Abstract
The genomes of living lungfishes can inform on the molecular-developmental basis of the Devonian sarcopterygian fish–tetrapod transition. We de novo sequenced the genomes of the African (Protopterus annectens) and South American lungfishes (Lepidosiren paradoxa). The Lepidosiren genome (about 91 Gb, roughly 30 times the human genome) is the largest animal genome sequenced so far and more than twice the size of the Australian (Neoceratodus forsteri)1 and African2 lungfishes owing to enlarged intergenic regions and introns with high repeat content (about 90%). All lungfish genomes continue to expand as some transposable elements (TEs) are still active today. In particular, Lepidosiren’s genome grew extremely fast during the past 100 million years (Myr), adding the equivalent of one human genome every 10 Myr. This massive genome expansion seems to be related to a reduction of PIWI-interacting RNAs and C2H2 zinc-finger and Krüppel-associated box (KRAB)-domain protein genes that suppress TE expansions. Although TE abundance facilitates chromosomal rearrangements, lungfish chromosomes still conservatively reflect the ur-tetrapod karyotype. Neoceratodus’ limb-like fins still resemble those of their extinct relatives and remained phenotypically static for about 100 Myr. We show that the secondary loss of limb-like appendages in the Lepidosiren–Protopterus ancestor was probably due to loss of sonic hedgehog limb-specific enhancers.
Lungfishes, together with the coelacanth, are the only remaining fish lineages of the Sarcopterygii (lobe-finned fishes) from within which tetrapods (amphibians, reptiles, birds and mammals), including humans, arose. It is now well established that lungfishes are more closely related to tetrapods than coelacanths1–3. In the Devonian about 425 million years ago (Ma), lungfishes exhibited their highest diversity, with about 70–100 species, and occupied initially marine and later freshwaters of the Gondwana supercontinent. After the major extinction event at the end of the Devonian, only a small number of sarcopterygian lineages, including lungfish, persisted. They were discovered in the nineteenth century4 and are found in Australia, Africa and South America. Their phylogenetic relationships mirror the pattern and timing of the Gondwana break-up. The two lungfish genera from Africa (Protopterus, with four species) and South America (Lepidosiren paradoxa) are more closely related to each other (family Lepidosirenidae). They last shared an ancestor in the Cretaceous, roughly 100 Ma. The Australian lungfish (Neoceratodus forsteri) had a last common ancestor with the two other lungfish lineages about 200 Ma5. Tetrapod ancestors conquered land with limbs that evolved from fins and were breathing air through lungs. These features probably predated the colonization of land. Only by studying the biology of the surviving lungfish lineages can we investigate the genomic basis and molecular-developmental mechanisms that facilitated the water–land transition of vertebrates.
Australian lungfish have large scales and internal gills as adults, just as the extinct lineages did6. Neoceratodus, with its sturdy large scales and limb-like fins, strongly resembles the external morphology of its close relative Ceratodus that went extinct more than 70 Ma. The Australian lungfish remained morphologically ‘static’, seemingly frozen in time, for about 100 Myr, whereas, concurrently, the African and South American lungfishes lost their scales almost completely, and also reduced their limb-like fins to thin filament-like threads that only barely aid in locomotion1.
Besides their significance in the evolutionary biology context, lungfish are notable because of their huge genomes. Genome sizes of eukaryotes vary greatly (over seven orders of magnitude; https://www.genomesize.com/statistics.php), but the significance and causes of genome size variation and evolutionary trends remain obscure. Large genomes generally have a higher content of repeats and transposable elements (TEs). The South American lungfish was estimated to have an even larger genome than the Australian and African lungfishes, which were previously the largest sequenced vertebrate genomes, being composed mainly of TEs and repetitive DNA1. The genomes of all lungfishes should allow us to address several important general questions about genome evolution in animals. More specifically, we were asking, what is the role of TE expansion in the evolution of genome size? Are TEs still actively expanding in extant lungfish species and why can they not control this? Does the overabundance of TEs lead to genome instability, driving erosion of synteny, and disrupt karyotype conservation? Can lungfish, as the closest relatives to tetrapods, help to reconstruct the ur-tetrapod karyotype?
When studying the relationship between molecular and phenotypic evolution during the conquest of land by vertebrates, knowledge of lungfish genome sequences permits us to address pertinent biological questions, specifically, how is the loss of genes and positive selection linked to particular adaptive features of lungfishes such as for their terrestrial lifestyle? What is the genetic basis of the Devonian sarcopterygian adaptations? How can the biological differences among the three last remaining lungfish lineages be explained? Which developmental differences explain the secondarily simplified fins in African and South American lungfishes?
Genome sequencing, assembly and annotation
Lungfish, some salamanders and Antarctic krill have the largest known animal genomes1,2,7,8. This extraordinary size makes such genomes interesting but also challenging to sequence, assemble and interpret.
We performed long-read sequencing to obtain chromosome-level genome assemblies of the South American and African lungfishes (Supplementary Information section 1). We generated 2,199 Gb from 103 Sequel II HiFi SMRT cells of the South American lungfish (25× coverage) and 2,850 Gb from 21 Sequel II CLR SMRT cells of the African lungfish (69× coverage). Hi-C chromosome conformation capture techniques aided in scaffolding in both species. In addition, 10x Genomics linked reads were generated for scaffolding and error correction in African lungfish. The 19 chromosomes of the South American lungfish and the 17 chromosomes of the African lungfish9,10 were assembled with scaffold N50 sizes of 4.3 Gb and 2.7 Gb, respectively (Supplementary Table 1). A high completeness was reached because 94.3% of the South American and 97.5% of the African lungfish assemblies could be assigned to full-length chromosomes. The total sizes of the South American and African lungfish genome assemblies are 87.2 Gb and 40.5 Gb, respectively. These roughly match the k-mer values (91.2 Gb and 47.5 Gb, respectively) and agree with estimates from flow cytometry and Feulgen photometry for the South American (80–120 Gb) and African lungfishes (40–60 Gb).
Based on transcriptome evidence, homologous proteins of vertebrates and ab initio gene prediction, we identified 19,777 protein-coding genes in South American lungfish and 19,181 in African lungfish (Supplementary Table 1). We also re-annotated the Australian lungfish genome1 using the improved strategy developed here for the two other giant genomes and retrieved 21,552 protein-coding genes (Supplementary Table 1). BUSCO analysis suggested that the annotated genes were less fragmented than in the previous version, which may explain the gene number discrepancy1.
The South American lungfish genome is more than double the size of the previously largest animal genome assemblies1,2,8, including those of the Australian and African lungfishes. Notably, 18 of the 19 South American lungfish chromosomes are each individually larger than the entire 3.055 Gb human genome11.
Including the new lungfish genomes in a phylogenomic analysis confirmed the position of lungfishes as closest living relatives of tetrapods3,12 (Supplementary Information section 2 and Extended Data Fig. 1b,c). A recent tip-dating analysis suggested that the divergence among the extant lineages coincided with the splitting up of the Gondwana supercontinent13.
Synteny and reconstruction of the ur-tetrapod karyotype
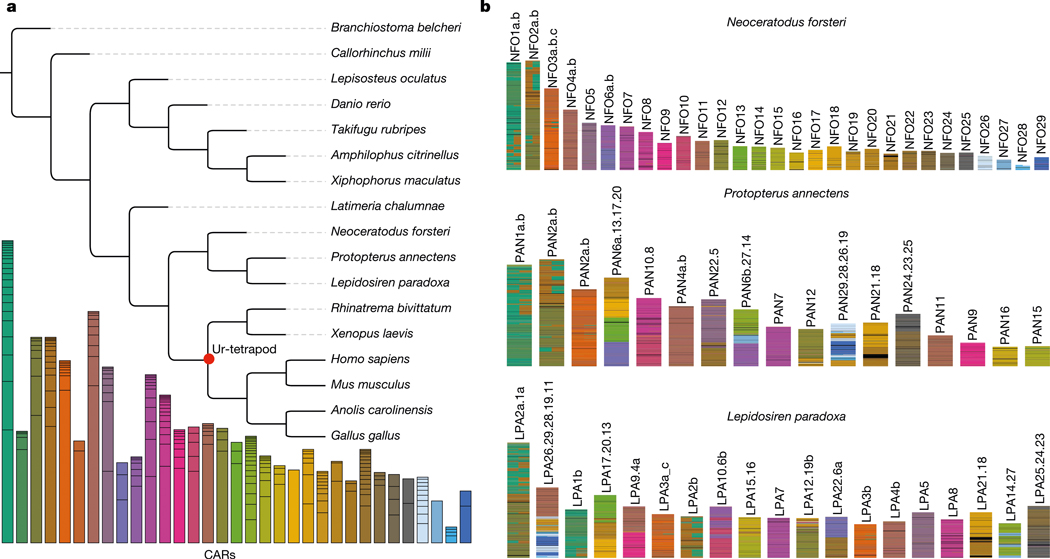
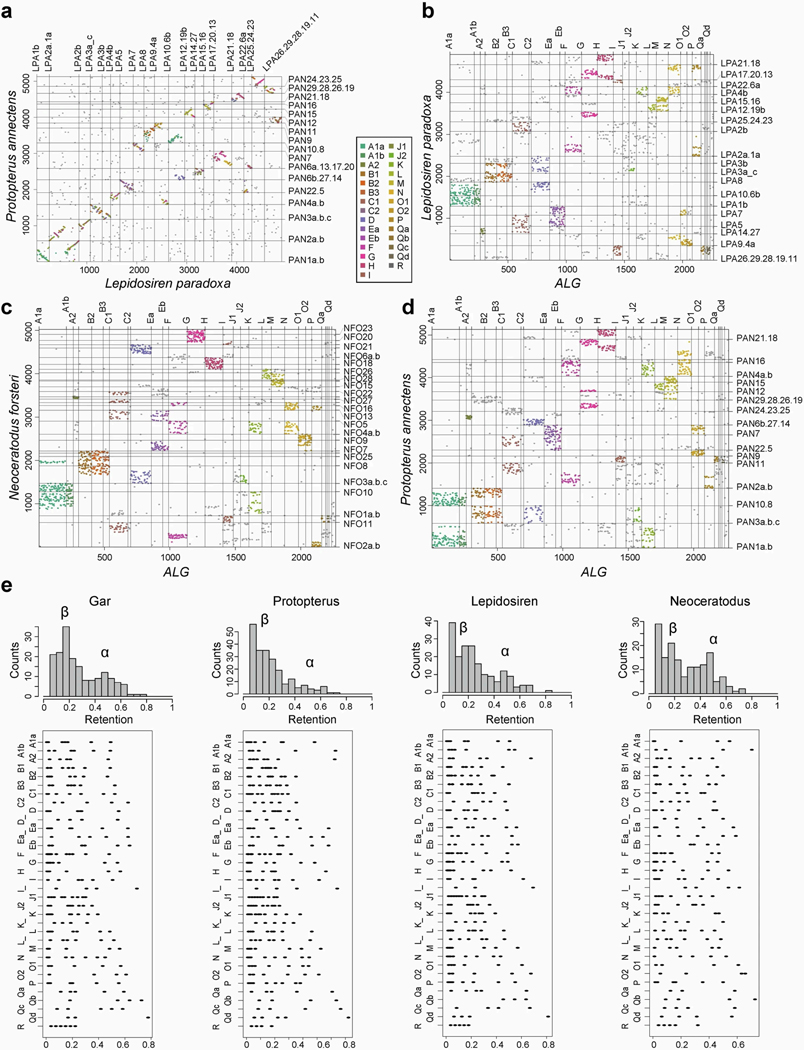
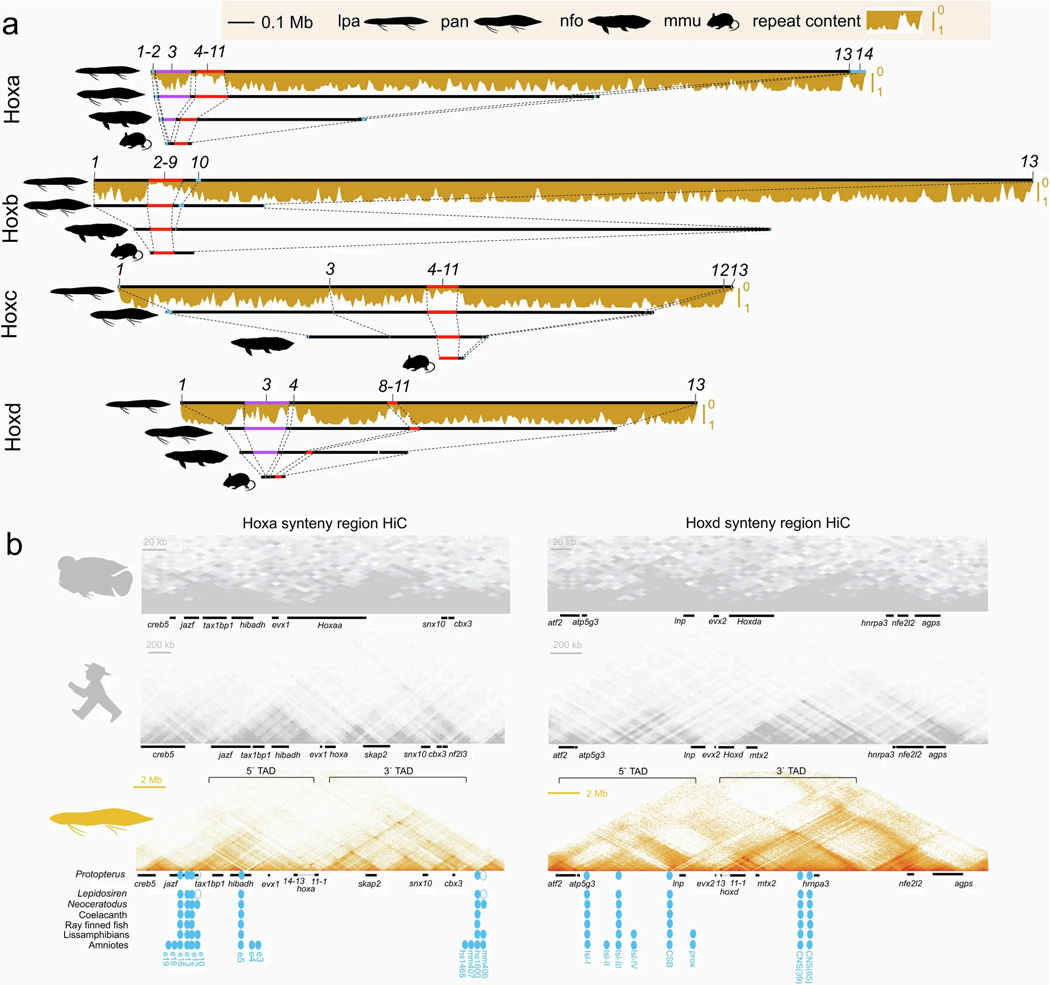
The 17 large chromosomal scaffolds plus 10 microchromosomes of the Australian lungfish can be completely represented as a combination of the ancestral linkage groups (ALGs)1,14, and the African and South American lungfish chromosomes exhibit a high degree of conserved synteny (Fig. 1a and Extended Data Fig. 2a,b). Moreover, the chromosomes of the three lungfish lineages show a high degree of collinearity (Fig. 1b and Supplementary Information section 3). This is notable, given the sizeable and independent expansions that these genomes underwent during the more than 200 Myr since they last shared a common ancestor. By comparing chromosomal synteny relationships, we find that entire lungfish chromosomes represent ancestral tetrapod linkage groups (Fig. 1c), which also correspond to one or several of the contiguous ancestral regions (CARs; Fig. 1c), providing support for the notion that whole chromosomes are retained and acted as distinct evolutionary units during early tetrapod evolution14–16. Moreover, we tracked the retention of CARs from the ancestors of lungfishes and tetrapods to the ur-tetrapod ancestor. More than 90% of the homologous regions stay in the same CAR, demonstrating a high retention rate. This is most clearly visible in the Australian lungfish genome that has the highest chromosome number, most of which (with the exception of chromosomes 2 and 5) preserve the ancestral tetrapod chromosome complement.
Fig. 1 |. Lungfish chromosomes help reconstruct the ur-tetrapod/vertebrate syntenic units.
a, AGORA reconstruction of CARs for different nodes on the vertebrate tree. The CARs of the ur-tetrapod are shown below the tree; each CAR represents one ALG or parts of ALGs. Individual CARs are grouped by Neoceratodus chromosomal homologies (Extended Data Fig. 2), showing that most of Neoceratodus chromosomes are often dominated by a single dominant reconstructed ur-tetrapod CAR, with other CARs likely to be part of the same ancestral ur-tetrapod chromosome. Black horizontal lines separate individual CARs that belong to an ur-tetrapod chromosome. b, Ancestral ‘ur-tetrapod’ CARs can be further traced in lungfish genomes, suggesting their additional mixing in Protopterus and Lepidosiren.
As well as this degree of conservation, we also identified several major chromosomal translocation and fusion events that happened during the early lungfish and tetrapod evolution. We inferred species-specific chromosomal fusions in all three lungfish lineages, for example, chromosome PAN10.8 in African lungfish, which is a recent fusion of ancestral chromosomes 10 and 8 (Fig. 1b and Supplementary Table 2). The Australian lungfish retains the highest degree of ancestral karyotype conservation, compared with the other lungfishes, as it has the fewest of such new chromosomal fusions, with most of the smaller chromosomes representing conserved microchromosomes1. These chromosomes fused to form larger macrochromosomes in the common ancestor of South American and African lungfishes, thus showing less ancestral chromosomal representation than the Australian lungfish (Fig. 1a and Extended Data Fig. 2a,b).
It has been argued that TEs contribute significantly to genome rearrangements17. By contrast, we find no significant difference (Mann–Whitney U-test (also known as the Wilcoxon rank-sum test) P value 0.2861) in the number of collinearity breaks between Neoceratodus and Lepidosiren or between Neoceratodus and Protopterus, even though the Lepidosiren genome has a much higher TE content (roughly 85 Gb) than Protopterus (about 40 Gb) and Neoceratodus (about 35 Gb) (see below).
Loss of duplicate genes
Compared with other vertebrates, lungfish chromosomes are more prone to losing genes in at least one of the paralogous chromosomes (‘homeologs’) that arose after the 2R ancient vertebrate whole-genome duplication (WGD). Two rounds of WGD resulted initially in four homeologs that are often still present in extant vertebrates15,18. But duplicate genes are not necessarily retained equally on all four chromosomes. Previous studies have identified a high retention (α) and a low retention (β) chromosome pair, arguing for an ancient vertebrate allotetraploidy event as cause for the WGD14. All lungfish genomes contain both α- and β-type chromosomes, identified on the basis of their degree of retention. However, at least some of the α chromosomes have lost a substantial proportion of the anciently duplicated genes (Extended Data Fig. 2c). Among lungfishes, only the Australian lungfish shows the ancestral two-peaked pattern of both α and β copies (Extended Data Fig. 2c). Given the higher retention of unmixed chromosomal units in Australian lungfish, compared with the other two lungfishes, these data support the notion that a comparatively more conserved ancestral state of the ur-tetrapod karyotype remains in the Australian lungfish, in terms of chromosomal synteny and gene complement, than in the more dynamic genomes of the other extant lungfish lineages.
Massive genome expansions
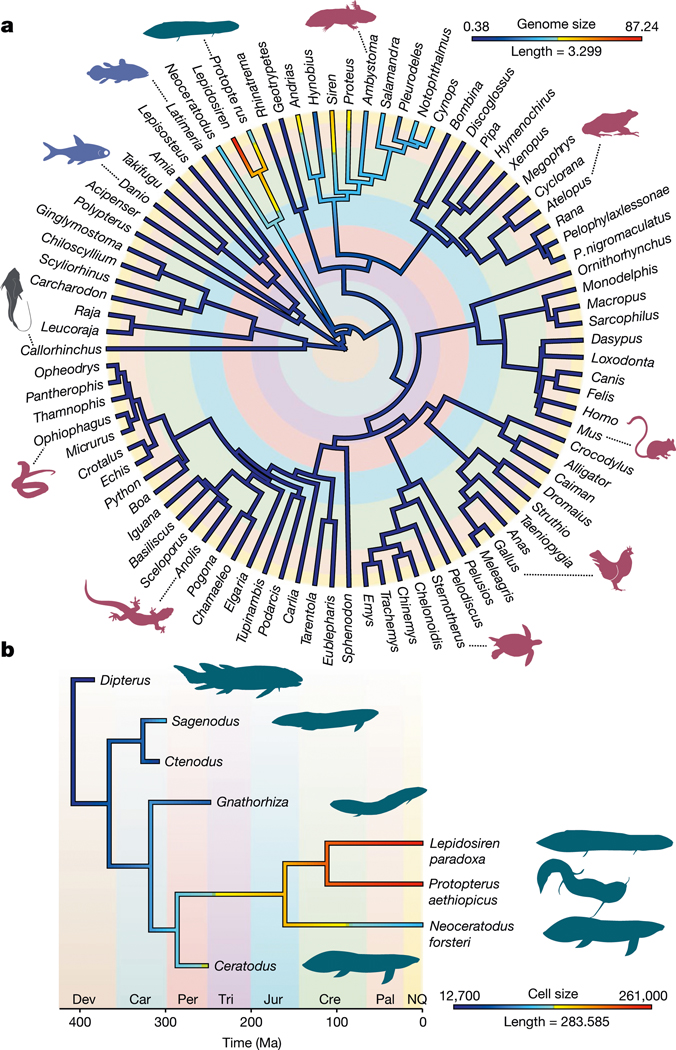
We inferred a Bayesian time-calibrated phylogeny (Supplementary Data 1), using a taxon-rich dataset3 and fossil calibrations13 to reconstruct the evolution of genome size. This showed that two evolutionarily independent bouts of large-scale genome expansion events happened in lungfishes and salamanders (Fig. 2a). The initial growth of lungfish genomes probably predates the age of the common ancestor of extant lungfishes (at a rate of 124 Mb per million years, assuming constant rates); it accelerated strongly in the lineage of the Lepidosirenidae (152 Mb every million years) and even more in the Lepidosiren lineage (371 Mb every million years or 3.71 Gb every 10 Myr). This is by far the fastest rate of diploid genome expansion reported, exceeding those known for any other vertebrate lineage by adding more than the equivalent of the human genome size every 10 Myr (Fig. 2a). A reconstruction of ancestral cell sizes using fossil and extant lungfishes19 and a new time-calibrated phylogeny13 now allowed a new view on genome size evolution (Fig. 2b). As cell and genome sizes are known to be strongly correlated20, we can approximate the dynamics of genome size evolution, including extinct lungfishes for which cell sizes are known19. It shows that their cell and consequently genome sizes expanded only after the split of modern lungfishes from their extinct ancestors. These analyses indicate that, not only the morphology, but also genome size seems to have remained comparatively static for long periods of time.
Fig. 2 |. Genome and cell size evolution.
a, Maximum likelihood reconstructions of the evolution of genome size in jawed vertebrates. Genome size evolution used a new Bayesian time-calibrated phylogeny and genome size values obtained from assembled genomes or the Genome Size Database (http://www.genomesize.com/search.php). b, Maximum likelihood reconstruction of cell size evolution in lungfish. Cell size reconstruction used the tip-dated phylogeny of ref. 13, including extinct lungfishes and cell size data from ref. 13. Branch lengths are in million years and colours denote genome size (in Gb) or cell volume (μl3). Major geological periods are highlighted with colours. Dev, Devonian; Car, Carboniferous; Per, Permian; Tri, Triassic; Jur, Jurassic; Cre, Cretaceous; Pal, Paleogene; NQ, Neogene–Quaternary.
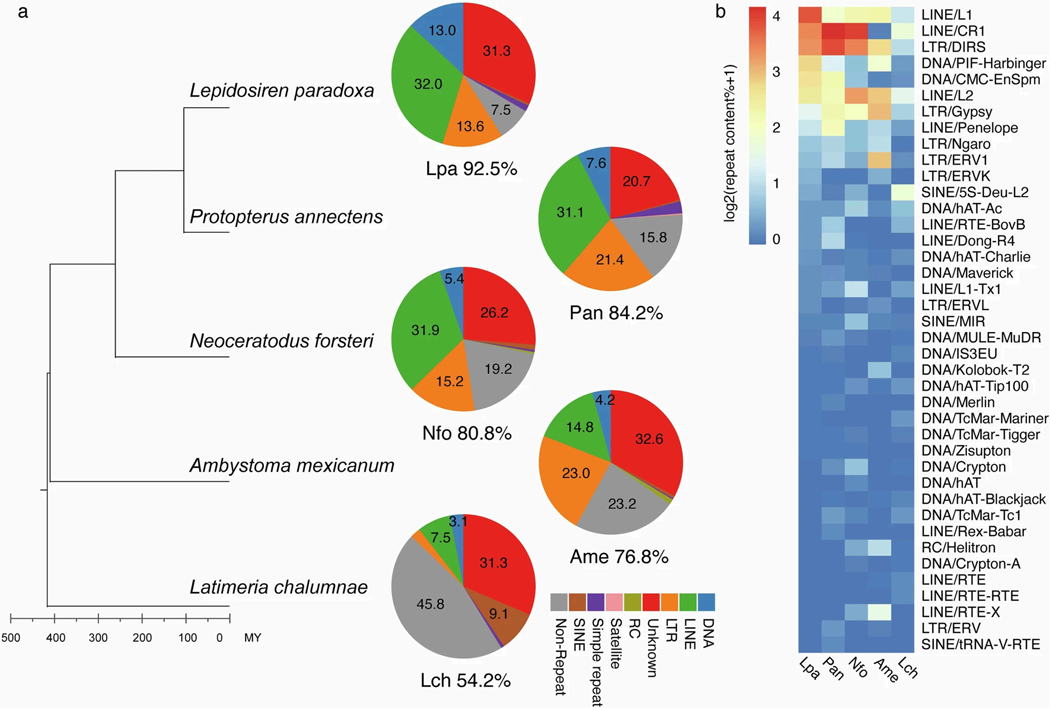
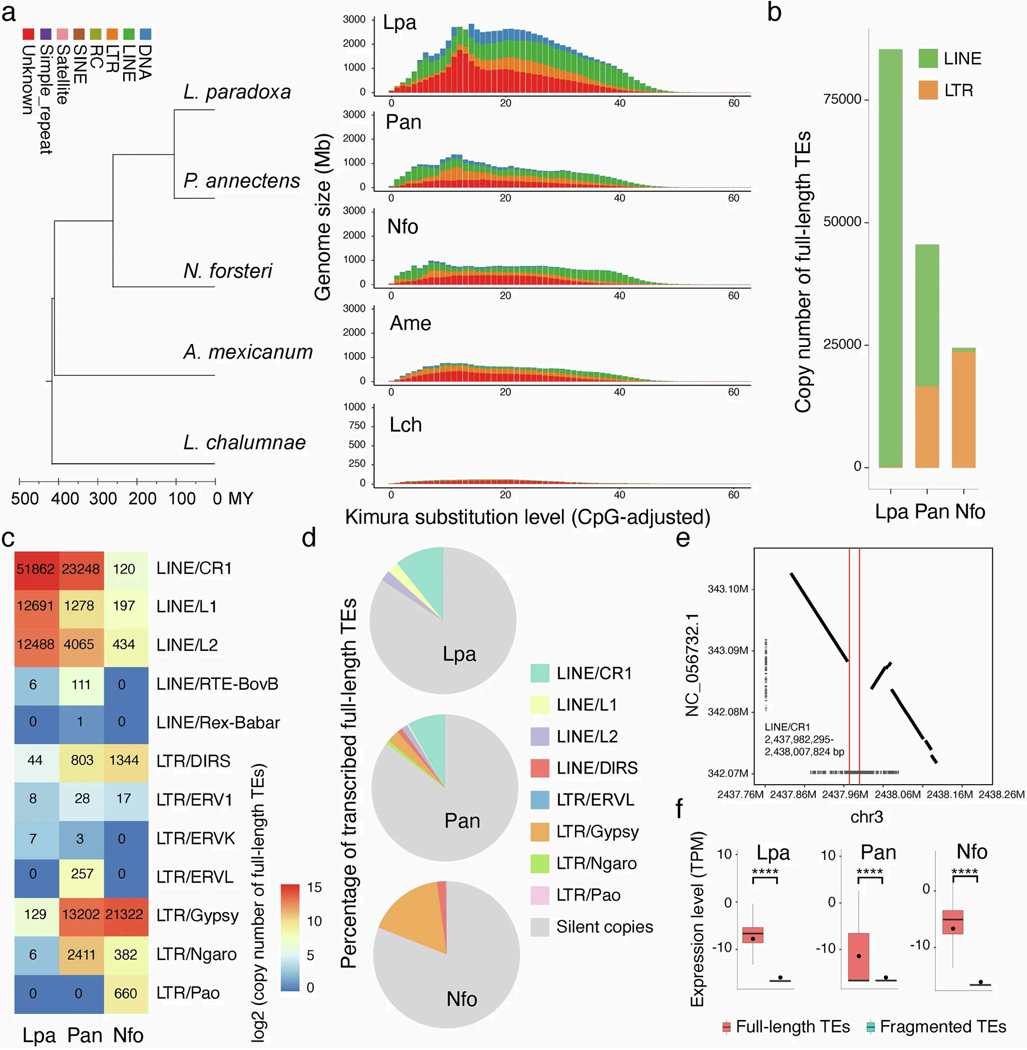
Extreme genome expansions can occur through the accumulation of sequence repeats and mobile elements1,2,7,8,21. It has been proposed that waves of TE expansion might coincide or even drive periods of phenotypic innovation22. We annotated the repeats including TEs of the three lungfish genomes, coelacanth and axolotl, by two rounds of standard repeat-masking procedures with default parameters. The massive genome expansions of both salamander and lungfish lineages were caused mainly by accumulation of TEs, but of different kinds—mainly long interspersed nuclear elements (LINEs) in lungfishes (but not uniformly in all three lineages) and long terminal repeats (LTRs) in axolotl (Extended Data Fig. 3).
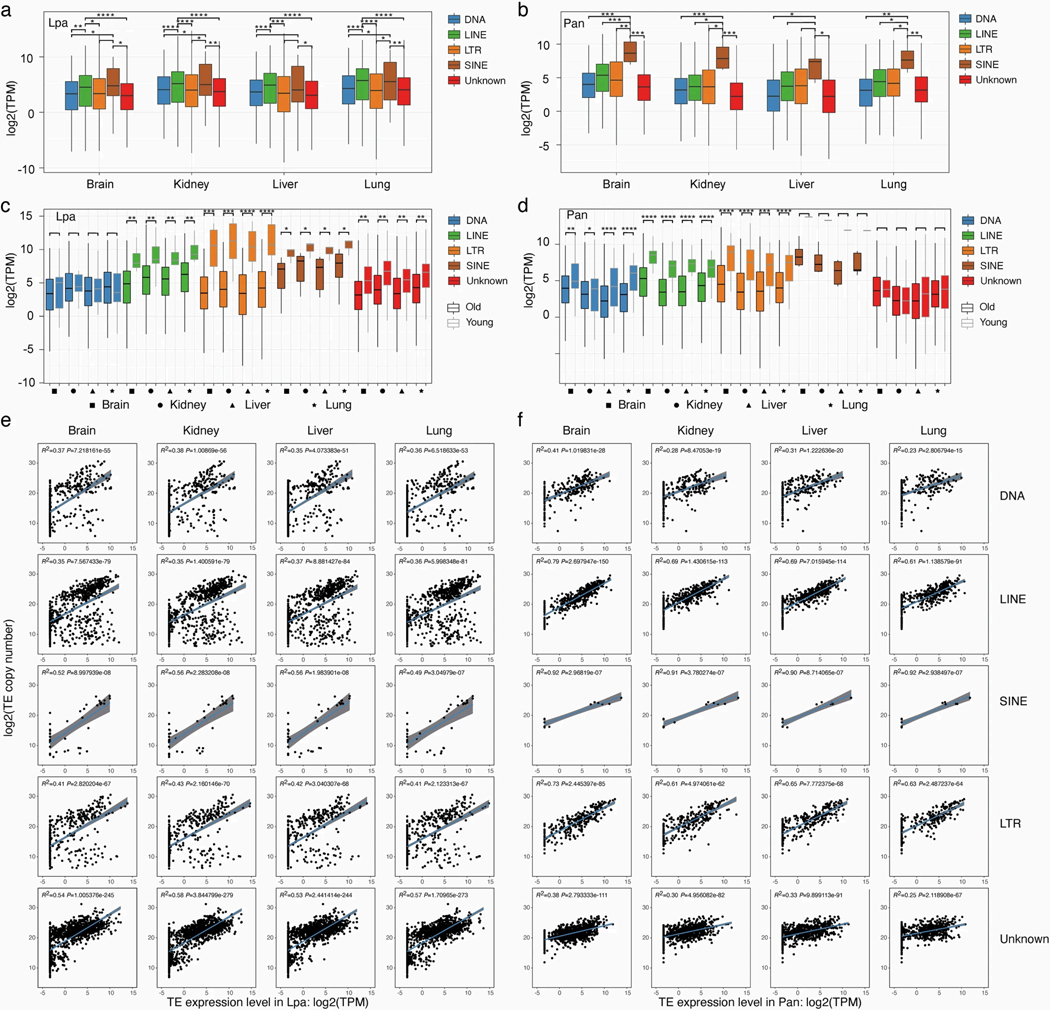
To determine whether or not TEs are expressed, and thus potentially still active, in lungfish genomes, we analysed transcriptome data of six different tissues from African and South American lungfishes. In all tissues, short interspersed nuclear elements (SINEs) are more strongly expressed than any other TE subclass in African lungfish, whereas both LINEs and, particularly, the SINE family are expressed disproportionately strongly (R2 > 0.5) in South American lungfish (Extended Data Fig. 4a,b,e,f).
Next, we divided the TEs into young or old copies on the basis of the distribution of their Kimura pairwise distances (Extended Data Fig. 5a). We found that young TE copies are significantly more highly expressed than old ones (Extended Data Fig. 4c,d), suggesting that these TEs are still active, and that TE expansion continues. This applies mostly for LINEs, LTRs and SINEs. To further investigate which TE families might still be active, we focused on full-length copies containing protein-coding and structural features necessary for autonomous (retro)transposition. This analysis showed a lack of full-length autonomous DNA transposons, suggesting that they now have low or no activity in lungfish. In the South American lungfish, a large number of full-length LINEs (greater than 75,000) are found. Among them, CR1 is the most highly expressed. The other two lungfish genomes contain fewer, but still extremely high, numbers of intact full-length TEs (Extended Data Fig. 5b,c). In African lungfish, both LINEs and LTR elements are the most numerous, whereas, in the Australian lungfish, LTR elements dominate. These complete copies are highly expressed, compared with fragmented ones, indicating that some TE families—particularly LINEs, LTRs and SINEs—might still be active in the giant lungfish genomes.
Expression of a full-length TE does not necessarily imply their activity. The available second African lungfish genome2 made it possible to identify intraspecific insertion polymorphisms by comparing the two independently sequenced genomes. The largest superfamily is LINE/CR1 (Extended Data Fig. 4b). Stringent filtering yielded 27 presumably complete LINE/CR1 copies in our African lungfish genome, compared with just 4 in the other individual. Of these 27 copies, 13 were syntenic, and 6 of these were exclusively present in our African lungfish genome and absent in the other individual’s genome (see examples in Extended Data Fig. 5e). This intraspecific variation provides direct evidence for current activity of TEs in lungfish.
Deficiencies in transposable element expansion control
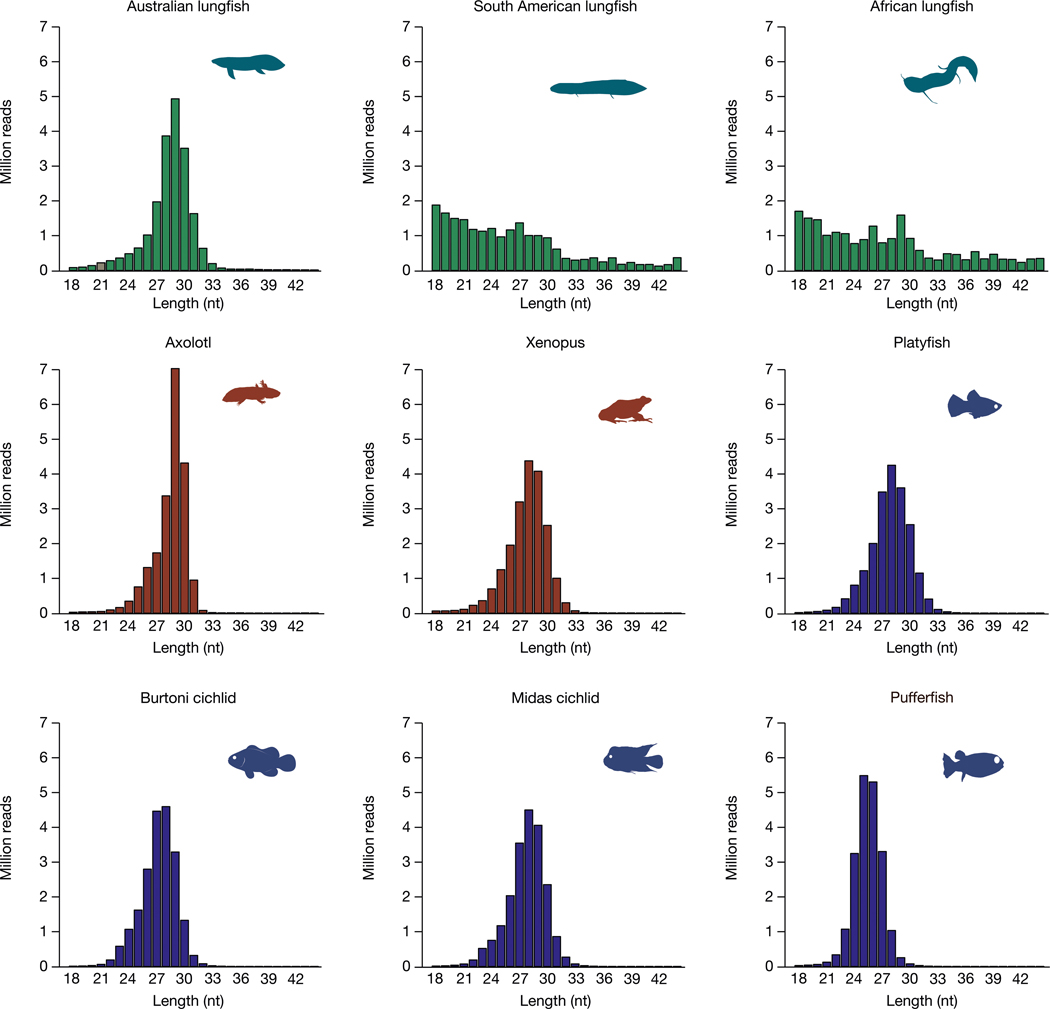
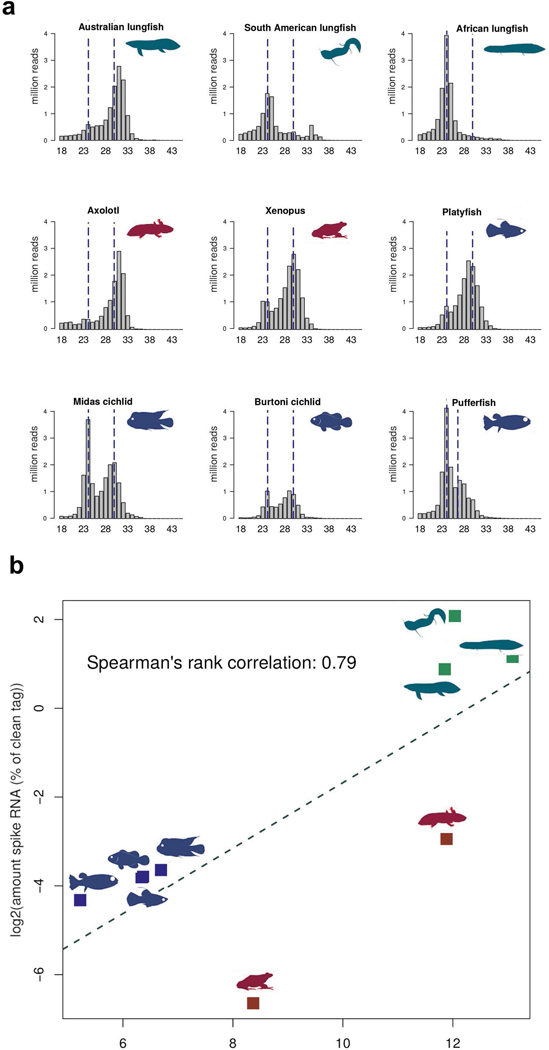
PIWI-interacting RNAs (piRNAs) are a class of non-coding small single-stranded RNAs with lengths of 23–31 bp. They have an important role in maintaining germline DNA integrity through silencing of TE transcription, thereby controlling copy number and activity of TEs, and thus genome size23–25. piRNAs carry a 3′ modification that makes them resistant to chemical oxidation and allows for their reliable identification. On the basis of sequenced small RNA libraries of oxidized RNA from the testes of 9 different species, we found the expected peak at around 28 nucleotides (Fig. 3 and Extended Data Fig. 6a) in the Australian lungfish. But in the African and South American lungfishes, most piRNAs seem to be degraded, as shorter reads were abundant and the 28-bp peak was missing (Fig. 3). Longer reads may be due to a failure in the trimming step during piRNA generation.
Fig. 3 |. Size distribution of clean reads of oxidized small RNA libraries from the three lungfish, amphibians and fish.
Except for the African and South American lungfish, all species have a clear peak at the expected size range of piRNAs.
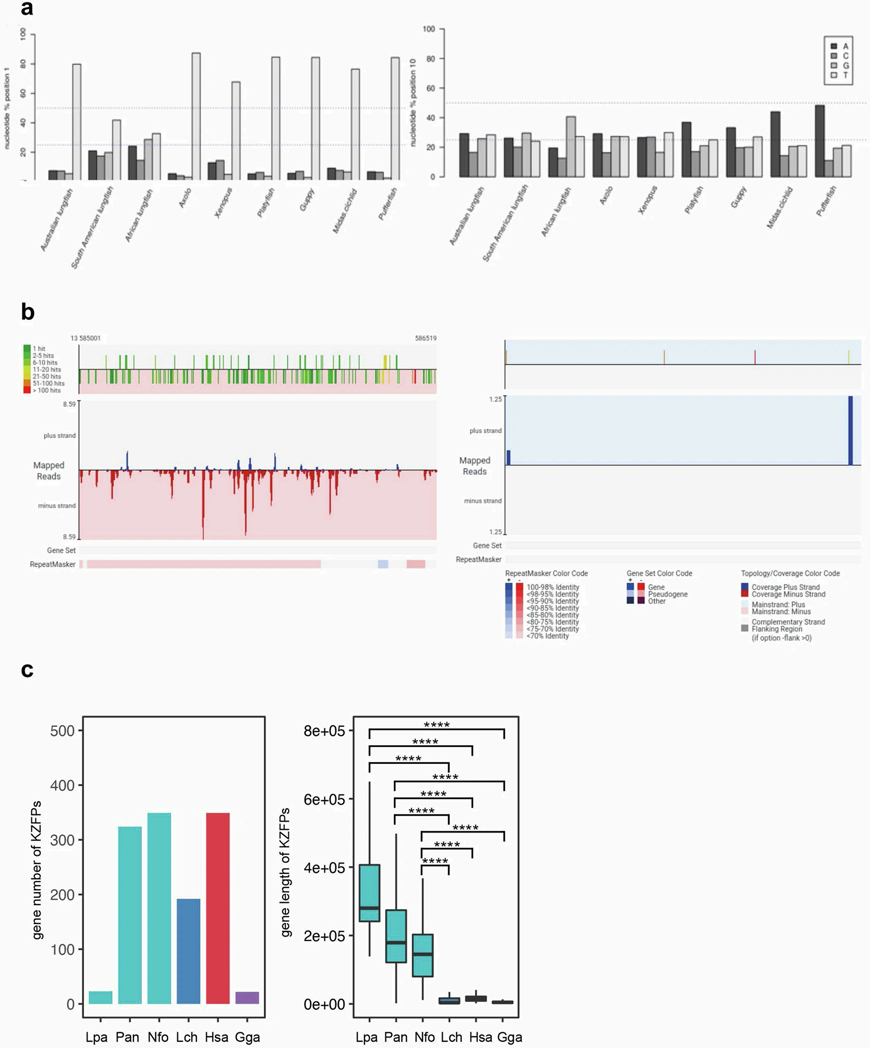
Moreover, Australian lungfish piRNAs, like those from amphibians and fish, had the expected 80% uracil nucleotide signature at position 1, as previously reported for other species26 (Extended Data Fig. 7a). In South American and African lungfishes, however, uracil at position 1 was under-represented, suggesting a corrupted processing of piRNA precursors. Only teleost fish were biased towards adenine at position 10, whereas, in lungfishes (and amphibians with much larger genomes), adenine was not over-represented, indicating a reduced ping-pong amplification27,28. Notably, the diversity of reads was strongly reduced in South American and African lungfishes. Here, the 25 most abundant reads covered 9.3% and 19.3% of all clean reads (Supplementary Table 3), indicating an overall low coverage of TE sequences by piRNAs (Fig. 3).
For quantification of piRNAs, a spike RNA was added to the input RNA during isolation. Reads were then mapped to the respective genomes and the spike sequence to obtain an estimate of the piRNA/total RNA proportion. All three lungfish species have only 5% piRNA content, compared with teleosts (Supplementary Table 3). Australian lungfish retained an apparently intact, but much reduced, abundance of piRNAs. Furthermore, axolotl has fewer piRNAs than the typically sized (0.5–2 Gb) teleost genomes9, suggesting a negative relationship between low piRNA content and large genome size (Extended Data Fig. 6b). As the piRNA pathway differs greatly between organisms29, the lower piRNA content of axolotl testis may be a result of different mechanisms.
Next, we identified and annotated piRNA clusters (Supplementary Table 4 and Extended Data Fig. 7b). Both piRNA cluster size and density in lungfish is reduced more than tenfold relative to genome size. Modelling showed that piRNA cluster size must exceed 0.2% of genome size to repress TE invasion efficiently30. N. forsteri just reaches 0.2%, but the piRNA clusters of the other two lungfishes are much smaller and also showed a reduced proportion of reads in the range of 24–32 nt (Supplementary Table 4). All other species measured have values greater than 4%. In addition, all lungfish had a proportion of main strand-encoded reads of roughly 100%. In accordance with the size distribution, the South American and African lungfishes had a reduced proportion of reads in the range of 24–32 nt (Supplementary Table 4).
Low levels of piRNA silencing of TEs is a mechanism that may partly account for the massive genome expansion during lungfish evolution31.All piRNA metabolism genes known from tetrapods and fish are present and expressed in the genomes of the three lungfish species (Supplementary Table 5). However, whether or not they are ‘normal’ with respect to expression level and protein structure needs further investigation because of the low conservation of some genes and the pathway in general.
C2H2 zinc-finger and Krüppel-associated box (KRAB)-containing zinc-finger protein (KZFP) genes have a principal role in recognition and transcriptional silencing of TEs32. Australian and African lungfish have more than 300 of these genes, just as humans do33. But the giant South American lungfish genome contains much fewer (n = 23), similar to the TE-poor chicken (Extended Data Fig. 7c) and other bird genomes33. The genome growth of the South American lungfish, largely explainable by further TE expansion, may be related to this lineage-specific loss of KZFP genes.
Gene and genome evolution
Retrogenes
Gene duplications provide raw material for new gene functions and evolutionary novelty, and thus can be important drivers of phenotypic evolution. New gene copies can emerge through DNA-mediated mechanisms (duplication of chromosomal segments), but also through the process of retroposition (retroduplication leading to intronless copies), whereby mRNAs are reverse-transcribed into DNA and inserted into the genome34. In mammals, retroposition is mainly mediated by LINE-1/L1 retrotransposons35. LINE-1/L1 is the most common repeat superfamily in South American lungfish and is also abundant in the two other lungfish species. Thus, relatively more retrogenes would be expected. Accordingly, although the overall genomic protein-coding gene content is similar, 1,847 parent–retrocopy pairs were identified in the South American lungfish, 1,201 in the African lungfish and 1,159 in the Australian lungfish. (Supplementary Table 6). In other non- mammalian vertebrates, the number of retrogenes is much smaller, ranging from about 50 to 400 (ref. 36).
Positive selection
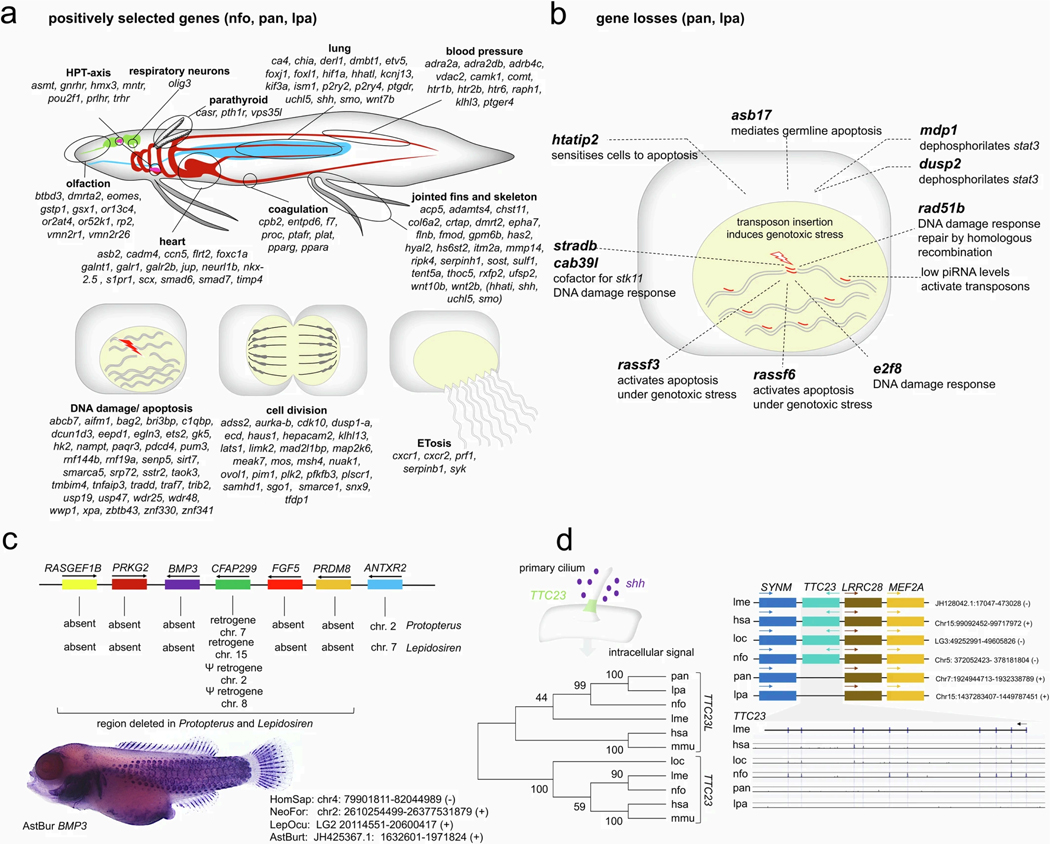
Positively selected genes were identified in all three lungfish genomes (site class 3, n = 49; site class 4, n = 474) (Supplementary Table 7) as they might be related to specific aspects of lungfish biology (Supplementary Information section 4 and Extended Data Fig. 8a), including a more terrestrial-oriented lifestyle with obligatory air breathing and incipient double circulatory system, enhanced olfaction and elaborately articulated fins. We found support for proposed alterations in the hypothalamic–pituitary–thyroid axis related to neotenic aspects of lungfish morphology. Genes related to lungfish immunity (ETosis) and to managing a giant genome during cell division and transcription were also identified (Supplementary Information section 4 and Extended Data Fig. 8a). Among those are many genes involved in the DNA damage response and apoptosis, likely to be related to the hyperactivity of transposons in the lungfishes’ genomes.
Further classes of positively selected genes have functions in lung, skeletal muscle, kidney and bone metabolism, with genes involved in ossification, calcium metabolism and the parathyreoid gland (Supplementary Information section 4 and Extended Data Fig. 8a). Calcium sensing and metabolism are crucial for dense bones of terrestrial animals, which is regulated by the parathyroid in tetrapods. The genes of the parathyroid gland evolved early in deuterostomes, but fish express them in the gills and only tetrapods have a true parathyroid2,37.
Gene losses
Ten identified gene losses are related to DNA damage response (Supplementary Information section 5 and Extended Data Fig. 8b), which would facilitate genome expansion by reducing somatic selection on genotoxic stress induced by transposon insertion. Other gene losses occurred around BMP3 (Supplementary Information section 5 and Extended Data Fig. 8c) including RASGEF1B, PRKG2, FGF5 and PRDM8. BMP3 is involved in the formation of scales38 (Extended Data Fig. 8c), which are secondarily almost completely reduced in the African and South American lungfishes39. Loss of PRKG2, RASGEF1B, TTC23 (Extended Data Fig. 8d) and hoxd12 are potentially related to the secondary reduction of the lepidosirenid fins (see below).
Hox cluster expansion in relation to genome growth
Lungfish hox clusters show marked expansion whereby the South American lungfish clusters are approximately 20 times larger than in the mouse (Supplementary Information section 5 and Extended Data Fig. 9a). Previous analysis of the hoxd clusters in giant genomes suggested that part of this cluster escaped expansion owing to purifying selection on gene regulatory constraints1. Comparison of all lungfish genomes identified similarly constrained sub-clusters in all four hox clusters (hoxa4–hoxa11, hoxb2–hoxb9, hoxc4–hoxc11 and hoxd8–hoxd11) (Extended Data Fig. 9a). Hox clusters have the lowest transposon content in vertebrate genomes40,41, and only squamates show some cluster expansion because of transposon invasion41,42. However, the South American lungfish hox clusters show massive presence of TEs in the expanded parts, whereas the size-constrained sub-cluster regions remain TE poor (Extended Data Fig. 9a). This confirms that constraints on the functional (sub-)clustering of hox genes and selection against transposon insertion remain strong, and are even noticeable in extraordinarily expanded genomes.
Hi-C analysis for the Midas cichlid, human and African lungfish hoxa and hoxd synteny regions showed that, in spite of the roughly 80-fold size difference, the flanking regulatory landscapes remain stable whereby both clusters are present on the intersection of a 3′ and 5′ topologically associated domain (Extended Data Fig. 9b and ref. 43). Known fin and limb enhancers (Extended Data Fig. 9b) are conserved except for hoxa cluster-related elements e10 and mm406 (ref. 44). Their loss is potentially related to the lepidosirenid vestige fin phenotype (see below). Thus, long-range regulatory landscapes remain preserved even under conditions of massive genome expansion.
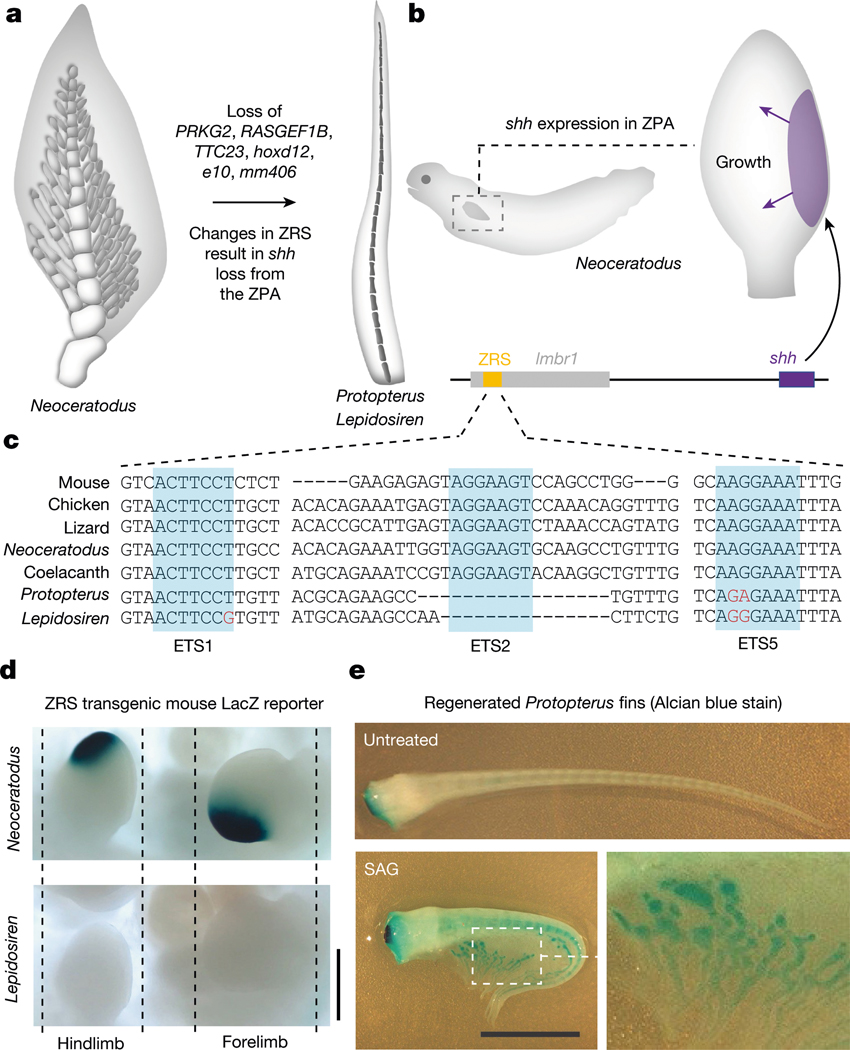
Sonic hedgehog modifications lead to fin reduction
Compared with the Australian lungfish, the African and South American lungfishes evolved secondarily simplified filament-like pectoral and pelvic fins caused by the absence of fin radials (a condition already found in Gnathoriza45) (Fig. 4a). In addition to loss of the hoxa fin/limb enhancers e10 and mm40644 (Extended Data Fig. 9b), we detected several gene losses associated with this fin reduction: PRKG2 (Extended Data Fig. 8c), whose loss results in reduced long bone size in mice, humans and cattle46; RASGEF1B (Extended Data Fig. 8c), a target of the sonic hedgehog (shh) pathway during limb development47; TTC23, which has an essential role in the transmission of an extracellular shh signal through primary cilia48; and hoxd12 (Extended Data Fig. 9a), a regulator of fin and limb development49. During embryogenesis, expansion of fins and limbs is driven by the activity of the Shh pathway, which determines digit and fin radial number in mouse and medaka50. Expression of shh in a conserved posterior fin or limb domain (‘zone of polarizing activity’ (ZPA)) is regulated by the zone of polarizing activity regulatory sequence (ZRS) enhancer, which is located within the intron of a 3′ distal gene, LMBR1 (ref. 50) (Fig. 4b). In Australian lungfish, whose fins resemble the tetrapod ancestral condition known from the fossil record, shh is expressed as expected in the ZPA51.
Fig. 4 |. Fin reduction in the Lepidosirenidae.
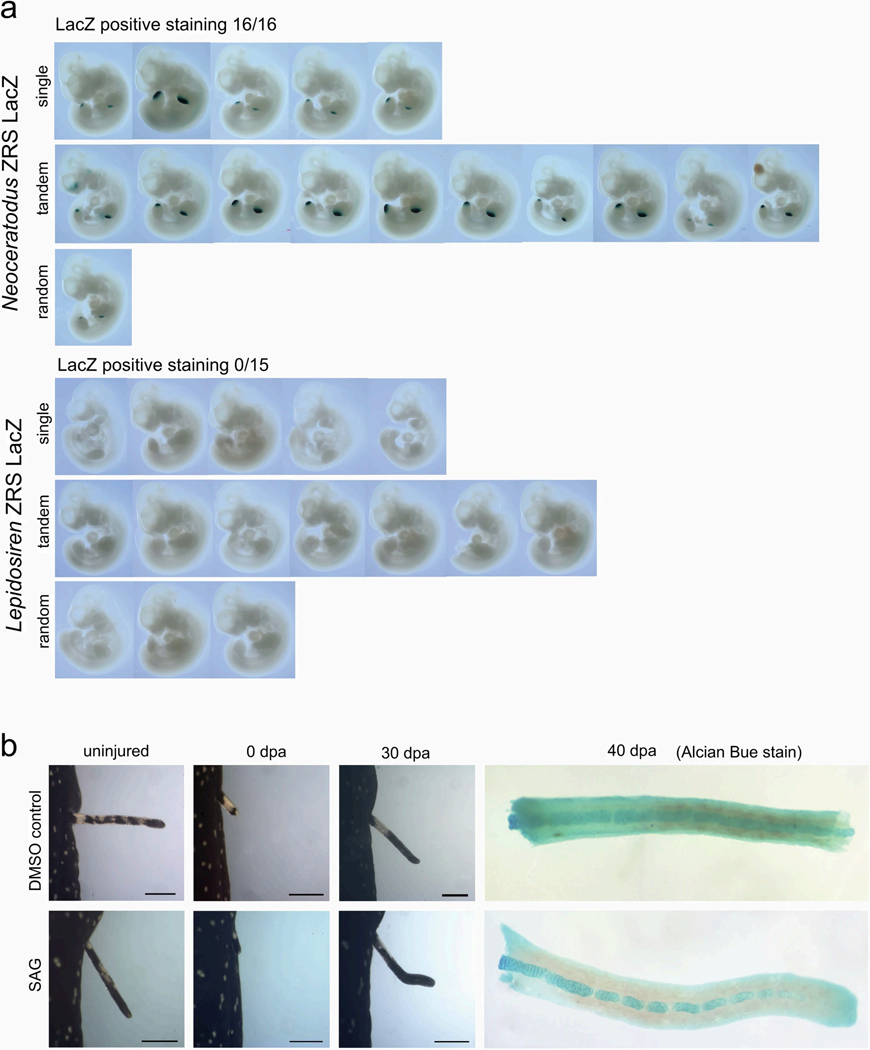
a, In comparison with the fins of the Australian lungfish, South American and African lungfish fins have absent or strongly reduced distal radials and gracile central radials, potentially related to loss of PRKG2, RASGEF1B, TTC23, hoxd12, e10 and mm406 and modification of the shh pathway. b, In the Australian lungfish, shh is expressed in a conserved posterior fin domain, the ZPA, which is driven by the ultraconserved long-range ZRS enhancer located in the LMBR1 gene. c, Genomic analysis of the ZRS enhancer indicates that South American and African lungfishes have modified and lost ETS transcription factor binding sites. d, Transgenic analysis in mouse limbs shows that the Australian lungfish ZRS drives the expected expression in the ZPA (16/16 embryos), whereas the South American lungfish ZRS does not show such activity (15/15 embryos). e, Stimulating regenerating African lungfish fins with the Shh agonist SAG results in the elaboration of post-axial radials (arrowheads) and partially rescues the ancestral phenotype (SAG-treated, n = 7; untreated, n = 7; representative image of one animal is shown). Scale bars, 0.5 cm (d), 1 cm (e).
The availability of all three lungfish genomes allowed us to comparatively evaluate alterations in the ZRS enhancer. Both South American and African lungfishes showed sequence changes indicative of disrupted ETS transcription factor binding sites (Fig. 4c), akin to those linked to limblessness in snakes52. The activity of lungfish ZRS elements was assayed in transgenic mouse limbs and, in contrast to the Australian lungfish, the South American lungfish ZRS element no longer drives limb expression (Fig. 4d and Extended Data Fig. 10a).
We further experimentally investigated the role of shh by using the Smoothened agonist (SAG) during pectoral fin regeneration in African and South American lungfish. Although South American lungfish fins do not regain any radial elements (Extended Data Fig. 10b), we observed elaboration of radial elements in African lungfish (Fig. 4e). There SAG stimulated fin regeneration with enlarged radials consisting of several segments that resemble the ancestral radials of Australian lungfish. Altogether, this suggests that modifications in the ZRS and further disruption of Shh signalling owing to loss of TTC23 contributed to reduction of lungfish fins and provides further evidence for the deep homology of digits and post-axial fin radials50,51. Different lineages such as snakes, caecilians and limbless lizards show partially divergent genetic signatures of limb reduction, whereby the ZRS enhancer is partially or completely lost in snakes and caecilians but seems to be unaffected in limbless lizards53,54. Our results indicate a specific role for modified Shh signalling by ZRS mutation in the reduction of the lepidosirenid distal fin radials as their fins closely resemble limbs with disrupted Shh signalling, which preserve only a central skeletal axis without distally articulating elements (the digits)55. The partial restoration of an ancestral phenotype in SAG-treated African lungfish suggests that the downstream network for fin radials is still partially responsive to Shh signalling (but probably eroded beyond functionality in South American lungfish). The inactive ZRS enhancer can thus be interpreted as a causal disruption in the original genotype–phenotype map and a driver of fin reduction (contra degeneration by relaxed selection after initial fin simplification). Notably, sall1, one of the genes previously implicated in the evolution of the sarcopterygian broadly articulated lobed-fin archetype1, is downstream of shh56. Failure to sufficiently activate this gene could therefore contribute to the reduced fin phenotype in the Lepidoserenidae.
Conclusion
Advances in DNA sequencing technologies and bioinformatics make it possible to sequence and assemble, at chromosome level, even macro-scale-sized genomes. The genomes of all three lineages of lungfish, because of their crucial phylogenetic position, hold the key to a better understanding of how molecular and developmental processes and genomic evolutionary changes contributed to the conquest of land and the evolution of tetrapods, one of the main transitions during vertebrate evolution. We show that massive bouts of genome expansion were driven by TEs, but were different in each of the three living lungfish lineages and the salamanders. TEs continue to actively spread through the already huge lungfish genomes, expanding them further. The twofold increase in size of the South American lungfish genome is paralleled by twice the amount of TEs, compared with the other two lungfishes. We identified potential molecular mechanisms, reduction or even complete lack of intact piRNAs that contributed to the massive expansion of lungfish genomes, particularly in the South American lungfish. There was, however, no relationship between TE abundance and genome stability, allowing the reconstruction of the ur-tetrapod karyotype. We also characterized molecular features that might explain some of the differences in morphological and physiological adaptations among extant lungfish. The Australian lungfish seems to have remained phenotypically largely unchanged for more than 100 Myr, whereas the lepidosirinid lungfishes changed markedly concurrently. For example, they lost their scales and almost all of the limb-like features of the fins. We unravelled modification of Shh signalling as the developmental basis of this secondary evolutionary simplification. Comparative genomic studies, analyses of gene losses and positively selected genes allowed us to infer the genomic basis of some important sarcopterygian features. The resource of chromosome-level genomes for all living lungfish lineages will now enable further research into lobe-finned ancestors of tetrapods who conquered land in the Devonian.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-024-07830-1.
Methods
Biological materials
Biopsy material for DNA and RNA isolation was obtained from three lungfish species (Neoceratodus forsteri, Protopterus annectens and Lepidosiren paradoxa), axolotl (Ambystoma mexicanum), Xenopus laevis and five teleost species (Tetraodon nigriviridis, Astatotilapia burtoni, Amphilophus amarillo, Xiphophorus maculatus and Poecilia reticulata). For details, see the accompanying reporting summary. Samples were collected in accordance with the regulations of the German Animal Welfare Law under permit T19/03, University of Konstanz, and authorization 568/300–1870/13, Würzburg, from the Veterinary Office of the District Government of Lower Franconia, Germany. The specimen of South American lungfish used can be assigned to the southern basin clade on the basis of an analysis of the mitochondrial DNA data57. Because four species of Protopterus are described, we ascertained that we were working with a specimen of P. annectens by analysis of the available comparative mitochondrial data.
Sequencing, assembly and annotation
The methods used for sequencing, assembly and annotation are described in detail in Supplementary Information section 1.
Repeats and transposable elements annotation
The repeat sequences of the coelacanth (Latimeria chalumnae), axolotl (Ambystoma mexicanum) and three lungfish (Protopterus annectens, Neoceratodus forsteri and Lepidosiren paradoxa) were predicted using standard RepeatMasker (v.4.1.2) procedures (RepeatMasker Open-4.0. 2013–2015), with the default TE Dfam (v.3.3) database58 and a de novo repeat library of each species constructed using RepeatModeler (v.2.0.2)59, including the TRF (v.4.10)60, RECON (v.1.0.8)61, RepeatScout (v.1.0.6)62 and rmblast (v.2.9.0)63, with default parameters. To further examine the remaining non-masked genome, we performed a second round of repetitive sequence prediction using the same workflow as mentioned before on the hard-masked genome by the first round of RepeatMasker. We retrieved repeat coverage information of other species analysed in this study from the literature64–66.
Repeat landscape plots of Kimura distance-based distribution analysis
Kimura two-parameter substitution levels between each repeat copy and its consensus sequence were calculated using a utility script calcDivergenceFromAlign.pl bundled in RepeatMasker (v.4.1.2) software. Repeat landscape plots were produced with an in-house-generated script draw.kimura.landscape.pl, using the divsum output from calcDivergenceFromAlign.pl.
Transposable element expression
Transposable element expression was first assessed at the family level with standard RediscoverTE (v.3.15)67 pipeline on brain, kidney, liver and lung poly(A+)-RNA data. Because of the large size of the lungfish genome, we split the genome into several 2 Gb chunks and ran the RediscoverTE pipeline separately and then merged them back for further downstream analyses. In addition, we used SQuiRE (v.0.9.9.93)68 to quantify locus-specific expression for the full-length TE expression analyses.
Full-length transposable element detection, abundance and transcription
The full-length TE copies of DNA transposons, LINEs and LTRs were identified following a method previously established for bird W chromosomes69. In brief, the DNA transposons and LINEs were identified by comparing the open reading frames (ORFs) in the insertions annotated by RepeatMasker (v.4.1.2)57 with a custom Pfam70 database (v.36.0) containing transposon-related proteins. ORFs from LINEs of at least 600 bp that spanned 90% of both endonuclease and reverse transcriptase domains were considered to be full-length elements. Likewise, ORFs belonging to DNA transposons of at least 1 kb that spanned 90% of the transposase protein domain were considered to be full length. For the full-length LTRs, we used LTRHARVEST (v.1.6.1)71 together with LTRDIGEST (v.1.6.1)72. LTRHARVEST results were filtered for false positives using LTRDIGEST in combination with hidden Markov model profiles of LTR retrotransposon-related proteins downloaded from Pfam70 and GyDB (v.2.0)73. To estimate the expression level of these full-length TEs as a proxy for activity, a copy (at a specific locus) was considered transcribed if at least 80% of its sequence was covered by uniquely mapped RNA-seq reads from combined tissues (brain, kidney, liver, lung). Otherwise, we defined them as silent copies.
For the detection of presence/absence patterns of LINE/CR1 insertions between the genomes of P. annectens sequenced in this study and the previously published study by Wang et al.2, we applied stringent filtering criteria, retaining only LINE/CR1 sequences exhibiting more than 99% identity with the consensus sequence and covering more than 80% of the consensus length. Next, we identified the syntenic region of the LINE/CR1 copies retrieved from our P. annectens genome relative to the other individual’s genome, using single-copy gene orthologue information. For those copies that exhibited synteny, we extracted the flanking regions, spanning ±100 kb around each syntenic sequence pair and subjected them to a polymorphic TE-finding pipeline GraffiTE74.
Annotation of KZFPs
The annotation of KZFPs was conducted with a validated method33. In detail, we first translated the whole genome of each species by six frames using EMBOSS (v.6.6.0) and then searched for domains relevant for KZFPs (C2H2 zinc-finger and KRAB) with HMMER2/HMMs from Pfam (v.36.0)70. The following score thresholds were used: C2H2, 0; KRAB, 13. Candidate KZFP genes were identified on the basis of the proximity of zinc-finger arrays and KRAB domains. For each genome, the maximum distance was defined by its annotation. Finally, existing annotation for protein-coding genes was incorporated whenever they overlapped with putative KZFP units.
Identification of retrocopies
Retrocopies were identified on the basis of the sequence similarity between a multi-exon gene (parent) and an intronless genome segment (retrocopy) following a similar strategy as in ref. 36. First, we retrieved all multi-exon genes from the genome and aligned their protein sequences onto the genome using GenblastA (v.1.0.4)75. Second, the aligned regions showing no intron were collected as retrocopy candidates and aligned back to the multi-exon genes using fasty36 (v.36.3.8h)76 to retrieve the best match. Third, these matches were aligned again using GeneWise (v.2–4)77 to confirm the loss of all introns in each retrocopy. Finally, those parent–retrocopy pairs with alignment coverage on parent of less than 70% or percentage identity smaller than 50% were discarded.
piRNA analyses
Small RNA from the testes of nine different species (Supplementary Table 8) was isolated. The lungfish samples were processed in parallel and with the same protocols as the other samples. Of note, they were not subjected to further polymerase chain reaction (PCR) cycles. Samples were stored at −80 °C immediately after animal dissection until downstream processing. Using a SPLIT RNA extraction kit (Lexogen) according to the manufacturer’s guidelines, 3–10 mg per sample was used as input to carry out RNA extraction. We performed RNA quantity and quality assessments with a NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Fisher) and a Fragment Analyzer System (Agilent Technologies), respectively. Sequencing-ready libraries were produced using a Nextflex Small RNA-Seq Kit v.3 (Perkin Elmer) following standard procedures. We performed indexed library preparation to allow for multiplexed sequencing. For library preparation, 200 ng of RNA per sample was used as input. Pooled libraries were purified from agarose gel using a PureLink Quick Gel Extraction Kit (Invitrogen). The pools were quality controlled on a Fragment Analyzer System and quantified using a Qubit 4.0 (Thermo Fisher). We performed high-throughput single-end (75 bp) sequencing on an Illumina NextSeq 500 platform at Lexogen GmbH.
Small RNA (less than 200 nt) from the same testis samples (Supplementary Table 8) was purified using mirVana microRNA (miRNA) isolation kit (Ambion/Life Technologies). RNA concentration and quality were evaluated by Qubit 4 Fluorometer (catalogue no. Q33238) using the Qubit RNA HS Assay-Kit (catalogue no. Q32852) following the protocol provided in the manufacturer’s instructions (Thermo Fisher Scientific). To reliably identify piRNAs in the small RNA fraction, we made use of the fact that the 3′ ends of piRNAs are protected from oxidation because of 2′-O-methylation. To be able to quantify the piRNA fraction and to assess the degree of amplification during next-generation sequencing library preparation, we added a 30-nt-long spike sequence with the same modified 3′ ends as piRNAs. On the basis of the amount of input RNA, 0.5% of artificial spike-in RNA was added (Custom RNA Oligos, Merck; sequence of the spike-in RNA: UAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUC with 2′-OMe-RNA at the 3′ end). To identify piRNAs in the small RNA fraction, we made use of the fact that the 3′ ends of piRNAs are protected from oxidation because of 2′-O-methylation. To enrich for oxidation-protected RNA sequences, small RNAs and spike-in RNAs were oxidized together as described in ref. 78. In brief, 25 μl of small RNA and 2 μl of spike RNA (diluted to 0.5% of total amount of small RNA), 8 μl of 5× Borate Buffer (pH 8.6) and 5 μl of 200 mM sodium periodate were incubated at 25 °C for 30 min. A total of 2 μl of glycerol was added to quench unreacted NaIO4 and incubated for 10 min at room temperature. Next, 229 μl of water, 30 μl of 3 M sodium acetate (pH 5.2) and 1 μl of glycogen were added to each tube, vortexed and spun briefly. A total of 900 μl (three volumes) of 100% ethanol were added, vortexed briefly and incubated on ice for 1 h. Next, the samples were spun at 17,000g for 30 min at 4 °C (vortex for 10 s after 15 min). After removal of the supernatant, 900 μl of 75% ethanol was added and the samples were spun at 17,000g for 5 min and 1 min at 4 °C, interrupted by the removal of the supernatant. Finally, the samples were air dried for 5 min and the pellets dissolved in 8 μl of water. To quantify the spike-in RNAs for testing, whether or not they are protected from oxidation, the TaqMan miR-221 assay kit and an Applied Biosystems 7900HT system was used. We followed the protocol provided in the manufacturer’s instructions (Life Technologies). The expression of small nuclear RNA RNU6b was used for normalization.
Sequencing of small RNAs, removing of adaptors and low-quality reads was done by BGI. Approximately 20 million clean reads were obtained for each sample (Supplementary Table 3). The amount of spike RNA (percentage of clean tag) was calculated as the percentage of reads mapping to the spike sequence out of all clean reads. RNA sequences were mapped to the respective genomes (Supplementary Table 8) using Bowtie2 (v.2.4.1, http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Remaining ribosomal RNA (rRNA) sequences were removed first using RiboDetector (ribodetector_cpu 0.2.7, https://github.com/hzi-bifo/RiboDetector) and highly abundant reads were blasted against the National Center for Biotechnology Information (NCBI) database. To find unique reads, these were collapsed using the fastx_toolkit (v.0.0.14, (ref. 79)) and further cleaned for remaining rRNA reads. Reads in features were counted by either HTseq count (v0.6.1, https://htseq.readthedocs.io/en/master/index.html) or the bedmap program from BEDOPS v.2.4.41 (https://bedops.readthedocs.io/en/latest/index.html) depending on chromosome size. piRNA signature of clean reads was detected using a shell script (grep -A1 “@” *_1.fq.gz | grep -v “@” | grep -v “\-\-” | cut -c <required position> | sort | uniq -c | sed -e ‘s/^[]*//’). Prediction and analyses of genomic piRNA clusters were done by the proTRAC software (v.2.4.4)80.
Sequence reads were BLASTed to hairpin sequences downloaded from miRBase (https://www.mirbase.org/) to calculate the proportion of reads mapping to miRNA sequences.
piRNA machinery genes
For species with genomes annotated by Ensembl, we retrieved the piRNA machinery genes by gene symbol using Ensembl API (https://rest.ensembl.org/). For lungfishes and axolotl, we identified those genes from each assembly using an ab initio method based on sequence similarity. First, the protein sequences of those genes retrieved from Ensembl were mapped onto the assembly using genblastA75. With the query proteins and their rough target region inferred by genblastA (v.1.0.4), we refined the alignments and parsed the gene intron/exon structures using GeneWise (v.2–4)77. Finally, protein sequences of the resulting gene predictions were retrieved and aligned back to the query sequences using blast (v.2.2.26)81 to access the alignment coverage and identity.
Orthology inference
Predicted proteins from the new lungfish genomes (Protopterus annectens and Lepidosiren paradoxa) were analysed together with genome-predicted proteins from representatives of major jawed vertebrate lineages, including Australian lungfish (Neoceratodus forsteri), coelacanth (Latimeria chalumnae), ray-finned fishes (Amphilophus citrinellus, Astatotilapia burtoni, Danio rerio, Lepisosteus oculatus, Tetraodon nigroviridis and Xiphophorus maculatus), chondryichthyans (Callorhinchus milii), amphibians (Ambystoma mexicanum and Xenopus laevis), diapsids (Anolis carolinensis and Gallus gallus) and synapsids (Homo sapiens and Mus musculus). Orthogroups were inferred with Orthofinder (v.2.4.0)82 using a species tree to refine orthology inference following ref. 3, but leaving unresolved the lungfish/coelacanth branch.
Phylogeny inference
Phylogeny was inferred using PhyloBayes MPI (v.1.9)83 under the site-heterogeneous CAT model that can overcome phylogenetic artefacts such as long branch attraction when reconstructing early sarcopterygian relationships12. Data were analysed by gene jackknifing3,84, that is, creating 100 independent sets of loci each with at least 200,000 aligned amino acid positions. A total of 100 independent Markov chain Monte Carlo chains were run until convergence (20,000 cycles, saving every tenth cycle), assessed a posteriori using PhyloBayes’ built-in functions (maxdiff = 1, meandiff = 0.00216271, effective sample size of more than 200 for all parameters after discarding the first 10% cycles as burn-in). Post-burn-in trees were summarized into a fully resolved majority-rule consensus tree (Supplementary Data 1).
Genome size evolution
We inferred a new time-calibrated phylogeny (Supplementary Data 1) using a phylogenomic dataset of 4,593 loci and 100 vertebrate taxa12, and 31 calibrations from recent studies1,13,84 using Bayesian inference (MCMCTree from PAML package v.4.9j) under best-fit amino acid replacement (JTT + G) and molecular clock (autocorrelated relaxed) models. The new time-calibrated tree was used to model the evolution of genome size by maximum likelihood using the ‘fastAnc’ function in the Phytools R package (v.1.19)85. Genome size data were approximated by the size of the assembled genome (when available) or by flow cytometry (haploid DNA content in Gb) obtained from the Animal Genome Size Database (http://www.genomesize.com). Ancestral genome sizes and branch lengths were used to calculate the rates of genome evolution for selected branches. To reconstruct the evolution of cell size, we used the data of osteocyte sizes from both living and fossil lungfishes86 and the time-calibrated phylogeny of ref. 13.
Macrosynteny analysis
Macrosyntenic relationships were profiled using 6,766 core orthologous groups that formed ALGs14. The orthologues to these orthogroups and between each of the lungfish species were computed by mutual best BLAST hit using NCBI BLAST suite v.2.13.0 and requiring mutual best-hit relationships to the chordate amphioxus, the jellyfish Rhopilema esculentum and the scallop Patinopecten yessoensis. This stringency enforced clear one-to-one orthologous group relationships. Published pipelines14,15 were used to construct macrosyntenic dotplots and chromosomal ALG composition plots. To estimate retention rates and gene loss on homologous chromosomes and in each of the ALG group, we first computed all putative paralogous sequences for the three lungfish and spotted gar genomes by requiring them to have higher sequence similarity (by BLAST) than the similarity to their closest amphioxus sequence. We then added these paralogs to the core 6,766 orthologous families. Any paralogs that did not fulfil such criteria were discarded.
To reconstruct the evolutionary history of ur-tetrapod chromosomes, we have investigated chromosomal homologies between the three lungfishes of this study, and the axolotl Ambystoma mexicanum (https://www.axolotl-omics.org/assemblies), the caecilian Rhinatrema bivittatum (NCBI aRhiBiv1.1), the epaulette shark Hemiscyllium ocellatum (NCBI sHemOce1.pat.X.cur), Silurana tropicalis, Mus musculus, Gallus gallus (NCBI, Build 6a), the spotted gar Lepisosteus oculatus (NCBI LepOcu1), the lamprey Petromyzon marinus (NCBI, kPetMar1. pri) and amphioxus Branchiostoma floridae; as a non-chromosomal genome, we included the current assembly of Latimeria chalumnae (NCBI, LatCha1). The chromosomes in each of the three lungfishes were then classified if they have ‘one-to-one’ homology (one and only one homologous chromosome) to each of the ingroup tetrapod and outgroup species. If at least one species in the ingroup and in the outgroup was found to contain a chromosome that fulfils this criterion, then this chromosome is inferred to be ancestral among lungfishes.
In parallel, we also used the Algorithm for Gene Order Reconstruction in Ancestors (AGORA) v.3.1 (ref. 16) for the reconstruction of ur-tetrapod chromosomes. This involved using genomes from the outgroup (Branchiostoma belcheri (annotation from ref. 87), Callorhinchus milii (GCF_018977255.1), Lepisosteus oculatus (GCF_000242695.1), Danio rerio (GCF_000002035.6), Takifugu rubripes (GCF_901000725.2), Amphilophus citrinellus (annotation from ref. 88), Xiphophorus maculatus (GCF_002775205.1) and Latimeria chalumnae (GCF_000225785.1)), the lungfish lineage (Neoceratodus forsteri, Protopterus annectens and Lepidosiren paradoxa) and the tetrapod lineage (Rhinatrema bivittatum (GCF_901001135.1), Xenopus laevis (GCF_017654675.1), Homo sapiens (GCF_000001405.40), Mus musculus (GCF_000001635.27), Anolis carolinensis (GCF_000090745.2) and Gallus gallus (GCF_016699485.2)). Orthogroups were inferred using Orthofinder based on these genomes, followed by providing the orthogroups, species tree and gene coordinates to AGORA for the reconstruction of the ancestral ur-tetrapod genome by using the AGORA vertebrate workflow16. A total of 25,967 orthogroups were inferred in the ur-tetrapod node, and 17,279 of them were assigned to the 811 CARs of the ur-tetrapod node. Among them, 33 CARs contained more than 100 genes and 273 CARs contained more than 10 genes. The AGORA-reconstructed CARs were further corresponded to the ALGs on the basis of their homology.
Positive selection
To estimate genes under positive selection in the lineage leading to the three lungfish species, the protein and complementary DNA (cDNA) fasta files for several fish species were downloaded from public databases (Supplementary Table 8). Orthologous proteins of all fish were identified using OrthoFinder v.2.5.482 with default settings. For each gene with a protein orthologue across all species, the corresponding protein and cDNA sequences were aligned and converted into a codon alignment using pal2nal v.14 (ref. 89). Resulting sequences were aligned by MUSCLE v.14 (ref. 90) (option: -fastaout) and non-conserved blocks were removed using Gblocks (v0.91b)91 (options: -b4 10 -b5 n –b3 5 –t = c). The Gblocks output was converted to PAML format. Trees were built using Phylip (v.3.696, https://phylipweb.github.io/phylip/) with Callorhinchus milii as the outgroup. For the phylogenetic analyses by maximum likelihood, the ‘Environment for Tree Exploration’ (ETE3 v.3.1.1) toolkit92 was used. For the detection of positive selection in lungfish, we calculated two branch-site-specific models, which involved model bsA1 (neutral) versus model bsA (positive selection) to identify sites under positive selection on a specific branch. Genes with a probability of greater than 0.95 for either site class 2a (positive selection in marked branch and conserved in rest) or site class 2b (positive selection in marked branch and relaxed in rest) were considered. Phylogenetic trees were drawn using Python scripts provided by ETE3.
Gene loss analysis
The criteria for gene loss were genes ‘present in coelacanth, spotted gar and Neoceratodus’ but ‘absent from both Lepidosiren and Protopterus’. A candidate gene list was compiled using Orthofinder (v.2.5.4)93 with default parameters. Candidates were filtered for obvious false positives by cross-checking against the existing Protopterus annotation in NCBI2. Remaining genes were manually followed up to identify genes with well-characterized physiological or developmental functions, hence the loss of which could directly inform on specific aspects of lungfish biology. The loss of these genes was subsequently confirmed manually by ViroBLAST (v.1.0)94 of Australian lungfish and coelacanth orthologues against the available transcriptomic and genomic databases for African and South American lungfish. Genes were considered absent if either no significant hits were obtained, or all significant hits could be assigned in cross-blastx analysis to paralogous genes. In two instances, namely of TTTC23 and BMP3/RASGEF1B/PRKG2, which are located in a highly conserved gene block, we performed a synteny analysis to confirm absence of these genes.
Hi-C and enhancer analysis
Conserved enhancer elements were identified by NCBI BLAST search (v.2.13.0) against the lungfish and other species genomes using the mouse sequences described in refs. 44,95. We performed Hi-C analysis as described in the Supplementary Information. Human Hi-C data were extracted from the embryonic stem cell dataset96 using the 3D genome browser97. Midas cichlid Hi-C genome-wide topological associated domain structure was called from Hi-C data using the program HiCExplorer v.3.6 (ref. 98). Hi-C contacts and topological associated domains were visualized by the UCSC Genome Browser.
Mouse transgenesis and SAG treatment of lungfish regenerating fins
For fin regeneration assays in the presence of an Shh pathway agonist, lungfish juveniles (P. annectens) of 11.6–41 cm in body length (n = 14) were obtained, maintained and treated in experimental conditions approved by IBAMA/SISBIO, internal control no. 47206–1, and the Ethics Committee for Animal Research and Experimentation CEUA-UFPA, protocol no. 037–2015 (Belém, Brazil), or IACUCAM-21–155 (Louisiana State University, Baton Rouge, LA, USA). Before any prolonged manipulation or amputation, animals were anesthetized with MS-222 at 0.2%. Animals had their pectoral fins amputated with sterile surgical steel blades at 0.1–0.5 cm distal from their body insertion and collected for skeletal staining as uninjured fins. Immediately after amputation and anaesthesia awakening (5–10 min), each animal was transferred to their individual tanks. On the second week post-amputation, the treatment with 200 nM of the SAG (Sigma Aldrich, catalogoue no. ML1314 or Adipogen catalogue no. AG-CR1–3585) started and was maintained until the seventh week. Control animals were treated with the equivalent volume amount of dimethylsulfoxide (DMSO). Treatment tanks were stored at room temperature (24–28 °C) in the dark (because of SAG photosensitivity) and solutions were changed once a day after animals had been fed (30–60 min, away from direct light). Fins were collected for skeletal staining at the eighth week.
For skeletal staining, fins were collected in ethanol 100% or buffered formalin (10%, pH of about 7.0). Subsequently, they were stained with Alcian blue (for cartilage staining) as described in ref. 99, with the following changes: after 24–48 h in ethanol, 70% samples were transferred to Alcian blue (1.2 mg ml−1, pH of about 2.0). After bleaching, clearing and glycerin series, fins were stored in 100% glycerin. All pictures for morphological/structural assessments were taken at time intervals specified on the panels with a Zeiss SteREO Discovery.V12 microscope with MRc5 camera.
For the ZRS sequence multiple alignment, ZRS orthologous sequences from chicken, lizard, coelacanth and Australian and African lungfishes were retrieved from the NCBI on the basis of a BLAST (v.2.13.0) search using a core of about 800 bp of the mouse ZRS enhancer sequence100. The South American lungfish ZRS orthologue sequence was retrieved from the genome assembly described in the present study. All the ZRS orthologue sequences were aligned using MAFFT (v.7) (https://www.ebi.ac.uk/jdispatcher/). Annotation/curation of the ETS transcription factor binding sites was manually entered with Adobe Illustrator.
For the ZRS enhancer–reporter mouse transgenic assays, DNA fragments corresponding to the L. paradoxa or N. forsteri ZRS sequences, flanked by the vector cloning site sequences (including a NotI restriction site), were synthesized by Twist Biosciences. The fragments were cloned into the PCR4-Shh::lacZ-H11 vector (Addgene_Plasmid#139098) using Gibson Assembly, as previously described in ref. 101. Enhancer–reporter transgenesis and mouse embryo X-gal staining were performed as previously described in ref. 101.
The mouse transgenic experiments were reviewed and approved by the University of California Irvine Laboratory Animal Resources (ULAR) under protocols AUP-20–001 and AUP-23–005. Mice were housed in the animal facility, where their conditions were electronically monitored 24/7 with daily visual checks by technicians. Mice were housed in BioBubble Clean Rooms, soft-walled enclosures powered by 80–100 air changes per hour of high-efficiency particulate air filtration under a light/dark cycle of 12:12 starting at 6 am, at 22–24.4 °C, and humidity 30–70%. All mice used in this study were of Mus musculus species and FVB strain.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1: Phylogenomics of lungfish.
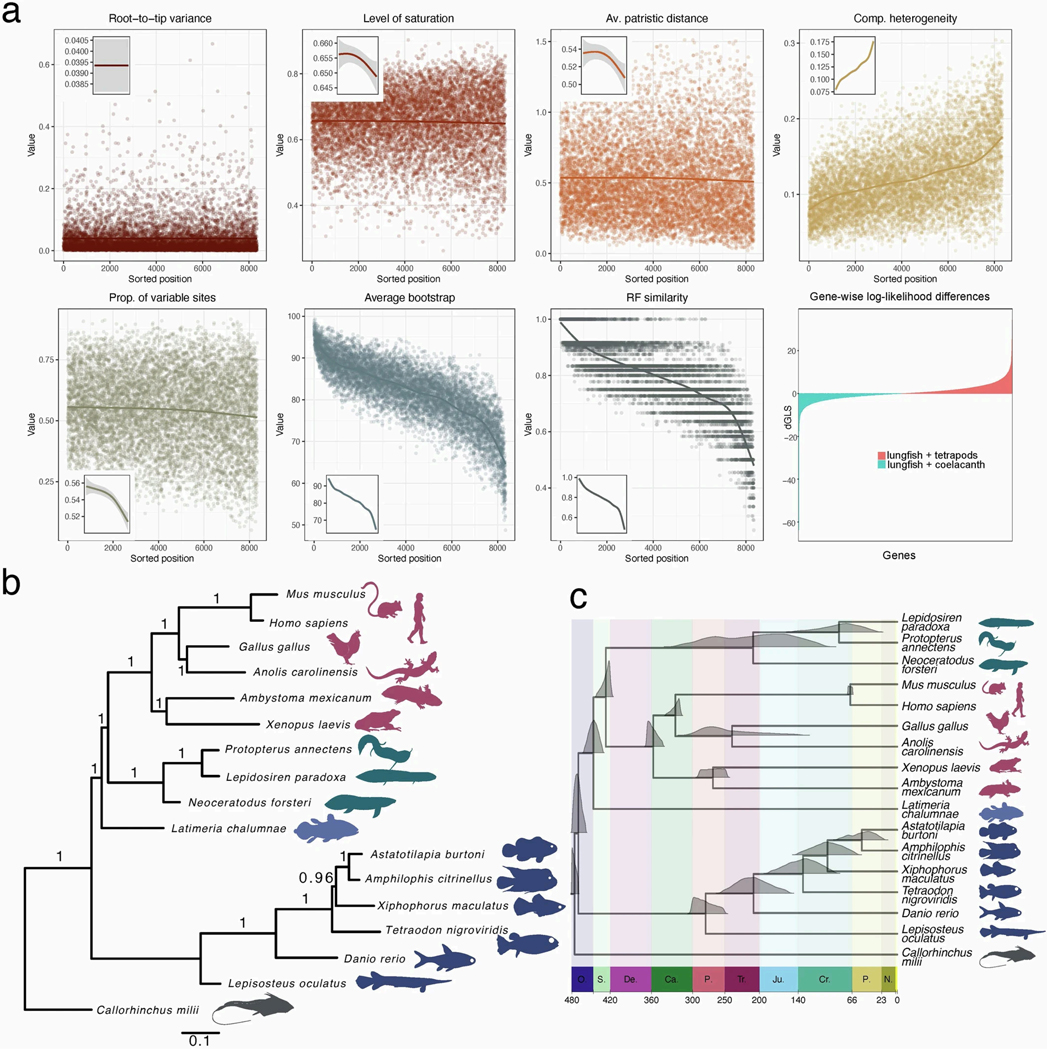
a, Loci selection for phylogenomics. Graphs show different properties (root-to-tip variance, level of saturation, average patristic distance, compositional heterogeneity, proportion of variable sites, average bootstrap support, Robinson-Foulds similarity) for the 8,339 loci as inferred by genesortR. The graph of gene-wise log-likelihood differences shows support of each locus for two relevant alternative hypotheses (see Supplementary Information 2). b, Bayesian phylogram showing the evolutionary relationships and relative rates of the three lungfish genomes within the context of vertebrate phylogeny. The phylogeny was reconstructed as the consensus of 100 Markov chains (MCMC) from 100 independent gene jackknife replicates analyzed by PhyloBayes-MPI under the CAT mixture model (indicated with numbers on the internal edges, 1 = 100 replicates). The scale bar is the expected amino acid replacements per site. c. Bayesian time-calibrated phylogeny inferred from the set of 8,323 orthologs. Posterior probability distributions of estimated ages of common ancestors are plotted on tree nodes. X axis is in million years and major geological periods are indicated (O. Ordovician, S. Silurian, De. Devonian, Ca. Carboniferous, P. Permian, Tr. Triassic, Ju. Jurassic, Cr. Cretaceous, P. Paleogene, N. Neogene).
Extended Data Fig. 2: High retention of ancestral linkage groups lungfish genomes.
a-d, Species-to-species dotplots showing high degree of retained collinearity in the African and South American lungfish genomes, despite their genome size. b-d, Oxford dotplots representing orthologous genes shared on the previously reported ancestral linkage groups (ALGs)15. Chromosome numbering corresponds to the homologous lungfish linkage groups which have independently fused in individual lineages. Neoceratodus with its 27 chromosomes represented the most ancestral (unfused) state. e, Retention rates of lungfish chromosomes. Often only one alpha copy is present in lungfishes, e.g. descendants of several chromosomal elements have two alpha chromosomes in gar and Australian lungfish but only one clear alpha chromosome remains in South American and African lungfish (with the alpha copies having lost genes). Retention rates were computed as the percentage of the retained (present) ohnologs of gene families that comprise a given ancestral linkage group. Total number of gene families per chromosome was counted and their position was not taken into account. Only chromosomes with at least 5% ancestral linkage group retention were counted. Lower plots show retention on individual chromosomes (represented by dots) grouped by their ancestral linkage group in different lungfishes and gar.
Extended Data Fig. 3: Genomic composition of repetitive elements.
a, Overall composition of repetitive elements from unmasked assemblies (two rounds of transposable element annotation) for the three lungfish (Lpa=Lepidosiren paradoxa, Pan=Protopterus annectens, Nfo=Neoceratodus forsteri), axolotl (Ame=Ambystoma mexicanum), and coelacanth (Lch=Latimeria chalumnae). The total TE coverage for each species is shown under each pie chart. RC, rolling-circle transposon; SINE, short interspersed element; LINE, long interspersed element; LTR, long terminal repeat; DNA, cut-and-paste DNA transposons. Total repeat coverage of other species analyzed in this study: Xenopus ~25%; Platyfish ~23%; Burtoni and Midas cichlids ~30%; and Pufferfish ~8%. b, Different repeat superfamilies expanded in lungfish genomes. Heatmap shows the repeat superfamily content of coelacanth (Lch=Latimeria chalumnae), axolotl (Ame=Ambystoma mexicanum) and three lungfish (Lpa=Lepidosiren paradoxa, Pan=Protopterus annectens, Nfo=Neoceratodus forsteri). The color is scaled to the genomic content across repeat superfamilies.
Extended Data Fig. 4: Expression of transposable element families.
a, b, Expression estimated for each transposable element family from poly (A)-enriched RNA-seq data. In all tissues, SINEs are more highly expressed than any other subclass in the African lungfish, while both LINEs and SINEs are slightly more expressed than any other subclass in the South American lungfish. n = 2029 (African lungfish) and 1897 (South American lungfish) transposable element families. Wilcoxon Signed Ranks Test (one-sided) was applied with * indicating p-value < 0.05, ** p-value < 0.005, *** p-value < 0.0005 and **** p-value < 0.00005. The box bounds the interquartile range divided by the median value, with the whiskers extending to a maximum of 1.5 times the interquartile range beyond the box. c, d, Higher expression of young transposable element families. When transposable element families are divided into young or old copies based on Kimura 2-parameter distance to consensus values (0–10% is young, >10% is old), young TEs are significantly higher expressed than old ones, suggesting that several types of TEs remain active and contribute to the ongoing expansion of the lungfish genomes. Out of the 13 SINE families of Protopterus annectens, only copies from the SINE/t-RNA-V-RTE are considered as young. e, f, | Correlation between expression of transposable element families and copy number. Expression was estimated for each transposable element family using poly (A)-enriched RNA-seq data. For all tissues and transposable element classes, a positive correlation is observed between expression level and copy number. When a transposable element family is highly expressed, this family tends to have more copies. All analyzed correlations are significantly positive (p-values < 0.001). A linear model estimated trend line and calculated 95% confidence interval around the trend (gray fill) are plotted (two-sided). Lpa, Lepidosiren paradoxa; Pan, Protopterus annectens.
Extended Data Fig. 5: Age estimation and comparison of full-length TEs across lungfish genomes.
a, Landscape of subclasses of transposable elements. Kimura substitution level (%) for each copy against its consensus sequence used as proxy for expansion history of the transposable elements. Older copies accumulated more nucleotide substitutions and show higher distance to the consensus sequences. The phylogeny depicts the estimation of divergence times among the five studied species. RC, rolling-circle transposon; SINE, short interspersed element; LINE, long interspersed element; LTR, long terminal repeat. b, Copy numbers of full-length TEs within orders. c, Copy numbers of full-length TEs within superfamilies, color scaled to copy number. d, Percentage of transcribed TEs. e. Example of synteny to show one full-length copy from LINE/CR1 exclusively present in our Protopterus genome and absent in the other individual’s genome. f, Comparison of expression between full-length and fragmented TEs. n = 122, 832, 031 (South American lungfish), 66, 736, 976 (African lungfish) and 58, 296, 831 transposable elements. Wilcoxon Signed Ranks Test (one-sided) was applied with **** indicating p-value < 0.00005. The box bounds the interquartile range divided by the median value, with the whiskers extending to a maximum of 1.5 times the interquartile range beyond the box and the middle dots indicate mean values. Lpa=Lepidosiren paradoxa, South American lungfish; Pan=Protoperus annectens, African lungfish; Nfo=Neoceratodus fosteri; Australian lungfish.
Extended Data Fig. 6: Size distribution and correlation between piRNA content and genome size.
a, Size distribution of clean reads of unoxidized small second distinct peak at the expected size range of piRNAs. b, Spearman rank RNA libraries of the same individuals as used for the piRNA analysis, with the correlation between genome size (log scale) and %RNA of clean tag) from the position of the peaks for miRNA and piRNA marked with dotted lines. In contrast oxidized testis small RNAs (silhouettes as in a). to the oxidized samples African and South American lungfish have a clear peak at the expected size range of miRNAs (~24 nts), but unlike the other species no second distinct peak at the expected size range of piRNAs. b, Spearman rank correlation between genome size (log scale) and %RNA of clean tag) from the oxidized testis small RNAs (silhouettes as in a).
Extended Data Fig. 7: Signature nucleotides of piRNAs, piRNA cluster structure and KZFP genes.
a, Proportion of nucleotides of the small RNA reads at the first position (left) and the tenth position (right) of the three lungfish, amphibian and fish samples. b, Graphical proTRAC output of a representative piRNA cluster for the pufferfish (left panel) and the South American lungfish (right panel). The top part visualizes the number of genomic hits produced by the query piRNA sequence. Dark green indicating that there is only one sequence hit in the genome, dark red indicating more than 1000 hits. Below is the sequence read coverage plot (blue: reads on the plus strand, red: reads on the minus strand). The RepeatMasker bar shows TEs annotated by RepeatMasker in this region. Lungfish clusters tend to have lower diversity and a higher read count. c, C2H2 zinc-finger and KRAB domain protein (KZFP) gene counts and genomic organization in sarcopterygians. Left, number of KZFP genes in indicated genomes. Right, gene length of KZFP genes in indicated species. n = 1168 KZFPs. Wilcoxon Signed Ranks Test (one-sided) was applied with **** indicating p-value < 0.00005. The box bounds the interquartile range divided by the median value, with the whiskers extending to a maximum of 1.5 times the interquartile range beyond the box. Lpa=Lepidosiren paradoxa; Pan=Protopterus annectens; Nfo=Neoceratodus forsteri; Lch=Latimeria chalumnae; Hsa=Homo sapiens; Gga=Gallus gallus.
Extended Data Fig. 8: Positively selected genes and gene losses.
a, Positively selected genes in all three lungfishes related to lungfish biology. b, Numerous gene losses in Lepidosiren paradoxa and Protopterus annectens indicate a cellular milieu that is permissive of transposon spreading due to a reduction in the DNA damage response and apoptosis. Due to low piRNA levels (through an as of yet unidentified mechanism) high activity of transposable elements is present in the germline resulting in frequent insertions and high levels of genotoxic stress due to double stranded DNA breaks which tend to result in G1 arrest and apoptosis as part of the DNA damage response which provides a mechanism for somatic selection against compromised cells. These gene losses are expected to reduce the levels of such selection and create a permissive environment for DNA transposition and helps explain the rapid expansion of the lungfishes’ genomes. c, The synteny block spanning RASGEF1B to ANTXR2 is widely preserved across vertebrates. The region containing RASGEF1B to PRDM8 has been deleted in Lepidosiren paradoxa and Protopterus annectens. The ciliary CFAP299 gene is still present in both species as an intronless retrogene. Loss of BMP3 can be linked to the reduced squamation of the derived Lepidoserenidae, while loss of PRKG2 and RASGEF1B can be linked to their derived fins. In the ray finned fish Astatotilapia burtoni, BMP3 is strongly expressed in the developing scales at 12 dpf. d, TTC23 is a component of the primary cilia and involved in the cellular perception of the shh signal transduction pathway. TTC23 is located in a highly conserved gene block which is also preserved in Lepidosiren paradoxa and Protopterus annectens, however without an identifiable TTC23 gene present. This “ghost locus” was further analyzed using Lagan Vista. Paired Lagan using the translated anchoring option and the Coelacanth sequence as baseline identifies the TTC23 exons in human, spotted gar and Neoceratodus forsteri, but not in Lepidosiren. paradoxa and Protopterus annectens.
Extended Data Fig. 9: Expanded hox clusters preserve regulatory landscape architecture.
a, In spite of a dramatic expansion of the lungfish Hox clusters whereby the Lepidosiren paradoxa clusters are approximately 20-fold enlarged compared to mouse, which is lower than the proportional difference in genome size. Consistent with this observation is that all four clusters preserve a conserved core subcluster (indicated in red) that has expanded relatively little and is low in repeat content. These regions are hoxa4-a11, hoxb2-b9, hoxc4-c11 and hoxd8-d11 indicating topological constraints on the expansion of these regions. In addition, hoxa3 and hoxd3 (purple) show expansion of their intronic region, which is similar to the expansion of the hoxa3 intron in the expanded axolotl Hoxa cluster7. An interesting difference is that the hoxa11-hoxa13 intergenic shows a tendency for expansion in lungfishes but not in axolotl, potentially related to additional constraints induced by the fin to limb transition. Furthermore, signatures of repeat insertion in the anterior Hoxc and posterior Hoxb clusters mirror those observed in anolis lizards41. b, HiC analysis for Midas cichlid, human and Protopterus annectens Hoxa and Hoxd clusters. Despite the approximate 70 times size difference between these species there is a remarkable conservation of the flanking regulatory landscapes whereby both clusters are present on the intersection of a 3’ and 5’ TAD. Known fin and limb enhancers (blue ovals) are conserved in an expected fashion (open ovals for Lepidosirenidae mm406 and e10 indicate secondary loss), altogether suggesting that long range regulatory landscapes remain preserved under conditions of genome expansion. Synteny regions shown encompass the following sizes: HoxA; Pan 3.2 Mb, Hsa 3.1 Mb Aci 0.31 Mb, Hoxd; Pan 28 Mb, Hsa 2.8 Mb, Aci 0.41 Mb. Species name abbreviations are the same as in the other figures.
Extended Data Fig. 10: Functional analysis of lungfishes ZRS and SAG treatment of Lepidosiren paradoxa regenerating fins.
a, Mouse transgenesis and LacZ staining for the Neoceratodus forsteri and Lepidosiren paradoxa ZRS sequences. Genotyping indicates whether insertion was either in a single or double copy at the targeted locus, or randomly integrated in the genome. Neoceratodus forsteri ZRS gives ZPA staining in 16/16 embryos, whereas the Lepidosiren paradoxa ZRS does not give staining in 15/15 embryos. b, Regeneration of pectoral fins in presence of the shh agonist SAG does not result in radial growth in Lepidosiren paradoxa (n = 3 for SAG treated animals, n = 3 for DMSO-treated animals; representative images of one animal per treatment are shown).
Supplementary Material
Acknowledgements
This work was supported by the German Research Foundation (DFG) through a grant to A.M., T. Burmester and M.S. (Me1725/24–1, Bu956/23–1, Scha408/16–1). O.S. was supported by the European Research Council’s Horizon 2020: European Union Research and Innovation Programme, grant no. 945026. Next-generation sequencing data production and data analysis were carried out at the DRESDEN-concept Genome Center, supported by the DFG Research Infrastructure Programme (project 407482635) and part of the Next Generation Sequencing Competence Network (project 423957469).
Footnotes
Code availability
Custom codes have been deposited at https://github.com/dukecomeback/lungfish, https://gitlab.mpi-cbg.de/assembly/programs/manualcurationhic, https://gitlab.mpi-cbg.de/assembly/programs/polishing and https://github.com/MartinPippel/DAmar.
Competing interests The authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586–024-07830–1.
Data availability
Genome assemblies and sequencing data are available from NCBI Bioprojects PRJNA808321, PRJNA808322, PRJNA813994, PRJNA813995 and PRJNA981572 and at BioSamples SAMN26083907 and SAMN26533844. Gene and repeat annotations are available at Figshare (https://figshare.com/articles/dataset/Lungfish_genome_annotation/24147732)102.
References
- 1.Meyer A et al. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 590, 284–289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K et al. African lungfish genome sheds light on the vertebrate water-to-land transition. Cell 184, 1362–1376.e1318 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Irisarri I et al. Phylotranscriptomic consolidation of the jawed vertebrate timetree. Nat. Ecol. Evol 1, 1370–1378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krefft JLG Description of a gigantic amphibian allied to the genus Lepidosiren from the Wide-Bay district, Queensland. Proc. Zool. Soc. Lond 1870, 221–224 (1870). [Google Scholar]
- 5.Meyer A & Dolven SI Molecules, fossils, and the origin of tetrapods. J. Mol. Evol 35, 102–113 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Kemp A The biology of the Australian lungfish, Neoceratodus forsteri (Krefft 1870). J. Morphol 190, 181–198 (1986). [Google Scholar]
- 7.Nowoshilow S et al. The axolotl genome and the evolution of key tissue formation regulators. Nature 554, 50–55 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Shao C et al. The enormous repetitive Antarctic krill genome reveals environmental adaptations and population insights. Cell 186, 1279–1294.e1219 (2023). [DOI] [PubMed] [Google Scholar]
- 9.Oliveira C et al. Chromosome formulae of neotropical freshwater fishes. Rev. Brasil. Genet 11, 577–624 (1988). [Google Scholar]
- 10.Suzuki A & Yamanaka K Chromosomes of an African Lungfish, Protopterus annectens. Proc. Jpn Acad. B Phys. Biol. Sci 64, 119–121 (1988). [Google Scholar]
- 11.Nurk S et al. The complete sequence of a human genome. Science 376, 44–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irisarri I & Meyer A The identification of the closest living relative(s) of tetrapods: phylogenomic lessons for resolving short ancient internodes. Syst. Biol 65, 1057–1075 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Brownstein CD, Harrington RC & Near TJ The biogeography of extant lungfishes traces the breakup of Gondwana. J. Biogeogr 50, 1191–1198 (2023). [Google Scholar]
- 14.Simakov O et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol 4, 820–830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simakov O et al. Deeply conserved synteny and the evolution of metazoan chromosomes. Sci. Adv 8, eabi5884 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muffato M et al. Reconstruction of hundreds of reference ancestral genomes across the eukaryotic kingdom. Nat. Ecol. Evol 7, 355–366 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourque G et al. Ten things you should know about transposable elements. Genome Biol. 19, 199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer A & Schartl M Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol 11, 699–704 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Thomson KS An attempt to reconstruct evolutionary changes in the cellular DNA content of lungfish. J. Exp. Zool 180, 363–371 (1972). [Google Scholar]
- 20.Gregory TR The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells Mol. Dis 27, 830–843 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Nystedt B et al. The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Falcon F, Tanaka EM & Rodriguez-Terrones D Transposon waves at the water-to-land transition. Curr. Opin. Genet. Dev 81, 102059 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Brennecke J et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Yi M et al. Rapid evolution of piRNA pathway in the teleost fish: implication for an adaptation to transposon diversity. Genome Biol. Evol 6, 1393–1407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J et al. Transposable element and host silencing activity in gigantic genomes. Front. Cell Dev. Biol 11, 1124374 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song J et al. Variation in piRNA and transposable element content in strains of Drosophila melanogaster. Genome Biol. Evol 6, 2786–2798 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aravin AA et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W et al. The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs. Mol. Cell 56, 708–716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquesi GIM et al. Vertebrate lineages exhibit diverse patterns of transposable element regulation and expression across tissues. Genome Biol. Evol 12, 506–521 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kofler R piRNA clusters need a minimum size to control transposable element invasions. Genome Biol. Evol 12, 736–749 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X et al. Transposable element expansion and low-level piRNA silencing in grasshoppers may cause genome gigantism. BMC Biol. 20, 243 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang P, Wang Y & Macfarlan TS The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet. 33, 871–881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imbeault M, Helleboid P-Y & Trono D KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Kaessmann H, Vinckenbosch N & Long M RNA-based gene duplication: mechanistic and evolutionary insights. Nat. Rev. Genet 10, 19–31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carelli FN et al. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 26, 301–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen M et al. Evolutionary patterns of RNA-based duplication in non-mammalian chordates. PLoS ONE 6, e21466 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okabe M & Graham A The origin of the parathyroid gland. Proc. Natl Acad. Sci. USA 101, 17716–17719 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C et al. Genome sequences reveal global dispersal routes and suggest convergent genetic adaptations in seahorse evolution. Nat. Commun 12, 1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerr T The scales of modern lungfish. Proc. Zool. Soc. Lond 125, 335–345 (1955). [Google Scholar]
- 40.Lander ES et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Di-Poï N, Montoya-Burgos JI & Duboule D Atypical relaxation of structural constraints in Hox gene clusters of the green anole lizard. Genome Res. 19, 602–610 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feiner N Accumulation of transposable elements in Hox gene clusters during adaptive radiation of Anolis lizards. Proc. Biol. Sci 283, 20161555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woltering JM., Noordermeer D., Leleu M. & Duboule D. Conservation and divergence of regulatory strategies at Hox loci and the origin of tetrapod digits. PLoS Biol. 12, e1001773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berlivet S et al. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 9, e1004018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kemp A, Cavin L & Guinot G Evolutionary history of lungfishes with a new phylogeny of post-Devonian genera. Palaeogeogr. Palaeoclimatol. Palaeoecol 471, 209–219 (2017). [Google Scholar]
- 46.Díaz-González F et al. Biallelic cGMP-dependent type II protein kinase gene (PRKG2) variants cause a novel acromesomelic dysplasia. J. Med. Genet 59, 28–38 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Lewandowski JP et al. Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Dev. Biol 406, 92–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breslow DK et al. A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat. Genet 50, 460–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L et al. Enlarged fins of Tibetan catfish provide new evidence of adaptation to high plateau. Sci. China Life Sci 66, 1554–1568 (2023). [DOI] [PubMed] [Google Scholar]
- 50.Letelier J et al. The Shh/Gli3 gene regulatory network precedes the origin of paired fins and reveals the deep homology between distal fins and digits. Proc. Natl Acad. Sci. USA 118, e2100575118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woltering JM et al. Sarcopterygian fin ontogeny elucidates the origin of hands with digits. Sci. Adv 6, eabc3510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kvon EZ et al. Comprehensive in vivo interrogation reveals phenotypic impact of human enhancer variants. Cell 180, 1262–1271.e1215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roscito JG et al. Convergent and lineage-specific genomic differences in limb regulatory elements in limbless reptile lineages. Cell Rep. 38, 110280 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Ovchinnikov V et al. Caecilian genomes reveal the molecular basis of adaptation and convergent evolution of limblessness in snakes and caecilians. Mol. Biol. Evol 40, msad102 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez-Rios J The many lives of SHH in limb development and evolution. Semin. Cell Dev. Biol 49, 116–124 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Farrell ER & Münsterberg AE csal1 is controlled by a combination of FGF and Wnt signals in developing limb buds. Dev. Biol 225, 447–458 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Carneiro J et al. Evidence of cryptic speciation in South American lungfish. J. Zool. Syst. Evol. Res 59, 760–771 (2021). [Google Scholar]
- 58.Storer J, Hubley R, Rosen J, Wheeler TJ & Smit AF The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob. DNA 12, 2 (2021); https://pubmed.ncbi.nlm.nih.gov/33436076/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flynn JM. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl Acad. Sci USA 117, 9451–9457 (2020); https://pubmed.ncbi.nlm.nih.gov/32300014/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benson G Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999); https://pubmed.ncbi.nlm.nih.gov/9862982/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bao Z, & Edyy SR Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12, 1269–1276 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price AL, Jones NC & Pevzner PA De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Camacho C et al. BLAST+: architecture and applications. BMC Bioinform. 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chalopin D, Naville M, Plard F, Galiana D & Volff J-N Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol 7, 567–580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conte MA et al. Chromosome-scale assemblies reveal the structural evolution of African cichlid genomes. Gigascience 8, giz030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brawand D et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513, 375–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong Y et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat. Commun 10, 5228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang WR, Ardeljan D, Pacyna CN, Payer LM & Burns KH SQuIRE reveals locus-specific regulation of interspersed repeat expression. Nucleic Acids Res. 47, e27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peona V et al. The avian W chromosome is a refugium for endogenous retroviruses with likely effects on female-biased mutational load and genetic incompatibilities. Philos. Trans. R. Soc. Lond. B Biol. Sci 376, 20200186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finn RD et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ellinghaus D, Kurtz S & Willhoeft U LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 9, 18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steinbiss S, Willhoeft U, Gremme G & Kurtz S Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37, 7002–7013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Llorens C et al. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 39, D70–D74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Groza C, Chen X, Wheeler TJ, Bourque G & Goubert C GraffiTE: a unified framework to analyzetransposable element insertion polymorphisms using genome-graphs. Preprint at bioRxiv 10.1101/2023.09.11.557209 (2023). [DOI] [Google Scholar]
- 75.She R, Chu JS, Wang K, Pei J & Chen N GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 19, 143–149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pearson WR Finding protein and nucleotide similarities with FASTA. Curr. Protoc. Bioinform 53, 3.9.1–3.9.25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Birney E, Clamp M & Durbin R GeneWise and Genomewise. Genome Res. 14, 988–995 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sellitto A et al. Molecular and functional characterization of the somatic PIWIL1/piRNA pathway in colorectal cancer cells. Cells 8, 1390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmieder R & Edwards R Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosenkranz D & Zischler H proTRAC-a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinform. 13, 5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camacho C et al. BLAST+: architecture and applications. BMC Bioinform. 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Emms DM & Kelly S OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lartillot N, Rodrigue N, Stubbs D & Richer J PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol 62, 611–615 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Delsuc F, Brinkmann H, Chourrout D & Philippe H Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 (2006). [DOI] [PubMed] [Google Scholar]
- 85.Revell LJ phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol 3, 217–223 (2012). [Google Scholar]
- 86.Thomson KS & Muraszko K Estimation of cell size and DNA content in fossil fishes and amphibians. J. Exp. Zool 205, 315–320 (1978). [Google Scholar]
- 87.Huang Z et al. Three amphioxus reference genomes reveal gene and chromosome evolution of chordates. Proc. Natl Acad. Sci. USA 120, e2201504120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kautt AF et al. Contrasting signatures of genomic divergence during sympatric speciation. Nature 588, 106–111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suyama M, Torrents D & Bork P PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Edgar RC MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Castresana J Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- 92.Huerta-Cepas J, Serra F & Bork P ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Emms DM & Kelly S OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Deng W, Nickle DC, Learn GH, Maust B & Mullins JI ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics 23, 2334–2336 (2007). [DOI] [PubMed] [Google Scholar]
- 95.Montavon T et al. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011). [DOI] [PubMed] [Google Scholar]
- 96.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19, 151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramírez F et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun 9, 189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taylor W & Van Dyke G Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium 9, 107–119 (1985). [Google Scholar]
- 100.Kvon EZ et al. Progressive loss of function in a limb enhancer during snake evolution. Cell 167, 633–642.e611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Osterwalder M et al. in Craniofacial Development Vol. 2403 (ed. Dworkin S) 147–186 (Humana, 2022). [Google Scholar]
- 102.Du K Lungfish genome annotation. figshare 10.6084/m9.figshare.24147732.v1 (2024). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies and sequencing data are available from NCBI Bioprojects PRJNA808321, PRJNA808322, PRJNA813994, PRJNA813995 and PRJNA981572 and at BioSamples SAMN26083907 and SAMN26533844. Gene and repeat annotations are available at Figshare (https://figshare.com/articles/dataset/Lungfish_genome_annotation/24147732)102.