Abstract
JC virus (JCV), along with other members of the polyomavirus family, encodes a class of highly conserved proteins, T antigens, that are capable of inducing aneuploidy in cultured cells. We have previously isolated T-antigen DNA variants of JCV from both colon cancer tissues and the corresponding nonneoplastic gastrointestinal tissues, raising new questions about the role of JCV in the development of chromosomal instability of the colon. Based on the sequence of the transcriptional control region (TCR), JCV can be classified as archetype or tandem repeat variants. Among the latter, Mad-1, the prototype virus first isolated from a patient with progressive multifocal leukoencephalopathy, is characterized by lacking the 23- and 66-bp sequences that are present in the archetype and by duplication of a 98-bp sequence. In this study, we evaluated differences in the TCR of JCV isolated from colon cancer tissues and nonneoplastic epithelium. To characterize JCV variants, we first treated eight pairs of DNA samples from colon cancers and noncancerous tissue with topoisomerase I and then amplified and cloned the JCV TCR. We obtained 285 recombinant clones from the JCV TCR, 157 from nonneoplastic samples, and 128 from colon cancer tissues. Of these clones, 262 spanned the length of the JCV Mad-1 TCR: 99.3% from nonneoplastic samples and 82.8% from colon cancer tissues. In sequencing 54 clones in both directions, we did not find archetype JCV either in the nonneoplastic tissue or in the cancer samples. From all colon cancer tissues, 18 clones had a deletion of one 98-bp tandem repeat. This deleted strain was not detected in any of the nonneoplastic tissues (14 versus 0% [χ2 = 23.6; P < 0.001]). Our study demonstrates that the only JCV strain present in the human colon is Mad-1, and the variant with a single 98-bp sequence is found exclusively in the cancer tissues. This strain may be involved in the development of chromosomal instability.
The human polyomavirus JC virus (JCV) infects a large proportion of the population worldwide (23). The initial infection is unapparent, but subsequently JCV establishes a latent infection, becoming reactivated when the immune system is impaired (8). The JCV genome is double-stranded, negatively supercoiled circular DNA that consists of early and late coding regions separated by two noncoding regions at both the 5′ and 3′ ends. Within the noncoding sequences is the transcriptional control region (TCR), which contains the promoter and enhancer for the early and late proteins; the intergenic region separates the 3′ regions of T antigen and the capsid protein, VP1.
T antigen is an oncogenic protein capable of transforming mammalian cells by interacting with cellular proteins such as p53 and the Rb family proteins (5, 21). T antigen also has ATPase and helicase activities, the latter of which may eventually contribute to chromosomal breakage and recombination (7). We have recently reported the presence of JCV T-antigen DNA sequences in nonneoplastic gastrointestinal tissues, colon cancer samples, and adjacent nonneoplastic mucosa, indicating that JCV might play a role in inducing chromosomal instability in the colonic cells (10, 20).
Based on the structure of its TCR, JCV can be classified into two forms: the archetype and tandem repeat variants. Although debatable, the archetype is presumed to be the circulating strain of the virus, which can be isolated from the kidneys and is excreted in urine. The tandem repeat variants are direct descendents of the archetype and result from deletions and duplications of sequences within the regulatory region (25). Among the tandem repeat variants of JCV, the best studied is Mad-1, which is the etiological agent of progressive multifocal leukoencephalopathy (PML) in immunocompromised individuals (17). The regulatory region of Mad-1 is characterized by two 98-bp repeats that feature a TATA box and by the absence of the 23- and 66-bp fragments that are present in the archetype. While tandem repeat variants have been found in the central nervous system of patients with PML, in brain tumors, and in peripheral lymphocytes of both PML and non-PML patients, the archetype has rarely been found in diseased brains. These findings suggest that the presence of a specific strain might reflect viral adaptation to the particular host tissue (4, 16, 19, 22).
Because the Mad-1 strain has the capacity for lytic infection of oligodendrocytes, JCV has always been considered strictly neurotropic. However, recent studies have shown that certain strains of JCV can replicate in lymphoid cells and that the capability of the virus to replicate in host cells is highly dependent on the structure of the TCR (2, 14). It has also been demonstrated that the ability of the virus to replicate in vitro depends upon the regulatory region. Paradoxically, the archetype appears incapable of replication in vitro in any cell, while strains characterized by tandem repeats display different capabilities and show higher rates of replication in glial cells (3).
In this study, we characterized the regulatory region in JCV isolates from colon cancer tissues and adjacent nonneoplastic colonic mucosa to better understand which strain might predominate. We found that the Mad-1 strain infects both colon cancer tissues and the nonneoplastic colonic mucosa. We also found that the archetype was absent in all samples, which suggests that archetype JCV cannot infect the colonic epithelium. Furthermore, a particular variant of the Mad-1 strain that lacks one of the 98-bp repeats is present in colon cancer tissues but absent in the adjacent nonneoplastic colonic epithelium. As previously demonstrated, this strain has different replication activity in vitro from the archetype and other tandem repeat variants and might be involved in initiating chromosomal instability in the colonic cells.
For the analysis of the TCR, we studied eight pairs of DNA samples that we found to be positive for JCV T-antigen DNA sequences in our earlier study. To enhance PCR amplification, we treated DNA with topoisomerase I, which, as we have previously demonstrated, relaxes the negatively supercoiled genome of the virus (10, 20) (L. Laghi, A. E. Randolph, and C. R. Boland, submitted for publication). We treated 500 ng of DNA with topoisomerase I (Life Technologies, Rockville, Md.) in the presence of 2 mM MgCl2, 14 mM Tris HCl (pH 8.4), and 32 mM KCl for 30 min at 37°C. To amplify the TCR, we chose primers spanning a part of the DNA sequence from T antigen to the agnoprotein, a strategy that ensured detection of both archetype and any tandem repeat variants (Fig. 1). The first round of DNA amplification was designed, based upon the JCV Mad-1 sequence, to target a fragment between nucleotides 4933 and 448. The nested reaction amplified a DNA sequence spanning nucleotides 4985 to 317, a sequence that included the entire JCV regulatory region.
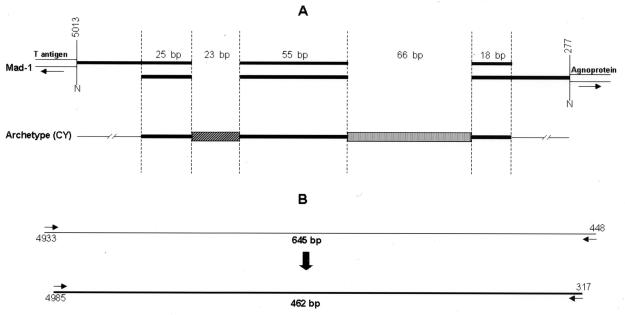
FIG. 1.
Map of the TCR of JCV Mad-1 and archetype. (A) Mad-1 is characterized by the absence of the 23- and 66-bp fragments and by duplications of the 25-, 55-, and 18-bp sequences, resulting in two identical 98-bp fragments, both featuring a TATA box. (B) For the PCR strategy, both first- and second-round reactions were targeted to amplify the complete TCR sequence spanning the highly conserved sequences of T antigen and agnoprotein. This design allowed both the rearranged and archetype forms to be amplified.
The primers for PCR amplifications were JPRRF1 (5′-AATGTTCCCCCATGCAGACC-3′ [nucleotides 4933 to 4952]) on the sense strand and an antisense primer binding at nucleotides 448 to 429 (15) for the first round. For the nested reaction, we used primer JPRNF1 (5′-GATTCCTCCCTATTCAGCACTTTG-3′ [nucleotides 4985 to 5003]) on the sense strand and an antisense primer binding at nucleotides 317 to 298 (15). The expected amplicons were 645 and 462 bp, respectively. All PCR amplifications included negative controls to rule out contamination.
The first-round amplification was carried out in a DNA Engine Thermal Cycler (MJ Research, Waltham, Mass.) in the presence of a 50 pM concentration of each primer, 20 mM Tris HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, and 1.5 U of Taq DNA polymerase (Life Technologies). The final volume was 50 μl, and hot-start PCR was performed using Ampliwax 50 gems (PE Applied Biosystems, Foster City, Calif.). Amplification included initial denaturation at 94°C for 3 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min; and a final extension at 72°C for 10 min.
The PCR products from the first-round amplification were separated by electrophoresis through a 1.2% agarose gel (Life Technologies) and transferred to a Hybond nylon membrane (Amersham Pharmacia Biotech, Piscataway, N.J.) in the presence of 0.2 M of NaOH for 3 h. The nylon filter was then prehybridized for 2 to 4 h in the presence of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.2% sodium dodecyl sulfate (SDS) at 42°C in a hybridizing oven. The hybridization was performed at 42°C for 2 h in the presence of 2× SSC, 0.2% SDS, and 5× Denhardt's solution, with a radiolabeled (end-labeled with 32P) nested primer as a probe. Following hybridization, the blot was washed in 2× SSC and 0.2% SDS three times for 5 min at room temperature, 1 h at 48°C, and 30 min at 50°C. Blots were exposed to autoradiography film overnight at −80°C. For the nested PCR, 2% of the volume of the first-round PCR product was reamplified in a 50-μl final volume containing 20 mM Tris HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, 1 U of Taq DNA polymerase, and a 40 pM concentration of each nested primer. The nested reaction included initial denaturation at 94°C for 3 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 45 s, and extension at 72°C for 45 s; and final extension at 72°C for 10 min. The resulting PCR products were then separated through 1.5% agarose gel, stained with ethidium bromide, and visualized under UV light.
We amplified JCV TCR sequences from all samples, and we confirmed the result by Southern blot analysis and nested PCR (Fig. 2).
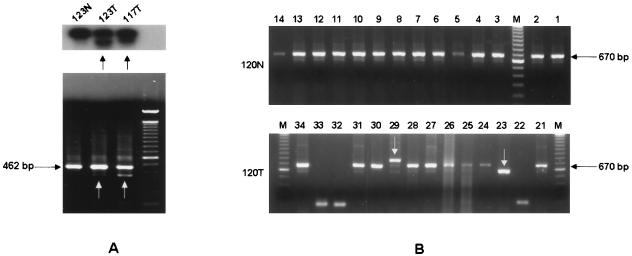
FIG. 2.
Detecting DNA rearrangements by Southern blot analysis and nested PCR. (A) Southern blot analysis of the first-round PCR of DNA extracted from two tumor samples and one nonneoplastic tissue is shown. As indicated by the arrows, colon cancers displayed multiple bands, which suggests the presence of DNA rearrangements. This result was confirmed by nested PCR (lower panel). (B) Clones generated by nested PCR product obtained from a nontumor sample and its cancerous counterpart were screened. The 670-bp fragment corresponds to the length of Mad-1 and the portion of plasmid DNA between the M13 primers (data confirmed by sequence analysis of 4 clones from 120N). In the 120T sample, two PCR products (indicated by arrows) showed higher (23) and lower (29) electrophoretic mobility compared with the 670-bp fragment, indicating the presence of DNA rearrangements. This result was also confirmed by sequence analysis (data not shown.).
Nested PCR products were subcloned into the pCR 2.1 vector (Invitrogen, Carlsbad, Calif.), and Escherichia coli were transformed by heat shock at 42°C for 30 s. After cloning the nested PCR products, we screened 340 recombinant colonies by PCR using M13 primers. The products were separated on a 1.2% agarose gel and visualized under UV light after ethidium bromide staining. Plasmids were then purified with the Flexiprep kit (Amersham Pharmacia Biotech), and clones underwent cycle sequencing in both directions with the Big Dye Terminator Sequencing kit (PE Applied Biosystems). Sequences were analyzed by an ABI 310 automated sequencer (PE Applied Biosystems) and aligned with those reported in GenBank by using MacVector Software (Oxford Molecular Group, Campbell, Calif.).
According to the sequence of JCV Mad-1, the expected size of our PCR products, including the partial plasmid DNA fragments comprised between the M13 primer sites, was 670 bp. After screening 340 clones by PCR, all clones showing shifts in band mobility and 31 clones of the expected size were sequenced in both directions. When nonspecific sequences were excluded due to inefficient ligation of the PCR product into the vector, 285 clones were obtained from the JCV TCR: 157 generated from nontumor samples and 128 from colon cancer tissues. Of these clones, 262 showed the predicted length of the JCV Mad-1 strain on agarose gel: 156 clones (99.3%) from nontumor samples and 106 (82.8%) from colon cancer tissues. The archetypal strain was not present in either colon cancer tissues or nontumor samples. Of the 54 clones sequenced in both directions (29 from colorectal cancer tissues and 25 from nontumor samples) those with the length of Mad-1 at gel electrophoresis were confirmed by sequence analysis.
In the colon cancer samples, 18 clones showed a deletion of one 98-bp sequence, while this strain was not detected in any of the nonneoplastic tissues (14 versus 0% [χ2 = 23.6; P < 0.001]). In four colon cancer samples, we sequenced one clone each having an extra 98-bp sequence. In colon cancer samples, a minimum of one to a maximum of four clones per sample were characterized by duplication or deletion of the 98-bp tandem repeat, while in one healthy tissue sample, 1 clone out of 20 displayed four copies of the 98-bp sequence ( 1 of 157 in nontumor samples versus 22 of 128 in colon cancer tissues: 0.6 versus 17.2% [χ2 = 26.03; P < 0.0001]). Proportions were compared by using the χ2 test. P values were considered to be statistically significant for P < 0.05. In all eight of the colon cancer samples, we amplified the strain characterized by the deletion of one 98-bp repeat. This strain was absent in the nonneoplastic counterpart, whereas in one nontumor sample, we detected a TCR with four copies of the 98-bp repeat. Among all sequenced clones, we did not find any other tandem repeat variants, which have been previously reported from studies of brains and peripheral lymphocytes of PML and immunocompetent patients (Table 1).
TABLE 1.
Distribution of JC virus TCRs in colon cancer tissues and corresponding nonneoplastic tissues
| Clone group | No. (%) of positive samples
|
|
|---|---|---|
| Colon cancer | Nonneoplastic epithelium | |
| Clones (n = 285) | 128 | 157 |
| Archetype strain | 0 | 0 |
| Mad-1 strain | 106 (82.5) | 156 (99.3) |
| Mad-1 variants | 22 (17.2) | 1 (0.6) |
| Single 98-bp sequence | 18 (14) | 0 |
| Triple 98-bp sequence | 4 (3.1) | 0 |
| Quadruple 98-bp sequence | 0 | 1 (0.6) |
| DNA samples (n = 16) | 8 | 8 |
| Mad-1 | 8 | 8 |
| Mad-1 variants | 8 | 1 |
We also found several point mutations in clones generated from both colon cancers and nontumor tissues. In some cases, these point mutations were randomly distributed, especially in PCR samples obtained from the nonneoplastic colon. However, in four clones generated from the colon cancer samples, the mutations, three of which were characterized by an adenine deletion and one of which was characterized by an adenine insertion at the A8 tract, were confined in the first A domain of the TCR (1) (Fig. 3). In the Mad-1 strain, the A region is duplicated and composed of a TATA box and a polyadenine tract. In eight more colon cancer samples, we found mutations confined between the E and the F domains of the TCR, which are immediately downstream of the late 98-bp repeat.
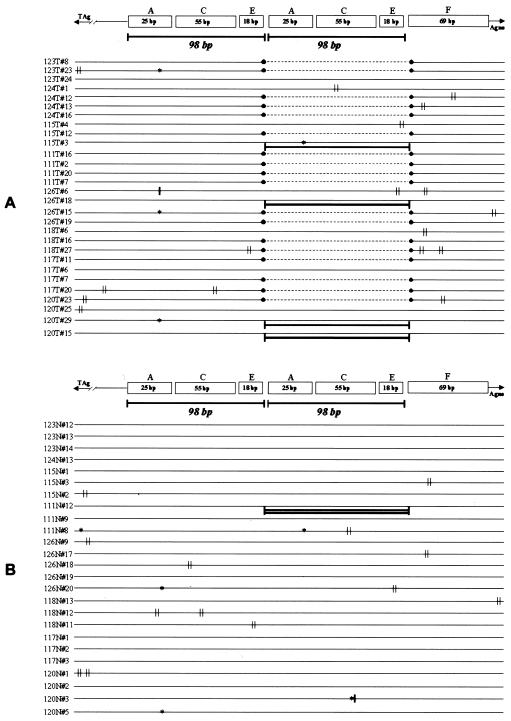
FIG. 3.
Schematic representation of DNA rearrangements and point mutations found in colon cancers (A) and matched nonneoplastic tissues (B). The nomenclature of motifs within the TCR is from Ault and Stoner (1). As shown, the deletion of a 98-bp sequence is an exclusive feature of colon cancers. In four clones generated from colon cancers, the 98-bp fragment was present in triplicate. In only one clone generated from a nontumor sample (111N#12), we found the 98-bp sequence repeated four times. In colon cancer samples, we found point mutations confined especially in the first TATA box and within the C and D regions. Symbols: ∥, point mutation, ∗, deletion; |, insertion.
The JCV archetype, assumed to be the circulating strain of the virus, can be isolated from the kidneys and is excreted with urine (25). The archetypal strain is seemingly unable to replicate in any cell type in vitro, while the Mad-1 strain can replicate in its natural host cells PHFG and also in POJ cells (both human fetal glial cells) (11, 12). The inability of the archetype strain to replicate in vitro is thought to result from the failure of the early promoter to express sufficient levels of transcripts to drive the DNA replication of the archetype under the control of T antigen (15). Furthermore, Mayreddy and coworkers have shown recently that the 23-bp fragment present in the archetype is a critical sequence that inactivates the late promoter, which controls the transcription of the capsid proteins, in glioma cells (13).
Many JCV strains characterized by tandem repeats have been detected in different cell types from immunocompetent and immunocompromised patients. Among these, the Mad-4 strain has been found to replicate partially in B lymphocytes, suggesting that viral replication depends upon its TCR and that by adapting itself, the host range of the virus might expand (14). According to a recent study, both archetypal and tandem repeat variants, such as Mad-1, Mad-4, and Mad-8, are present in tonsillar stromal cells and tonsillar lymphocytes, suggesting that tonsillar tissue might be the first site for viral replication (15). Monaco and coworkers have described detecting 8 clones with the archetype sequence among the 21 clones obtained from tonsillar lymphocytes (38%).
In our study, we amplified JCV TCR sequences from all samples. Interestingly, in colon cancer samples, the autoradiographs and the results of nested PCR showed multiple bands, which suggested the presence of multiple strains generated by DNA rearrangements in the TCR. We did not find archetypal TCR sequence either in colorectal cancers or nonneoplastic epithelium. However, we did find the Mad-1 strain or strains characterized by a single 98-bp sequence without any archetypal feature, such as the presence of the 23-bp fragment. This finding suggests that the archetype JCV is not present in the gastrointestinal tract and that the colonic cells may allow replication of JCV Mad-1 DNA. Moreover, we did not find Mad-4 and Mad-8 strains, which indicates that the DNA fragments we amplified were not the result of lymphocyte contamination.
Several hypotheses can be offered to explain the infection of the gastrointestinal tract by the Mad-1 strain of JCV. (i) First, rearrangements might possibly occur in nonlymphoid tissue, and lymphocytes might simply transport the virus to secondary sites, such as the gastrointestinal tract. Once transported to the secondary site, the viral strain selected for replication would be determined by the specific host tissue. As previously suggested, B lymphocytes undergo genomic rearrangements necessary for the production and modification of antibodies. JCV could adapt itself to different cell types by modifying its TCR through the same (or a similar) system useful to accomplish these genomic rearrangements (1, 15). (ii) Second, the absence of the archetype in colonic epithelium might be the result of rearrangements of the archetypal regulatory region in situ, in order to proliferate in the colonic tissue. (iii) Finally, the exclusive Mad-1 infection of the colon might also reflect the presence of multiple circulating variants of the virus (e.g., archetype and tandem repeat variants) that might be excreted by a different mechanism (urine and alimentary tract).
Recently, Elsner and Deorries showed the presence of the Mad-1 strain in brains and kidneys of persistently infected patients in Germany, with the absence of archetypal sequences even in kidneys (6). In the United States data also reflect the results from Europe, indicating that the most prominent strain is Mad-1 (24).
As suggested by these data and our previous study in which we detected a high frequency of JCV infection in the upper and lower gastrointestinal tract of immunocompetent patients (20), the presence of Mad-1 in the gastrointestinal tract could be a good source for diffusion of the infection; this could justify the prominence of Mad-1 strains compared with the archetype.
Our earlier work showed the presence of JCV T-antigen sequences from colorectal cancer tissues and corresponding nonneoplastic tissues, with a viral copy number of at least 10-fold higher in colon cancers compared with the matched nontumor tissues (10). In this study, we used some of the JCV samples that we had determined to be positive for T antigen in our previous study. All colon cancer specimens, but not the adjacent nonneoplastic tissues, were characterized by the presence of a strain with the sequence of the Mad-1 TCR but with the deletion of one 98-bp sequence. This variant was previously found in kidneys and brains of infected patients (6), and in bone marrow and brain tissue from one PML patient (9). According to previous reports, an artificial mutant strain, pM1Δ98, that contains one 98-bp repeat obtained by deleting the late 98-bp sequence from Mad-1, consistently demonstrates less viability and lower replication rates than the Mad-1 strain in PHFG cells (3). However, when introduced into Rat-2 fibroblasts, the pM1Δ98 artificial mutant is capable of transforming the host cells more efficiently than the prototype Mad-1 (2). This confirms the ability of the virus, although in vitro, to adapt to different hosts through modification of its regulatory region sequence. The presence of a 98-bp-deletion variant of Mad-1 in colorectal cancer tissues only, which already displayed a higher viral load, could reflect the higher replication rate of the virus in actively proliferating colon cancer cells. At this time we are investigating the expression of T antigen in healthy colon and colon cancer tissues. Our preliminary data indicate the expression of T antigen in both tissues (unpublished data).
In our study, we also detected many point mutations that in some cases were randomly distributed, which suggests the possibility that some of these represent errors introduced by Taq DNA polymerase. However, in clones generated from colon cancers, mutations were distributed in the A domain (four clones) and in the area between the E and F domains (eight clones): The reproducibility and consistency of these data suggest the presence of true mutations. Mutations in the A domain might change the specificity of the regulatory region for expressing T antigen in different cell types, since this area usually confers glial specificity and is important for directing transcription under the presence of factors such as Tst-1, that are expressed in Schwann cells. Furthermore, as has been reported (1), the area between the E and F domains represents a common site for strand breaks, which might permit deletions or duplications.
Finally, we can suggest two models for the involvement of JCV in the onset of genomic instability. First, the Mad-1 strain might be selected for replication and persistently infect the gastrointestinal tract. With impairment of the local immunological system, the virus can actively replicate and select for a specific variant (lacking in one of the 98-bp repeat sequences) that is capable of transforming the host tissue. Ongoing replication of the virus might also lead to alterations in the coding regions for T antigen that could participate in tumorigenesis. Alternatively, the activation of the Mad-1 strain of the JCV might be secondary to preexisting genomic instability. The higher rates of cellular proliferation could result in the presence of the strain lacking in one of the 98-bp repeat sequences after integration and recombination of the viral DNA with the host genome.
Although the Mad-1 JCV variant is believed to be neurotropic, the early promoter in the TCR interacts with numerous cellular transcription factors, such as NF-κB, NF-1, and Sp1 (for a review, see reference 18). These factors are expressed in many cell types. Future research should be directed toward understanding how these transcription factors might regulate JCV replication in colonic epithelial cells and the eventual transformation of the host.
Acknowledgments
We thank Eugene Major for helpful suggestions and critical reading of the manuscript. We thank Pamela Edmunds for valuable assistance during the preparation of the manuscript and Amy Pagel for technical help.
This work was funded by the California Research Program, grant 1II0048 (to C.R.B.).
REFERENCES
- 1.Ault G S, Stoner G L. Human polyomavirus JC promoter/enhancer rearrangement patterns from progressive multifocal leukoencephalopathy brain are unique derivatives of a single archetypal structure. J Gen Virol. 1993;74:1499–1507. doi: 10.1099/0022-1317-74-8-1499. [DOI] [PubMed] [Google Scholar]
- 2.Bollag B, Chuke W F, Frisque R J. Hybrid genomes of the polyomaviruses JC virus, BK virus, and simian virus 40: identification of sequences important for efficient transformation. J Virol. 1989;63:863–872. doi: 10.1128/jvi.63.2.863-872.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel A M, Swenson J J, Mayreddy R P, Khalili K, Frisque R J. Sequences within the early and late promoters of archetype JC virus restrict viral DNA replication and infectivity. Virology. 1996;216:90–101. doi: 10.1006/viro.1996.0037. [DOI] [PubMed] [Google Scholar]
- 4.Deorries K, Vogel E, Geunther S, Czub S. Infection of human polyomaviruses JC and BK in peripheral blood leukocytes from immunocompetent individuals. Virology. 1994;198:59–70. doi: 10.1006/viro.1994.1008. [DOI] [PubMed] [Google Scholar]
- 5.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, Vandyke T, Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elsner C, Deorries K. Human polyomavirus JC control region variants in persistently infected CNS and kidney tissue. J Gen Virol. 1998;79:789–799. doi: 10.1099/0022-1317-79-4-789. [DOI] [PubMed] [Google Scholar]
- 7.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 8.Frisque R J, White F A. The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy. In: Roos R, editor. Molecular neurovirology. Totowa, N.J: Humana Press; 1992. pp. 25–158. [Google Scholar]
- 9.Jensen P N, Major E O. Viral variant nucleotide sequences help expose leukocytic positioning in the JC virus pathway to the CNS. J Leukoc Biol. 1999;65:428–438. doi: 10.1002/jlb.65.4.428. [DOI] [PubMed] [Google Scholar]
- 10.Laghi L, Randolph A E, Chauhan D P, Marra G, Major E O, Neel J V, Boland C R. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci USA. 1999;96:7484–7489. doi: 10.1073/pnas.96.13.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandl C W, Frisque R J. Characterization of cells transformed by the human polyomavirus JC virus. J Gen Virol. 1986;67:1733–1739. doi: 10.1099/0022-1317-67-8-1733. [DOI] [PubMed] [Google Scholar]
- 11.Mandl C, Walker D L, Frisque R J. Derivation and characterization of POJ cells, transformed human fetal glial cells that retain their permissivity for JC virus. J Virol. 1987;61:755–763. doi: 10.1128/jvi.61.3.755-763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayreddy R P, Safak M, Razmara M, Zoltick P, Khalili K. Transcription of the JC virus archetype late genome: importance of the kappa B and the 23-base-pair motifs in late promoter activity in glial cells. J Virol. 1996;70:2387–2393. doi: 10.1128/jvi.70.4.2387-2393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monaco M C, Shin J, Major E O. JC virus infection in cells from lymphoid tissue. Dev Biol Stand. 1998;94:115–122. [PubMed] [Google Scholar]
- 15.Monaco M C, Jensen P N, Hou J, Durham L C, Major E O. Detection of JC virus DNA in human tonsil tissue: evidence for site of initial viral infection. J Virol. 1998;72:9918–9923. doi: 10.1128/jvi.72.12.9918-9923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman J T, Frisque R J. Detection of archetype and rearranged variants of JC virus in multiple tissues from a pediatric PML patient. J Med Virol. 1997;52:243–252. doi: 10.1002/(sici)1096-9071(199707)52:3<243::aid-jmv2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Padgett B L, Walker D L, ZuRhein G M, Hodach A E, Chou S M. JC papovavirus in progressive multifocal leukoencephalopathy. J Infect Dis. 1976;133:686–690. doi: 10.1093/infdis/133.6.686. [DOI] [PubMed] [Google Scholar]
- 18.Raj G V, Khalili K. Transcriptional regulation: lessons from the human neurotropic polyomavirus JCV. Virology. 1995;213:283–291. doi: 10.1006/viro.1995.0001. [DOI] [PubMed] [Google Scholar]
- 19.Rencic A, Gordon J, Otte J, Curtis M, Kovatich A, Zoltick P, Khalili K, Andrews D. Detection of JC virus DNA sequence and expression of the viral oncoprotein, tumor antigen, in brain of immunocompetent patient with oligoastrocytoma. Proc Natl Acad Sci USA. 1996;93:7352–7357. doi: 10.1073/pnas.93.14.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricciardiello L, Laghi L, Ramamirtham P, Chang C L, Chang D K, Randolph A E, Boland C R. JC virus DNA sequences are frequently present in the human upper and lower gastrointestinal tract. Gastroenterology. 2000;119:1228–1235. doi: 10.1053/gast.2000.19269. [DOI] [PubMed] [Google Scholar]
- 21.Staib C, Pesch J, Gerwig R, Gerber J K, Brehm U, Stangl A, Grummt F. p53 inhibits JC virus DNA replication in vivo and interacts with JC virus large T-antigen. Virology. 1996;219:237–246. doi: 10.1006/viro.1996.0241. [DOI] [PubMed] [Google Scholar]
- 22.Tornatore C, Berger J R, Houff S A, Curfman B, Meyers K, Winfield D, Major E O. Detection of JC virus DNA in peripheral lymphocytes from patients with and without progressive multifocal leukoencephalopathy. Ann Neurol. 1992;31:454–462. doi: 10.1002/ana.410310426. [DOI] [PubMed] [Google Scholar]
- 23.Walker, D. L., and B. L. Padgett. The biology and molecular biology of JC virus, p. 327–377. In N. P. Salzman (ed.), The Papovaviridae. Plenum Press, New York, N.Y.
- 24.White F A, Ishaq M, Stoner G L, Frisque R J. JC virus DNA is present in many human brain samples from patients without progressive multifocal leukoencephalopathy. J Virol. 1992;66:5726–5734. doi: 10.1128/jvi.66.10.5726-5734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]