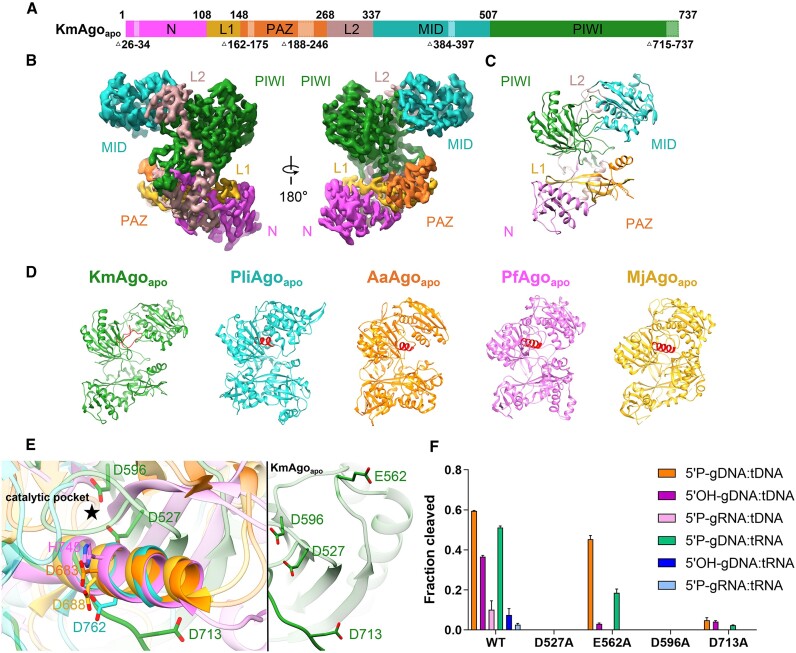
We first determined the structure of the apo-form KmAgo (KmAgoapo) to 2.85 Å overall resolution (Figure 1A–C, Table 1, Supplementary Figure S3a, S7a, S8a). KmAgo shares 10–23% sequence identity with the Ago proteins from eukaryotes, bacteria, and archaea whose structures have already been determined (Supplementary Table S3). KmAgo possesses four conserved domains (N, PAZ, MID, PIWI) and two linker regions (L1, L2) found in other Ago proteins. The overall bi-lobed molecular architecture is typical for the family of Ago proteins, and the arrangement of the two lobes is comparable to other pAgos (Figure 1D). Some regions of KmAgo appear to have intrinsic flexibility. This concerns particular regions in the PAZ domain and the C-terminal region (CTR) of the PIWI domain (Figure 1A–D). For other Agos, it has been shown that these regions are involved in nucleic acid interactions and undergo conformational changes upon nucleic acid binding (25,31–33).
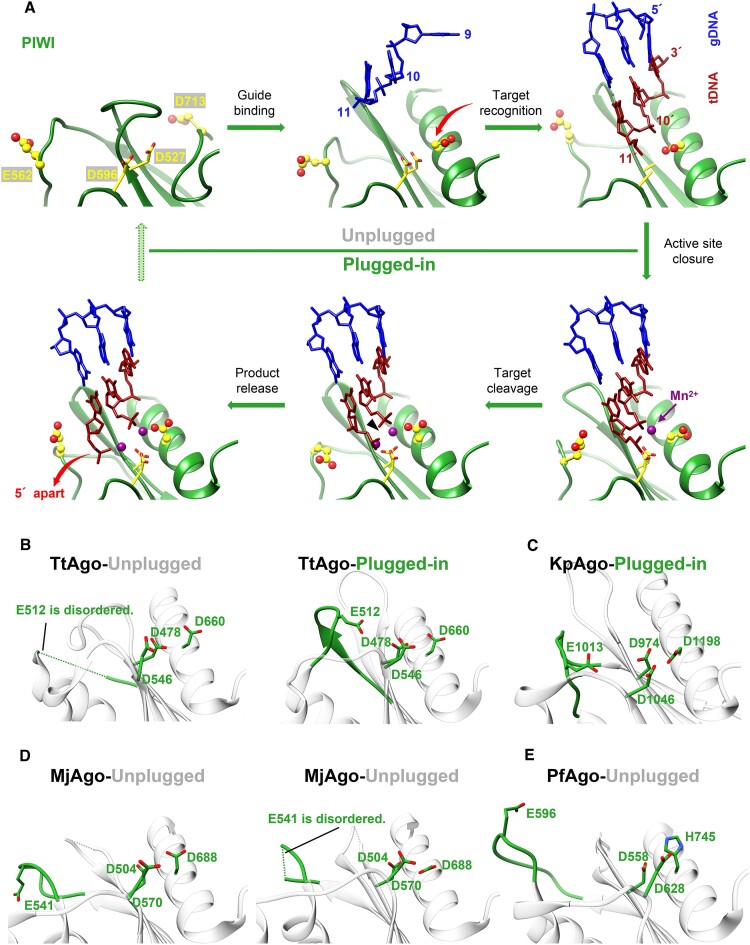
Figure 1.
The overall structure of KmAgoapo. (A) The primary structure of KmAgoapo with amino acid numbering. Domains and their unstructured residues are colored and labeled. (B, C) Cryo-EM density map (B) and ribbon diagrams (C) of KmAgoapo, with domains colored as in (A). (D) Structural comparison of the apo form of KmAgo, PliAgo (PDB: 7R8F), AaAgo (PDB: 1YVU), PfAgo (PDB: 1U04) and MjAgo (PDB: 5G5S). The structures were superimposed onto each other, focusing on their C-terminal regions, which were subsequently highlighted in red. (E) (left) A magnified view comparing the C-terminal regions of KmAgoapo with other pAgo structures, as depicted in (D). (right) A magnified view of catalytic tetrad in KmAgoapo from the same perspective. Color schemes are indicated by the labels. (F) Cleavage activity of the catalytic site mutants of KmAgo. Average cleavage efficiencies were plotted from three technical replicates. Error bars represent the standard deviations.
Structural investigations of TtAgo (Thermus thermophilus Ago) have revealed that the catalytic pocket is comprised of three Asp residues, with an inserted Glu residue completing the catalytic tetrad, indicating the establishment of a cleavage-competent state (33). The PIWI domain of KmAgo carries the catalytic tetrad D527, E562, D596 and D713 (Supplementary Figure S1). When comparing KmAgoapo to the other apo-form Agos, we observed striking differences in the catalytic pocket of the PIWI domain (Figure 1E). In KmAgo, both E562 within the glutamic acid finger and D713 situated in the CTR are observed in an ‘unplugged’ position. D713 is located on a loop away from the catalytic pocket. While in other Agos, the corresponding catalytic residues of D713 are situated within an alpha helix, and the side chain of those residues are positioned in the catalytic pocket. Thus, we introduced single alanine mutations at these four positions to verify the catalytic residues (Figure 1F, Supplementary Figure S9). A single mutation at positions D527, D596 or D713 almost abolished cleavage activity, suggesting that D713 is the catalytic aspartate. Surprisingly, although mutation of glutamate finger (E562) abolished guide RNA mediated cleavage activity, it still had good guide DNA mediated cleavage activity. Furthermore, the cleavage activity of E562A mediated by guide DNA targeting RNA decreased by over 50%. However, it still exhibited robust cleavage activity when targeting DNA. Studies on D-R pAgos have shown that target selection is mainly achieved during the cleavage step (16). We hypothesize that the potential interaction between KmAgo's E562 residue and target DNA may not be essential for accurate targeting and cleavage of DNA in the active site, whereas it is pivotal for target RNA recognition and processing. The ‘unplugged’ state of both D713 and E562 suggests that the catalytic center of KmAgo in the apo state is immature, thereby lacking cleavage ability. The structure of apo KmAgo reveals how the concerted action of ‘unplugged’ D713 and E562 keeps Ago in an inactive state.
The KmAgo-guide DNA structure featuring a distinctive kink in the gDNA
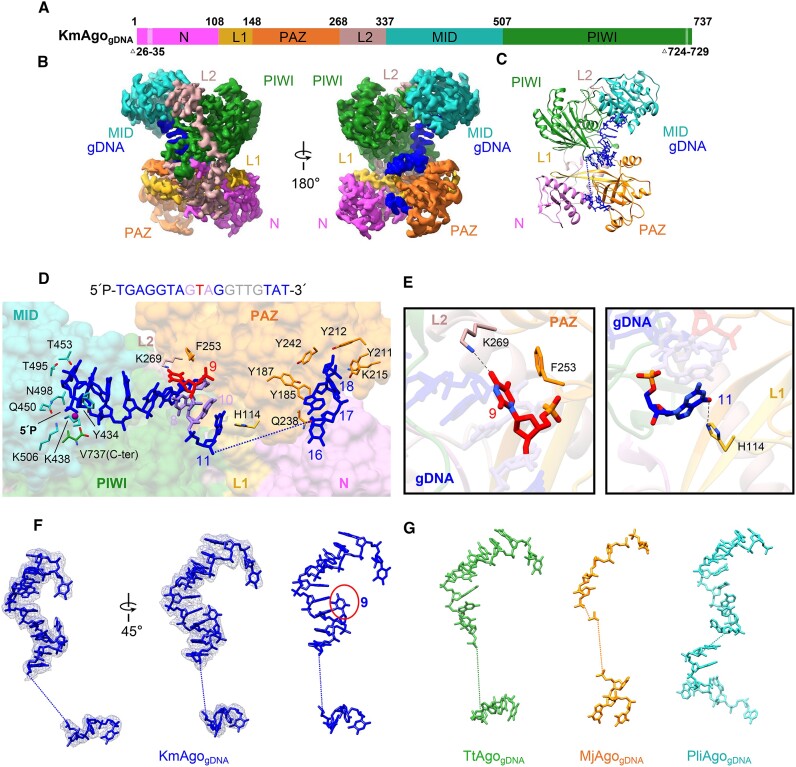
To provide structural insights into guide binding, we determined the 2.66 Å cryo-EM structure of KmAgo bound to 18 nt 5′P-gDNA (Figure 2A–C, Table 1, Supplementary Figure S3b, S7b, S8b). The bound 5′P-gDNA is shown in blue, with the chain traceable for nucleotides 1–11 and 16–18. Nucleotides 12 through 15 are poorly ordered (Figure 2D, F), typical of guide-bound Ago structures (25,28). The gDNA strand passes through a central basic channel located between the PAZ-containing (N, L1 and PAZ) and PIWI-containing (MID and PIWI) lobes of the bi-lobal scaffold of KmAgo (Figure 2B, C). This allows the gDNA to contact all domains and linkers, ultimately defining the nucleic acid-binding channel within the protein. The 5′- and 3′-terminal nucleotides of the guide DNA strand are anchored in the MID and PAZ domains, respectively. To stabilize the guide 5′-phosphate, KmAgo shares a conserved four-amino acid motif X-K-Q-K (X = K, R, H and Y) in the MID domain (KmAgo: Y434, K438, Q450, K506; Supplementary Figure S1). A Mn2+ ion is coordinated by the first and third phosphates of the guide DNA strand and by the carboxyl group of V737. The first base is flipped out and stacked over the side chain of a tyrosine Y434. The insertion at the 3′-end of the guide enables the terminal two nucleotides (17,18) to engage in interactions with multiple aromatic side chains, including Y212, Y187, Y185, Y211 and Y242. This interaction significantly augments the stability of the PAZ domain compared to its unbound state. Mutation of R41, Y187, Y211 or Y242 to alanine significantly enhances the cleavage activity, particularly towards RNA targets. Notably, the R41A and Y211A mutants displayed remarkably higher cleavage activity compared to DNA, when guided with 5′P-gDNA (Figure 2C, D, Supplementary Figures S10 and S11).
Figure 2.
The overall structure of KmAgogDNA. (A) The primary structure of KmAgogDNA with amino acid numbering. Domains and their unstructured residues are colored and labeled. (B, C) Cryo-EM density map (B) and ribbon diagrams (C) of KmAgogDNA, with domains colored as in (A). The density of guide DNA is indicated by a blue color. (D) (Top) 5′P-gDNA sequence within the binary complex. Unmodelled segments were highlighted in gray. (Bottom) A magnified representation of the interactions between KmAgo and 5′P-gDNA. KmAgo is represented as a semi-transparent surface. The amino acid residues crucial for guide positioning, along with the nucleotides, are labeled and represented using stick models. (E) Detailed interactions at the guide strand T9 and G11. (F) Structure of the guide DNA in KmAgo-gDNA, displayed with cryo-EM density shown in mesh. The kink at position 9 is marked with a red circle. (G) Structure of the guide DNA in TtAgo-gDNA (PDB: 3DLH), MjAgo-gDNA (PDB: 5G5T) and PliAgo-gDNA (PDB: 7R8H) binary complex.
In Agos bound to guide DNA, there is usually no noticeable kink (Figure 2F, G) (16,25,28). Surprisingly, a distinct kink is observed in the binary structure of KmAgo, specifically within the guide nucleotides 8–10 (Figure 2D, F). In detail, the nucleotides G2 to G8 are continuously stacked, while the nucleotide T9 is flipped out. The flipped nucleotide T9 is stabilized through pi-stacking with F253 within the PAZ domain. K269, which extends from the L2 linker, makes base-specific contacts with nucleotide T9 (Figure 2E). The unexpected kink leads to the direct stacking of nucleotide G8 with the nucleotide A10 base. Additionally, prior to the disordering of the guide, we observed hydrogen bonding between G11 and H114 (Figure 2E). Mutational analysis showed that F253A, K269A and H114A remarkably increased cleavage activity (Supplementary Figures S10b and S11). These results indicated that this unique kink may serve as a species-specific readout to modulate cleavage efficiency.
Guide DNA binding induces D713 plugged in the catalytic pocket
Comparison of the apo KmAgo with the gDNA-loaded binary complex structure reveals that gDNA binding induces a significant conformational change, resulting in the repositioning of the domains in the two lobes and the opening of the binding channel (Supplementary Figure S12a). Among them, MID domain shifts around 11° and PAZ domain rotates by 33°, both with respect to PIWI domain. In the MID domain, the residues (K429-T453) undergo significant upward movement upon the entry of the guide, preventing steric clashes between KmAgo and the 5′-end of the guide DNA (Supplementary Figure S12d). The residues (S383-T415) become more organized relative to its apo form (Supplementary Figure S12e).
An important difference in the KmAgo binary complex with 18 nt guide DNA (relative to the apo KmAgo) lies in the insertion of D713 into the catalytic pocket, forming a triad with the other two catalytic Asp residues. The formation of this triad may indicate the presence of a pre-mature catalytic pocket, wherein E562 remains positioned outside the catalytic pocket in an unplugged state (Supplementary Figure S12 and S13a). We observed conformational changes on proceeding from the apo form to the binary complex with 18 nt guide DNA. Specifically, large structural transitions are observed within CTR, loop 1 and loop 2 of KmAgo (Supplementary Figure S13a), which is different from all previously characterized Agos. In PliAgo and MjAgo, no significant conformational changes are observed in these regions (Supplementary Figure S13b, c) (16,25). In contrast, in KmAgo the CTR undergoes a conformational change, transitioning from a loop structure to a helical conformation, thereby enabling the insertion of D713 into the catalytic pocket (Supplementary Figure S12b). Loop 1 extends outward by 20°, resulting in the formation of an antiparallel β-sheet within the binary complex. Loop 2 rises by 8°, indicating a conformational change in this region (Supplementary Figure S12c).
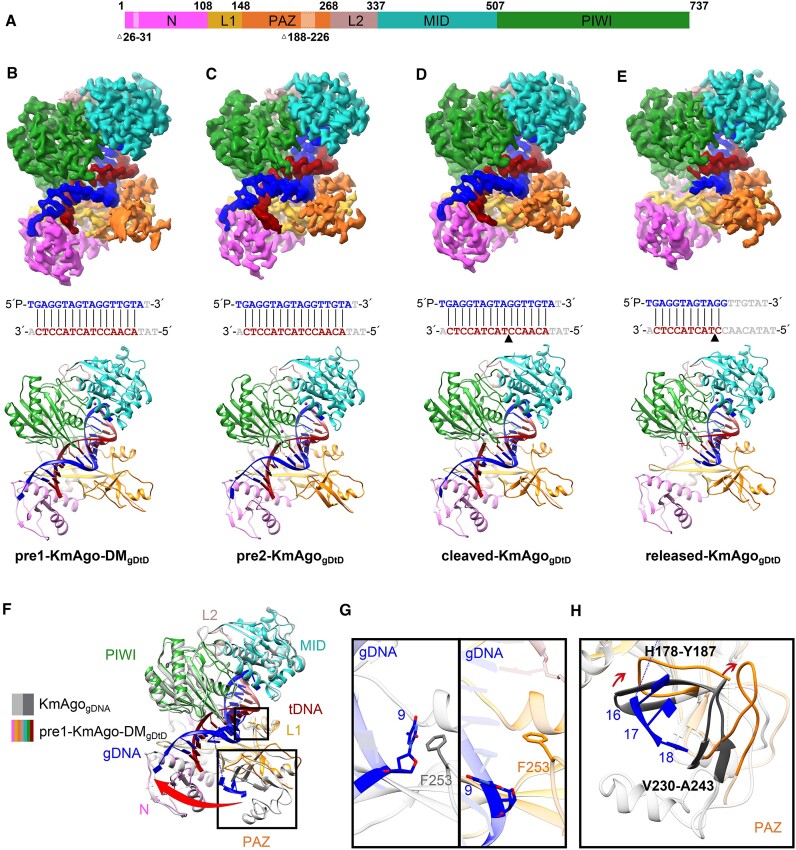
Ago ternary complex with guide DNA and target DNA
To gain structural insights into KmAgo guide mediated target recognition, catalytic inactive KmAgo-DM (D527A/D596A) was used to observe conformations after target recognition but before target cleavage. We determined the cryo-EM structure at 2.68 Å resolution of KmAgo-DM in complex with an 18 nt 5′P-gDNA and 19 nt target DNA (pre1-KmAgo-DMgDtD) (Figure 3a, 3b, Table 1, Supplementary Figures S4a, S7c, S8c). Then, by promptly preparing samples after adding the target DNA, we obtained a 2.96 Å wild-type KmAgo ternary complex structure (pre2-KmAgogDtD) containing intact target DNA (Figure 3A, C, Table 1, Supplementary Figures S4b, S7d, S8d). By extending the incubation time to 60 min, we captured the structures containing cleaved target DNA (Figure 3A, D, E). Initial 3D reconstructions indicated the presence of multiple DNA configurations within the KmAgo, leading to the refinement of two structures of the ternary KmAgo–guide–target complex at 2.76 Å overall resolution (Table 1, Supplementary Figures S5, S7e, S8e, S8f). In the first structure (cleaved-KmAgogDtD) (Figure 3A, D), we observed intact guide-target duplex segments on either side of the cleaved target DNA strand. In the second structure (released-KmAgogDtD) (Figure 3A, E), not only did the structure of this complex show cleavage of the target DNA strand at the 10′–11′ step but also duplex formation was retained only on the side containing the seed segment, with the guide strand disordered beyond position 12. Moreover, the density of the target chain (C12’-T19’) is absent, resulting in the downward flipping of the 3′ end of the guide strand at a specific angle.
Figure 3.
The overall structures of KmAgo-DMgDtD and KmAgogDtD. (A) Domain architecture of ternary complexes with amino acid numbering. Domains and their unstructured residues are colored and labeled. (B–E) Cryo-EM density map (Top) and ribbon diagrams (Bottom) of pre1-KmAgo-DMgDtD (B), pre2-KmAgogDtD (C), cleaved-KmAgogDtD (D), released-KmAgogDtD (E). (Middle) The sequence and pairing of gDNA (blue) and tDNA (red) strands in the ternary complex. Nucleotides that were not modelled are represented in gray. The black triangle in (D) and (E) denotes the location of the cleavage site. (F) Superposition of pre1-KmAgo-DMgDtD (colored as in B) and KmAgogDNA (gray). (G) Enlarged view of the interaction between position 9 and F253. The left panel is the binary structure, and the right panel is the pre1-KmAgo-DMgDtD. (H) Significant conformational changes in the PAZ domain are indicated by red arrows.
The overall protein structures of these four ternary complexes are almost identical, and the DNA target (in red) can be traced from positions 2′ to 16′, with guide-target pairing spanning 15 bp from base-pair position 2–2′ to 16–16′ within the nucleic acid-binding channel (Figure 3B–D), except for the released-KmAgogDtD (Figure 3E). The 5′-phosphate of the guide DNA is still anchored in the MID pocket, but the 3′-end (position 18 is disordered and cannot be traced) is released from the PAZ pocket to the N domain on formation of the 15-bp guide-target duplex (Figure 3B-D). We observed conformational changes on proceeding from the binary complex to the ternary complex, and these changes can be visualized after superimposing the PIWI-containing (Mid and PIWI) lobe as indicated by the red arrow in Figure 3F. These changes involve a pivotal rotation of the PAZ domain and large structural transitions within it (Figure 3F). To enable the pairing of gDNA-tDNA, the protruding T9 residue in the binary structure undergoes an inward flip, while the side chain of F253 relocates to create ample space for unhindered base pairing (Figure 3G). Besides, two loops (H178-Y187, V230-A243) within the PAZ domain moves up to tighten the binding channel (Figure 3H).
Dynamic changes of E562 during the catalytic cycle
Ago-mediated cleavage typically requires the embedding of a glutamate residue (referred to as the ‘glutamate finger’) into the catalytic pocket, resulting in the formation of a complete DEDX catalytic tetrad (30). In TtAgo, the loop region harboring the glutamate finger (E512) necessitates a substantial backbone conformational change to facilitate insertion into the pocket and subsequent cleavage (33). Intriguingly, in KmAgo, upon superimposing the glutamate finger (E562) across various states, we observed tDNA cleavage despite minor fluctuations in the backbone of E562, indicating a heightened flexibility within the loop region encompassing E562 (Supplementary Figure S14). In all ternary complexes, the glutamate finger of KmAgo lacks the stabilizing hydrogen bond network typically observed in other Ago proteins (30) (62–64).
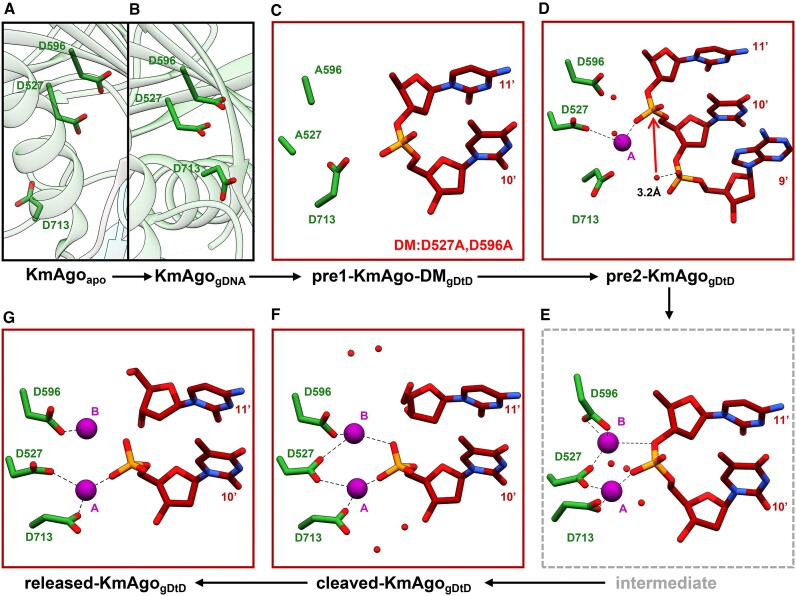
Subsequently, we conducted an individual analysis of the catalytic pocket in each state of KmAgo, revealing that the positions of D527 and D596 within the catalytic tetrad remain relatively stable. D713 inserts during the transition from the apo to the binary state, thereby promoting the pre-maturation of the catalytic pocket. E562 undergoes dynamic alterations throughout the catalytic cycle, resulting from the swinging motion of the backbone (Figure 4A). Further comparing the relative positions of the catalytic tetrad in different states of other Agos (Figure 4B–E), we find that the catalytic pocket of KmAgo when ions enter shares the most similarity with the inserted state of KpAgo's pocket (Figure 4C) (30). Based on our observations, we hypothesize that KmAgo transitions from an unplugged state to a plugged-in state upon ion entry (Figure 4A). The active site of pre1-KmAgo-DMgDtD exhibited no observable metal ion density (Figure 5C), likely due to catalytic mutations that distorted the optimal geometry for divalent cation coordination, thereby suggesting a conformation incompatible with cleavage.
Figure 4.
Comparison of the catalytic tetrad in KmAgo with other species. (A) The conformational changes within the active site of KmAgo during target recognition and catalysis. The residues located within the active site are highlighted in yellow, with E562 and D713 represented as ball-and-stick models. In the initial stages of guide binding and target recognition, the active site is unplugged (upper row). This is followed by the ‘plugged-in’ of the glutamate finger, binding of the catalytic metal, and activation of catalysis (lower row). The accompanying variations in D713 and product release are denoted by red arrows. The PDB accession numbers are (from the upper left corner, clockwise): 8XHS, 8XHV, 8XJX, 8XK0, 8XK3, 8XK4. (B–E) States whose conformations can be deduced from the other known structures are marked as green (plugged-in) and gray (unplugged). (B) TtAgo (left panel, 3DLH; right panel, 4NCB). (C) KpAgo (4F1N). (D) MjAgo (left panel, 5G5S; right panel, 5G5T). (E) PfAgo (1U04).
Figure 5.
Conformational changes of the catalytic center. Cryo-EM structure snapshots (A–D, F–G) and a molecular dynamics simulation model of the intermediate state (E) in the cleavage cycle of KmAgo are presented. The pair of Mn2+ cations are labeled and depicted as magenta spheres. Water molecules are represented as red spheres. The three catalytic residues D527, D596 and D713 are shown in stick representation. The red arrow in (D) indicates the nucleophilic attack. 3.2 Å refers to the distance from the attacking water molecule to the non-bridging phosphate oxygen.
A pair of Mn2+ cations mediate cleavage chemistry
The PIWI domain of Ago adopts an RNase H fold, with catalytic D527, D596 and D713 residues lining the active site of the KmAgo. Two Mn2+ cations have been shown to facilitate target hydrolysis during catalytic cleavage by RNase-H-containing nucleases, with cation A assisting nucleophilic attack by positioning and activating a water molecule and cation B stabilizing the transition state and leaving group (33,65). Utilizing the resolved structure of the KmAgo ternary complex in the plugged-in state, we aim to elucidate the molecular mechanism governing the cleavage of tDNA mediated by 5′P-gDNA.
In pre2-KmAgogDtD, Mn2+ A is seen, but Mn2+ B is missing (Figure 5D). Despite D713 not yet coordinated with Mn2+ A, Mn2+ A is directly coordinated with D527 and the scissile phosphate in a configuration resembling the catalytic centers of TtAgo and bacterial RNase H (33,65). Mn2+ A is surrounded by three water molecules, one prepared for in-line attack on the backbone phosphate at the cleavage site. These results indicated that this structure represents a cleavage-compatible conformation.
In cleaved-KmAgogDtD, a pair of Mn2+ cations (A and B), separated by 3.9 Å, are located on either side of the scissile phosphate between nucleotides T10’ and C11’ (Figure 5F). Mn2+ A is coordinated with D527 and D713 to a nonbridging phosphate oxygen of the newly generated 5′-phosphate. D713 coordinates with Mn2+ A by slightly deflecting its side chain. Mn2+ B is coordinated with D527 and D596 to one nonbridging oxygen of the newly generated 5′-phosphate. In released-KmAgogDtD, Mn2+ A is still coordinated with D527 and D713 to non-bridging phosphate oxygen of the newly generated 5′-phosphate, while Mn2+ B is only coordinated with D596 (Figure 5G). In TtAgo, the plugged-in glutamate finger is coordinated with the divalent metal B through a pair of bridging water molecules and may act as a structural anchor to impart the catalytic specificity of Ago (Supplementary Figure S15a) (30,33). Notably, upon the entry of Mn2+ into the catalytic center, the side chain of E562 shifted towards the catalytic pocket, resulting in a plugged-in state of KmAgo. However, our observations did not indicate that E562 was involved in the coordination of the Mn2+ cation B. (Supplementary Figure S15b–d).
To further elucidate this dynamic aspect of E562 and delineate the potential intermediate state between pre2-KmAgogDtD and cleaved-KmAgogDtD, molecular dynamics (MD) simulations starting from the cleaved complex with repaired scissile phosphate were performed (Supplementary Figure S16). The root mean square deviation (RMSD) analysis of protein C-alpha atoms showed the system equilibrated during the simulation (Supplementary Figure S16b). The results revealed that Mn2+ B stably coordinates within the catalytic pocket, forming a stable interaction with D527 and D596, alongside the departing group (Figure 5E, Supplementary Figure S16c, d, f). The E562 residue remains external to the catalytic pocket throughout the simulations, underscoring its non-essential role in the catalytic coordination (Supplementary Figure S16e, g). These observations demonstrate a non-traditional DEDX tetrad catalytic mechanism.
Comparison of Ago ternary complexes with target DNA and RNA
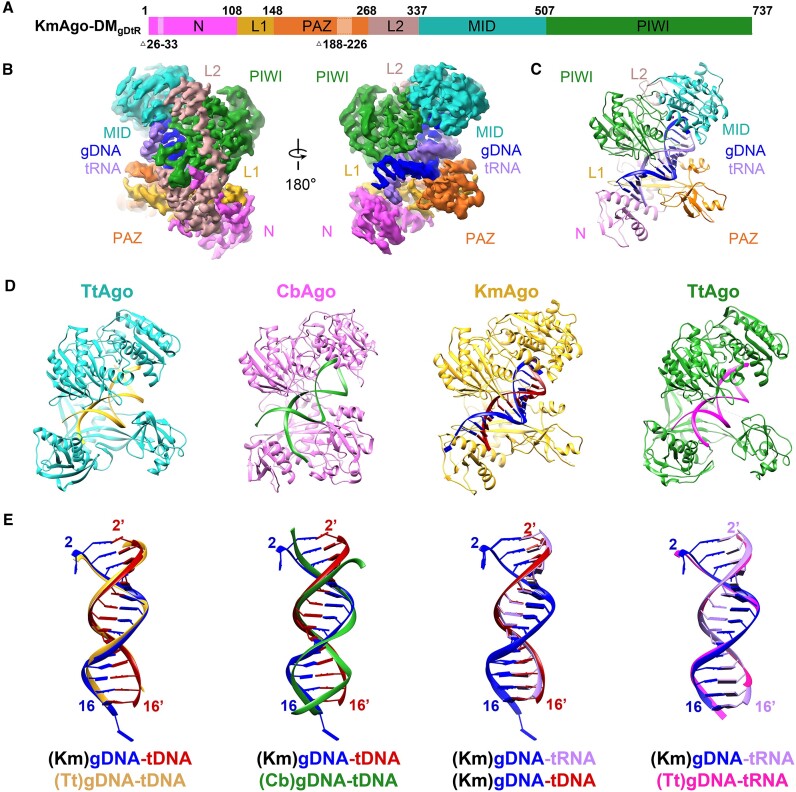
Since KmAgo also employs guide DNA to cleave RNA targets at moderate temperatures, we determined the structure of KmAgo-DM in complex with an 18 nt 5′P-gDNA and a 19 nt target RNA (KmAgo-DMgDtR) at 2.84 Å resolution (Figure 6A–C, Table 1, Supplementary Figures S6, S7f, S8g). The guide DNA strand (in blue), which can be traced from positions 1–17, is anchored at its 5′end. The target RNA strand (in purple) can be traced from positions 2′ to 16′, with guide-target pairing spanning 15 bp between base-pair positions 2–2′ to 16–16′. The overall structures of these KmAgo ternary complexes are identical (Supplementary Figure S17a).
Figure 6.
The overall structure of KmAgo-DMgDtR. (A) Domain architecture of KmAgo-DMgDtR with amino acid numbering. Domains and their unstructured residues are colored and labeled. (B, C) Cryo-EM density map (B) and ribbon diagrams (C) of KmAgo-DMgDtR, with domains colored as in (A). (D) Structural representation of KmAgo and other ternary complexes. (Left) TtAgogDtD (PDB: 4NCB); (middle left) CbAgogDtD (PDB: 6QZK); (middle right) KmAgo-DMgDtD; (right) TtAgogDtR (PDB: 3HK2). (E) Superposition of the guide DNA–target DNA and guide DNA–target RNA duplex depicted in (D) pairwise. The color schemes are designated by the corresponding labels.
KmAgo is phylogenetically closely related to mesophilic CbAgo (sequence identity 22.26%). However, our structural homology search using Dali servers showed that the overall structure of KmAgo exhibits distant similarities with thermophilic TtAgo, which is inconsistent with their distant sequence identity (sequence identity 17.95%). The guide-target duplex within the KmAgo ternary complex adopts an A-form (Supplementary Figure S17b), and exhibits good superposition with TtAgo, whereas it does not align well with CbAgo, particularly towards the termini of the duplex (Figure 6D, E). The structural observations are in agreement with the functional roles of these Agos. KmAgo and TtAgo exhibit efficient cleavage activity towards both DNA and RNA (6,11,28), whereas CbAgo is primarily effective against DNA (14).
Mutational analysis of residues interacting with targets
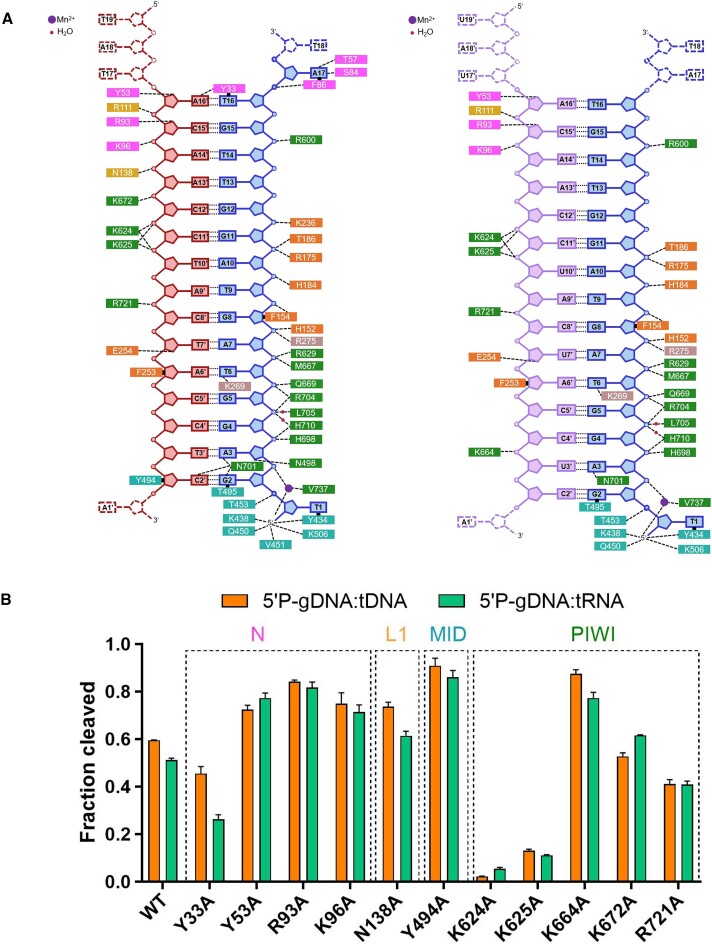
Both the DNA–RNA and DNA–DNA duplex within KmAgo are stabilized by a network of intermolecular interactions, with the entire set shown schematically in Figure 7A, and a subset of which are shown in Supplementary Figure S18. Although the amino acid residues involved in the interaction between DNA and proteins are similar in both complexes, more amino acid residues are interacting with target DNA compared to target RNA (Figure 7A). This may be one of the reasons why KmAgo has a higher efficiency in DNA cleavage. To ascertain the amino acid residues that play a crucial role in the cleavage activity of KmAgo, we mutated the amino acid residues interacting with target DNA and target RNA into alanine and measured the cleavage activity of these mutants (Figure 7B, Supplementary Figure S19).
Figure 7.
Comparison of the major contacts in KmAgo-DM ternary complexes. (A) (Left) Schematic representation of the primary interactions between KmAgo-DM and gDNA (blue) and tDNA (red). (Right) Schematic representation of the primary interactions between KmAgo-DM and gDNA (blue) as well as tRNA (purple). The residues are colored according to their domains, similar to the representation in Figure 6A. (B) Mutational analysis of amino acid residues interacting with targets. Average cleavage efficiencies were plotted from three technical replicates. Error bars represent the standard deviations.
Apart from K624A and K625A, which lead to almost complete loss of cleavage, Y33A and R721A also reduced cleavage. Notably, we observed that mutation of K672, which interacts with DNA target nucleotide A13’ (Figure 7B, Supplementary Figure S18c), increased 5′P-gDNA mediated RNA cleavage activity but slightly decreased 5′P-gDNA mediated DNA cleavage activity. Interestingly, the mutation of Y53, R93, K96, N138, Y494 or K664 to alanine increased various types of cleavage activity, especially Y494A and K664A, which significantly increased gDNA mediated cleavage activity. The activity of Ago nucleases involves multiple stages, and the activity is influenced by many factors. In addition to guide/target binding affinity, the guide/target binding rate and the products release rate also have a significant impact on their activity (14). We speculate that although mutating these sites to alanine may disrupt potential hydrogen bonds with the guide/target, thereby reducing binding affinity to some extent, it may also promote the rapid release of cleavage products or facilitate guide/target binding, ultimately exhibiting higher activity. These findings identify numerous putative mutation sites that could enhance the cleavage activity of KmAgo, thereby facilitating the advancement of nucleic acid manipulation tools reliant on KmAgo.
Discussion
In this study, we determined seven high-resolution cryo-EM structures of KmAgo: an apo-form KmAgo (2.85 Å), a guide DNA loaded KmAgo (2.66 Å), three guide DNA/target DNA loaded KmAgo with distinct catalysis step (2.96, 2.76 and 2.76 Å), a guide DNA/target DNA loaded KmAgo-DM (2.68 Å), and a guide DNA/target RNA loaded KmAgo-DM (2.84 Å). These structures encompass the stages of apo-form, guide binding, target recognition, cleavage, and release, enabling us to gain a comprehensive understanding of the conformational changes in the KmAgo catalysis process.
Surprisingly, the structural and biochemical data consistently suggest that KmAgo utilizes a non-traditional DEDX tetrad catalytic mechanism. Notably, E562 remains in a plugged-in state while efficiently executing cleavage reactions, despite not maintaining continuous coordination with divalent metal ions. In contrast, the last catalytic residue, D713, which coordinates with Mn2+ A in catalysis, may act as a structural anchor to impart the catalytic specificity of KmAgo. In the absence of a guide, D713 is positioned away from the catalytic center. After guide binding, D713 enters the catalytic center, resulting in the formation of a pre-mature catalytic pocket.
In addition, we have identified many key sites that significantly affect the cleavage activity through structural analysis and mutation analysis, laying the foundation for modifying KmAgo to adapt to a particular application. This structural analysis of KmAgo advances our understanding of the diversity of molecular mechanisms by Ago proteins. From an application perspective, our mechanism discovery provides a foundation for designing and optimizing DNA and RNA manipulation tools. Mesophilic pAgos, such as KmAgo, have the potential to be engineered as effective tools for probing RNA structures and detecting nucleic acids. A 2021 study showed that KmAgo could perform efficient RNA cleavage at elevated temperatures, which facilitates the melting of secondary structures and enables unbiased RNA processing (43). In 2022, a study addressed the need for high-temperature reaction conditions in Ago-based methods by utilizing mesophilic Agos, specifically KmAgo, to achieve CRISPR-like isothermal detection. Nonetheless, there remains potential for enhancing detection sensitivity, necessitating the augmentation of enzymatic activity in mesophilic Agos (66).
Supplementary Material
Acknowledgements
We thank the Cryo-EM facility of Hubei University for single particle cryo-EM data collection and computation support.
Author contributions: Lixin Ma(Conceptualization, Formal analysis, Validation, Writing—review & editing); Yang Liu (Conceptualization, Formal analysis, Visualization, Writing—original draft, review & editing); Shan Wu (Conceptualization, Formal analysis, Visualization, Writing—review & editing); Xin Tao (Formal analysis, Methodology, Visualization, Writing—original draft); Hui Ding (Formal analysis, Methodology, Visualization, Writing—original draft); Shaowen Wu (Formal analysis, Methodology, Visualization, Writing—original draft); Fei Wang (Formal analysis, Writing—review & editing); Hu Xu (Formal analysis, Methodology); Jie Li (Formal analysis, Methodology); Chao Zhai (Formal analysis, Writing—review & editing); Shunshun Li (Formal analysis, Methodology) and Kai Chen (Formal analysis, Methodology).
Contributor Information
Xin Tao, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Hui Ding, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Shaowen Wu, State Key Laboratory of Swine and Poultry Breeding Industry, Agro-biological Gene Research Center, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, China.
Fei Wang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Hu Xu, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Jie Li, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Chao Zhai, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Shunshun Li, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Kai Chen, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Shan Wu, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Yang Liu, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Lixin Ma, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Data availability
The cryo-EM maps have been deposited into the Electron Microscopy Data Bank (accession numbers: EMD-38354 for KmAgoapo, EMD-38355 for KmAgogDNA, EMD-38409 for pre1-KmAgo-DMgDtD, EMD-38408 for KmAgo-DMgDtR, EMD-38412 for pre2-KmAgogDtD, EMD-38414 for cleaved-KmAgogDtD, EMD-38415 for released-KmAgogDtD).
The coordinates have been deposited into the Protein Data Bank (accession numbers: 8XHS for KmAgoapo, 8XHV for KmAgogDNA, 8XJX for pre1-KmAgo-DMgDtD, 8XJW for KmAgo-DMgDtR, 8XK0 for pre2-KmAgogDtD, 8XK3 for cleaved-KmAgogDtD, 8XK4 for released-KmAgogDtD).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Key Research and Development Program of China [2023YFA0913700]; National Natural Science Foundation of China [32300517 to Y.L., 22307022 to S.W.]; China Postdoctoral Science Foundation [2022M721074]; Distinguished Young Scholars of Hubei Province [2022CFA078]; Knowledge Innovation Program of Wuhan-Shuguang Project [2023020201020418]. Funding for open access charge: Ministry of Science and Technology of the People's Republic of China.
Conflict of interest statement. None declared.
References
- 1. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 2. Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V., Patel D.J., van der Oost J.. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014; 21:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hutvagner G., Simard M.J.. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008; 9:22–32. [DOI] [PubMed] [Google Scholar]
- 4. Ghildiyal M., Zamore P.D.. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009; 10:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakanishi K. Anatomy of four human Argonaute proteins. Nucleic Acids Res. 2022; 50:6618–6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J.et al.. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuzmenko A., Oguienko A., Esyunina D., Yudin D., Petrova M., Kudinova A., Maslova O., Ninova M., Ryazansky S., Leach D.et al.. DNA targeting and interference by a bacterial Argonaute nuclease. Nature. 2020; 587:632–637. [DOI] [PubMed] [Google Scholar]
- 8. Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., van der Oost J.. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015; 43:5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S., Stockl S., Tinnefeld P., Schneider S., Klingl A.et al.. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017; 2:17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A.. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Li W., Jiang X., Wang Y., Zhang Z., Liu Q., He R., Chen Q., Yang J., Wang L.et al.. A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 2021; 49:1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Olovnikov I., Chan K., Sachidanandam R., Newman D.K., Aravin A.A.. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013; 51:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuzmenko A., Yudin D., Ryazansky S., Kulbachinskiy A., Aravin A.A.. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019; 47:5822–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hegge J.W., Swarts D.C., Chandradoss S.D., Cui T.J., Kneppers J., Jinek M., Joo C., van der Oost J.. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019; 47:5809–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W., Liu Y., He R., Wang L., Wang Y., Zeng W., Zhang Z., Wang F., Ma L.. A programmable pAgo nuclease with RNA target preference from the psychrotolerant bacterium Mucilaginibacter paludis. Nucleic Acids Res. 2022; 50:5226–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lisitskaya L., Shin Y., Agapov A., Olina A., Kropocheva E., Ryazansky S., Aravin A.A., Esyunina D., Murakami K.S., Kulbachinskiy A.. Programmable RNA targeting by bacterial Argonaute nucleases with unconventional guide binding and cleavage specificity. Nat. Commun. 2022; 13:4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koopal B., Potocnik A., Mutte S.K., Aparicio-Maldonado C., Lindhoud S., Vervoort J.J.M., Brouns S.J.J., Swarts D.C.. Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA. Cell. 2022; 185:1471–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zaremba M., Dakineviciene D., Golovinas E., Zagorskaite E., Stankunas E., Lopatina A., Sorek R., Manakova E., Ruksenaite A., Silanskas A.et al.. Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD(+) depletion. Nat. Microbiol. 2022; 7:1857–1869. [DOI] [PubMed] [Google Scholar]
- 19. Zeng Z., Chen Y., Pinilla-Redondo R., Shah S.A., Zhao F., Wang C., Hu Z., Wu C., Zhang C., Whitaker R.J.et al.. A short prokaryotic Argonaute activates membrane effector to confer antiviral defense. Cell Host Microbe. 2022; 30:930–943. [DOI] [PubMed] [Google Scholar]
- 20. Jolly S.M., Gainetdinov I., Jouravleva K., Zhang H., Strittmatter L., Bailey S.M., Hendricks G.M., Dhabaria A., Ueberheide B., Zamore P.D.. Thermus thermophilus Argonaute Functions in the Completion of DNA Replication. Cell. 2020; 182:1545–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fu L., Xie C., Jin Z., Tu Z., Han L., Jin M., Xiang Y., Zhang A.. The prokaryotic Argonaute proteins enhance homology sequence-directed recombination in bacteria. Nucleic Acids Res. 2019; 47:3568–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee K.Z., Mechikoff M.A., Kikla A., Liu A., Pandolfi P., Fitzgerald K., Gimble F.S., Solomon K.V.. NgAgo possesses guided DNA nicking activity. Nucleic Acids Res. 2021; 49:9926–9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ryazansky S., Kulbachinskiy A., Aravin A.A.. The expanded universe of prokaryotic Argonaute proteins. mBio. 2018; 9:e01935-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lisitskaya L., Aravin A.A., Kulbachinskiy A.. DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 2018; 9:5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D., Schneider S.. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2017; 2:17035. [DOI] [PubMed] [Google Scholar]
- 26. Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L.. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004; 305:1434–1437. [DOI] [PubMed] [Google Scholar]
- 27. Miyoshi T., Ito K., Murakami R., Uchiumi T.. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016; 7:11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J.. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008; 456:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J.. Structure of yeast Argonaute with guide RNA. Nature. 2012; 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J.. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J.. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005; 19:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anzelon T.A., Chowdhury S., Hughes S.M., Xiao Y., Lander G.C., MacRae I.J.. Structural basis for piRNA targeting. Nature. 2021; 597:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao X., Shang K., Zhu K., Wang L., Mu Z., Fu X., Yu X., Qin B., Zhu H., Ding W.et al.. Nucleic acid-triggered NADase activation of a short prokaryotic Argonaute. Nature. 2023; 625:822–831. [DOI] [PubMed] [Google Scholar]
- 37. Shen Z., Yang X.Y., Xia S., Huang W., Taylor D.J., Nakanishi K., Fu T.M.. Oligomerization-mediated activation of a short prokaryotic Argonaute. Nature. 2023; 621:154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiao Y., Maeda S., Otomo T., MacRae I.J.. Structural basis for RNA slicing by a plant Argonaute. Nat. Struct. Mol. Biol. 2023; 30:778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsumoto N., Nishimasu H., Sakakibara K., Nishida K.M., Hirano T., Ishitani R., Siomi H., Siomi M.C., Nureki O.. Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA. Cell. 2016; 167:484–497. [DOI] [PubMed] [Google Scholar]
- 40. Makarova K.S., Wolf Y.I., van der Oost J., Koonin E.V.. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hegge J.W., Swarts D.C., van der Oost J.. Prokaryotic Argonaute proteins: novel genome-editing tools?. Nat. Rev. Micro. 2018; 16:5–11. [DOI] [PubMed] [Google Scholar]
- 42. Graver B.A., Chakravarty N., Solomon K.V.. Prokaryotic Argonautes for in vivo biotechnology and molecular diagnostics. Trends Biotechnol. 2023; 42:61–73. [DOI] [PubMed] [Google Scholar]
- 43. Kropocheva E., Kuzmenko A., Aravin A.A., Esyunina D., Kulbachinskiy A.. A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis. Nucleic Acids Res. 2021; 49:4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carey M.F., Peterson C.L., Smale S.T.. PCR-mediated site-directed mutagenesis. Cold Spring Harb. Protoc. 2013; 2013:738–742. [DOI] [PubMed] [Google Scholar]
- 45. Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 47. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 48. Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Žídek A., Nelson A.W.R., Bridgland A.et al.. Improved protein structure prediction using potentials from deep learning. Nature. 2020; 577:706–710. [DOI] [PubMed] [Google Scholar]
- 49. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 50. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B.et al.. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2017; 27:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Webb B., Sali A.. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016; 54:5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jo S., Kim T., Iyer V.G., Im W.. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008; 29:1859–1865. [DOI] [PubMed] [Google Scholar]
- 54. Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E.. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015; 1:19–25. [Google Scholar]
- 55. Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmuller H., MacKerell A.D. Jr.. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017; 14:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deift P., Zhou X.. A steepest descent method for oscillatory Riemann–Hilbert problems. Asymptotics for the MKdV equation. Ann Math. 1993; 137:295–368. [Google Scholar]
- 57. Parrinello M., Rahman A.. Crystal structure and pair potentials: a molecular-dynamics study. Phys. Rev. Lett. 1980; 45:1196. [Google Scholar]
- 58. Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A Gen Phys. 1985; 31:1695–1697. [DOI] [PubMed] [Google Scholar]
- 59. Hess B., Bekker H., Berendsen H.J., Fraaije J.G.. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997; 18:1463–1472. [Google Scholar]
- 60. Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G.. A smooth particle mesh Ewald method. J. Chem. Phys. 1995; 103:8577–8593. [Google Scholar]
- 61. McGibbon R.T., Beauchamp K.A., Harrigan M.P., Klein C., Swails J.M., Hernandez C.X., Schwantes C.R., Wang L.P., Lane T.J., Pande V.S.. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015; 109:1528–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Boland A., Huntzinger E., Schmidt S., Izaurralde E., Weichenrieder O.. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10466–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J.. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W.. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005; 121:1005–1016. [DOI] [PubMed] [Google Scholar]
- 66. Li X., Dong H.R., Guo X., Huang F., Xu X.Y., Li N.L., Yang Y., Yao T.B., Feng Y., Liu Q.. Mesophilic Argonaute-based isothermal detection of SARS-CoV-2. Front. Microbiol. 2022; 13:957977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps have been deposited into the Electron Microscopy Data Bank (accession numbers: EMD-38354 for KmAgoapo, EMD-38355 for KmAgogDNA, EMD-38409 for pre1-KmAgo-DMgDtD, EMD-38408 for KmAgo-DMgDtR, EMD-38412 for pre2-KmAgogDtD, EMD-38414 for cleaved-KmAgogDtD, EMD-38415 for released-KmAgogDtD).
The coordinates have been deposited into the Protein Data Bank (accession numbers: 8XHS for KmAgoapo, 8XHV for KmAgogDNA, 8XJX for pre1-KmAgo-DMgDtD, 8XJW for KmAgo-DMgDtR, 8XK0 for pre2-KmAgogDtD, 8XK3 for cleaved-KmAgogDtD, 8XK4 for released-KmAgogDtD).