Abstract
Background
Phylogeny, combined with trait-based measures, offers insights into parasite sharing among hosts. However, the specific traits that mediate transmission and the aspects of host community diversity that most effectively explain parasite infection rates remain unclear, even for the Bartonella genus, a vector-borne bacteria that causes persistent blood infections in vertebrates.
Methods
This study investigated the association between rodent host traits and Bartonella infection, as well as how rodent community diversity affects the odds of infection in the Atlantic Forest, using generalized linear models. Additionally, we assessed how host traits and phylogenetic similarities influence Bartonella infection among mammal species in Brazil. To this end, rodents were sampled from ten municipalities in Rio de Janeiro, southeastern Brazil. Then, we calculated several diversity indices for each community, including Rényi’s diversity profiles, Fisher’s alpha, Rao’s quadratic entropy (RaoQ), Functional Diversity (FDis), Functional Richness (FRic), and Functional Evenness (FEve). Finally, we compiled a network encompassing all known interactions between mammal species and Bartonella lineages recorded in Brazil.
Results
We found no significant relationship between diversity indices and the odds of Bartonella infection in rodent communities. Furthermore, there was no statistical support for the influence of individual-level traits (e.g., body length, sex, and age) or species-level ecological traits (e.g., locomotor habitat, dietary guild, and activity period) on Bartonella infection in rodents. A country-scale analysis, considering all mammal species, revealed no effect of host traits or phylogeny on Bartonella infection.
Conclusions
This study highlighted wild mammals that share Bartonella lineages with livestock, synanthropic, and domestic animals, underscoring the complexity of their maintenance cycle within the One Health framework. A key question arising from our findings is whether molecular host–cell interactions outweigh host body mass and ecological traits in influencing Bartonella infection, potentially opening new avenues for understanding host–parasite relationships and infection ecology.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-024-06523-y.
Keywords: Bartonella, Functional diversity, Host trait, Interaction network, Mammal, Phylogeny
Background
Host evolutionary history, combined with trait-based measures, is associated with parasite spread among hosts [1, 2]. Variations in host traits modulate vector exposure, parasite encounters, and pathogen spread within local host communities [3–5]. Therefore, functional diversity measures, which encompass the variety and distribution of ecological, morphological, and physiological traits within a community, can serve as an adequate proxy for the structural role of host communities [6, 7]. However, it remains unclear whether and which functional diversity indices can serve as an indicator of infection rates [5, 6, 8–10].
The Bartonella genus consists of facultative intracellular alphaproteobacteria and vector-borne pathogens that can cause persistent hemotropic infections in their vertebrate hosts [11]. From an ecological perspective, Bartonella infects a broad diversity of host species, with varying levels of specificity across host phylogeny. Even host-specific species, such as B. henselae, which are commonly found in domestic cats, have also been identified in dogs and wild mammals in Brazil [12, 13]. Furthermore, these bacteria have been detected in various mammalian orders and ectoparasites worldwide, including Argentina [14], Chile [15], Colombia [16], Israel [17], the USA [18], Italy [19], Thailand [20], Japan, Russia, Korea, and Taiwan [21].
Bartonella spp. exhibit a phylogenetic pattern that separates them into lineages, each displaying a distribution of virulence factors that contribute to their persistence and pathogenicity [22]. These virulence factors show evolutionary patterns in host specificity, with certain lineages closely associated with specific mammal orders, such as lineage 2 with ruminants and lineage 4 with rodents [22–24]. Thus, these bacteria provide a suitable system for studying disease–diversity relationships.
Rodent species are frequently reported as hosts for many zoonotic agents [25] and also harbor a high diversity of Bartonella species [15, 26, 27]. Certain traits of rodent species, such as age at sexual maturity, short gestation periods, and large litter sizes, traits associated with fast life history strategies, can influence the risk of infection by zoonotic agents [25, 28]. Therefore, a trait profile approach to known Bartonella host species may allow us to forecast which species are likely to act as reservoirs for these bacteria.
In this context, the aim of this study was threefold. First, data on Bartonella infection in rodent communities were used to (i) investigate which host traits at the individual level are most associated with the odds of Bartonella infection and (ii) examine how community diversity affects the odds of infection in Atlantic Forest areas. Second, for host species of different mammalian orders that occur in Brazil, (iii) our objective was to assess whether the probability of Bartonella infection is influenced by traits and phylogenetic similarities between these host species.
In the individual and species-level analyses, we hypothesized that infection would be more frequent among hosts with similar traits. Morphological traits such as body length, body mass, and tail length are related to individual age and lifetime exposure to parasites [29, 30]; therefore, we expected these individual measures to be positively related to infection probability. Furthermore, since male rodents tend to increase their mobility during reproductive periods and may experience hormone-induced immunosuppression [31], we anticipated that males would have a higher probability of Bartonella infection compared with females. We also hypothesized that scansorial or semi-scansorial locomotor habitats and an invertebrate diet would increase the probability of infection, as these traits increase the likelihood of encountering a vector [32, 33]. Finally, because activity period is associated with resource sharing and is evolutionarily related to other ecological traits such as foraging strata [34], we expected cathemeral rodents, which are active both day and night, to face a higher risk of exposure due to overlap activity periods with vectors and other hosts.
In the community analysis, we expected that at greater functional divergence [i.e., Functional Evenness index (FEve)], which may be associated with a reduced abundance of highly competent species, would result in lower odds of Bartonella infection. Conversely, higher functional diversity [that is, Functional Diversity (FDis), Functional Richness (FRic), and Rao’s quadraticentropy (RaoQ) indices] was expected to increase the chance of Bartonella infection by enhancing trait diversity and the abundance of potential host species. Definitions of these functional diversity indices are provided in the Methods subsection titled Host community structure.
Finally, considering that Bartonella lineages have functional factors related to pathogenicity [22], we also aimed to examine whether host traits at the species level influence the sharing of Bartonella lineages among hosts of different mammalian orders. By integrating analyses across these scales, we can achieve a comprehensive understanding of how individual- or species-level traits, community diversity, and phylogenetic relationships affect parasite infection [6, 7]. This integrated approach facilitates the development of more effective monitoring, management, and control strategies tailored to specific ecological contexts and host community structures [35].
Methods
Two distinct datasets were used to investigate our hypotheses on three scales: individual, community, and species. For the first set of analyses, we used data from several rodent communities sampled across the Brazilian Atlantic Forest, including individual-level information on life history traits and Bartonella infection. This biome was chosen due to the higher prevalence of these bacteria in rodents compared with other Brazilian biomes [36]. For the second set of analyses, we expanded the scale to include all other mammalian orders occurring in Brazil. Consequently, we analyzed traits at the species level and Bartonella infection, as there is no individual-level database available.
Study area and rodent community data source
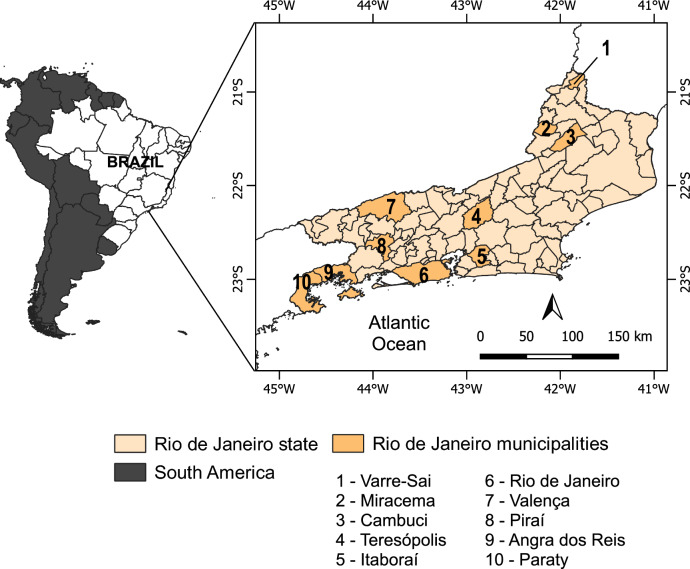
We used geo-located data from previous studies on individual rodents, infected or not with Bartonella, in ten municipalities in the state of Rio de Janeiro, southeastern Brazil (see Fig. 1) [37, 38]. Additional data were gathered from a research project on the biodiversity of Atlantic Forest, covering rodent captures from 2004 to 2019 in the same areas. A unified dataset was compiled, containing information on 398 rodent individuals. Rodent species were identified by morphological examination, karyotyping, and molecular analyses by experts from the Laboratory of Biodiversity and Parasitology of Wild Mammal Reservoirs, following the methodology outlined by D’Andrea et al. [39]. The number of individuals sampled in each community ranged from 12 to 138, with 3–8 rodent species. All procedures involving rodents were approved in advance by the Institutional Ethics Committee of Animal Research at the Oswaldo Cruz Foundation under the license number LW39/14.
Figure 1.
Study areas for wild rodents in the state of Rio de Janeiro, Brazil (2004–2019)
Bartonella was detected using polymerase chain reaction (PCR) methods and DNA sequencing. Bacterial DNA was extracted from liver and spleen samples and screened by PCR targeting the gltA, rpoB, ftsZ, and groEL genes. In total, 32 rodent individuals with the Bartonella gene were detected, with the number of infected individuals in each community ranging from 0 to 8. Detailed information on the study area, sampling methods, and molecular assays can be found in Rozental et al. [37] and Gonçalves-Oliveira et al. [38].
Host trait data
Data on morphological and ecological traits associated with host life history were obtained from our database, the EltonTraits database [40], and from Paglia et al. [41]. The morphological data from our database were used for individual-level analyses, while for species-level analyses, we used the body mass available in EltonTraits and Paglia. Missing data on species-level traits were estimated using mode values (the most commonly occurring value) from closely related species within the same genus, except for body mass, which was estimated using the mean value of the genus. The estimated data are indicated in the data file at figshare [42]: https://doi.org/10.6084/m9.figshare.25838281. An overview of the scale of factors for each trait and the number of replications can be found in the Table 1.
Table 1.
Overview of the scale at which key parameters were measured and their respective replication
| Scale of inference for the chance of infection by the Bartonella bacteria | Scale at which the factor of interest is applied | Number of replicates at the appropriate scale |
|---|---|---|
| Rodent communities | Communities | Eight communities for each diversity index |
| Rodent individuals | Species | Body length: 17 species |
| Rodent individuals | Species | Tail length/body length: 17 species |
| Rodent individuals | Species | Age: 16 adult, 8 young |
| Rodent individuals | Species | Sex: 14 feminine, 15 masculine |
| Rodent individuals | Species | Locomotor habitat: 1 scansorial, 3 semi-scansorial, 15 ground, 1 semi-aquatic, 2 arboreal |
| Rodent individuals | Species | Dietary guild: 8 insectivore, 13 herbivore, 2 omnivore |
| Rodent individuals | Species | Activity period: 3 diurnal, 11 nocturnal, 9 cathemeral |
| Rodent species | Species | Body mass: 17 species |
| Rodent species | Species | Locomotor habitat: 1 scansorial, 2 semi-scansorial, 11 ground, 1 semi-aquatic, 2 arboreal |
| Rodent species | Species | Dietary guild: 5 insectivore, 11 herbivore, 1 omnivore |
| Rodent species | Species | Activity period: 2 diurnal, 9 nocturnal, 6 cathemeral |
| Mammal species | Species | Body mass: 108 species |
| Mammal species | Species | Dietary guild: 27 insectivore, 55 herbivore, 18 omnivore, 8 carnivore |
| Mammal species | Species | Activity period: 12 diurnal, 76 nocturnal, 20 cathemeral |
Individual hosts are subsamples that contribute to the precision of the estimates at the species level but are not independent replicates. Therefore, we included the species name as a random effect in the statistical models to avoid pseudoreplication. Further details on the data used in the models can be found in the Additional file: Table S1, and the raw data is available on figshare [40]: https://doi.org/10.6084/m9.figshare.25838281
Host community structure
Diversity indices representing different aspects of community structure and composition were calculated for each rodent community using Rényi’s diversity profiles, Fisher’s alpha [43], Rao’s quadratic entropy [44], FDis [45], FRic, and FEve [46]. The Rényi diversity profile is a technique for ordering diversity [47] that produces curves indicating the richness and evenness of each community. Fisher’s alpha is a scale-independent biodiversity indicator based on the curvature of the species abundance distribution. Rao’s quadratic entropy is a dissimilarity metric used within a functional space that measures the abundance-weighted sum of pairwise functional distances between all species. FDis is another dissimilarity metric that incorporates the abundance-weighted distance of species trait values from the community centroid. FRic measures the volume of the multidimensional space occupied by all species, while FEve assesses the regularity of the trait distribution and relative species abundance. All diversity metrics were calculated using the packages Vegan [48], BiodiversityR [49], and FD [50] available in R version 4.1.0 [51].
Brazilian mammal host and Bartonella lineage network
We compiled a second dataset using existing GenBank data. All metadata for Bartonella sequences, including host name, gene, locality, GenBank accession number, and reference, where the host species was identified, were downloaded. This information was obtained using Geneious software from the NCBI database [52], and all Bartonella sequences recorded in Brazil were recovered. For sequences with no scientific host name provided, we obtained information on Bartonella hosts from the articles in which the sequences were published (available in the Data Sources section). Data on all mammal species that tested negative for Bartonella DNA were also gathered from these articles.
In the present study, the term “genotype” refers specifically to the variant forms of the partial sequences of the genes gltA, rpoB, ftsZ, groEL, nuoG, ITS, pap31, 16S, ribC, and htrA carried by Bartonella bacteria. Bartonella genotypes were classified into lineages on the basis of the phylogenies available in the peer-reviewed literature from which the sequences were published (see the Data Sources section). Specifically, Bartonella genotypes closely related to species that occur in Brazil, such as Bartonella coopersplainsensis, B. bovis, B. clarridgeiae, B. rochalimae, B. quintana, B. henselae, B. koehlerae, and B. vinsonii, were classified into lineages as indicated by Wagner and Dehio [22]. This classification facilitates the analysis of how Bartonella lineages are shared among hosts, as these lineages exhibit patterns of distribution of virulence factors that affect their pathogenicity and host adaptation [22].
Host phylogenetic distance
At the rodent individual scale, we used phylogenetic distances estimated from the Atlantic Forest non-volant small mammal tree [53]. We incorporated cytochrome B sequences from three rodent species not included in this tree to complete the final phylogenetic distance matrix. Sequence alignment and pairwise distances for each species were calculated using the maximum likelihood method with 500 bootstrap replications, assuming Gamma distribution with invariant sites and a very strong branch swap filter in MEGA© software (www.megasoftware.net). The phylogenetic tree was built using the Kimura 2-parameter method, which is commonly used for estimating genetic distances and phylogenetic relationships [54].
At the species scale, we used the cophenetic.philo function of the ape package [55] to calculate the phylogenetic distances between Brazilian mammal species with PCR-positive or negative Bartonella detections. We sampled 10,000 equally plausible mammal phylogenetic trees from the posterior distribution published by Upham et al. [56], covering 108 species. We then created a rooted consensus tree using TreeAnnotator v1.10, summarizing it with a burn-in of the first 1000 trees and a cutoff of 0.7 posterior probability. The most representative tree was rooted at the midpoint of the clusters. One mammal species detected with Bartonella, Bubalus bubalis (Cetartiodactyla), was absent from the supertree, as were two species that tested negative for Bartonella DNA: Aotus infulatus (Primates) and Sphiggurus villosus (Rodentia).
Statistical analyses
Diversity indices and odds of Bartonella infection for each community
To investigate whether the odds of Bartonella infection were affected by the functional diversity of the host community, we used generalized linear models (GLM) from the package lme4 [57] with a binomial family and a logit link function. The significance of the model was evaluated by simulating a null model to test the absence of an effect of functional diversity indices on Bartonella infection and evaluated using analysis of variance (ANOVA) at the probability level of 0.05.
Rodent traits as predictors of Bartonella infection
To assess whether rodent sex, age, body length, dietary guild, locomotor habitat, and activity period may drive Bartonella infection, we used data from all individuals sampled in the ten communities. Note that, because body length and tail length are highly correlated (Pearson correlation coefficient, r = 0.7, P < 0.05), we evaluated whether the ratio of these two variables would be a better predictor than body length alone. Conversely, the Spearman correlations between age, sex, and body length were statistically not significant. The exotic synanthropic species Rattus rattus and Mus musculus were excluded from this analysis.
To test whether host traits influenced infection status while accounting for rodent phylogenetic relatedness, candidate phylogenetic generalized linear mixed models (GLMM) were fitted using the brms package with default priors and infection status as a Bernoulli-distributed response. The identity of rodent species and the phylogenetic covariance matrix were included as random effects [58]. Four chains of 10,000 iterations each were run with a burn-in period of 5000 and thinned every 10 steps, resulting in a total of 4000 samples. GLMMs were compared using leave-one-out cross-validation (LOOIC), and goodness of fit was assessed with Bayesian R2, which includes the total modeled variance attributed to fixed effects [59, 60]. Fixed effects [means and 95% highest density intervals (HDI)] were estimated from the posterior distributions of each predictor in the best phylogenetic GLMM.
Network analysis of all PCR-positive hosts for Bartonella in Brazil
To highlight species notable for sharing multiple lineages, we created a bipartite network in which the nodes represent mammalian host species from different orders or Bartonella lineages, and the edges among the nodes represent associations between host species and Bartonella lineages. We calculated the degree centrality (number of edges connected to a node) and the betweenness centrality (number of shortest paths passing through a node) using the igraph package [61]. These measures describe each host’s role in sharing Bartonella lineages. For instance, hosts with high degree centrality play a significant role in spreading parasite diversity, while hosts with high betweenness centrality act as bridges between different groups of hosts, particularly if they have greater contact with other hosts due to similarities in their ecological traits.
The GLMs were fitted to examine whether the host trait patterns affected the host centralities in the network. We used different statistical methods to analyze network centrality measures. For degree centrality, we applied a GLM assuming a Poisson distribution of the data. For betweenness centrality, we transformed the data using a logit function and then used a GLM assuming a normal distribution (Gaussian errors).
Trait and phylogenetic similarities as predictors
The effects of host ecological and evolutionary similarity on interaction patterns were evaluated using multiple regression on distance matrices [62] with the ecodist package [63]. We examined how phylogenetically similar hosts or those with similar traits share Bartonella compared with dissimilar ones. Three pairwise matrices were created for rodents sampled from the Atlantic Forest, which were tested for the detection of Bartonella DNA: phylogenetic distance, trait profile distance, and a distance matrix for positivity. The same three pairwise matrices were also created for all mammals tested for the detection of Bartonella DNA in Brazil. The distance matrix for positivity, derived from the presence or absence of Bartonella in each host species, was constructed using Jaccard’s qualitative index [64].
Data source and R code
A list of data and R code used in the study are provided at figshare [42]: https://doi.org/10.6084/m9.figshare.25838281. A list of all included studies can be found in the Data Sources section.
Results
Structures of the rodent community and the chances of infection with Bartonella
Six host species with Bartonella DNA were detected in eight of the ten analyzed communities, as follows: Akodon cursor (87 individuals caught, 75 tested, 15 PCR positive), Akodon montensis (21 caught, 3 tested, 2 PCR positive), Delomys dorsalis (62 caught, 14 tested, 3 PCR positive), Euryoryzomys russatus (9 caught, 7 tested, 6 PCR positive), Nectomys squamipes (33 caught, 30 tested, 3 PCR positive), and Oxymycterus dasytrichus (20 caught, 14 tested, 3 PCR positive). A summary of the data collected used in our models is provided in the Additional file: Table S1. The Rényi diversity profile (Additional file: Fig. S1) showed that the communities’ richness indices (alpha = 0), Shannon (alpha = 1), and Simpson (alpha = 2) had overlapping confidence intervals, making comparison using classical diversity metrics difficult.
Using data from eight study sites, our findings show that Fisher’s alpha ranged between 1.64 and 2.52, and functional diversity varied among communities as follows: FRic (0.007–0.26), FEve (0.41–0.91), RaoQ (0.05–0.17), and FDis (0.17–0.41). Our models did not indicate significant relationship between functional diversity and the chances of Bartonella infection in the rodent communities investigated (Additional file: Table S2).
Host trait: predictors at the individual level
The best phylogenetic GLMM was fitted with binary data on the presence or absence of Bartonella as dependent variable, with dietary guild and activity period as fixed effects. We excluded the locomotor habitat from the models due to overfitting. The complete ranking of the candidate models is shown in the Additional file: Table S3. We observed a weak effect of the dietary guild and the activity period on the odds of Bartonella infection in Atlantic Forest rodents, with this effect characterized by considerable uncertainty, as indicated by the credible interval overlapping zero (Fig. 2). There was no statistical support for the influence of individual-level morphological traits or species-level ecological traits on Bartonella infection, after accounting for species identity and phylogeny as random effects (Additional file: Table S3, Fig. 2).
Figure 2.
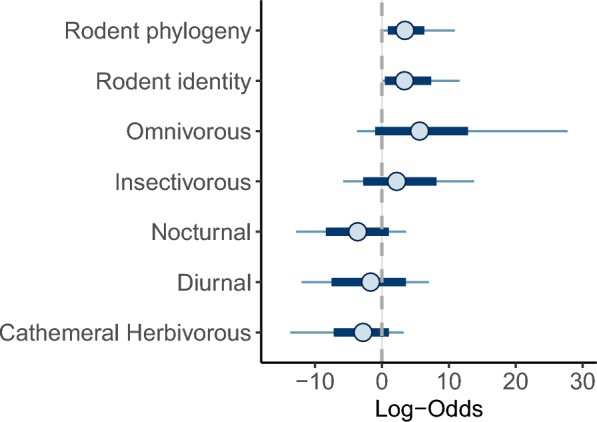
Predictors of Bartonella infection in rodent. The graph illustrates the effect of the dietary guild and the activity period on the odds of Bartonella infection in Atlantic Forest rodents. The uncertainty of these effects is represented by the width of the credible interval. Posterior means of the odds of Bartonella infection are shown, with 80% HDI (thick segments) and 95% HDI (thinner outer lines) from the most parsimonious phylogenetic GLMM. The reference levels for the chances of Bartonella infection are cathemeral and herbivorous rodents
Influence of traits and phylogenetic distance at the species level
Although we expected that trait and phylogenetic similarities would increase opportunities for contact with susceptible species and thus enhance Bartonella infection, no significant effect was detected at the species level (Additional file: Table S4). This finding remained consistent when considering phylogenetic proximity in the Bartonella infection patterns among all mammal hosts listed for Brazil (Additional file: Table S4).
Brazilian host-Bartonella lineage network
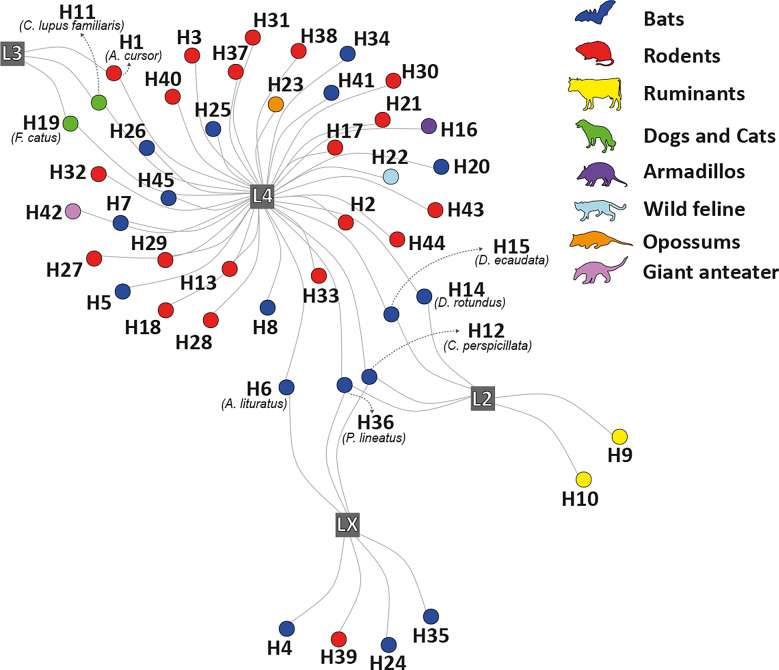
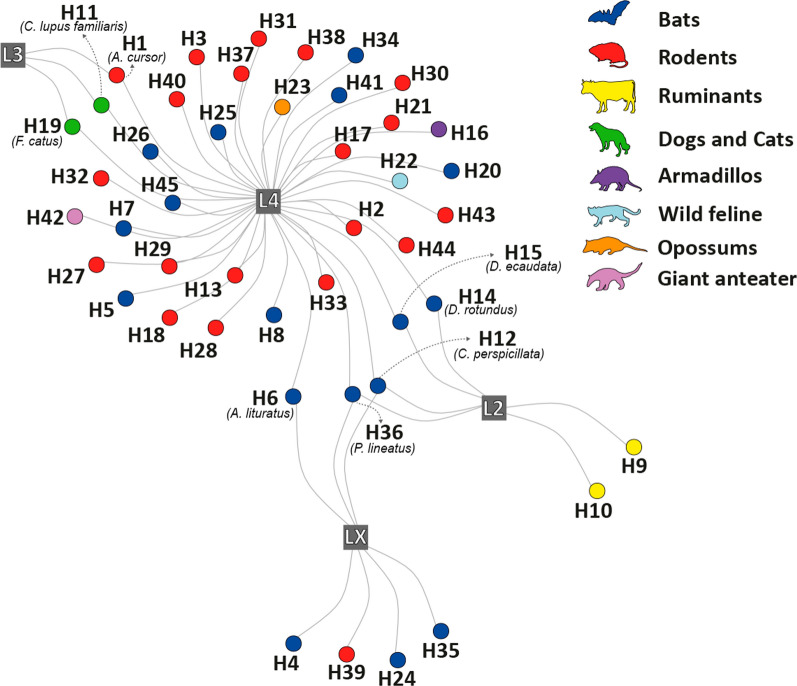
Considering Bartonella lineages instead of genotypes allowed us to explore a broader aspect of the association between these proteobacteria and its hosts. In general, Bartonella was detected in at least one species of six mammal orders in Brazil, with the bacteria most commonly found in rodent and bat species. The Bartonella lineage could not be determined for eight bat species and three rodent species. This network illustrates which Bartonella lineages have more widespread host distributions, and the network node properties suggest that the Chiroptera order likely plays a major role in sharing the Bartonella lineage with other host orders, due to its higher betweenness centrality. The degree centrality was highest for the common vampire bat (Desmodus rotundus; H14) and the hairy-hegged vampire bat (Diphylla ecaudata; H15), the cursor grass mouse (A. cursor; H1), the great fruit-eating bat (Artibeus lituratus; H6), the domestic dog (Canis lupus familiaris; H11), and the domestic cat (Felis catus; H19), indicating that these host species interacted with two Bartonella lineages in the host–parasite network (Fig. 3). Seba’s short-tailed bat (Carollia perspicillata; H12) and white-lined broad-nosed bat (Platyrrhinus lineatus; H36) also presented the highest degree (3) and the betweenness centrality values (Additional file: Table S5). Although host traits were not associated with network centralities (see Additional file: Tables S6-S7), bats with higher centrality had overlapped activity periods with A. cursor, and domestic dogs and cats, since these species are nocturnal and cathemeral. D. rotundus and D. ecaudata are carnivorous, similar to domestic host species. C. perspicillata, P. lineatus, and A. lituratus are herbivorous, but like A. cursor, they also include invertebrates in their diet.
Figure 3.
Bipartite network of interactions between mammal species (circles) and Bartonella lineages (squares) found in Brazil. One node represents a lineage with no phylogenetic tree classification (LX) identified in seven host species reported in the studies used to build the interaction network. Lineage 4 (L4) is the most promiscuous, detected in 36 of the 45 mammal species known as hosts of Bartonella in Brazil. Lineage 2 (L2) has been detected in six species, including two ruminants and four bats. Canis lupus familiaris (H11), Felis catus (H19), and Akodon cursor (H1) share L3 and L4. Carollia perspicillata (H12) and Desmodus rotundus (H14), Diphylla ecaudata (H15), and Platyrrhinus lineatus (H36) share L2 and L4. Other host species share L4, as follows: H2. Akodon montensis; H3. Akodon sp.; H4. Anoura caudifer; H5. Artibeus fimbriatus; H6. Artibeus lituratus; H7. Artibeus obscurus; H8. Artibeus planirostris; H9. Bos taurus; H10. Bubalus bubalis; H13. Delomys dorsalis; H16. Euphractus sexcinctus; H17. Euryoryzomys macconnelli; H18. Euryoryzomys russatus; H20. Glossophaga soricina; H21. Hylaeamys megacephalus; H22. Leopardus geoffroyi; H23. Marmosops ocellatus; H24. Myotis izecksohni; H25. Myotis riparius; H26. Myotis sp.; H27. Neacomys spinosus; H28. Necromys lasiurus; H29. Nectomys squamipes; H30. Oecomys mamorae; H31. Oligoryzomys nigripes; H32. Oxymycterus dasytrichus; H33. Oxymycterus nasutus; H34. Phyllostomus discolor; H35. Phyllostomus sp.; H37. Proechimys gardneri; H38. Rattus norvegicus; H39. Rattus rattus; H40. Rhipidomys macrurus; H41. Sturnira lilium; H42. Tamandua tetradactyla; H43. Thrichomys fosteri; H44. Thrichomys laurentius; H45. Uroderma bilobatum. Three sequences were not used in the network, as their lineage classifications were either not found in the primary article or because the host’s scientific name was not reported, namely KY356756 (host not reported), MG878887, and MG878888 (Glossophaga soricina as host)
Discussion
In Brazil, eco-epidemiological studies on Bartonella infection in rodents have been mainly concentrated in the Atlantic Forest region [36–38]. Here, we anticipated that the functional diversity of rodent communities sampled across the Atlantic Forest would affect the odds of Bartonella infection. However, variations in functional diversity measures—namely FRic, FEve, RaoQ, and FDis—did not explain the odds of Bartonella infection in these rodent communities. This could be due to the high functional redundancy found in non-volant small mammal communities with more than five species in the Atlantic Forest [53]. With several species exhibiting similar trait values, the impact of functional diversity on transmission dynamics may be mitigated, as multiple species can fulfill similar ecological roles. Another hypothesis is that the density of host populations and their contact rates with vectors play a more critical role in Bartonella transmission than community structure itself. This indicates that control measures should prioritize reducing humans-vector contact rather than focusing solely on community composition.
Other components of community diversity, such as species richness and Shannon index, have previously been linked to the prevalence of Bartonella infections in various regions of the USA [65]. However, it was unclear whether these indices accounted for the different sampling efforts across the analyzed communities, which is crucial for inter-site biodiversity comparisons. In our study, we included the sampling effort as a covariate in a GLM model and found no association between species richness or the Shannon index and the chances of Bartonella infection. Additionally, our findings may be influenced by environmental factors that were not account in our analyses, as well as the presence of vectors. Vectors is a key determinant in the spread of Bartonella within host communities [66]. Environmental factors, such as temperature and humidity, also influence the abundance of fleas [67]. Therefore, more comprehensive studies that integrate environmental parameters with vector presence are essential to elucidate the prevalence of Bartonella in host communities.
In this study, we anticipated that cathemeral and insectivorous hosts would be associated with increased Bartonella infection. However, we found no statistical support for the influence of morphological and ecological traits on Bartonella infection in rodents. These expected relationships may be obscured by the complex interactions between host behaviour, vector activity, and habitat use. Host exposure is related to encounter rates between hosts and vectors. Fleas that transmit Bartonella are most active during the daytime [68], making cathemeral (partially daytime) and diurnal (fully daytime) hosts more susceptible to infection. Additionally, while oral transmission of Bartonella in rodents is less effective in causing bacteremia [69], insectivorous rodent species may have increased exposure to Bartonella vectors in their foraging habitats. For instance, ticks are commonly found ectoparasites in environments where insectivorous rodents live, such as forests and grasslands [32, 33]. Therefore, surveillance efforts should also consider the diversity of habitats that rodents inhabit, including natural, altered, and agricultural areas, as these environments can affect both vector and host populations.
Regarding the interaction network between mammal host species and Bartonella lineages, lineage 4 (L4) appears to spread among a diverse range of hosts compared with other lineages. This suggests that L4 has a broad potential to infect hosts that are phylogenetically distant and occupy different ecological niches. While Bartonella genotypes often cluster by mammal taxonomic orders [70], this can obscure high host specificity when investigating at the lineage level, where genotypes closely related to a particular lineage are grouped together. The specificity of Bartonella genotypes suggests greater biological restrictions at this level, but the classification of lineages, as described by Wagner and Dehio [22], reveals important patterns in the distribution of pathogenicity-related virulence factors among lineages. It is worth noting that the unknown Bartonella lineages (represented in the network as LX) may not constitute a single lineage and may not fit into known lineages. In Brazil, most Bartonella infection records are at the genotype level, with limited strain isolation or species delimitation. This limits our ability to classify Bartonella species and trace ecological drivers in mammal hosts. Additionally, some hosts and vectors can harbor multiple genotypes, and strains may recombine, affecting genetic similarity among genotypes [71].
Our network analyses aimed to characterize the centrality of mammal species in the sharing of Bartonella lineages recorded in Brazil. Some bat species share Bartonella lineages with cattle and other bats (see, e.g., [72, 73]), while synanthropic rodent species and domestic animals share two Bartonella lineages (L3 and L4). The results highlight key rodent and bat species that should be targeted in eco-epidemiological studies due to their propensity to circulate among domestic animals and share their habitats.
Our findings also suggest that bats may possess traits not examined in this study that make them more prone to various Bartonella lineages. It is possible that other ecological traits, such as occupying a greater diversity of roosting habitats, increase the risk of Bartonella exposure between bat species [74]. Although Bartonella lineages encompass a diverse array of species and genotypes, these bacteria are functionally convergent, possessing factors related to pathogenicity and the capacity to infect host cells [75]. For instance, specific sets of Bep repertoires have adapted to three different Bartonella lineages, demonstrating remarkable host adaptation [22]. Consequently, factors such as body mass and ecological traits of the host may have a weaker effect on Bartonella infections compared with molecular–host cell interactions.
Conclusions
Understanding the transmission pathways that drive Bartonella infection has significant methodological and zoonotic disease management implications. In Brazil, a continental-sized country, the number of host communities investigated for these bacteria is still very limited. Functional diversity indices require uniformity in comparative data. Although there was no sufficient statistical support to establish trait-based indices, analyses in different biomes should assess the effect of these diversity indices on pathogen infection. The absence of accurate specimen identification and inventory data, such as sampling effort and information on all hosts tested for the detection of Bartonella, including those negative for the pathogen, complicates meta-analyses at the scale of host communities. Consequently, study programs focusing on the role of host trait diversity in modulating pathogen infection could provide a semi-quantitative tool for indicator-based surveillance, species management, and control strategies. Regarding all mammal species, no effect of host traits and phylogeny was observed on the sharing of Bartonella lineage. An open question is whether molecular–host cell interactions play a more significant role in infection dynamics than host body mass and ecological traits. Identifying key species in Bartonella transmission will help guide policies in the human and animal health sectors by informing cross-sectional surveillance efforts. This work highlights wild host species that share Bartonella lineages with livestock, synanthropic rodent species, and domestic animals, underscoring the complexity of the maintenance cycle of these proteobacteria within the One Health framework.
Data sources
André, M. R., Denardi, N. C. B., de Sousa, K. C. M., Gonçalves, L. R., Henrique, P. C., Ontivero, C. R. G. R., Ontivero, C. R. G. R., Gonzalez, I. H. L., Nery, C. V. C., Chagas, C. R. F., Monticelli, C., Santis, A. C. G. A., & Machado, R. Z. (2014). Arthropod-borne pathogens circulating in free-roaming domestic cats in a zoo environment in Brazil. Ticks and Tick-borne Diseases, 5(5), 545-551. https://doi.org/10.1016/j.ttbdis.2014.03.011
André, M. R., Dumler, J. S., Herrera, H. M., Gonçalves, L. R., de Sousa, K. C., Scorpio, D. G., Santis, A. C. G. A., Domingos, I. H., De Macedo, G. C., & Machado, R. Z. (2016). Assessment of a quantitative 5'nuclease real-time polymerase chain reaction using the nicotinamide adenine dinucleotide dehydrogenase gamma subunit (nuoG) for Bartonella species in domiciled and stray cats in Brazil. Journal of Feline Medicine and Surgery, 18(10), 783-790. https://doi.org/10.1177/1098612X15593787
André, M. R., Canola, R. A. M., Braz, J. B., Perossi, I. F. S., Calchi, A. C., Ikeda, P., Machado, R. Z., Vasconcelos, R. O., & Camacho, A. A. (2019). Aortic valve endocarditis due to Bartonella clarridgeiae in a dog in Brazil. Revista Brasileira de Parasitologia Veterinária, 28, 661-670. https://doi.org/10.1590/S1984-29612019078
André, M. R., Gutiérrez, R., Ikeda, P., Amaral, R. B., Sousa, K. C. M., Nachum‐Biala, Y., Lima, L., Teixeira, M. M. G., Machado, R. Z., & Harrus, S. (2019). Genetic diversity of Bartonella spp. in vampire bats from Brazil. Transboundary and Emerging Diseases, 66(6), 2329-2341. https://doi.org/10.1111/tbed.13290
Bonato, L., Figueiredo, M. A. P., Gonçalves, L. R., Machado, R. Z., & André, M. R. (2015). Occurrence and molecular characterization of Bartonella spp. and hemoplasmas in neotropical primates from Brazilian Amazon. Comparative Immunology, Microbiology and Infectious Diseases, 42, 15-20. https://doi.org/10.1016/j.cimid.2015.09.001
Braga, I. A., Dias, I. S. D. O., Chitarra, C. S., Amude, A. M., & Aguiar, D. M. (2015). Molecular detection of Bartonella clarridgeiae in domestic cats from Midwest Brazil. Brazilian Journal of Infectious Diseases, 19, 451-452. https://doi.org/10.1016/j.bjid.2015.05.002
Calchi, A. C., Vultão, J. G., Alves, M. H., Yogui, D. R., Desbiez, A. L. J., do Amaral, R. B., de Santi, M., Teixeira, M. M. G., Werther, K., Machado, R. Z., & André, M. R. (2020). Multi‐locus sequencing reveals a novel Bartonella in mammals from the Superorder Xenarthra. Transboundary and Emerging Diseases, tbed.13545. https://doi.org/10.1111/tbed.13545
de Paiva Diniz, P. P. V., Maggi, R. G., Schwartz, D. S., Cadenas, M. B., Bradley, J. M., Hegarty, B., & Breitschwerdt, E. B. (2007). Canine bartonellosis: serological and molecular prevalence in Brazil and evidence of co-infection with Bartonella henselae and Bartonella vinsonii subsp. berkhoffii. Veterinary Research, 38(5), 697-710. http://doi.org/10.1051/vetres:2007023
de Sousa, K. C. M., do Amaral, R. B., Herrera, H. M., Santos, F. M., Macedo, G. C., de Andrade Pinto, P. C. E., Barros-Battesti, D. M., Machado, R. Z., & André, M. R. (2018). Genetic diversity of Bartonella spp. in wild mammals and ectoparasites in Brazilian Pantanal. Microbial Ecology, 76(2), 544-554. https://doi.org/10.1007/s00248-017-1138-0
Ferreira, M. S., Guterres, A., Rozental, T., Novaes, R. L. M., Vilar, E. M., Oliveira, R. C. D., Fernandes, J., Forneas, D., Alvino Junior, A., Brandão, M. L., Cordeiro, J. L. P., Alvarez, M. R. D. V., Althoff, S. L., Moratelli, R., Cordeiro-Estrela, P., da Silva, R. C. & Lemos, E. R. S. D. (2018). Coxiella and Bartonella spp. in bats (Chiroptera) captured in the Brazilian Atlantic Forest biome. BMC veterinary research, 14(1), 1-10. https://doi.org/10.1186/s12917-018-1603-0
Gonçalves-Oliveira, J., Rozental, T., Guterres, A., Teixeira, B. R., Andrade-Silva, B. E., Costa-Neto, S. F. da, Furtado, M. C., Moratelli, R., D’Andrea, P. S., & Lemos, E. R. S. (2020). Investigation of Bartonella spp. in brazilian mammals with emphasis on rodents and bats from the Atlantic Forest. International Journal for Parasitology: Parasites and Wildlife, 13, 80-89. https://doi.org/10.1016/j.ijppaw.2020.07.004
Gonçalves, L. R., Favacho, A. R. de M., Roque, A. L. R., Mendes, N. S., Fidelis Junior, O. L., Benevenute, J. L., Herrera, H. M., D’Andrea, P. S., de Lemos, E. R. S., Machado, R. Z., & André, M. R. (2016). Association of Bartonella species with wild and synanthropic rodents in different Brazilian biomes. Applied and Environmental Microbiology, 82(24), 7154-7164. https://doi.org/10.1128/AEM.02447-16
Gonçalves, L. R., Harrus, S., Gutiérrez, R., Herrera, H. M., Souza Ramos, I. A., Porfírio, G. E. de O., Nachum‐Biala, Y., Sousa, K. C. M., Silva, T. M. V., Campos, J. B. V., Lemos, W., Moraes Barros-Battesti, D., Machado, R. Z., & André, M. R. (2020). Molecular detection and genetic diversity of Bartonella species in large ruminants and associated ectoparasites from the Brazilian Cerrado. Transboundary and Emerging Diseases, tbed.13517. https://doi.org/10.1111/tbed.13517
Gonçalves, L. R., Harrus, S., Herrera, H. M., Gutierrez, R., Pedrassani, D., Nantes, W. A. G., Santos, F. M., Porfírio, G. E. O., Barreto, W. T. G., Macedo, G. C., Assis, W. O., Campos, J. B. V., Silva, T. M. V., Biolchi, J., Sousa, K. C. M., Nachum-Biala, Y. N., Barros-Battesti, D. M., Machado, R. Z., & André, M. R. (2020). Low occurrence of Bartonella in synanthropic mammals and associated ectoparasites in peri-urban areas from Central-Western and Southern Brazil. Acta Tropica, 207, 105513. https://doi.org/10.1016/j.actatropica.2020.105513
Hayman, D. T., McDonald, K. D., & Kosoy, M. Y. (2013). Evolutionary history of rat-borne Bartonella: the importance of commensal rats in the dissemination of bacterial infections globally. Ecology and Eolution, 3(10), 3195-3203. https://doi.org/10.1002/ece3.702
Ikeda, P., Seki, M. C., Carrasco, A. O. T., Rudiak, L. V., Miranda, J. M. D., Gonçalves, S. M. M., Hoppe, E. G. L., Albuquerque, A. C. A., Teixeira, M. M. G., Passos, C. E., Werther, K., Machado, R. Z., & André, M. R. (2017). Evidence and molecular characterization of Bartonella spp. and hemoplasmas in neotropical bats in Brazil. Epidemiology and Infection, 145(10), 2038-2052. https://doi.org/10.1017/S0950268817000966
Ikeda, P., Marinho Torres, J., Perles, L., Lourenço, E. C., Herrera, H. M., de Oliveira, C. E., Zacarias Machado, R., & André, M. R. (2020). Intra- and inter-host assessment of Bartonella diversity with focus on non-hematophagous bats and associated ectoparasites from Brazil. Microorganisms, 8(11), 1822. https://doi.org/10.3390/microorganisms8111822
Miceli, N. G., Gavioli, F. A., Gonçalves, L. R., André, M. R., Sousa, V. R. F., Sousa, K. C. M. D., & Machado, R. Z. (2013). Molecular detection of feline arthropod-borne pathogens in cats in Cuiabá, state of Mato Grosso, central-western region of Brazil. Revista Brasileira de Parasitologia Veterinária, 22, 385-390. https://doi.org/10.1590/S1984-29612013000300011
Pedrassani, D., Biolchi, J., Gonçalves, L. R., Mendes, N. S., Zanatto, D. C. D. S., Calchi, A. C., Machado, R. Z., & André, M. R. (2019). Molecular detection of vector-borne agents in cats in Southern Brazil. Revista Brasileira de Parasitologia Veterinária, 28, 632-643. https://doi.org/10.1590/S1984-29612019077
Rozental, T., Ferreira, M. S., Guterres, A., Mares-Guia, M. A., Teixeira, B. R., Gonçalves, J., Bonvicino, C. R., D’Andrea, P. S., & de Lemos, E. R. S. (2017). Zoonotic pathogens in Atlantic Forest wild rodents in Brazil: Bartonella and Coxiella infections. Acta Tropica, 168, 64-73. https://doi.org/10.1016/j.actatropica.2017.01.003
Silva, B. T. G. D., Souza, A. M. D., Campos, S. D. E., Lemos, E. R. S. D., Favacho, A. R. D. M., & Almosny, N. R. P. (2018). Presence of Bartonella spp. in domestic cats from a state park in Rio de Janeiro, Brazil. Revista do Instituto de Medicina Tropical de São Paulo, 60. https://doi.org/10.1590/S1678-9946201860014
Souza, A. M., Almosny, N. R. P., Favacho, A. R. M., Almeida, D. N. P., Ferreira, R. F., Ferreira, E. O., Moreira, N; S., & Lemos, E. R. S. (2017). Bartonella spp. and hematological changes in privately owned domestic cats from Rio de Janeiro, Brazil. The Journal of Infection in Developing Countries, 11(08), 591-596. https://doi.org/10.3855/jidc.8152
Souza, U. A., Webster, A., Dall’Agnol, B., Morel, A. P., Peters, F. B., Favarini, M. O., Mazim, F. D., Soares, J. B. G., Tirelli, F. P., Tortato, M. A., de Lemos, E. R. S., Trigo, T. C., Soares, J. F., & Reck, J. (2021). Molecular and serological survey of the cat-scratch disease agent (Bartonella henselae) in free-ranging Leopardus geoffroyi and Leopardus wiedii (Carnivora: Felidae) from Pampa biome, Brazil. Microbial Ecology, 81(2), 483-492. https://doi.org/10.1007/s00248-020-01601-x
Staggemeier, R., Venker, C. A., Klein, D. H., Petry, M., Spilki, F. R., & Cantarelli, V. V. (2010). Prevalence of Bartonella henselae and Bartonella clarridgeiae in cats in the south of Brazil: a molecular study. Memórias do Instituto Oswaldo Cruz, 105, 873-878. https://doi.org/10.1590/S0074-02762010000700006
Supplementary Information
Additional file 1: Fig. S1 Rényi’s diversity profiles of the rodent communities sampled in ten municipalities in Rio de Janeiro state, Brazil. Table S1 Summary of the number of communities, individuals and species studied at the three different levels of the analyses. Table S2 Functional diversity indices models using the odds ratio of Bartonella infection (logit-transformed) as the response variable between rodent communities in the state of Rio de Janeiro, Brazil. Table S3 Phylogenetic generalized linear mixed models predicting the status of Bartonella infection between rodent individuals of the Atlantic Forest (n = 192 after removing missing values). Table S4 Multiple regression coefficients for species interaction distance matrices, considering the presence and absence of Bartonella per host species and their phylogenetic and trait profile distances. Table S5 Properties of network node: values of degree and betweenness centralities. Table S6 Full ranking of candidate generalized linear models predicting degree centrality. Table S7 Full ranking of candidate generalized linear models predicting betweenness centrality.
Acknowledgement
We are grateful to Dr. Shimon Harrus for his suggestions and criticism of the manuscript.
Author contributions
G.L.T.C., J.O.G., and C.S.A. conceived the ideas; G.L.T.C., J.O.G., and C.S.A. designed methodology; G.L.T.C, J.O.G., and C.S.A. collected the data; G.L.T.C., J.O.G., and C.S.A. analyzed the data; P.S.D.A. coordinated the research project on on the biodiversity of Atlantic Forest, investigating taxonomic and ecological aspects; E.R.S.L. coordinated the investigation of parasitological aspects; and G.L.T.C., J.O.G., E.R.S.L., P.S.D.A., and C.S.A. led the writing of the manuscript. G.L.T.C. and J.O.G. should be considered joint first author. All authors contributed critically to the drafts and gave final approval for publication.
Funding
This study was funded by Programa Fiocruz de Fomento à Inovação – INOVA Fiocruz, grant no.: VPPCB-008-FIO-18; Serrapilheira Institute, grant no.: 1912–32354 and 6435–13754.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the figshare repository [42]: https://doi.org/10.6084/m9.figshare.25838281. Additionally, a list of data sources used in the study are provided in the Data sources section.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gabriella Lima Tabet Cruz and Jonathan Gonçalves-Oliveira have contributed equally to this work.
References
- 1.Becker DJ, Speer KA, Brown AM, Fenton MB, Washburne AD, Altizer S, et al. Ecological and evolutionary drivers of haemoplasma infection and bacterial genotype sharing in a neotropical bat community. Mol Ecol. 2020;29:1534–49. 10.1111/mec.15422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKee CD, Bai Y, Webb CT, Kosoy MY. Bats are key hosts in the radiation of mammal-associated Bartonella bacteria. Infect Genet Evol. 2021;89:104719. 10.1016/j.meegid.2021.104719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker DJ, Streicker DG, Altizer S. Using host species traits to understand the consequences of resource provisioning for host-parasite interactions. J Anim Ecol. 2018;87:511–25. 10.1111/1365-2656.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osorio-Canadas S, Arnan X, Bassols E, Vicens N, Bosch J. Seasonal dynamics in a cavity-nesting bee-wasp community: shifts in composition, functional diversity and host-parasitoid network structure. PLoS ONE. 2018;13:e0205854. 10.1371/journal.pone.0205854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreazzi CS, Martinez-Vaquero LA, Winck GR, Cardoso TS, Teixeira BR, Xavier SC, et al. Vegetation cover and biodiversity reduce parasite infection in wild hosts across ecological levels and scales. Ecography. 2023;2023:e06579. 10.1111/ecog.06579. [Google Scholar]
- 6.Llopis-Belenguer C, Balbuena JA, Lange K, de Bello F, Blasco-Costa I. Towards a unified functional trait framework for parasites. Trends Parasitol. 2019;35:972–82. 10.1016/j.pt.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Huang ZY, Halliday FW, Becker DJ. Host functional traits as the nexus for multilevel infection patterns. Trends Ecol Evol. 2023;38:1125–8. 10.1016/j.tree.2023.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Zhou S. A combination of species evenness and functional diversity is the best predictor of disease risk in multihost communities. Am Nat. 2015;186:755–65. 10.1086/683774. [DOI] [PubMed] [Google Scholar]
- 9.Morris A, Guégan JF, Benbow ME, Williamson H, Small PL, Quaye C, et al. Functional diversity as a new framework for understanding the ecology of an emerging generalist pathogen. EcoHealth. 2016;13:570–81. 10.1007/s10393-016-1140-x. [DOI] [PubMed] [Google Scholar]
- 10.Fecchio A, Lima MR, Bell JA, Schunck F, Corrêa AH, Beco R, et al, Repenning M, Braga ÉM. Loss of forest cover and host functional diversity increases prevalence of avian malaria parasites in the Atlantic Forest. Int J Parasitol. 2021;51:719–28. 10.1016/j.ijpara.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Goodrich I, McKee C, Kosoy M. Longitudinal study of bacterial infectious agents in a community of small mammals in New Mexico. Vector Borne Zoonotic Dis. 2020;20:496–508. 10.1089/vbz.2019.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Paiva Diniz PPV, Maggi RG, Schwartz DS, Cadenas MB, Bradley JM, Hegarty B, et al. Canine bartonellosis: serological and molecular prevalence in Brazil and evidence of co-infection with Bartonella henselae and Bartonella vinsonii subsp. berkhoffii. Vet Res. 2007;38:697–710. 10.1051/vetres:2007023. [DOI] [PubMed] [Google Scholar]
- 13.Souza UA, Webster A, Dall’Agnol B, Morel AP, Peters FB, Favarini MO, et al. Molecular and serological survey of the cat-scratch disease agent (Bartonellahenselae) in free-ranging Leopardusgeoffroyi and Leoparduswiedii (Carnivora: Felidae) from Pampa biome Brazil. Microbial Ecol. 2021;81:483–92. 10.1007/s00248-020-01601-x. [DOI] [PubMed] [Google Scholar]
- 14.De Salvo MN, Hercolini C, Arístegui E, Bruno A, Brambati DF, Cicuttin GL. Bartonella spp. associated with rodents in an urban protected area, Buenos Aires (Argentina). Comp Immunol Microb Infect Dis. 2020;72:101515. 10.1016/j.cimid.2020.101515. [DOI] [PubMed] [Google Scholar]
- 15.Müller A, Gutiérrez R, Seguel M, Monti G, Otth C, Bittencourt P, et al. Molecular survey of Bartonella spp. in rodents and fleas from Chile. Acta Trop. 2020;212:105672. 10.1016/j.actatropica.2020.105672. [DOI] [PubMed] [Google Scholar]
- 16.Silva-Ramos CR, Ballesteros-Ballesteros JA, Chala-Quintero SM, Matiz-González JM, Herrera-Sepúlveda MT, Faccini-Martínez ÁA, et al. Genetic diversity of Bartonella spp. among cave-dwelling bats from Colombia. Acta Trop. 2024;29:107370. 10.1016/j.actatropica.2024.107370. [DOI] [PubMed] [Google Scholar]
- 17.Gutiérrez R, Morick D, Gross I, Winkler R, Abdeen Z, Harrus S. Bartonella in domestic and stray cats from Israel: comparison of bacterial cultures and high-resolution melt real-time PCR as diagnostic methods. Vector Borne Zoonotic Dis. 2013;13:857–64. 10.1089/vbz.2013.1308. [DOI] [PubMed] [Google Scholar]
- 18.Sato S, Brinkerhoff RJ, Hollis E, Funada S, Shannon AB, Maruyama S. Detection of zoonotic Bartonella pathogens in rabbit fleas, Colorado, USA. Emerg Infect Dis. 2020;26:778–81. 10.3201/eid2604.191161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greco G, Zarea AAK, Sgroi G, Tempesta M, D’Alessio N, Lanave G, et al. Zoonotic Bartonella species in Eurasian wolves and other free-ranging wild mammals from Italy. Zoonoses Pub Health. 2021;68:316–26. 10.1111/zph.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poofery J, Narapakdeesakul D, Riana E, Arnuphapprasert A, Nugraheni YR, Ngamprasertwong T, et al. Molecular identification and genetic diversity of Bartonella spp. in 24 bat species from Thailand. Transbound Emerg Dis. 2021;69:e717–33. 10.1111/tbed.14389. [DOI] [PubMed] [Google Scholar]
- 21.Kim KS, Inoue K, Kabeya H, Sato S, Takada T, Pangjai D, et al. Prevalence and diversity of Bartonella species in wild small mammals in Asia. J Wildl Dis. 2016;52:10–21. 10.7589/2015-01-015. [DOI] [PubMed] [Google Scholar]
- 22.Wagner A, Dehio C. Role of distinct type-IV-secretion systems and secreted effector sets in host adaptation by pathogenic Bartonella species. Cell Microbiol. 2019;21:e13004. 10.1111/cmi.13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buffet JP, Kosoy M, Vayssier-Taussat M. Natural history of Bartonella-infecting rodents in light of new knowledge on genomics, diversity and evolution. Future Microbiol. 2013;8:1117–28. 10.2217/fmb.13.77. [DOI] [PubMed] [Google Scholar]
- 24.Segers FH, Kešnerová L, Kosoy M, Engel P. Genomic changes associated with the evolutionary transition of an insect gut symbiont into a blood-borne pathogen. ISME J. 2017;11:1232–44. 10.1038/ismej.2016.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han BA, Schmidt JP, Bowden SE, Drake JM. Rodent reservoirs of future zoonotic diseases. PNAS. 2015;112:7039–44. 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutiérrez R, Krasnov B, Morick D, Gottlieb Y, Khokhlova IS, Harrus S. Bartonella infection in rodents and their flea ectoparasites: an Overview. Vector Borne Zoonotic Dis. 2015;15:27–39. 10.1089/vbz.2014.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krügel M, Król N, Kempf VA, Pfeffer M, Obiegala A. Emerging rodent-associated Bartonella: a threat for human health? Parasites Vectors. 2022;31:113. 10.1186/s13071-022-05162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plourde BT, Burgess TL, Eskew EA, Roth TM, Stephenson N, Foley JE. Are disease reservoirs special? Taxonomic and life history characteristics. PLoS ONE. 2017;12:e0180716. 10.1371/journal.pone.0180716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosoy M, Mandel E, Green D, Marston E, Childs J. Prospective studies of Bartonella of rodents. Part I. Demographic and temporal patterns in population dynamics. Vector Borne Zoonotic Dis. 2004;4:285–95. 10.1089/vbz.2004.4.285. [DOI] [PubMed] [Google Scholar]
- 30.Costa F, Porter FH, Rodrigues G, Farias H, de Faria MT, Wunder EA, et al. Infections by Leptospira interrogans, Seoul Virus, and Bartonella spp. among Norway rats (Rattus norvegicus) from the urban slum environment in Brazil. Vector Borne Zoonotic Dis. 2014;14:33–40. 10.1089/vbz.2013.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krasnov BR, Morand S, Hawlena H, Khokhlova IS, Shenbrot GI. Sex-biased parasitism, seasonality and sexual size dimorphism in desert rodents. Oecologia. 2005;146:209–17. 10.1007/s00442-005-0189-y. [DOI] [PubMed] [Google Scholar]
- 32.Luza AL, Gonçalves GL, Pillar VD, Hartz SM. Processes related to habitat selection, diversity and niche similarity in assemblages of non-volant small mammals at grassland–forest ecotones. Nat Conserv. 2016;14:88–98. 10.1016/j.ncon.2016.09.003. [Google Scholar]
- 33.Hansford KM, Wheeler BW, Tschirren B, Medlock JM. Urban woodland habitat is important for tick presence and density in a city in England. Ticks Tick Borne Dis. 2022;13:101857. 10.1016/j.ttbdis.2021.101857. [DOI] [PubMed] [Google Scholar]
- 34.Cox DTC, Gardner AS, Gaston KJ. Diel niche variation in mammals associated with expanded trait space. Nat Commun. 2021;12:1–10. 10.1038/s41467-021-22023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacher TE, Kennerley R, Long B, McCay S, Roach NS, Turvey ST, et al. Support for rodent ecology and conservation to advance zoonotic disease research. Conserv Biol. 2021;35:1061–2. 10.1111/cobi.13763. [DOI] [PubMed] [Google Scholar]
- 36.Gonçalves LR, de Favacho ARM, Roque ALR, Mendes NS, Fidelis Junior OL, Benevenute JL, et al. Association of Bartonella species with wild and synanthropic rodents in different Brazilian biomes. Appl Environ Microbiol. 2016;82:7154–64. 10.1128/AEM.02447-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozental T, Ferreira MS, Guterres A, Mares-Guia MA, Teixeira BR, Gonçalves J,et al. Zoonotic pathogens in Atlantic Forest wild rodents in Brazil: Bartonella and Coxiella infections. Acta Trop. 2017;168:64–73. 10.1016/j.actatropica.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Gonçalves-Oliveira J, Rozental T, Guterres A, Teixeira BR, Andrade-Silva BE, da Costa-Neto SF, et al. Investigation of Bartonella spp. in brazilian mammals with emphasis on rodents and bats from the Atlantic Forest. Int J Parasitol Parasites Wildl. 2020;13:80–9. 10.1016/j.ijppaw.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Andrea PS, Teixeira BR, Gonçalves-Oliveira J, dos Dias D, Val Vilela R, dos Lucio CS, et al. A Coleção Mastozoológica do Laboratório de Biologia e Parasitologia de Mamíferos Silvestres Reservatórios—Fundação Oswaldo Cruz. Braz J Mammal. 2021;e90:e90202119. 10.32673/bjm.vie90.19. [Google Scholar]
- 40.Wilman H, Belmaker J, Simpson J, de la Rosa C, Rivadeneira MM, Jetz W. EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals: ecological archives E095–178. Ecology. 2014;95:2027–2027. 10.1890/13-1917.1. [Google Scholar]
- 41.Paglia AP, Da Fonseca GA, Rylands AB, Herrmann G, Aguiar LM, Chiarello AG, et al. Lista Anotada dos Mamíferos do Brasil 2ª Edição/annotated checklist of Brazilian mammals. Occasional Papers Conserv Biol. 2012;6:1–82. [Google Scholar]
- 42.Cruz GLT, Gonçalves-Oliveira J, de Lemos ERS, D’Andrea PS, Andreazzi CS. From host individual traits to community structure and composition: Bartonella infection insights. 2024. Figshare. Dataset. 10.6084/m9.figshare.25838281.v1.
- 43.Fisher RA, Corbet AS, Willians CB. The relation between the number of species and the number of individuals in a random sample of an animal population. J Anim Ecol. 1943;12:42–58. 10.2307/1411. [Google Scholar]
- 44.Botta-Dukát Z. Rao’s quadratic entropy as a measure of functional diversity based on multiple traits. J Veg Sci. 2005;16:533–40. 10.1111/j.1654-1103.2005.tb02393.x. [Google Scholar]
- 45.Laliberté E, Legendre P. A distance-based framework for measuring functional diversity from multiple traits. Ecology. 2010;91:299–305. 10.1890/08-2244.1. [DOI] [PubMed] [Google Scholar]
- 46.Villéger S, Mason NWH, Mouillot D. New multidimensional functional diversity indices for a multifaceted framework in functional ecology. Ecology. 2008;89:2290–301. 10.1890/07-1206.1. [DOI] [PubMed] [Google Scholar]
- 47.Tóthmérész B. Comparison of different methods for diversity ordering. J Veg Sci. 1995;6:283–90. 10.2307/3236223. [Google Scholar]
- 48.Oksanen J, Simpson G, Blanchet F, Kindt R, Legendre P, Minchin P, et al. Vegan: community ecology package. R package version 2.6–4; 2022. [Google Scholar]
- 49.Kindt R, Coe R. Tree diversity analysis: a manual and software for common statistical methods for ecological and biodiversity studies. Nairobi: World Agroforestry Centre; 2005. [Google Scholar]
- 50.Laliberté E, Legendre P, Shipley B, Laliberté ME. FD: measuring functional diversity from multiple traits, and other tools for functional ecology R package version 1.0–12.1. Austria: R Core Team Vienna; 2014. [Google Scholar]
- 51.R Core Team. R: A language and environment for statistical computing. Vienna: R foundation for Statistical Computing; 2021. [Google Scholar]
- 52.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bovendorp RS, Brum FT, McCleery RA, Baiser B, Loyola R, Cianciaruso MV, et al. Defaunation and fragmentation erode small mammal diversity dimensions in tropical forests. Ecography. 2019;42:23–35. 10.1111/ecog.03504. [Google Scholar]
- 54.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:11–120. 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 55.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–8. 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 56.Upham NS, Esselstyn JA, Jetz W. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 2019;17:e3000494. 10.1371/journal.pbio.3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. 10.18637/jss.v067.i01. [Google Scholar]
- 58.Bürkner PC. brms: an R package for Bayesian multilevel models using Stan. J Stat Softw. 2017;80:1–28. [Google Scholar]
- 59.Vehtari A, Gelman A, Gabry J. Practical Bayesian model evaluation using leave-one-out cross-validation and WAIC. Stat Comput. 2017;27:1413–32. 10.1007/s11222-016-9696-4. [Google Scholar]
- 60.Gelman A, Goodrich B, Gabry J, Vehtari A. R-squared for Bayesian regression models. Am Stat. 2019;73:307–9. 10.1080/00031305.2018.1549100. [Google Scholar]
- 61.Csardi G, Nepusz T. The igraph software package for complex network research. Int J Complex Syst. 2006;1695:1–9. [Google Scholar]
- 62.Lichstein JW. Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant Ecol. 2007;188:117–31. 10.1007/s11258-006-9126-3. [Google Scholar]
- 63.Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Statist Softw. 2007;22:1–19. 10.18637/jss.v022.i07. [Google Scholar]
- 64.Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat. 1908;44:223–70. [Google Scholar]
- 65.Bai Y, Kosoy MY, Calisher CH, Cully JF Jr, Collinge SK. Effects of rodent community diversity and composition on prevalence of an endemic bacterial pathogen-Bartonella. Biodiversity. 2009;10:3–11. 10.1080/14888386.2009.9712856. [Google Scholar]
- 66.Kedem H, Cohen C, Messika I, Einav M, Pilosof S, Hawlena H. Multiple effects of host-species diversity on coexisting host-specific and host-opportunistic microbes. Ecology. 2014;95:1173–83. 10.1890/13-0678.1. [DOI] [PubMed] [Google Scholar]
- 67.Krasnov BR, Shenbrot GI, Khokhlova IS. Dark diversity of flea assemblages of small mammalian hosts: effects of environment, host traits and host phylogeny. Int J Parasitol. 2022;52:157–67. 10.1016/j.ijpara.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Bossard RL, Broce AB, Dryden MW. Effects of circadian rhythms and other bioassay factors on cat flea (Pulicidae: Siphonaptera) susceptibility to insecticides. J Kans Entomol Soc. 2000;73:21–9. [Google Scholar]
- 69.Marignac G, Barrat F, Chomel B, Vayssier-Taussat M, Gandoin C, Bouillin C, et al. Murine model for Bartonella birtlesii infection: new aspects. Comp Immunol Microb Infect Dis. 2010;33:95–107. 10.1016/j.cimid.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 70.Frank HK, Boyd SD, Hadly EA. Global fingerprint of humans on the distribution of Bartonella bacteria in mammals. PLoS Negl Trop Dis. 2018;12:e0006865. 10.1371/journal.pntd.0006865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gutiérrez R, Cohen C, Flatau R, Marcos-Hadad E, Garrido M, Halle S, et al. Untangling the knots: co-infection and diversity of Bartonella from wild gerbils and their associated fleas. Mol Ecol. 2018;27:4787–807. 10.1111/mec.14906. [DOI] [PubMed] [Google Scholar]
- 72.Ikeda P, Seki MC, Carrasco AOT, Rudiak LV, Miranda JMD, Gonçalves SMM, et al. Evidence and molecular characterization of Bartonella spp. and hemoplasmas in neotropical bats in Brazil. Epidemiol Infect. 2017;145:2038–52. 10.1017/S0950268817000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.André MR, Gutiérrez R, Ikeda P, Amaral RB, Sousa KCM, Nachum-Biala Y, et al. Genetic diversity of Bartonella spp. in vampire bats from Brazil. Transbound Emerg Dis. 2019;66:2329–41. 10.1111/tbed.13290. [DOI] [PubMed] [Google Scholar]
- 74.McKee CD, Krawczyk AI, Sándor AD, Görföl T, Földvári M, Földvári G, et al. Host phylogeny, geographic overlap, and roost sharing shape parasite communities in European bats. Frontiers Ecol Evol. 2019;7:69. 10.3389/fevo.2019.00069. [Google Scholar]
- 75.Fromm K, Dehio C. The impact of Bartonella VirB/VirD4 type IV secretion system effectors on eukaryotic host cells. Frontiers Microbiol. 2021;12:762582. 10.3389/fmicb.2021.762582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Cruz GLT, Gonçalves-Oliveira J, de Lemos ERS, D’Andrea PS, Andreazzi CS. From host individual traits to community structure and composition: Bartonella infection insights. 2024. Figshare. Dataset. 10.6084/m9.figshare.25838281.v1.
Supplementary Materials
Additional file 1: Fig. S1 Rényi’s diversity profiles of the rodent communities sampled in ten municipalities in Rio de Janeiro state, Brazil. Table S1 Summary of the number of communities, individuals and species studied at the three different levels of the analyses. Table S2 Functional diversity indices models using the odds ratio of Bartonella infection (logit-transformed) as the response variable between rodent communities in the state of Rio de Janeiro, Brazil. Table S3 Phylogenetic generalized linear mixed models predicting the status of Bartonella infection between rodent individuals of the Atlantic Forest (n = 192 after removing missing values). Table S4 Multiple regression coefficients for species interaction distance matrices, considering the presence and absence of Bartonella per host species and their phylogenetic and trait profile distances. Table S5 Properties of network node: values of degree and betweenness centralities. Table S6 Full ranking of candidate generalized linear models predicting degree centrality. Table S7 Full ranking of candidate generalized linear models predicting betweenness centrality.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the figshare repository [42]: https://doi.org/10.6084/m9.figshare.25838281. Additionally, a list of data sources used in the study are provided in the Data sources section.