Abstract
Background
The genus Robinsonia DC. (tribe Senecioneae, Asteraceae) endemic to the Juan Fernández Islands in Chile is one of the most conspicuous insular plant groups in the world. Unlike typical herbaceous Asteraceae plants, these plants demonstrate spectacular and unusual rosette tree growth forms as shown by the alpine giant senecios (genus Dendrosenecio, tribe Senecioneae) endemic to the East African mountains. However, monophyly of the genus and phylogenetic relationships among species of Robinsonia as well as their plastome evolution remain elusive. This study aims to explore their phylogeny, species diversification, and molecular evolution based on the complete plastome sequences in the context of adaptive radiation on oceanic islands.
Results
The insular Robinsonia plastomes are highly conserved in their structures and organization of contents. Five divergence hotspots as potential chloroplast markers and five positively selected coding genes (accD, ndhF, rpoA, ycf1, and ycf2) are identified. Robinsonia plastomes has an overall nucleotide diversity higher than that of the sky island Dendrosenecio, but much lower than herbaceous Senecio. Phylogenetic analysis demonstrates the monophyly of Robinsonia and identifies two major infrageneric lineages. Both Robinsonia and Dendrosenecio are deeply nested within large genus Senecio.
Conclusions
While plastid genomes of Robinsonia are highly conserved, their sequences strongly demonstrated the monophyly of the genus and inferred robust interspecific relationships, including herbaceous Senecio and woody Dendrosenecio. Different sets of positively selected chloroplast genes, five for Robinsonia and two for Dendrosenecio, may play an important role in the adaptation strategies of these fascinating woody species in insular and continental sky island habitats. Overall phylogenetic positions and sister lineages of Robinsonia and Dendrosenecio require additional study based on broader sampling of Senecio.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05711-3.
Keywords: Adaptive radiation, Asteraceae, Critically endangered, Insular woodiness, Juan Fernández Islands, Plastome evolution
Introduction
Oceanic islands offer the opportunity to understand the evolutionary processes underlying rapid species diversification subsequent to the island colonization of immigrant populations by means of long-distance dispersal. The most commonly described evolutionary process in island biogeography is speciation associated with adaptive radiation called cladogenesis [1, 2]. Adaptive radiation connotes the entry of a single ancestor into a variety of habitats followed by species diversification and ecological shift. An initial immigrant population splits into morphologically and ecologically distinct evolutionary lineages through adaptation to divergent habitats available on an island (or an archipelago) free from competition and predation followed by inherent genetic variability release [1]. There have been numerous documented examples of island adaptive radiation, including the Lobelia complex (Campanulaceae) [3] and silversword alliance [4] in Hawaii; Scalesia (Asteraceae) in the Galápagos Islands [5–7]; Echium (Boraginaceae) [8, 9], Aeonium (Crassulaceae) [10, 11], and the woody Sonchus alliance (Asteraceae) [12–15] in the Canary Islands.
The Juan Fernández Islands, located 667 km off the west coast of Chile, are of volcanic origin and comprise two major islands and a much smaller island: Robinson Crusoe Island (also known as Isla Masatierra, 48 km2); Alejandro Selkirk Island (Isla Masafuera, 50 km2), and Santa Clara Island (2 km2, close to Robinson Crusoe Island). Robinson Crusoe Island is estimated to be approximately four million years old and Alejandro Selkirk Island is estimated one million years old [16]. The older island, Robinson Crusoe, is closer to Chile and the younger Alejandro Selkirk is 181 km west of Robinson Crusoe. The archipelago harbors a small, but unique flora with high levels of endemic flowering plants, including one family (Lactoridaceae), 10 genera and 105 species (approximately 14% and 67% at the generic and specific levels, respectively) [17, 18]. The Juan Fernández Islands are well suited for addressing evolutionary questions due to having only two major islands, their small sizes, the small number of endemic species, and their proximity to the major source areas in continental South America [17]. The most spectacular radiations in the Juan Fernández Islands have been in the family Asteraceae, having evolved tree-like insular habits called rosette trees in three groups of genera from three different tribes: Centaurodendron Johow and Yunquea Skottsb. of Cardueae, Dendroseris D. Don of Lactuceae, and Robinsonia DC. of Senecioneae. Whereas the first two genera contained only three species, the other two genera comprised 11 species for Dendroseris and eight for Robinsonia. Genus Robinsonia includes eight unique rosette dioecious shrub (or tree) species with extensive morphological and ecological diversity [19, 20].
Robinsonia species are found mostly on the older and nearer Robinson Crusoe Island, with only R. masafuerae Skottsb. found on Alejandro Selkirk Island. Morphological [19] and genetic [21, 22] data indicate that R. masafuerae originated following dispersal from Robinson Crusoe of an R. evenia-like ancestor. Robinsonia exhibits a range of morphological variation accompanied by adaptation to diverse habitats (Fig. 1). Despite a suite of biological features shared by all species, such as rosette tree habit, dioecious breeding system and same chromosome numbers (n = 20) [23–25], species vary considerably in habit (from a subshrub 1 ~ 2 m tall to a true tree, 5 ~ 6 m or more), and their habitats vary from epiphytism on tree ferns in very moist forests (R. evenia) to growth on montane scrubland and open cliff faces (R. gayana) [1, 19, 26].
Fig. 1.
Representative species of Robinsonia. A, R. berteroi; B, C, R. evenia; D, R. gracilis; E, R. masafuerae; F, R. thurifera; G, R. gayana. In C-E, heads from female (left) and male (right) plants for comparisons. Photo credit: Tod F. Stuessy, The Ohio State University, USA
As shown on Robinsonia species, one of the most prominent convergent aspects of island floras is the relatively high proportion of woody species in otherwise herbaceous lineages, a phenomenon known as insular woodiness [1, 27–29]. Insular woodiness, the evolutionary transition from herbaceous toward the woody condition on islands, are well exemplified by the Hawaiian silverswords (Dubautia and Argyroxiphium) and woody violets (Viola), as well as the Macaronesian tree lettuces (Sonchus) and viper buglosses (Echium) [4, 8, 9, 12–15, 30]. The spectra of growth forms have been considered adaptive modes, and woody insular representatives of predominantly herbaceous plants have likely evolved woodiness to develop tree-or shrub-like habits on islands. However, this tendency can also be observed in continental areas, most conspicuously in the equatorial highlands of Africa, Malaysia, and South America [1]. Insular woodiness and its evolutionary drivers remain poorly understood because the evidence is diverse for different island lineages, resulting in several hypotheses, such as aseasonal climate, competition for sunlight, drought resistance, and/or lack of large native herbivores [1, 31, 32]. The tribe Senecioneae (Asteraceae) encompasses several genera including the largest genus Senecio (ca. 1,250 species) [33, 34], and exhibits enormous variation in life-history strategies and morphology [35]. The tendency of Asteraceae to be represented on oceanic islands by woody species is well demonstrated by Senecioneae species with prominent examples in Robinsonia on the Juan Fernández Islands and Dendrosenecio on East African sky islands. The woody habit (accumulation of considerable secondary xylem) might be achieved in herbaceous Senecioneae when uniform conditions, typical of some oceanic islands, release plants from their seasonal cycles of growth [36].
Chloroplasts in plant cells play a crucial role in sustaining life on earth through the process of photosynthesis and oxygen release. They encode house-keeping and photosynthesis-associated proteins, serving as the active metabolic centers in cellular reactions to their environment [37]. Metabolites that are synthesized in chloroplasts are important for plant adaptation to environmental stress, such as drought [38], salinity [39], extreme temperature [40], high light [41] and heavy metal stress [42]. Plastid genomes are widely used to infer plant phylogeny and evolutionary history, as the reduced cost of next-generation sequencing and improved genomic analyses have facilitated the inclusion of massive amounts of data. These data revealed considerable genome-wide variation, which increased the phylogenetic resolution, especially at lower taxonomic levels, recent divergence, and rapid radiation. In contrast, conservative genome evolution yields limited sequence variation, which hinders phylogenetic resolution; therefore, plastome sequencing is now an efficient option for increasing phylogenetic resolution at lower taxonomic levels in plant phylogenetic and population genetic analyses [43].
In this study, we sequenced and assembled the whole plastid genomes of the genus Robinsonia to better understand their organization and evolution, as well as to reevaluate phylogenetic relationships inferred from previous studies. Specifically, we inferred the phylogenetic relationships within Robinsonia to hypothesize subgeneric, sectional, and species relationships, with inclusion of currently available complete plastome sequences of several Senecio and Dendrosenecio species. We also performed comparative plastome analyses to determine the structure, gene content, and rearrangements of plastid genomes of Robinsonia. Their genomic characteristics were subsequently compared with those of tropical alpine herbaceous Senecio and woody Dendrosenecio species in East Africa to understand the genomic similarities and differences among herbaceous and woody Senecioneae plastomes evolved on oceanic islands and continental mountainous highlands (sky islands). Lastly, this study identified positively selected genes of plastid genomes that can give us some insights into adaptation strategies in insular and continental sky island habitats. In addition, several highly variable chloroplast regions, as useful markers for population genetic or phylogeographic studies of Robinsonia and Senecio, are identified.
Materials and methods
Plant materials and DNA extraction
Plant materials of Robinsonia species used in this study were previously collected in the field during four expeditions of the Universidad de Concepción, Chile and Ohio State University, USA to the Juan Fernández Islands. All but one species of Robinsonia were included, representing the two subgenera, Rhetinodendron and Robinsonia. The materials of R. macrocephala of subg. Robinsonia section Symphyochaeta were not available, as it is now thought to be extinct. As acknowledged in previous studies [44–47], the Robinsonia samples are representatives of the collections from the Robinson Crusoe National Park under the permission issued by CONAF (Corporación National Forestal). The fresh leaves were either dried (placed in sealable plastic bags with silica gel) or placed on ice and retained at 4 °C until extracted in the laboratory at The Ohio State University, Columbus, Ohio, USA. Total genomic DNAs were extracted using the DNeasy Plant Mini kit (Qiagen, Valencia, CA, United States) at Sungkyunkwan University laboratory following the manufacturer’s instruction. The vouchers were deposited in the OS (Herbarium of Ohio State University) and WU (Herbarium of Universität Wien) (Table 1).
Table 1.
Genomic features of the complete plastid genomes of seven Robinsonia species sequenced and analyzed in this study
| Taxa | Collection no. | GenBank accession no. | Total plastid size (bp) / GC content (%) | LSC size (bp) / GC content (%) | IR size (bp) / GC content (%) | SSC size (bp) / GC content (%) | No. of genes | No. of protein-coding genes | No. of tRNA genes | No. of rRNA genes | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Robinsonia Subgenus Rhetinodendron | |||||||||||
| R. berteroi | 11,238 | NC085195 | 151,239/37.2 | 83,334 /35.3 | 24,824/42.9 | 18,257/30.4 | 130 | 87 | 37 | 6 | |
| Robinsonia Subgenu s Robinsonia | |||||||||||
| Section Symphyochaeta | |||||||||||
| R. macrocephala | not available due to extinction | ||||||||||
| S ection Robinsonia | |||||||||||
| R. gayana | 19,251 J | NC085197 | 151,330/37.2 | 83,429/35.3 | 24,817/42.9 | 18,267/30.5 | 130 | 87 | 37 | 6 | |
| R. saxatilis | 11,186 | NC085200 | 151,247/37.6 | 83,352/35.3 | 24,815/42.9 | 18,265/30.5 | 130 | 87 | 37 | 6 | |
| R. thurifera | 11,161 | NC085201 | 151,330/37.2 | 83,429/35.3 | 24,817/42.9 | 18,267/30.5 | 130 | 87 | 37 | 6 | |
| S ection Eleutherolepis | |||||||||||
| R. evenia | 19,283 F | NC085196 | 151,246/37.2 | 83,352/35.3 | 24,816/42.9 | 18,262/30.4 | 130 | 87 | 37 | 6 | |
| R. gracilis | 19,138 | NC085198 | 151,278/37.2 | 83,377/35.3 | 24,816/42.9 | 18,269/30.4 | 130 | 87 | 37 | 6 | |
| R. masafuerae | 19,637 | NC085199 | 151,328/37.2 | 83,428/35.3 | 24,816/42.9 | 18,268/30.4 | 130 | 87 | 37 | 6 | |
LSC: large single copy region; SSC: small single copy region; IR: inverted repeat
Plastome sequencing, assembly, and annotation
Illumina paired-end (PE) genomic libraries with a fragment size of 550 base pairs (bp) were prepared and sequenced using the Illumina HiSeq platform (Illumina, Inc., San Diego, Ca, USA) at Macrogen Corporation (Seoul, Korea). The sequence contigs were assembled using the de novo genomic assembler, Velvet 1.2.10 [48], and annotation was performed using GeSeq [49], ARAGORN v1.2.36 [50], and RNAmmer 1.2 Server [51]. Each draft annotation was then manually investigated and corrected whenever necessary using Geneious v8.1.6 (Biomatters Ltd., Auckland, New Zealand), performing a BLAST search by comparing with homologous genes in Nicotiana tabacum (NC001879) and Senecio vulgaris (NC046693) from the GenBank database at the National Center for Biotechnology Information (NCBI) as references. The complete plastome sequences of seven Robinsonia species were registered in GenBank under the accession numbers NC085195 (R. berteroi, Collection # 11238); NC085197 (R. gayana, Collection # 19251 J); NC085201 (R. thurifera, Collection # 11161); NC085198 (R. gracilis, Collection # 19138); NC085196 (R. evenia, Collection # 19283 F); NC085199 (R. masafuerae, Collection # 19637); and NC085200 (R. saxatilis, Collection # 11186). OGDRAW was used to draw circular plastid genome maps [52].
Comparative plastome analyses and identification of highly divergent regions
We performed several comparative plastome analyses of Robinsonia species on the Juan Fernández Islands in the Pacific Ocean, comparing with herbaceous Senecio and woody Dendrosenecio plastomes from highland East Africa. This comparison aimed to investigate the similarities and differences among herbaceous and woody plastomes that convergently evolved as woody growth forms on both of oceanic islands and continental sky islands. The sequences of six Dendrosenecio and five Senecio plastomes reported by Gichira et al. [53] were obtained from GenBank for genomic comparison; i.e., D. battiscombei, D. brassiciformis, D. elgonensis, D. johnstonii, D. keniodendron, and D. meruensis for woody Dendrosenecio and S. moorei, S. keniophytum, S. purtschelleri, S. schweinfurthii, and S. roseiflorus for herbaceous Senecio plastomes (see Table 1 in Gichira et al. for their genomic features) [53]. The level of codon usage bias was determined by the relative synonymous codon usage (RSCU; the relative frequency of occurrence of the synonymous codon for a specific amino acid) value calculated from the codon usage frequency using MEGA7 [54]. To evaluate the pressure of natural selection on the protein-coding genes of the seven plastomes, site-specific models implemented in EasyCodeML [55] were used in the preset running mode based on the CodeML algorithms. Selective pressure has been inferred by the ratio of nonsynonymous and synonymous substitution rates (denoted as ω = dN/dS), with ω = 1 indicating neutral mutations; ω < 1, purifying selection; and ω > 1, diversifying positive selection. Positively selected sites were presented based on the fit of seven codon substitution models (M0, M1a, M2a, M3, M7, M8, and M8a) with heterogeneous ω values across sites implemented in EasyCodeML using likelihood ratio tests (LRT) [56, 57]. Overall sequence divergence was estimated using the mVISTA online program [58] (https://genome.lbl.gov/vista/mvista/submit.shtml) employing the LAGAN alignment mode [59]. Nucleotide diversity (Pi) was calculated using sliding window analysis (window length = 1000 bp and step size = 200 bp excluding sites with alignment gaps) to detect the most divergent regions (i.e., mutation hotspots) in DnaSP [60].
Phylogenetic analysis
Phylogenetic relationships of the newly sequenced accessions of Robinsonia were investigated in the context of their relationships with the complete plastid sequences of other closely related species obtained from GenBank, including Senecio (nine species), Dendrosenecio (12 species), and two other related genera in subtribe Senecioninae (Jacobaea and Pericallis). Two species of Ligularia from subtribe Tussilagininae were used as outgroups. In total, 32 plastid genomes were used to generate maximum likelihood (ML) trees based on both the complete plastid genome sequences and concatenated sequences of protein coding genes with 1000 replicate bootstrap (BS) analyses using IQ-TREE [61] after alignment using MAFFT v.7 [62]. The complete plastid genome sequences were partitioned for genic (protein, tRNAs, and rRNAs coding genes) and non-coding intergenic regions. The best fit evolutionary models were chosen as TVM + F + I for the concatenated sequences of 80 protein coding genes, HKY + F + I for tRNA, HKY + F for rRNAs, and K3Pu + F + I + G4 for noncoding intergenic regions, which were scored according to Bayesian information criterion scores and weights by testing 88 DNA models using ModelFinder [63] implemented in IQ-TREE.
Results
Gene content, order, and organization of the plastomes of Robinsonia
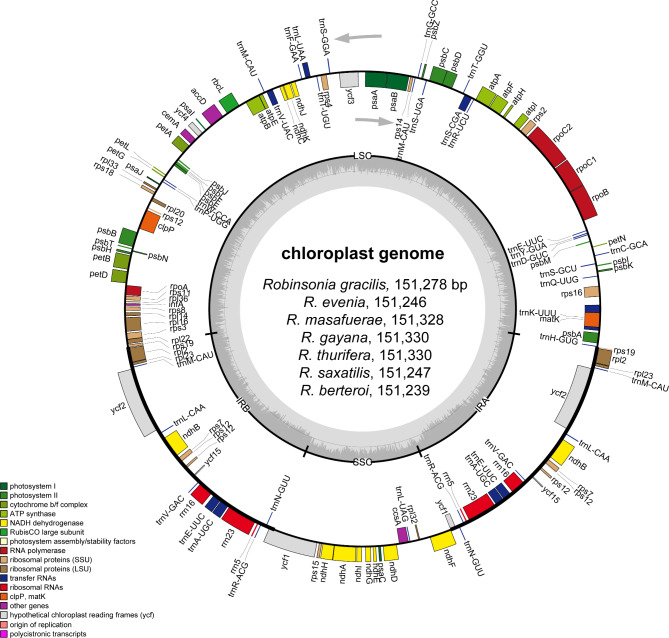
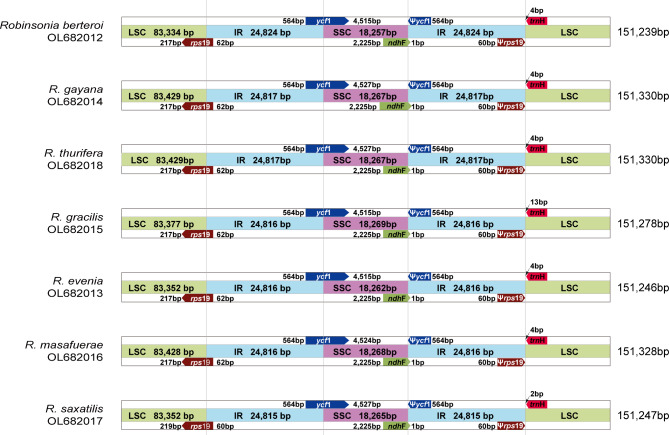
The seven plastomes of Robinsonia species (R. berteroi, R. evenia, R. gayana, R. gracilis, R. masafuerae, R. saxatilis and R. thurifera) were highly conserved in gene content and arrangement, displaying 99.7% pairwise sequence similarity despite the morphological and ecological differences among them. The total lengths of the seven Robinsonia plastomes ranged from 151,239 (R. berteroi, subg. Rhetinodendron) to 151,330 bp (R. gayana and R. thurifera, subg. Robinsonia), which were slightly longer than Dendrosenecio (average 150 bp shorter) species [53]. Each of the seven Robinsonia plastomes consisted of four typical plastid regions: LSC (83,334 − 83,429 bp), SSC (18,257 − 18,269 bp), and IR regions (24815 − 24,824 bp), sharing the same genes and similar gene contents at all adjacent junctions among the four regions (Figs. 2 and 3). The overall guanine-cytosine (GC) content of each plastid genome was 37.2%, with LSC, SSC, and IRs regions having 35.3%, 30.4 − 30.5%, and 42.9% GC contents, respectively. Each of the seven cp. genomes contained 130 genes, including 87 protein-coding genes (excluding pseudogenes), six rRNA genes, and 37 tRNA genes (Tables 1 and 2). Eighteen genes, including seven tRNA genes contained introns. Three genes, clpP, rps12, and ycf3, had two introns. The trnK-UUU tRNA gene harbored the largest intron, containing the matK gene. In total, 17 genes were duplicated in the IR regions, including seven tRNAs, three rRNAs, and seven protein genes. The trans-splicing gene rps12, consisting of three exons, was located in the LSC region of exon 1, whereas exons 2 and 3 were imbedded in the IR regions. Incompletely duplicated parts of ycf1 and rps19 in the IR regions were considered pseudogenes in all the cp genomes sequenced in this study.
Fig. 2.
Gene maps of the plastid genomes of seven Robinsonia species sequenced and analyzed in this study. The genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to different functional groups are shown in different colors. The thick lines indicate the extent of the inverted repeats (IRs) that separate the genomes into small single copy (SSC) and large single copy (LSC) regions
Fig. 3.
Comparison of the border positions of the large single copy (LSC), small single copy (SSC), and inverted repeat (IR) regions among seven Robinsonia plastid genomes. Gene names are indicated in boxes, and their lengths in the corresponding regions are displayed above the boxes. Ψ indicates a pseudogene
Table 2.
Genes present in the complete plastid genomes of seven Robinsonia species sequenced in this study
| Category | Group | Genes | |
|---|---|---|---|
| Photosynthesis | photosystem_I | psaA, psaB, psaC, psaI, and psaJ | |
| photosystem_II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, and psbZ | ||
| NADH_dehydrogenase |
ndhA*, ndhB( 2)*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, and ndhK 2)*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, and ndhK |
||
| cytochrome_b/f_complex | petA, petB*, petD*, petG, petL, and petN | ||
| ATP_synthase | atpA, atpB, atpE, atpF*, atpH, and atpI | ||
| Large_subunit_of_Rubisco | rbcL | ||
| Self-replication | Large_subunits_of_ribosome |
rpl2( 2)*, rpl14, rpl16*, rpl20, rpl22, rpl23( 2)*, rpl14, rpl16*, rpl20, rpl22, rpl23( 2), rpl32, rpl33, and rpl36 2), rpl32, rpl33, and rpl36 |
|
| Small_subunits_of_ribosome |
rps2, rps3, rps4, rps7( 2), rps8, rps11, rps12 ( 2), rps8, rps11, rps12 ( 2)**, rps14, rps15, rps16*, rps18, and rps19 2)**, rps14, rps15, rps16*, rps18, and rps19 |
||
| DNA-dependent_RNA_polymerase | rpoA, rpoB, rpoC1*, and rpoC2 | ||
| translation initiation factor | infA | ||
| Ribosomal_RNAs |
rrn5( 2), rrn16( 2), rrn16( 2), and rrn23( 2), and rrn23( 2) 2) |
||
| Transfer_RNAs |
trnA-UGC( 2)*, trnC-GCA, trnD-GUC, trnE-UUC( 2)*, trnC-GCA, trnD-GUC, trnE-UUC( 3)*, trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU*, trnL-CAA( 3)*, trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU*, trnL-CAA( 2)*, trnL-UAA, trnL-UAG, trnM-CAU( 2)*, trnL-UAA, trnL-UAG, trnM-CAU( 4), trnN-GUU( 4), trnN-GUU( 2), trnP-UGG, trnQ-UUG, trnR-ACG( 2), trnP-UGG, trnQ-UUG, trnR-ACG( 2), trnR-UCU, trnS-CGA, trnS-GCU*, trnS-GGA, trnS-UGA, trnS-GGU, trnT-UGU, trnV-GAC( 2), trnR-UCU, trnS-CGA, trnS-GCU*, trnS-GGA, trnS-UGA, trnS-GGU, trnT-UGU, trnV-GAC( 2), trnV-GCA, trnW-CCA, and trnY-GUA 2), trnV-GCA, trnW-CCA, and trnY-GUA |
||
| Other genes | Maturase | matK | |
| Protease | clpP** | ||
| Envelope_membrane_protein | cemA | ||
| Acetyl-CoA_carboxylase | accD | ||
| C-type_cytochrome_synthesis_gene | ccsA | ||
| Genes of unknown function | Proteins_of_unknown_function |
ycf1, ycf2( 2), ycf3**, ycf4, ycf15( 2), ycf3**, ycf4, ycf15( 2) 2) |
( N) indicates the genes that have N copies. * and ** indicate genes containing one and two introns, respectively.
N) indicates the genes that have N copies. * and ** indicate genes containing one and two introns, respectively.
Comparative plastome analyses: codon usage , positive selection, sequence divergence and mutation hot spot
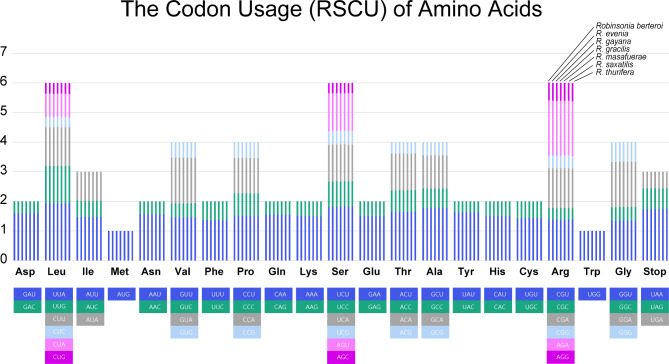
The total size of all protein coding genes (single copies, excluding repeated genes) in the seven Robinsonia plastomes was 68,214 − 68,232 bp, encoding 22,738 − 22,744 codons including stop codons. Their average number of codon usage was smaller than that of Dendrosenecio (22,815 − 22840 for single copies of coding genes; 26,293 − 26,382 for all coding genes), but slightly larger than Senecio (22,640 − 22,689; 26,067 − 26,177) in Gichira et al.’s [53]. The patterns of frequently used codons and their RSCU values were consistent among the seven Robinsonia plastomes (Fig. 4). The highest RSCU value was indicated in the usage of UUA codon for leucine (1.94) followed by AGA for arginine (1.85 − 1.86), while the lowest ones, CUC for leucine (0.34 − 0.35) and AGC (0.34 − 0.35) for Serine followed by CUG for leucine (0.36). The codons AUG (M) and UGG (W), which encode methionine and tryptophan, respectively, showed no bias (RSCU = 1) (Supplementary table S1).
Fig. 4.
The relative synonymous codon usage (RSCU) of the protein-coding genes in chloroplast genomes of seven Robinsonia species. The codon usages of amino acids are plotted along the x-axis, while the stacked RSCU values in each bar column are plotted along the y-axis respectively. Each amino acid contains seven clustered bar columns representing seven species; 1st column through 7th column for R. berteroi, R. evenia, R. gayana, R. gracilis, R. masafuerae, R. saxatilis, and R. thurifera
From seven Robinsonia plastomes, we identified five coding genes potentially evolved under positive selection, accD, ndhF, rpoA, ycf1, and ycf2 using the pairwise comparison of codon substitution models, M7 (beta) vs. M8 (beta and ω > 1). They were presented with a significant posterior probability (p) more than 0.95 indicated with an asterisk, * (P ≥ 0.95) or ** (P ≥ 0.99), calculated using the Bayes empirical Bayes (BEB) test [57]. On the other hand, two coding genes, atpA and rpoC2 were found in the six Dendrosenecio plastomes undergoing positive selection on continental sky islands, whereas none were found in the five sympatric Senecio plastomes occurring in Mt. Kenya (Table 3).
Table 3.
Positively selected sites having dN/dS values > 1 detected in seven Robinsonia and six Dendrosenecio plastomes
| Gene name | Models | np | ln L | Model compared | Likelihood ratio test p-value | Positively selected sites | |
|---|---|---|---|---|---|---|---|
| Robinsonia plastomes (R. berteroi, R. evenia, R. gayana, R. gracilis, R. masafuerae, R. saxatilis, and R. thurifera) | |||||||
| accD | M8 | 16 | -1964.763594 | M7 vs. M8 | 0.667566011 | 39 S 0.962*, 116 I 0.962* | |
| M7 | 14 | -1965.167711 | |||||
| ndhF | M8 | 16 | -2893.891812 | M7 vs. M8 | 0.001549017 | 491 F 0.970* | |
| M7 | 14 | -2900.361947 | |||||
| rpoA | M8 | 16 | -1327.780011 | M7 vs. M8 | 0.628296557 | 95 V 0.962*, 280 R 0.962* | |
| M7 | 14 | -1328.244754 | |||||
| ycf1 | M8 | 16 | -6754.608911 | M7 vs. M8 | 0.010754360 | 432 N 0.991** | |
| M7 | 14 | -6759.141355 | |||||
| ycf2 | M8 | 16 | -8899.330842 | M7 vs. M8 | 0.517702255 | 794 D 0.976*, 831 G 0.976*, 1234 E 0.976* | |
| M7 | 14 | -8899.989197 | |||||
| Dendrosenecio plastomes (D. battiscombei, D. brassiciformis, D. elgonensis, D. johnstonii, D. keniodendron, and D. meruensis) | |||||||
| atpA | M8 | 14 | -2016.260099 | M7 vs. M8 | 0.557743556 | 123 I 0.977*, 477 S 0.977*, 508 G 0.977* | |
| M7 | 12 | -2016.843955 | |||||
| rpoC2 | M8 | 14 | -5520.953083 | M7 vs. M8 | 0.447862506 | 278 R 0.984*, 717 I 0.984*, 1039 L 0.984*, 1040 N 0.984* | |
| M7 | 12 | -5521.756352 | |||||
Significant posterior probability value for positively selected sites are indicated with * for > 0.95 and ** >0.99
np is number of parameters, lnL, the log-likelihod values, and LRT p-value, Likelihood Ratio Test p-value
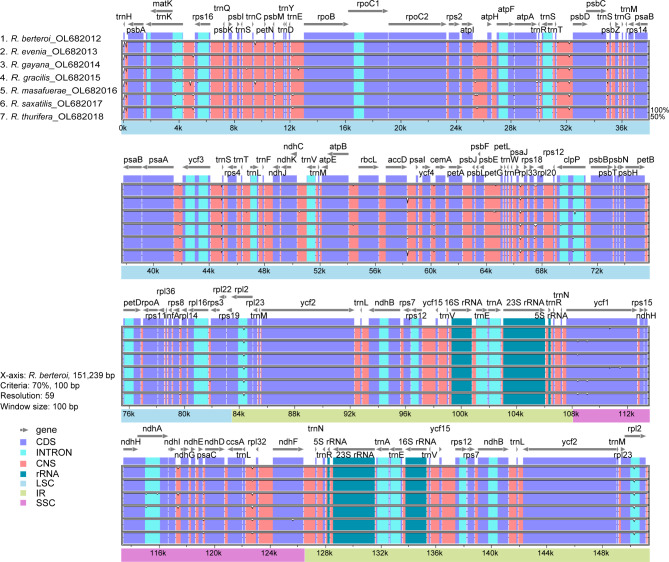
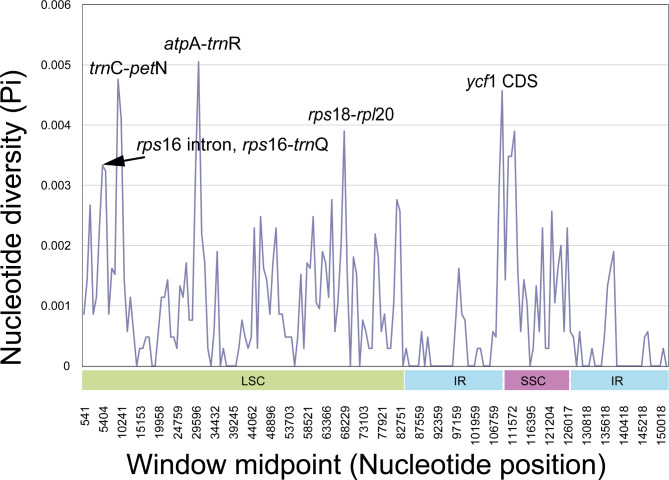
The divergence of the seven Robinsonia plastomes was assessed using the mVISTA platform [58], with the annotated sequences of Robinsonia berteroi as a reference. The mVISTA graph exhibited a high degree of synteny and gene order conservation among the plastomes of Robinsonia, sharing almost identical coding regions (except ycf1) and more variable noncoding and intron regions (Fig. 5). However, the results of mVISTA analysis including six Dendrosenecio and five Senecio together with Robinsonia plastomes indicated different patterns of polymorphic sites among the three groups, mostly from noncoding and intron regions, even though all of them revealed similar synteny in the coding regions (except petB and ycf1) (Supplementary fig. S1). The overall nucleotide diversity value (Pi) of the seven Robinsonia plastomes (average 0.00092, ranging from 0 to 0.00505) calculated using DnaSP [60] was higher than that of the six Dendrosenecio plastomes (average 0.00058, ranging from 0 to 0.0112). However, their diversity was much lower than five Senecio plastomes (average 0.00357, ranging from 0 to 0.0142). Among the seven Robinsonia plastomes, the SSC region containing ycf1, which is known to have high diversity in plants [47, 64–66], showed the highest nucleotide diversity (0.001668), whereas the lowest value was observed in the IR boundary regions (0.00032). Five divergence hotspots among the Robinsonia plastomes have been suggested as potential plastid markers for the phylogenetic studies of Robinsonia species and closely related Senecio groups. Four intergenic hotspot regions (rps16-trnQ, trnC-petN, atpA-trnR, and rps18-rpl20), and one protein coding region (ycf1) were identified in LSC and SSC regions (Fig. 6).
Fig. 5.
Comparison of the plastid genomes of seven Robinsonia species, against R. berteroi by mVISTA. Grey arrows indicate genes with their orientation and position. Genome regions are color-coded as blue blocks for the conserved coding genes (exon), aqua blue blocks for introns, and orange blocks for the conserved non-coding sequences in intergenic regions (CNS). Thick lines below the alignment indicate the quadripartite regions of genomes; LSC region is in light blue, IR regions, in beige, and SSC region, in pink
Fig. 6.
Five hotspot regions in the seven Robisonia plastomes
Phylogenetic analysis
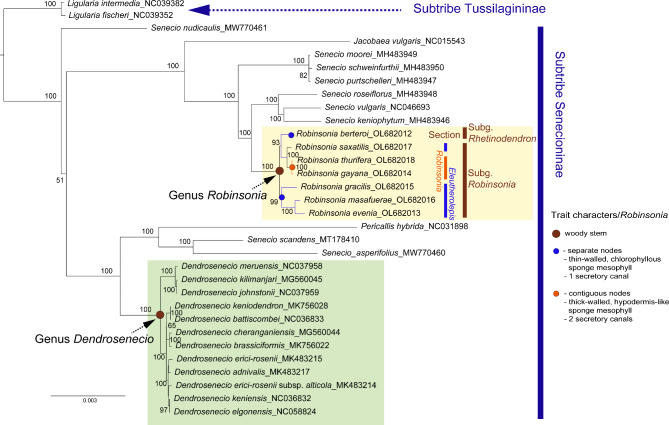
ML analyses were conducted based on sequences of complete plastomes (132,037 aligned nucleotide bp) and protein coding genes (68,573 bp) of the 32 representative Asteraceae plastomes, including Senecio, Dendrosenecio, and Robinsonia species. Both ML phylogenetic trees provided good resolution of inter-generic and inter-specific relationships, sharing congruent topology with slight variances in BS values (Fig. 7and Supplementary fig. S2). Robinsonia and Dendrosenecio were monophyletic with robust support (100% BS, respectively), whereas Senecio was not monophyletic. Within the ingroup species of the subtribe Senecioninae against the outgroup species of the subtribe Tussilagininae (Ligularia intermedia and L. fischeri), two major clades were revealed, with Robinsonia and Dendrosenecio embedded in different clades, suggesting that Robinsonia is not closely related to Dendrosenecio. Interestingly, Robinsonia clustered with Senecio species collected from East Africa, and the monophyletic clade of Dendrosenecio was more closely related to other Senecio species occurring primarily in Asia than to the sympatric African Senecio species. These relationships, however, hinge on a broader sampling of Senecio species in a future study. Within the monophyletic clade of all extant Robinsonia species, the plastome sequences did not support the current subgeneric classification of Robinsonia into two subgenera: Rhetinodendron and Robinsonia. Robinsonia berteroi of monotypic subg. Rhetinodendron was in a sister relationship with section Robinsonia of subg. Robinsonia (98% of BS) rather than a sister to whole subg. Robinsonia; two subgenera are not reciprocally monophyletic. Within subg. Robinsonia, two sections of Robinsonia (R. gayana, R. thurifera, and R. saxatilis) and Eleutherolepis (R. gracilis, R. masafuerae and R. evenia) were resolved into distinct clades. R. saxatilis initially described as being the closest to R. evenia and treated under the section Eleutherolepis [20] was nested in the section Robinsonia instead of Eleutherolepis (100% BS) on both the full sequences and coding genes of plastomes (Fig. 7and Supplementary fig. S2).
Fig. 7.
Maximum likelihood tree based on the complete plastome sequences of 32 representatives of Asteraceae including species of Robinsonia, Dendrosenecio and Senecio. Numbers above nodes are bootstrap values with 1000 replicates. Trait characters are marked with brown circle representing woody stem; blue, separate nodes spacing, typically thin-walled, chlorophyllous leaf sponge mesophyll, and one secretory canal per vascular bundle; and orange, nearly contiguous nodes, thick-walled, strongly hypodermis-like mesophyll, and two secretory canals. Trait data taken from Skottsberg (1921) and Carlquist (1974) [1, 26], but not available for R. saxatilis
Discussion
Phylogenetic relationships of Robinsonia
Robinsonia species have been variously recognized at the genus level: R. berteroi (DC.) R.W. Sanders, Stuessy & Martic was initially recognized under distinct genera, that is, initially described as Balbisia berteroi DC [67–69]. and then as Rhetinodendron berteroi (Dcne.) Hemsl [70–72]. Skottsberg (1921) also recognized the monotypic genus Rhetinodendron Meisn. with Rhetinodendron berteroi and the genus Robinsonia DC. with six species [26]. He divided Robinsonia into subgenera and sections as follows: subg. Symphyochaeta (DC.) Skottsb. (R. macrocephala Dcne.) and subg. Eleutherochaeta (DC.) Skottsb. with two sections, i.e., section Symphyolepis Skottsb. (Robinsonia gayana Dcne. and R. thurifera Dcne.) and section Eleutherolepis DC. (R. evenia Phil., R. masafuerae Skottsb. and R. gracilis Dcne.). Later, Sanders et al. (1987) provided an explicit phylogenetic hypothesis using primarily morphological characteristics and suggested that all species of Robinsonia species should be recognized as a monophyletic genus resulting from a single introduction [19]. The genus Rhetinodendron was merged with Robinsonia as the monotypic subg. Rhetinodendron (along with R. berteroi) based on its strong similarity to Robinsonia species in terms of flavonoid chemistry [73] and morphology [19]. The remaining six species were placed under subg. Robinsonia, which is divided into sections Symphyochaeta (R. macrocephala), Robinsonia (R. gayana and R. thurifera) and Eleutherolepis (R. evenia, R. gracilis, and R. masafuerae). Danton (2006) described Robinsonia saxatilis Danton from Robinson Crusoe Island and referred it to section Eleutherolepis DC. of subg. Robinsonia [20]. Unfortunately, several species of Robinsonia are highly threatened by extinction because of their restricted ranges and small population sizes. Robinsonia macrocephala was once known from several locations in Masatierra [26], but has not been found elsewhere despite repeated searches on several expeditions and is now thought to be extinct [74]. Robinsonia berteroi was once reported from eight localities [26], but is now known from only one mature individual found in 2015 and other individuals (between one and 15) in inaccessible places where prospecting efforts should be made. Thus, R. berteroi is categorized as Critically Endangered (CR, DS 06/2017 MMA) instead of Extinct (EX) on the IUCN Red List of Threatened Species [75].
Molecular phylogenetic analyses of Robinsonia generally supported the relationships based on the morphological characters suggested by Sanders et al. (1987) [19]. Although the phylogenetic study using restriction site mutations in cpDNA and IGS of nuclear rDNA failed to resolve the relationships among Robinsonia species [76], internal transcribed spacer (ITS) phylogeny supported the monophyly of Robinsonia species (including R. berteroi) [22], implying a single introduction of the ancestor of Robinsonia to the Islands. However, later studies [35, 77] which included more extensive sampling of Senecio species, reported that R. berteroi appeared to be more closely related to Senecio species than to the other Robinsonia species on ITS or ITS-the external transcribed spacer (ETS) combined phylogenies. However, in Pelser et al.’s [77], the combined plastid data (ndhF gene; trnL intron; psbA-trnH, psbJ-petA, 5’ and 3’ trnK, and trnL-F intergenic spacers) supported the monophyly of Robinsonia species (including R. berteroi), but the ITS-ETS combined data set failed to resolve Robinsonia as monophyletic, placing R. berteroi outside of the Robinsonia clade. Pelser et al. concluded that “Thus, the monophyly of Robinsonia remains inconclusive despite additional data and analyses.” However, Pelser et al. [77] proposed merging Robinsonia into one of the largest genera of angiosperms Senecio because all species of the former were nested within the latter.
Although the results of the present study are highly concordant with those of previous molecular studies, the whole plastome sequences provided additional insights into questions that have remained unanswered despite several earlier studies. The sister lineages of Robinsonia and Dendrosenecio have yet to be determined due to the limited taxonomic sampling of the plastomes of the broadly occurring Senecio, however, the plastome phylogeny provided strong support for the monophyly of Robinsonia, with R. berteroi resolved within the same clade as other species (Fig. 7and Supplementary fig. S2). In addition, R. berteroi was resolved as a sister to a clade consisting of R. thurifera, R. gayana and R. saxatilis, rather than a sister to all other Robinsonia species, where it had commonly been placed in other molecular phylogenies, supporting its recognition as subg. Rhetinodendron [22, 77]. To the best of our knowledge, this is the first time that this relationship has been suggested for R. berteroi. Regarding the taxonomic treatment of R. berteroi, we continue to support its recognition as a separate subgenus (subg. Rhetinodendron), particularly because of its smaller flowering heads and narrowly elongated, thyrsoid capitulescences (Fig. 1A). Robinsonia saxatilis was initially viewed as being closely related to R. evenia of sect. Eleutherolepis [20]. However, Takayama et al. [21] reported that this new species was more properly placed in section Robinsonia near R. gayana or possibly R. thurifera based on amplified fragment length polymorphism (AFLP) and simple sequence repeat (SSR) analyses. The complete plastome phylogeny in the current study also suggests that R. saxatilis shares its most recent common ancestor with a common ancestor of R. gayana-R. thurifera, further corroborating earlier AFLP and SSR results.
Plastome structure and evolution
The whole cp. genomes of the seven Robinsonia species reported for the first time in this study, were highly conserved, sharing common genomic features such as sequence similarity, gene content, and gene number (Table 1). We compared their genomic features with the plastomes of Dendrosenecio and Senecio species previously reported by Gichira et al. [53] and found that the plastomes of the three genera shared similar genomic features despite their morphological and distributional divergences (i.e., the woody insular endemics of Robinsonia in the Juan Fernández Islands, the woody highland endemics of Dendrosenecio in East Africa, and the herbaceous endemics of Senecio in East Africa). Minor differences were found in their total lengths; with Dendrosenecio (150 kb) being slightly shorter than Robinsonia and Senecio (151 kb) with differences mainly in SSC sizes. Generally, the lengths of the chloroplast genome and its quadripartite regions vary among plant lineages because of the contraction and expansion of IR regions. Evaluating their contraction and expansion by comparing the location of the boundaries among the four chloroplast regions (two IRs, LSC and SSC) can provide insights into plastome evolution [78]. All contained the functional protein-coding gene of ycf1 at IR/SSC with its pseudogene copy, ycf1Ψ at SSC/IR, and functional rps19 at LSC/IR with pseudogene copy rps19Ψ at IR/LSC endpoints (Fig. 3).
Codon usage bias and positive selection were compared to investigate selective pressure on protein-coding genes in the plastomes of Robinsonia, Dendrosenecio, and Senecio. The patterns of frequently used codons and the average number of codon usage were very similar, with the highest RSCU value indicated in the usage of UUA(L) and AGA(R), whereas the lowest were AGC(S), CUC(L), UAC(Y), and CUG(L). Additionally, codons AUG (M) and UGG (W) showed no bias (RSCU = 1) in any of the plastomes of the three genera (Supplementary table S1). However, several genes that potentially evolved under positive selection were identified in the woody plastomes of Robinsonia and Dendrosenecio (Table 3), whereas no positive selection sites were detected in the herbaceous Senecio plastomes. Seven Robinsonia plastomes contained five coding genes, accD, ndhF, rpoA, ycf1, and ycf2, under positive selection, however, only two genes, atpA and rpoC2, were detected in the six Dendrosenecio plastomes. Most of these genes encode proteins related to photosynthesis and self-replication of plastids, which are extremely important for plant survival, including the NADH dehydrogenase gene (ndhF), DNA-dependent RNA polymerase genes (rpoA and rpoC2), ATP subunit gene (atpA), Acetyl-CoA carboxylase gene (accD), and genes of unknown function (ycf1 and ycf2). These findings suggest that a substantial amount of positive selection in these gene regions may play an important role in the adaptation strategies of woody endemics on oceanic islands (Robinsonia) and the tropical mountains of East Africa (Dendrosenecio). The equatorial highlands contain an ecological spectrum similar to that of oceanic islands, and woodiness increases in both regions. However, typical oceanic islands are distinct from equatorial highlands in that the former contain a wider gamut of habitats (shore to alpine, aquatic to xeric) in contrast to the latter with a single extreme habitat and relatively uniform climates throughout the year (‘summer every day, winter every night’) [1]. Plastid genes broadly adapt to changing ecological conditions [79, 80], and the positive selection of different sets of genes may have resulted in the adaptive radiation of Robinsonia and Dendrosenecio in their specific habitats.
Seven Robinsonia plastomes were highly conserved in the mVISTA and DnaSP analyses, but diverged from six Dendrosenecio and five Senecio plastomes, revealing differences in the pattern of polymorphic sites and overall nucleotide diversity. The nucleotide diversity (Pi) of Robinsonia (average 0.00092) was higher than that of Dendrosenecio (0.00058), but lower than that of Senecio (0.00357). Robinsonia’s plastome diversity was comparable to that of other insular woody endemics such as Dendroseris (0.00061) on the Juan Fernández Islands and Dendrosonchus (0.00090) on the Canary Islands [47, 81]. Furthermore, the overall patterns for highly variable regions were similar among the three genera with the highest diversity in the SSC region and the lowest in the IR regions. Although the chloroplast genomes were highly conserved among the seven Robinsonia species, we identified five divergence hotspots from the LSC and SSC regions as potential plastid markers for phylogenetic studies of Robinsonia and closely related taxa (Fig. 6): four intergenic hotspot regions (rps16-trnQ, trnC-petN, atpA-trnR, and rps18-rpl20), and one protein coding region (ycf1).
Adaptive radiation of Robinsonia
The Juan Fernández Islands seems too limited to promote adaptive radiation due to its small size of two islands (Robinson Crusoe Island and Alejandro Selkirk Island), and outlying islets (Santa Clara Island). Nevertheless, Robinsonia represents early or limited adaptive radiation, and the species occupy very specific discontinuous niches over a range of mesophytic to xerophytic habitats, indicating a remarkable degree of correlation between habits and habitats [1]. Their habitats vary from obligate epiphytism on tree ferns (R. evenia) in very moist forest to growth on montane scrubland and open cliff faces (R. gayana). Carlquist (1974) has commented that the adaptive radiation on the Juan Fernández Islands seems to be in response to intense niche diversity and separation and that the species originated by in situ adaptation to local conditions [1]. Selection for adaptation to different ecological conditions drives adaptive radiation, accelerating speciation through the processes of genetic differentiation among populations within species, acquisition of reproductive isolation among populations, and the rise of ecological differentiation among such populations [82]. Consequently, oceanic island flora displays the differences in traits representation of dispersal capacity, pollination syndrome and life history strategy between island and mainland floras [83]. The phylogenetic inference in this study also suggests that the diversification of Robinsonia species could be the results of adaptive radiation associated with correlated habits (i.e., functional traits) and habitats on the Juan Fernández Islands.
The clade composing three species of R. evenia, R. gracilis, and R. masafuerae on the plastome phylogenetic tree (Fig. 7and Supplementary fig. S2) supports the section Eleutherolepis of subgenus Robinsonia, and those three species share the trait characters of separate nodes spacing, typically thin-walled, chlorophyllus leaf mesophyll, and one secretory canal per vascular bundle. On the other hand, R. gayana and R. thurifera (section Robinsonia) display nearly contiguous nodes, thick-walled hypodermis-like mesophyll, and two secretory canals [1, 19, 26]. Two groups of Robinsonia species are ecologically differentiated with habitats being mesophytic (section Eleutherolepis) or xerophytic (section Robinsonia). Hypodermis, a characteristic of brightly illuminated situations, tends to increase with exposed situation, and there is a relative tendency of conversion of spongy tissue to hypodermis observed in section Robinsonia. Increased presence of secretory canal with greater illumination and xeromorphy, although not uniform, seems evident as seen in Asteraceae plants of dry localities [1].
Crawford et al. (2018) synthesized information from several studies [1 pp. 206–211, 19, 84] and interpreted the possible factors driving the radiation of Robinsonia in the Juan Fernández Islands and maintaining species distinctions [85]. The mesomorphy in leaf anatomy of R. evenia, R. gracilis, and R. masafuerae (section Eleutherolepis of Robinsonia) is attributed to their epiphytism on cloud-forest tree fern; i.e., R. evenia (obligate epiphytism), R. gracilis (facultative epiphytism), and R. masafuerae (associated with ferns), all within mesic habitats of cloud forest. R. evenia is considered as an obligate epiphyte found on tree ferns [1, p. 206], although some specimens are known to occur on rock surfaces, i.e., eplithic lithophytes (Patricio López-Sepúlveda, personal observation). R. gracilis is not often epiphytic, but seedlings may start as epiphytes. R. gracilis may be found near to R. gayana (section Robinsonia), but the former occurs on ridges with open scrub, whereas the latter, often in crevices of cliffs in montane scrub. R. masafuerae, the only Robinsonia species found on Alejandro Selkirk Island and closely related with R. evenia on Robinson Crusoe Island, prefers fern-rich gullies and is sometimes epiphytic.
Contrastingly, the xeromorphy of R. gayana and R. thurifera (section Robinsonia of Robinsonia) is correlated with the growth on cliff faces and emergence in large size (to 5–6 m tall) from the fern-forest canopy, respectively [1, p. 210]. The closely related R. gayana and R. thurifera appeared to prefer different habitats, although they may occasionally occur together [19]. The former species grows primarily on exposed, rocky, xerophytic habitats, while the latter occurs primarily on forest edges in more mesic environments. Another factor isolating the two species is the later flowering of R. thurifera than its sympatric sister species, R. gayana [19]. R. saxatilis, which has been suggested to have evolved from a common ancestor of R. gayana-R. thurifera in this study, also exbibits strictly rock and non-forest ecology without epiphytic seedlings on tree fern trunks [20]. R. berteroi currently classified under monotypic subg. Rhetinodendron exhibits similar trait characters as section Eleutherolepis of Robinsonia species, however, it occurs from less mesomorphic habitats in open brushwood and forest along the ridge [1, 19]. The plastome phylogeny in this study resolved R. berteroi closer to the clade of xeromorphic Robinsonia species rather than to the clade of mesomorphic Eleutherolepis species with similar trait characters (Fig. 7).
Conclusions
This study, for the first time, assembled the complete plastome sequences of seven Robinsonia species, woody insular endemics on the Juan Fernández Islands in Chile, and performed phylogenetic and phylogenomic analyses to better understand the plastome evolution of Robinsonia. The analyses included woody Dendrosenecio and herbaceous Senecio from the tropical mountains in eastern Africa. Despite their morphological and distributional divergence, the whole cp genomes of the three genera were highly conserved, sharing common genomic features such as sequence similarity, gene content, and gene numbers. However, plastome sequences provide high resolution and strong support for the phylogenetic relationships among Robinsonia and closely related taxa including several Senecio and Dendrosenecio species. The monophyly of Robinsonia was robustly confirmed based on both ML trees of the complete plastome sequences and 80 concatenated coding genes. Robinsonia is divided into two major lineages with R. berteroi included within the same clade as other species, but nested within Senecio. Robinsonia berteroi currently classified under subg. Rhetinodendron was unexpectedly resolved as a sister to the clade consisting of R. thurifera, R. gayana and R. saxatilis (section Robinsonia of subg. Robinsonia) rather than being a sister to all remaining Robinsonia species (subg. Robinsonia). However, we support the current taxonomic treatment of R. berteroi as a separate subgenus (subg. Rhetinodendron), particularly because of its morphological distinction from the other Robinsonia species. Robinsonia saxatilis, which was initially described as being closely related to R. evenia of sect. Eleutherolepis, has been suggested to have evolved from a common ancestor of R. gayana-R. thurifera, supporting the findings based on AFLP and SSR analyses. Given the limited taxonomic sampling of the broadly occurring Senecio in this study, the sister lineages of Robinsonia and Dendrosenecio have yet to be determined. In the adaptive radiation procedure of Robinsonia species in the Juan Fernández Islands, the possible factors driving their radiation and maintaining species distinction could be their preference for different habitats and different flowering times even between species occasionally occurring together. The results of comparative phylogenomic analyses of the plastomes of Robinsonia, Dendrosenecio and Senecio species indicated that different sets of positively selected chloroplast genes, five in Robinsonia and two in Dendrosenecio, may have contributed to the adaptive radiation of these fascinating woody species in insular and continental sky island habitats. The overall patterns for the highly variable regions were similar among the three genera with the highest diversity in the SSC region and the lowest in the IR regions. Five mutation hotspots (rps16-trnQ, trnC-petN, atpA-trnR, rps18-rpl20, and ycf1) in the LSC and SSC regions were identified as potential chloroplast markers for future phylogenetic and phylogeographic studies on Robinsonia and related Senecio groups.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Supplementary Figure S1. Comparison of the plastid genomes of seven Robinsonia, six Dendrosenecio and five Senecio species, against R. berteroi by mVISTA.
Supplementary Material 2: Supplementary Figure S2. Maximum likelihood tree based on the protein-coding gene sequences of 32 representative Asteraceae species in Robinsonia, Dendrosenecio and Senecio. Trait characters are marked with brown circle representing woody stem; blue, separate nodes spacing, typically thin-walled, chlorophyllous leaf sponge mesophyll, and one secretory canal per vascular bundle; and orange, nearly contiguous nodes, thick-walled, strongly hypodermis-like mesophyll, and two secretory canals. Trait data taken from Skottsberg (1921) and Carlquist (1974) [1, 26], but not available for R. saxatilis
Supplementary Material 3: Supplementary Table S1. Relative synonymous codon usage (RSCU) of protein-coding genes in the chloroplast genomes of seven Robinsonia species
Acknowledgements
We thank the anonymous reviewers for their insightful comments and suggestions. We also thank the Cambridge University Press for the permission of using the photographs in Fig. 1, originally published by Stuessy et al. (Plants of Oceanic Islands: Evolution, Biogeography, and Conservation of the Flora of the Juan Fernández (Robinson Crusoe) Archipelago, 2018: C58, C59, C61-C67).
Abbreviations
- AFLP
Amplified fragment length polymorphic
- BS
Bootstrap
- cp
Chloroplast
- cpDNA
Chloroplast DNA
- DnaSP
DNA sequence polymorphism
- ETS
External transcribed spacer
- IGS
Intergenic spacer
- ITS
Internal transcribed spacer
- IR
Inverted repeat
- LSC
Large single copy
- ML
Maximum likelihood
- Pi
Nucleotide diversity
- Plastome
Plastid genome
- RSCU
Relative synonymous codon usage
- SSC
Small single copy
- SSR
Simple sequence repeat
Author contributions
Conceptualization, M-S.C., JY.Y., S-C.K.; Methodology, M-S.C., JY.Y., S-H.K.; Software and analysis, M-S.C., JY.Y., S-H.K.; Resources, T.F.S., D.J.C., P.L-S.; Data curation, M-S.C., JY.Y.; Writing−original draft preparation by M-S.C., and review and editing by T.F.S., D.J.C., S-C.K.; Visualization, M-S.C., JY.Y., S-H.K.; Supervision and project administration, S-C.K.; Funding acquisition, M-S.C. All the authors have read and approved the final version of the manuscript.
Funding
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education to M-S Cho (2022R1I1A2063355).
Data availability
The datasets generated and/or analyzed in the current study are available from the NCBI GenBank repository [https://www.ncbi.nlm.nih.gov/].
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Carlquist S. Island biology. New York and London: Columbia University; 1974. [Google Scholar]
- 2.Stuessy TF, Jakubowsky G, Gómez RS, Pfosser M, Schlüter PM, Fer T, Sun BY, Kato H. Anagenetic evolution in island plants. J Biogeogr. 2006;33:1259–65. [Google Scholar]
- 3.Givnish TJ, Millam KC, Mast AR, Paterson TB, Theim TJ, Hipp AL, Henss JM, Smith JF, Wood KR, Sytsma KJ. Origin, adaptive radiation and diversification of the hawaiian lobeliads (Asterales: Campanulaceae). Proc R Soc B. 2009;276:407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlquist S, Baldwin BG, Carr GD. Tarweeds & silverswords: evolution of the Madiinae (Asteraceae). St. Louis: Missouri Botanical Garden; 2003. [Google Scholar]
- 5.Eliasson U. Studies in Galapagos Plants. XIV. The genus Scalesia Arn. Opera Bot. 1974;36:1–117. [Google Scholar]
- 6.Fernández-Mazuecos M, Vargas P, McCauley RA, Monjas D, Otero A, Chaves JA, Andino JEG, Rivas-Torres G. The radiation of Darwin’s giant daisies in the Galápagos Islands. Curr Biol. 2020;30(24):4989–98. [DOI] [PubMed] [Google Scholar]
- 7.Schilling EE, Panero JL, Eliasson UH. Evidence from chloroplast DNA restriction site analysis on the relationships of Scalesia (Asteraceae: Heliantheae). Am J Bot. 1994;81:248–54. [Google Scholar]
- 8.Böhle UR, Hilger HH, Martin WF. Island colonization and evolution of the insular woody habit in Echium L. (Boraginaceae). Proc Natl Acad Sci USA. 1996;93:11740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcıa-Maroto F, Manas-Fernández A, Garrido-Cárdenas JA, Alonso DL, Guil-Guerrero JL, Guzmán B, Vargas P. D6-Desaturase sequence evidence for explosive pliocene radiations within the adaptive radiation of Macaronesian Echium (Boraginaceae). Mol Phylogenet Evol. 2009;52:563–74. [DOI] [PubMed] [Google Scholar]
- 10.Jorgensen TH, Olesen JM. Adaptive radiation of island plants: evidence from Aeonium (Crassulaceae) of the Canary Islands. Perspect Plant Ecol Evol Sys. 2001;4:29–42. [Google Scholar]
- 11.Mort ME, Soltis DE, Soltis PS, Francisco-Ortega J, Santos-Guerra A. Phylogenetics and evolution of the macaronesian clade of Crassulaceae inferred from nuclear and chloroplast sequence data. Syst Bot. 2002;27:271–88. [Google Scholar]
- 12.Kim S-C, Crawford DJ, Francisco-Ortega J, Santos-Guerra A. A common origin for Woody Sonchus and five related genera in the macaronesian islands: molecular evidence for extensive radiation. Proc Natl Acad Sci USA. 1996a;93:7743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S-C, Crawford DJ, Jansen RK. Phylogenetic relationships among the genera of subtribe Sonchinae (Asteraceae): evidence from ITS sequences. Syst Bot. 1996b;221:417–32. [Google Scholar]
- 14.Kim S-C, McGowen MR, Lubinsky P, Barber JC, Mort ME, Santos-Guerra A. Timing and tempo of early and successive adaptive radiations in Macaronesia. PLoS ONE. 2008;3:e2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santiago LS, Kim S-C. Correlated evolution of leaf shape and physiology in the Woody Sonchus alliance (Asteraceae: Sonchinae) in Macaronesia. Int J Plant Sci. 2009;170:83–92. [Google Scholar]
- 16.Stuessy TF, Foland KA, Sutter JF, Sanders RW, Silva M. Botanical and geological significance of potassium-argon dates from the Juan Fernandez Islands. Science. 1984;225:49–51. [DOI] [PubMed] [Google Scholar]
- 17.Stuessy TF, Swenson U, Crawford DJ, Anderson G. Plant conservation in the Juan Fernandez archipelago. Chile Aliso. 1998b;16:89–101. [Google Scholar]
- 18.Takayama K, Crawford DJ, López-Sepúlveda P, Greimler J, Stuessy TF. Factors driving adaptive radiation in plants of oceanic islands: a case study from the Juan Fernández Archipelago. J Pl Res. 2018;131:469–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders RW, Stuessy TF, Marticorena C, Silva MO. Phytogeography and evolution of Dendroseris and Robinsonia, tree-Compositae of the Juan Fernandez islands. Opera Bot. 1987;92:195–215. [Google Scholar]
- 20.Danton P. Contribution à la flore de l’archipel Juan Fernández (Chili). Description de deux taxons nouveaux: Nicotiana cordifolia subsp. sanctaeclarae subsp. nov. (Solanaceae), Robinsonia saxatilis sp. nov. (Asteraceae). Acta Bot Gallica. 2006;153:249–255.
- 21.Takayama K, López-Sepúlveda P, Greimler J, Crawford DJ, Peñailillo P, Baeza M, Ruiz E, Kohl G, Tremetsberger K, Gatica A, Letelier L. Relationships and genetic consequences of contrasting modes of speciation among endemic species of Robinsonia (Asteraceae, Senecioneae) of the Juan Fernández Archipelago, Chile, based on AFLPs and SSRs. New Phytol. 2015;205:415–28. [DOI] [PubMed] [Google Scholar]
- 22.Sang T, Crawford DJ, Stuessy TF, Silva M. ITS sequences and the phylogeny of the genus Robinsonia (Asteraceae). Syst Bot. 1995;20:55–64. [Google Scholar]
- 23.Anderson GJ, Bernardello G, Stuessy TF, Crawford DJ. Breeding system and pollination of selected plants endemic to Juan Fernández Islands. Am J Bot. 2001;88:220–33. [PubMed] [Google Scholar]
- 24.Bernardello G, Anderson GJ, Stuessy TF, Crawford DJ. A survey of floral traits, breeding systems, floral visitors, and pollination systems of the angiosperms of the Juan Fernández islands (Chile). Bot Rev. 2001;67:255–308. [Google Scholar]
- 25.Sanders RW, Stuessy TF, Rodriguez R. Chromosome numbers from the flora of the Juan Fernandez islands. Am J Bot. 1983;70:799–810. [Google Scholar]
- 26.Skottsberg C. The phanerogams of the Juan Fernandez islands. In: Skottsberg C, editor. The natural history of Juan Fernandez and Easter Island. Volume 2. Uppsala: Almqvist and Wiksells; 1921. pp. 95–240. [Google Scholar]
- 27.Lens F, Davin N, Smets E, del Arco M. Insular woodiness on the Canary Islands: a remarkable case of convergent evolution. Int J Plant Sci. 2013;174:992–1013. [Google Scholar]
- 28.Whittaker RJ, Fernández-Palacios JM, Matthews TJ, Borregaard MK, Triantis KA. Island biogeography: taking the long view of nature’s laboratories. Science. 2017;357(6354):eaam8326. [DOI] [PubMed] [Google Scholar]
- 29.Zizka A, Onstein RE, Rozzi R, Weigelt P, Kreft H, Steinbauer MJ, Bruelheide H, Lens F. The evolution of insular woodiness. P Natl Acad Sci. 2022;119:e2208629119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballard HE Jr, Sytsma KJ. Evolution and biogeography of the Woody Hawaiian violets (Viola, Violaceae): Arctic origins, herbaceous ancestry and bird dispersal. Evolution. 2000;54:1521–32. [DOI] [PubMed] [Google Scholar]
- 31.Choat B, Brodribb TJ, Brodersen CR, Duursma RA, López R, Medlyn BE. Triggers of tree mortality under drought. Nature. 2018;558:531–9. [DOI] [PubMed] [Google Scholar]
- 32.Lens F, Tixier A, Cochard H, Sperry JS, Jansen S, Herbette S. Embolism resistance as a key mechanism to understand adaptive plant strategies. Curr Opin Plant Biol. 2013;16:287–92. [DOI] [PubMed] [Google Scholar]
- 33.Nordenstam B. Taxonomic studies in the tribe Senecioneae (Compositae). Opera Bot. 1978;44:1–84. [Google Scholar]
- 34.Nordenstam B. Tribe Senecioneae. In: Kadereit JW, Jeffrey C, editors. Flowering plants. Eudicots. Asterales. Vol. 8 of Kubitzki J, editor. The families and genera of vascular plants. Berlin: Springer; 2007. pp. 208–241.
- 35.Pelser PB, Nordenstam B, Kadereit JW, Watson LE. An ITS phylogeny of tribe Senecioneae (Asteraceae) and a new delimitation of Senecio L. Taxon. 2007;56:1077–104. [Google Scholar]
- 36.Carlquist S. Wood anatomy of Senecioneae (Compositae). Aliso. 1962;5:123–46. [Google Scholar]
- 37.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao C, Haigh AM, Holford P, Chen ZH. Roles of chloroplast retrograde signals and ion transport in plant drought tolerance. Int J Mol Sci. 2018;19(4):963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krasensky J, Broyart C, Rabanal FA, Jonak C. The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid Redox Signal. 2014;21(9):1289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo S, Kim C. Current understanding of temperature stress-responsive chloroplast FtsH metalloproteases. Int J Mol Sci. 2021;22(22):12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phee BK, Cho JH, Park S, Jung JH, Lee YH, Jeon JS, Bhoo SH, Hahn TR. Proteomic analysis of the response of Arabidopsis chloroplast proteins to high light stress. Proteomics. 2004;4(11):3560–8. [DOI] [PubMed] [Google Scholar]
- 42.Khan M, Nawaz N, Ali I, Azam M, Rizwan M, Ahmad P, Ali S. Regulation of photosynthesis under metal stress. In: Ahmad P, Ahanger MA, Alyemeni MN, Alam P, editors. Photosynthesis productivity environ stress. Hoboken: Wiley; 2019. p. 95–105.
- 43.Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crawford DJ, Stuessy TF, Silva MO. Allozyme divergence and the evolution of Dendroseris (Compositae: Lactuceae) on the Juan Fernandez islands. Syst Bot. 1987;12:435–43. [Google Scholar]
- 45.Crawford DJ, Stuessy TF, Cosner MB, Haines DW, Silva MO, Baeza M. Evolution of the genus Dendroseris (Asteraceae: Lactuceae) on the Juan Fernandez islands: evidence from chloroplast and ribosomal DNA. Syst Bot. 1992;17:676–82. [Google Scholar]
- 46.Pacheco P, Crawford DJ, Stuessy TF, Silva MO. Flavonoid evolution in Dendroseris (Compositae, Lactuceae) from the Juan Fernandez Islands, Chile. Am J Bot. 1991;78:534–43. [Google Scholar]
- 47.Cho M-S, Kim S-H, Yang JY, Crawford DJ, Stuessy TF, López-Sepúlveda P, Kim S-C. Plastid phylogenomics of Dendroseris (Cichorieae; Asteraceae): insights into structural organization and molecular evolution of an endemic lineage from the Juan Fernandez Islands. Front Plant Sci. 2020;11:594272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 2008;18:821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, FischerA., Bock R, Greiner S. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 2007;35:3100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:W575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gichira AW, Avoga S, Li Z, Hu G, Wang Q, Chen J. Comparative genomics of 11 complete chloroplast genomes of Senecioneae (Asteraceae) species: DNA barcodes and phylogenetics. Bot Stud. 2019;60:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao F, Chen C, Arab DA, Du Z, He Y, Ho SY. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evolut. 2019;9:3891–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z, PAML. A program package for phylogenetic analysis by maximum likelihood. Bioinformatics. 1997;13:555–6. [DOI] [PubMed] [Google Scholar]
- 57.Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–18. [DOI] [PubMed] [Google Scholar]
- 58.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. NISC comparative sequencing program. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–302. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2014;32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, Cheng T, Guo J, Zhou S. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015;5:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cho M-S, Yang JY, Mejías JA, Kim S-C. Phylogenomic insight into dysploidy, speciation, and plastome evolution of a small Mediterranean Genus Reichardia (Cichorieae; Asteraceae). Sci Rep. 2022;12:11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim S-H, Yang JY, Cho M-S, Stuessy TF, Crawford DJ, Kim S-C. Chloroplast Genome provides insights into molecular evolution and species relationship of Fleabanes (Erigeron: Tribe Astereae, Asteraceae) in the Juan Fernández Islands, Chile. Plants. 2024;13:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Candolle AP. Genres nouveaux appartenant à La Famille Des Composées Ou Synanthérées: Première decade. In: Guillemin JBA, editor. Archives de Botanique, V2. Paris: Au Bureau des Archives; 1833. pp. 330–4.
- 68.de Candolle AP. Prodromus Systematis Naturalis Regni Vegetabilis. Volume 7. Paris: Treuttel et Würtz; 1838. [Google Scholar]
- 69.Decaisne J. Monographie des genres Balbisia et robinsonia, de la famille des Composées. Ann Sci Nat Bot Sér 2. 1834;1:16–30.
- 70.Meisner CF. Plantarum vascularium genera secundum ordines naturales digesta. Leipzig: Libraria Weidmannia; 1839. p. 136. [Google Scholar]
- 71.Hemsley WB. Report on the Botany of Juan Fernandez, the South-Eastern moluccas, and the Admiralty Islands. In: Thompson CW, Murray J, editors. Report on the scientific results of the voyage of H.M.S. Challenger during the years 1873–1876, Botany, vol. 1, part 4. London: Her Majesty’s Stationary Office; 1884. pp. 1–96. [Google Scholar]
- 72.Johow F. Estudios Sobre la flora de Las Islas De Juan Fernandez. Santiago: Gobierno de Chile; 1896. [Google Scholar]
- 73.Pacheco P, Crawford DJ, Stuessy TF, Silva MO. Flavonoid evolution in Robinsonia (Compositae) of the Juan Fernandez islands. Am J Bot. 1985;72:989–98. [Google Scholar]
- 74.Stuessy TF, Crawford DJ, Marticorena C, Rodríguez R. Island biogeography of angiosperms of the Juan Fernandez archipelago. In: Stuessy TF, Ono M, editors. Evolution and speciation of island plants. Cambridge: Cambridge University Press; 1998a. pp. 121–38. [Google Scholar]
- 75.Novoa P. Robinsonia Berteroi. IUCN Red List Threatened Species. 2020;eT158536719A158673805. 10.2305/IUCN.UK.2020-1.RLTS.T158536719A158673805.en.Accessed 29 Nov 2023 .
- 76.Crawford DJ, Stuessy TF, Cosner MB, Haines DW, Silva MO. Ribosomal and chloroplast DNA restriction site mutations and the radiation of Robinsonia (Asteraceae: Senecioneae) on the Juan Fernandez islands. Pl Syst Evol. 1993;184:233–9. [Google Scholar]
- 77.Pelser PB, Tepe EJ, Kennedy AH, Watson LE. The fate of Robinsonia (Asteraceae): sunk in Senecio, but still monophyletic. Phytotaxa. 2010;5:31–46. [Google Scholar]
- 78.Menezes APA, Resende-Moreira LC, Buzatti RSO, Nazareno AG, Carlsen M, Kalapothakis LFP, Lovato E. Chloroplast genomes of Byrsonima species (Malpighiaceae): comparative analysis and screening of high divergence sequences. Sci Rep. 2018;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Piot A, Hackel J, Christin PA, Besnard G. One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta. 2018;247:255–66. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, Wen F, Hong X, Li Z, Mi Y, Zhao B. Comparative chloroplast genome analyses of Paraboea (Gesneriaceae): insights into adaptive evolution and phylogenetic analysis. Front Pl Sci. 2022;13:1019831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cho M-S, Yang JY, Yang TJ, Kim S-C. Evolutionary comparison of the chloroplast genome in the Woody Sonchus Alliance (Asteraceae) on the Canary Islands. Genes. 2019;10:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Givnish TJ. Ecology of plant speciation. Taxon. 2010;59(5):1326–66. [Google Scholar]
- 83.König C, Weigelt P, Taylor A, Stein A, Dawson W, Essl F, Pergl J, Pyšek P, Van Kleunen M, Winter M, Chatelain C. Source pools and disharmony of the world’s island floras. Ecography. 2021;44(1):44–55. [Google Scholar]
- 84.Stuessy TF, Swenson U, Crawford DJ, Silva M. Isolating mechanisms and modes of speciation in endemic angiosperms of the Juan Fernandez islands. In: Stuessy TF, Ono M, editors. Evolution and speciation of island plants. Cambridge: Cambridge University Press; 1998c. pp. 79–96. [Google Scholar]
- 85.Crawford DJ, Stuessy TF, Takayama K, López-Sepúlveda P, Anderson G, Bernardello G. Speciation. In: Stuessy TF, Crawford DJ, López-Sepúlveda P, Baeza CM, Ruiz EA, editors. Plants of oceanic islands: evolution, biogeography, and conservation of the flora of the Juan Fernández (Robinson Crusoe) Archipelago. Cambridge: Cambridge University Press; 2018. pp. 308–31. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Supplementary Figure S1. Comparison of the plastid genomes of seven Robinsonia, six Dendrosenecio and five Senecio species, against R. berteroi by mVISTA.
Supplementary Material 2: Supplementary Figure S2. Maximum likelihood tree based on the protein-coding gene sequences of 32 representative Asteraceae species in Robinsonia, Dendrosenecio and Senecio. Trait characters are marked with brown circle representing woody stem; blue, separate nodes spacing, typically thin-walled, chlorophyllous leaf sponge mesophyll, and one secretory canal per vascular bundle; and orange, nearly contiguous nodes, thick-walled, strongly hypodermis-like mesophyll, and two secretory canals. Trait data taken from Skottsberg (1921) and Carlquist (1974) [1, 26], but not available for R. saxatilis
Supplementary Material 3: Supplementary Table S1. Relative synonymous codon usage (RSCU) of protein-coding genes in the chloroplast genomes of seven Robinsonia species
Data Availability Statement
The datasets generated and/or analyzed in the current study are available from the NCBI GenBank repository [https://www.ncbi.nlm.nih.gov/].