Abstract
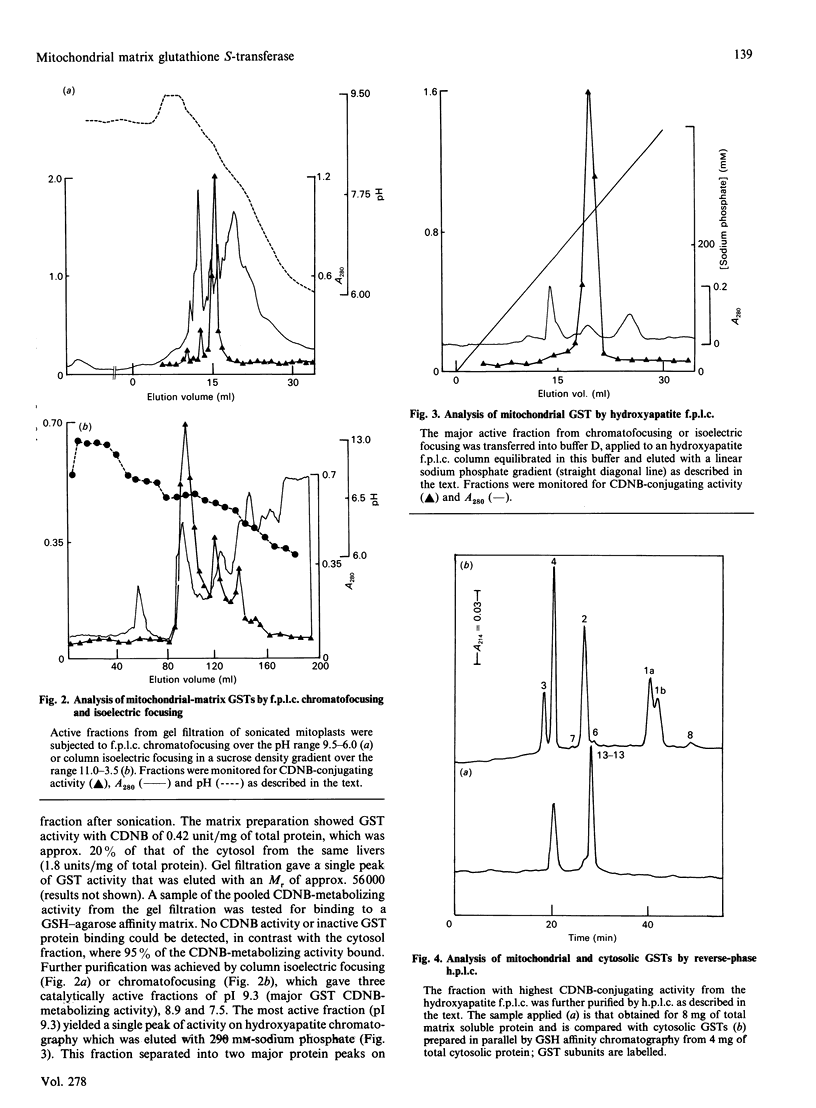
A rat liver mitochondrial-matrix fraction was prepared and shown to have 1-chloro-2,4-dinitrobenzene(CDNB)-metabolizing glutathione transferase (GST) activity. Further fractionation by sequential gel filtration, isoelectric focusing or chromatofocusing and hydroxyapatite chromatography yielded three GSTs of pI 9.3, 8.9 and 7.5, none of which bound to a GSH-agarose affinity matrix. Most of the activity was associated with the pI-9.3 form, which was selected for further study. Its activity was tested with the following potential substrates in addition to CDNB: 1,2-dichloro-4-nitrobenzene, p-nitrobenzyl chloride, trans-4-phenylbut-3-en-2-one, 1,2-epoxy-3-(p-nitrophenoxy)propane, ethacrynic acid, menaphthyl sulphate, cumene hydroperoxide, linoleic acid hydroperoxide and 4-hydroxynon-2-enal. Appreciable activity was obtained only with CDNB and ethacrynic acid (82 and 26 mumol/min per mg of protein respectively). The apparent Km for GSH, using 1 mM-CDNB, was 1.9 mM. The enzyme is a dimer of subunit Mr 26,500. It has a free N-terminus, which has enabled the first 33 amino acids to be sequenced. This portion of primary structure has a sequence in common with members of the Theta class of GSTs (eg. 36% identity with subunit 12) and also a sequence which might function as a mitochondrial import signal. It is novel and has been named 'GST 13-13'.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alin P., Danielson U. H., Mannervik B. 4-Hydroxyalk-2-enals are substrates for glutathione transferase. FEBS Lett. 1985 Jan 7;179(2):267–270. doi: 10.1016/0014-5793(85)80532-9. [DOI] [PubMed] [Google Scholar]
- Allen J. A., Coombs M. M. Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA. Nature. 1980 Sep 18;287(5779):244–245. doi: 10.1038/287244a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Backer J. M., Weinstein I. B. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980 Jul 11;209(4453):297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- Beale D., Meyer D. J., Taylor J. B., Ketterer B. Evidence that the Yb subunits of hepatic glutathione transferases represent two different but related families of polypeptides. Eur J Biochem. 1983 Dec 1;137(1-2):125–129. doi: 10.1111/j.1432-1033.1983.tb07805.x. [DOI] [PubMed] [Google Scholar]
- Botti B., Ceccarelli D., Tomasi A., Vannini V., Muscatello U., Masini A. Biochemical mechanism of GSH depletion induced by 1,2-dibromoethane in isolated rat liver mitochondria. Evidence of a GSH conjugation process. Biochim Biophys Acta. 1989 Sep 15;992(3):327–332. doi: 10.1016/0304-4165(89)90092-5. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Coles B., Ketterer B. The role of glutathione and glutathione transferases in chemical carcinogenesis. Crit Rev Biochem Mol Biol. 1990;25(1):47–70. doi: 10.3109/10409239009090605. [DOI] [PubMed] [Google Scholar]
- Di Ilio C., Aceto A., Piccolomini R., Allocati N., Faraone A., Cellini L., Ravagnan G., Federici G. Purification and characterization of three forms of glutathione transferase from Proteus mirabilis. Biochem J. 1988 Nov 1;255(3):971–975. doi: 10.1042/bj2550971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egilsson V., Evans I. H., Wilkie D. Toxic and mutagenic effects of carcinogens on the mitochondria of Saccharomyces cerevisiae. Mol Gen Genet. 1979 Jul 2;174(1):39–46. doi: 10.1007/BF00433303. [DOI] [PubMed] [Google Scholar]
- Greenawalt J. W. The isolation of outer and inner mitochondrial membranes. Methods Enzymol. 1974;31:310–323. doi: 10.1016/0076-6879(74)31033-6. [DOI] [PubMed] [Google Scholar]
- Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974 Nov 25;249(22):7130–7139. [PubMed] [Google Scholar]
- Hiratsuka A., Sebata N., Kawashima K., Okuda H., Ogura K., Watabe T., Satoh K., Hatayama I., Tsuchida S., Ishikawa T. A new class of rat glutathione S-transferase Yrs-Yrs inactivating reactive sulfate esters as metabolites of carcinogenic arylmethanols. J Biol Chem. 1990 Jul 15;265(20):11973–11981. [PubMed] [Google Scholar]
- Iizuka M., Inoue Y., Murata K., Kimura A. Purification and some properties of glutathione S-transferase from Escherichia coli B. J Bacteriol. 1989 Nov;171(11):6039–6042. doi: 10.1128/jb.171.11.6039-6042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jocelyn P. C., Cronshaw A. Properties of mitochondria treated with 1-chloro-2,4-dinitrobenzene. Biochem Pharmacol. 1985 May 1;34(9):1588–1590. doi: 10.1016/0006-2952(85)90706-3. [DOI] [PubMed] [Google Scholar]
- Kispert A., Meyer D. J., Lalor E., Coles B., Ketterer B. Purification and characterization of a labile rat glutathione transferase of the Mu class. Biochem J. 1989 Jun 15;260(3):789–793. doi: 10.1042/bj2600789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P. Resolution, purification and some properties of three glutathione transferases from rat liver mitochondria. Hoppe Seylers Z Physiol Chem. 1980 Jan;361(1):9–15. doi: 10.1515/bchm2.1980.361.1.9. [DOI] [PubMed] [Google Scholar]
- La Roche S. D., Leisinger T. Sequence analysis and expression of the bacterial dichloromethane dehalogenase structural gene, a member of the glutathione S-transferase supergene family. J Bacteriol. 1990 Jan;172(1):164–171. doi: 10.1128/jb.172.1.164-171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Alin P., Guthenberg C., Jensson H., Tahir M. K., Warholm M., Jörnvall H. Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7202–7206. doi: 10.1073/pnas.82.21.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith M. J., Reed D. J. Status of the mitochondrial pool of glutathione in the isolated hepatocyte. J Biol Chem. 1982 Apr 10;257(7):3747–3753. [PubMed] [Google Scholar]
- Meyer D. J., Coles B., Pemble S. E., Gilmore K. S., Fraser G. M., Ketterer B. Theta, a new class of glutathione transferases purified from rat and man. Biochem J. 1991 Mar 1;274(Pt 2):409–414. doi: 10.1042/bj2740409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern R., Guthenberg C., Depierre J. W. Microsomal glutathione S-transferase. Purification, initial characterization and demonstration that it is not identical to the cytosolic glutathione S-transferases A, B and C. Eur J Biochem. 1982 Nov;128(1):243–248. [PubMed] [Google Scholar]
- Morgenstern R., Lundqvist G., Andersson G., Balk L., DePierre J. W. The distribution of microsomal glutathione transferase among different organelles, different organs, and different organisms. Biochem Pharmacol. 1984 Nov 15;33(22):3609–3614. doi: 10.1016/0006-2952(84)90145-x. [DOI] [PubMed] [Google Scholar]
- Myers K. A., Saffhill R., O'Connor P. J. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis. 1988 Feb;9(2):285–292. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- Niranjan B. G., Wilson N. M., Jefcoate C. R., Avadhani N. G. Hepatic mitochondrial cytochrome P-450 system. Distinctive features of cytochrome P-450 involved in the activation of aflatoxin B1 and benzo(a)pyrene. J Biol Chem. 1984 Oct 25;259(20):12495–12501. [PubMed] [Google Scholar]
- O'Brien P. J. Intracellular mechanisms for the decomposition of a lipid peroxide. I. Decomposition of a lipid peroxide by metal ions, heme compounds, and nucleophiles. Can J Biochem. 1969 May;47(5):485–492. doi: 10.1139/o69-076. [DOI] [PubMed] [Google Scholar]
- Prohaska J. R., Ganther H. E. Selenium and glutathione peroxidase in developing rat brain. J Neurochem. 1976 Dec;27(6):1379–1387. doi: 10.1111/j.1471-4159.1976.tb02619.x. [DOI] [PubMed] [Google Scholar]
- Roise D., Theiler F., Horvath S. J., Tomich J. M., Richards J. H., Allison D. S., Schatz G. Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 1988 Mar;7(3):649–653. doi: 10.1002/j.1460-2075.1988.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryle C. M., Mantle T. J. Studies on the glutathione S-transferase activity associated with rat liver mitochondria. Biochem J. 1984 Sep 1;222(2):553–556. doi: 10.1042/bj2220553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. T., Evans C. G., Doane-Setzer P., Castro V. M., Tahir M. K., Mannervik B. Denitrosation of 1,3-bis(2-chloroethyl)-1-nitrosourea by class mu glutathione transferases and its role in cellular resistance in rat brain tumor cells. Cancer Res. 1989 May 15;49(10):2621–2625. [PubMed] [Google Scholar]
- Tan K. H., Meyer D. J., Gillies N., Ketterer B. Detoxification of DNA hydroperoxide by glutathione transferases and the purification and characterization of glutathione transferases of the rat liver nucleus. Biochem J. 1988 Sep 15;254(3):841–845. doi: 10.1042/bj2540841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert N. E. Isolation of subcellular organelles of metabolism on isopycnic sucrose gradients. Methods Enzymol. 1974;31:734–746. doi: 10.1016/0076-6879(74)31077-4. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Jagt D. L., Hunsaker L. A., Garcia K. B., Royer R. E. Isolation and characterization of the multiple glutathione S-transferases from human liver. Evidence for unique heme-binding sites. J Biol Chem. 1985 Sep 25;260(21):11603–11610. [PubMed] [Google Scholar]
- Wilkinson R., Hawks A., Pegg A. E. Methylation of rat liver mitochondrial deoxyribonucleic acid by chemical carcinogens and associated alterations in physical properties. Chem Biol Interact. 1975 Mar;10(3):157–167. doi: 10.1016/0009-2797(75)90109-x. [DOI] [PubMed] [Google Scholar]
- Wunderlich V., Schütt M., Böttger M., Graffi A. Preferential alkylation of mitochondrial deoxyribonucleic acid by N-methyl-N-nitrosourea. Biochem J. 1970 Jun;118(1):99–109. doi: 10.1042/bj1180099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich V., Tetzlaff I., Graffi A. Studies on nitrosodimethylamine: preferential methylation of mitochondrial DNA in rats and hamsters. Chem Biol Interact. 1972 Jan;4(2):81–89. doi: 10.1016/0009-2797(72)90001-4. [DOI] [PubMed] [Google Scholar]



