Abstract
Alzheimer’s disease (AD) is a multifactorial disease with both genetic and environmental factors contributing to its etiology. Previous evidence has implicated disturbed insulin signaling as a key mechanism that plays a role in both neurodegenerative diseases such as AD and comorbid somatic diseases such as diabetes mellitus type 2 (DM2). In this study, we analysed available genome-wide association studies (GWASs) of AD and somatic insulin-related diseases and conditions (SID), i.e., DM2, metabolic syndrome and obesity, to identify genes associated with both AD and SID that could increase our insights into their molecular underpinnings. We then performed functional enrichment analyses of these genes. Subsequently, using (additional) GWAS data, we conducted shared genetic etiology analyses between AD and SID, on the one hand, and blood and cerebrospinal fluid (CSF) metabolite levels on the other hand. Further, integrating all these analysis results with elaborate literature searches, we built a molecular landscape of the overlap between AD and SID. From the landscape, multiple functional themes emerged, including insulin signaling, estrogen signaling, synaptic transmission, lipid metabolism and tau signaling. We also found shared genetic etiologies between AD/SID and the blood/CSF levels of multiple metabolites, pointing towards “energy metabolism” as a key metabolic pathway that is affected in both AD and SID. Lastly, the landscape provided leads for putative novel drug targets for AD (including MARK4, TMEM219, FKBP5, NDUFS3 and IL34) that could be further developed into new AD treatments.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13195-024-01609-2.
Introduction
Alzheimer’s disease (AD) is a multifactorial, neurodegenerative disease that involves gradually progressing dementia over the course of multiple years. AD etiology is thought to be caused by a complex interplay of genetic and environmental risk factors, and it has been demonstrated that the typical AD-linked pathophysiological brain characteristics – involving amyloid beta and tau – start many years before the onset of clinical symptoms [13, 94]. AD prevalence increases with age and affects approximately 4% of the world population age 60 and over [5]. Since the size of the elderly population continues to grow across the world, AD constitutes a significant and increasing societal burden and is more prevalent in women than men [44]. Moreover, about 5% of people with AD develop symptoms at a younger age, i.e., before 65. Further, approved available pharmacotherapeutic approaches for AD include N-methyl-D-aspartate receptor antagonism and acetylcholinesterase inhibition, but the efficacy of the currently available drugs is limited and disease progression cannot be meaningfully halted or reversed yet [40, 59, 62].
Although some AD-causing mutations in a few familial risk genes (APP, PS1, PS2) that cause an early onset form of the diseases have been established [102], the vast majority of patients have a sporadic, late onset form of AD that is a polygenic disease, involving multiple common genetic factors that differ between affected and unaffected individuals [101]. Apolipoprotein E (APOE) is an important lipid carrier in the brain and common variation in the APOE gene constitutes a major risk factor for sporadic AD, in that individuals carrying the APOE4-allele have a greatly increased risk of developing AD, and APOE2-allele carriers have a decreased risk compared to individuals with the (most common) APOE3-allele [30, 2]. Furthermore, (neuro)inflammation is thought to play a role in AD [14], and variation in the major histocompatibility complex (MHC) locus – constituting a highly polymorphic genetic region that codes for the vast range of cell surface proteins involved in antigen presentation for (adaptive) immunity – has been linked to many diseases, including AD and diabetes [54, 101]. In addition, amyloid precursor protein (APP) – precursor of amyloid beta, which aggregates into toxic, extraneuronal amyloid plaques in AD – and microtubule-associated protein tau (MAPT) – which is hyperphosphorylated and accumulates into intraneuronal neurofibrillary tangles in AD – are key molecules in the pathogenesis of AD [30].
Furthermore, AD has been found to have multiple somatic comorbidities, including (peripheral) insulin-related diseases and conditions, i.e., diabetes mellitus type 2 (DM2), metabolic syndrome (MES) and obesity (OBS) [70]. In addition, it is increasingly recognized that AD also involves some form of disrupted insulin signaling in the central nervous system (CNS), more specifically brain insulin resistance [38]. While insulin signaling is most commonly known for its role in glucose homeostasis and typical somatic insulin-related diseases (SID), such as diabetes mellitus, insulin and the insulin receptor (INSR) are also expressed (and functional) in the CNS, and AD and SID may therefore share disease mechanisms [38]. In non-brain tissues such as muscle, hepatic or adipose tissue, the INSR B-isoform – that can bind both insulin and insulin-like growth factor (IGF) 1 – is expressed, whereas in neurons in the brain, the INSR A-isoform – lacking exon 11 in the hormone-binding domain and able to bind insulin and IGF2 – is expressed [28]. The INSR (A-isoform) is also commonly overexpressed in cancer [73], and IGF2 has also been linked to neurodevelopment and neurodegeneration through downstream signaling via the IGF2 receptor (IGF2R) [1]. Several studies have linked AD to insulin signaling, and some have postulated AD as a ‘type 3 diabetes’ [47]. In this regard, insulin signaling has also been demonstrated to play an important role in the formation of both amyloid plaques and neurofibrillary tangles, and APOE-E4 may adversely affect neuronal insulin signaling leading to cellular dysfunction and death [38, 105]. In the brain, insulin is also involved in regulating neuronal growth and development [33, 78]. In addition, similar to its effects on peripheral glucose, insulin stimulates neuronal glucose uptake by promoting the translocation of glucose transporters 3 and 4 (GLUT3/SLC2A3 and GLUT4/SLC2A4) to the neuronal cell membrane. Moreover, CNS insulin regulates peripheral glucose homeostasis through hypothalamic feedback on hepatic glucose release [55, 78].
Although a number of molecules and pathogenic processes implicated in the disease have been identified, a significant part of AD etiology remains unresolved. In addition, currently available treatments lack efficacy and these are not disease-modifying treatments (DMTs) [67]. Therefore, in the current study, we aimed to improve the current knowledge of the molecular mechanisms and processes involved in AD and to identify potential novel drug targets for AD that could be further developed into DMTs. To achieve these aims, we used the ‘molecular landscape’ building approach that we have used previously for other neurological and psychiatric diseases, such as amyotrophic lateral sclerosis [43], Parkinson’s disease [42], obsessive–compulsive disorder [95] and Tourette’s [100]. A unique feature of our approach is that we start in an unbiased manner, from the (gen)omics perspective. Indeed, our current disease-level ‘hypothesis’ is that there is some genetic and molecular overlap between AD and the above-mentioned SID, which is in part based on our own findings that there are local [24] rather than global [23] genetic correlations between AD and SID. To this end, we integrated the latest available genome-wide association studies (GWASs) of AD as well as D2M, MES and OBS, and compiled a list of genes relevant for both AD and SID, based on gene-based analyses, functional annotation and expression quantitative trait loci (eQTL) mapping. Then, we performed functional enrichment analyses on these AD/SID-related genes to identify potentially enriched functional themes, pathways and upstream regulator molecules. Furthermore, for unbiased screening purposes and using additional GWAS data, we performed shared genetic etiology analyses to investigate the presence, extent and direction of genetic sharing between AD/SID on the one hand and 237 blood and 338 cerebrospinal fluid (CSF) metabolite levels on the other hand. Subsequently, integrating the genes and themes/pathways/upstream regulators from our genomic data analyses with an elaborate literature search, we built a molecular landscape of the overlap between AD and SID. Lastly, this landscape provides (novel) insights into the molecular mechanisms underlying AD etiology, as well as ‘leads’ for drug targets that may be further developed into DMTs for AD.
Methods
GWAS data of Alzheimer’s disease (AD) and somatic insulin-related diseases (SID)
We collected summary statistics of the largest and latest available genome-wide association studies (GWASs) of AD [8, 101] and three major SID, i.e., type 2 diabetes mellitus (DM2) [54], metabolic syndrome (MES) [50] and obesity (OBS) [98]. Sample sizes were 86,531 (proxy) cases and 676,386 (proxy) controls for AD, 74,124 cases and 824,006 controls for DM2, 59,677 cases and 231,430 controls for MES, and 9,805 cases and 235,085 controls for OBS. Further details are available in the (supplementary information of the) respective GWASs of AD/SID. The GWAS summary statistics were filtered based on allele frequencies (between 1 and 99%), imputation quality (INFO > = 0.8), sample size (> = 67% for cases and controls), type of genetic variation (keeping only bi-allelic single nucleotide polymorphisms (SNPs)) and strand-ambiguity (keeping only non-ambiguous SNPs).
Primary analyses of AD and SID GWAS data
Gene-based analyses in MAGMA
First, we used the AD and SID GWAS data to perform gene-based analyses in MAGMA [48] (version 1.09), and this to investigate aggregated genetic association signals at the gene-level. For each of the 4 GWASs of AD, DM2, MES and OBS, we calculated gene-level statistics (using the aggregate of SNP-wise mean and top model) for all protein-coding genes. The 1000 Genomes [4] (phase 3, European samples) reference panel was used to obtain information about linkage disequilibrium (LD). Then, we performed cross-trait association analyses on the gene-level statistics of AD with the 3 SID (i.e., AD with DM2, AD with MES, and AD with OBS), similar to previous studies [41, 61]. In short, we used the gene-based meta-analysis, as implemented in MAGMA, to aggregate the AD and SID gene-level statistics [41, 61]. We considered a gene as relevant for both AD and SID if it showed: a cross-trait gene-level P-value < 2.50E-06 (i.e., ‘gene-wide’ significance), a cross-trait gene-level P-value that is at least an order of magnitude smaller than its single disorder P-value, and a single disorder P-value in both AD and SID < 0.05 [41, 61]. The primary MAGMA analyses were run by aggregating the SNP variation within the gene boundaries (i.e., without an annotation window around the gene), because we found that applying an annotation window resulted in less accurate cross-trait results for certain genetic loci with extensive linkage disequilibrium (LD) structures and strong association signals in the GWAS (e.g., the APOE- or MHC-locus). The genes that were found to be relevant for both AD and at least one SID were included in the list of input genes for the molecular landscape. Nonetheless, auxiliary MAGMA analyses (normal gene-level and cross-trait), using a 100kb up- and downstream annotation window around each gene were performed, as SNPs outside the gene boundaries may affect (the expression of) the gene [93]. However, the genes from these analyses were used for additional functional gene enrichment analyses (see below) only.
Functional annotation and gene mapping (FUMA)
In addition to MAGMA, we performed functional annotation and gene mapping in FUMA [97] on the 4 GWASs of AD, DM2, MES and OBS (using the online interface at https://fuma.ctglab.nl/, last accessed on April 1st 2024). First, we identified genomic risk loci using FUMA default parameters for identifying statistical significance (i.e., SNP P-value of genetic association cut-off at 5.00E-08) and independency between SNPs (i.e., an r2 threshold of 0.6 for first identifying ‘independent’ significant SNPs, and a second r2 threshold of 0.1 for ‘lead SNPs’ that represent each cluster of significant SNPs). Genomic risk loci were identified based on 250kb LD blocks. Information about LD was obtained from the default 1000 Genomes [4] (phase 3, European samples) reference panel.
Functional annotation of SNPs within each genomic risk locus was performed using the available ANNOVAR (http://annovar.openbioinformatics.org/en/latest/), CADD (http://cadd.gs.washington.edu/) and RegulomeDB (http://regulomedb.org/index) databases, as well as chromatin states. For the subsequent gene mapping, we filtered SNPs based on CADD (> 12.37, to include more deleterious SNPs) and RegulomeDB score (< = 7, to include SNPs that are potentially regulating gene function). Gene mapping was performed in FUMA based on genomic position, expression quantitative trait locus (eQTL) and 3D chromatin interaction (3CI) information. For eQTL and 3CI mapping, we used only the available brain tissue databases to investigate whether significant SNPs for AD and SID are specifically regulating gene expression in the brain. In this way, the mapped eQTL/3CI genes are brain-expressed genes that are potentially affected by the SNPs from the AD/SID GWASs.
We used the FUMA default FDR cut-offs of 0.05 for eQTL and 1.00E-06 for 3CI mapping. Brain databases used for eQTL mapping were: eQTL Catalogue (BrainSeq DLPFC, Schwartzentruber 2018 sensory neuron; https://www.ebi.ac.uk/eqtl/), PsychENCODE (PFC, TC, CB; http://resource.psychencode.org/), xQTL (DLPFC; http://mostafavilab.stat.ubc.ca/xqtl/), CommonMind Consortium (CMC; https://www.synapse.org//#!Synapse:syn5585484), BRAINEAC (10 regions; http://www.braineac.org/), and GTEx ver. 8 (13 regions; http://www.gtexportal.org/home/). For 3CI mapping, we used the following brain databases: dorsolateral prefrontal cortex and hippocampus, adult and fetal cortex, PsychENCODE (prefrontal cortex; http://resource.psychencode.org/) and FANTOM5 (http://fantom.gsc.riken.jp/5/). We required SNPs to be overlapping with the one end of a chromatin interaction loop, while the other end of the loop had to be overlapping with transcription start sites. In addition, SNPs had to be overlapping with enhancer regions and gene transcription start sites had to overlap with promoter regions, using the Roadmap Consortium database (E053 to E082 brain datasets; http://egg2.wustl.edu/roadmap/web_portal/DNase_reg.html), thereby effectively only considering enhancer-promoter interactions. The genes that were mapped for both AD and SID (i.e., AD on the one hand and DM2, MES and/or OBS on the other hand) were included as input genes for the molecular landscape.
Functional enrichment analyses of AD/SID-related genes
Using the list of input genes for the molecular landscape that we derived from the MAGMA and FUMA analyses, we performed functional enrichment analyses with Ingenuity Pathway Analysis [46] (IPA,QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). We used the total set of all MAGMA and FUMA AD/SID-relevant landscape input genes, as well as the subset of MAGMA genes that overlapped between all 3 AD/SID cross-trait analyses (see above) as input datasets for IPA. Finally, we also investigated the AD/SID-relevant genes, that were obtained through the auxiliary MAGMA analyses (applying a 100kb annotation window, see above), with IPA (again, using all genes, and the ones overlapping between all 3 AD/SID cross-trait analyses). We used IPA to obtain information about enriched molecular pathways (metabolic and cell signaling cascades), upstream regulator molecules (regulating the expression of the input genes), interacting protein networks (networks of proteins interacting with the input proteins to identify network hubs and connectivity patterns of biological relevance) and ‘diseases and cellular functions’ (expected causal effects of genes/proteins on the disease/function). Because oncology research is overrepresented in the literature, and our primary interest in building our landscape is in non-oncological diseases (AD/SID), we do not mention the enriched annotations for any type of cancer-related diseases or functions in the results.
IPA performs enrichment analyses is based on the ‘Ingenuity Knowledge Base’, a large, curated database that itself is based on existing literature (https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). We applied the IPA default Fisher’s exact test (right-tailed) with subsequent Benjamini Hochberg (BH) correction (BH cut-off 0.05) to identify significant enrichment signals.
Genetic sharing analyses for AD/SID-related genes and blood and CSF metabolite levels
After the MAGMA and FUMA analyses, we collected summary statistics data from GWASs of an extensive range of blood [69] and CSF [66] metabolite levels. Sample sizes ranged from 703 to 1802 for the blood metabolite GWASs and the sample size was (up to) 291 for the GWAS of CSF metabolites (all continuous traits). All metabolite GWAS summary statistics were filtered similar to the AD/SID GWAS data (i.e., regarding: allele frequency (between 5 and 95%), sample size, imputation quality, type of variation, and strand ambiguity). Further details are available in the (supplementary information of the) respective GWASs.
Polygenic score (PGS)-based analyses
To investigate the presence, extent and direction of genetic sharing between AD/SID and blood/CSF metabolite levels, we performed polygenic score (PGS)-based analyses using PRSice [22] (version 1.25, optimized for summary statistics based analyses, which uses the R package ‘GTX’ (https://github.com/tobyjohnson/gtx)) using AD/SID and the metabolite GWAS data (237 blood and 338 CSF metabolites). Because of sample overlap between the largest available GWASs for AD, DM2, and the blood metabolite levels GWASs, we used the publicly available FINNGEN (R10; data and info available at https://www.finngen.fi/en) GWAS data of AD (10,520 cases and 402,662 controls) and DM2 (65,085 cases and 335,112 controls) as ‘base’ phenotype for the PGS-based analyses with the blood metabolite levels as ‘target’ phenotypes. The 1000 Genomes [4] (phase 3, European samples) reference panel was used to obtain information about LD. LD was addressed by clumping SNPs within a 500kb window with an R2 threshold of 0.25. PGSs were estimated based on 7 broad GWAS P-value thresholds for SNP inclusion into the score (SNP P ≤ 0.001, 0.05, 0.1, 0.2, 0.3, 0.4 and 0.5). A PGS represents a weighted sum of the SNPs’ effects of the ‘base’ phenotype, which is then regressed onto the second or ‘target’ phenotype. Bonferroni correction was applied to identify statistically significant results for blood (Bonferroni P-value cut-off 0.05 / (237 metabolites * 7 P-thresholds) = 3.01E-05) and CSF (Bonferroni P-value cut-off 0.05 / (338 metabolites * 7 P-thresholds) = 2.11E-05) metabolic traits.
Molecular landscape of the overlap between AD and SID
We built the molecular landscape using an approach that we have applied multiple times previously [42, 43, 95, 100]. Firstly, we filtered the list of putative input genes for the molecular landscape (as obtained from the MAGMA and FUMA analyses, see above) for protein-coding genes that are expressed in the brain (using the Human Protein Atlas [89] (https://www.proteinatlas.org/), Uniprot Knowledge Base [87] (https://www.uniprot.org/) and existing literature through searching PubMed (https://pubmed.ncbi.nlm.nih.gov/)). Then, we used the Uniprot Knowledge Base [87] and existing literature to obtain functional and structural information about each of the proteins encoded by the (filtered) input genes. The subcellular localization of proteins was investigated through the use of multiple sources, including the Uniprot Knowledge Base [87], Ingenuity Pathway Analysis [46] (IPA,QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis), the Human Protein Atlas [89] and through manually reviewing the literature. We investigated protein–protein interactions between all proteins encoded by the input genes using existing literature, the Ingenuity Knowledge Base, and multiple protein interaction databases including BioGRID (https://thebiogrid.org/), STRING (https://string-db.org/) and IntAct (https://www.ebi.ac.uk/intact/home) to identify high-quality interactions based on experimental evidence (i.e., no predicted interactions). Together, these databases capture a comprehensive range of protein–protein interactions, both in number and type of interactions (i.e., physical binding and all other functional relationships, such as activation, inhibition, degradation, expression regulation, etc.) across the protein interactome. For each protein–protein interaction, the nature (e.g. activation through phosphorylation) as well as the plausibility (based on physical proximity, such as the same subcellular compartment) was determined. All filtered and curated interactions between the proteins encoded by the input genes were included in the molecular landscape. We also added a number of enriched interacting proteins or upstream regulators from the functional enrichment analyses to the landscape, given their role and likely biological significance in relation to the input genes/proteins for the landscape. Furthermore, blood or cerebrospinal fluid metabolites that emerged from the genetic sharing analyses were added to the landscape. The protein-metabolite interactions were investigated and curated/filtered by using the same approach and databases as used for protein–protein interactions. The landscape was drawn using Serif DrawPlus software (version X8). For visual convenience, we avoided crossing arrows as much as possible, and tried to restrict redundancy in drawing proteins and their interactions. Color and symbolic coding were used for proteins and protein interactions and these are described in detail in the Results section and the corresponding landscape figure.
Expression analyses of landscape genes in AD brains
Subsequently and to corroborate the built landscape, we used the results from a previously published study [82] – based on publicly available RNA sequencing data from the AMP-AD knowledge portal on the Synapse platform (project SynID: syn2580853) – to investigate differential gene expression in 7 brain regions from AD patients compared to healthy controls. For all genes encoding proteins in the landscape, we investigated whether they are significantly differentially expressed (FDR P < 0.05, regardless of fold change) in one or more of the 7 brain regions from AD patients. The (differential) expression of these genes was also considered when assessing the different aspects of specificity for identifying potentially novel drug targets from the molecular landscape (see below).
Identification of putative novel drug targets
From the molecular landscape of the overlap between AD and SID, we identified a number of putative (novel) drug targets for AD. This was not an analysis based on a single tool or bioinformatics pipeline, but an assessment based on extensive literature searches for all landscape proteins for different aspects of drug target specificity, complemented by the gene expression analyses mentioned above. In this regard, an ideal drug target for AD should adhere to five broad aspects of target specificity. Firstly, the target should have ‘regional specificity’, in that it should be expressed in the brain and be affected/functional in relevant cell types (such as neurons, astrocytes and microglial cells), and preferably also be differentially expressed in AD compared to healthy controls. For the latter, we assessed the differential expression of all landscape proteins in the brain of AD patients versus controls, as described above. Secondly, an excellent AD target needs to show ‘temporal specificity’, i.e., it is linked in time with AD onset and/or its expression pattern changes with AD symptoms over time. A third important aspect is ‘symptomatic specificity’: the target should be linked to one or more disease symptoms or signs. Given the role of insulin signaling in both AD and SID, we considered both cognitive and neurological symptoms associated with AD and insulin resistance, together with related endocrine/metabolic traits, as disease symptoms/signs for assessing the symptomatic specificity of potential drug targets. Fourthly, a good AD drug target should have ‘molecular specificity’, i.e., it needs to play an important role in disease etiology, as is reflected by its involvement in multiple protein interactions and functional themes in the molecular landscape of AD-SID overlap. Finally and importantly, an ideal target should have ‘modulatory specificity’: it should be possible to modulate the putative drug target to modify – in a beneficial way – the progression of AD and reduce AD symptoms. In our consideration of novel targets, we did not consider some ‘well-known’ landscape proteins – such as tau (MAPT) and APOE – which have already been extensively studied.
Results
Input genes for the landscape resulting from primary analyses of AD and SID GWAS data
Based on the MAGMA analyses of the latest available GWAS summary statistics for AD and SID, a total of 211 genes passed inclusion criteria from the AD x DM2, AD x MES and AD x OBS cross-trait analyses and were included as input genes for the landscape. A total of 15 genes emerged from all three cross-trait analyses (Supplementary Table 1). The auxiliary MAGMA analyses (using a 100kb up- and downstream annotation window around each gene) resulted in 492 genes from the AD x DM2, AD x MES and AD x OBS cross-trait analyses, of which 42 genes emerged from all three cross-trait analyses (Supplementary Table 1).
The functional gene mapping with FUMA resulted in 135 mapped genes for AD and 904 mapped genes for SID (661 for DM2, 185 for MES, and 14 for OBS). In total, 19 genes overlapped between the FUMA analyses for AD and (at least one) SID, and these were also included as input genes for the landscape (Supplementary Table 2).
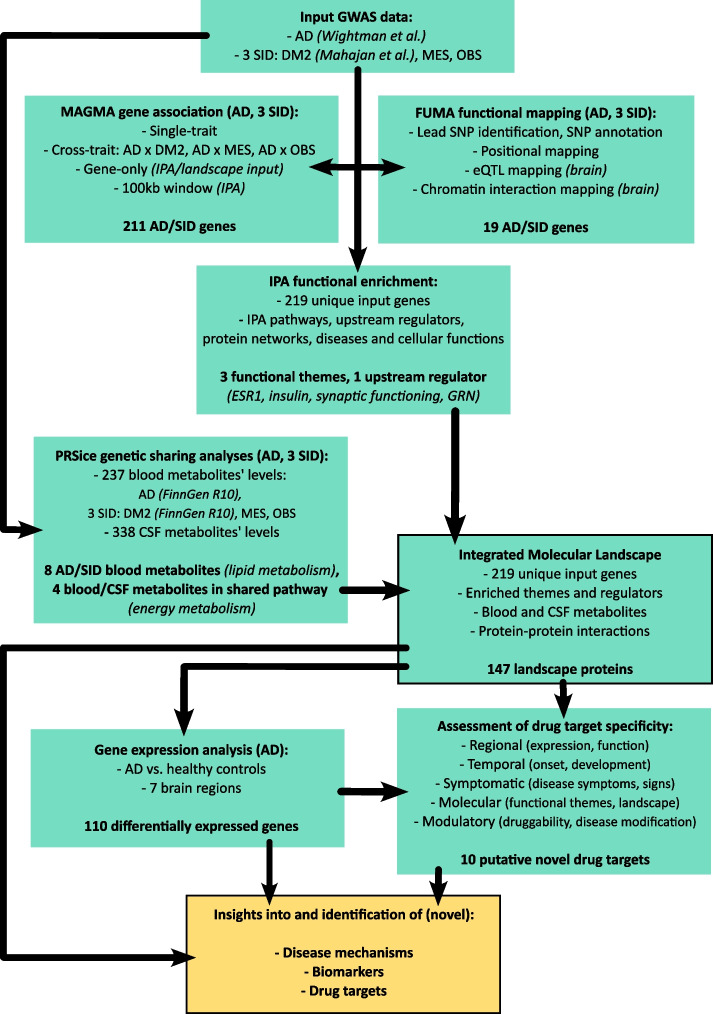
Combining the 211 MAGMA-derived genes and 19 FUMA-derived genes resulted in 219 unique input genes for the landscape (Supplementary Table 3). In Fig. 1 we have provided an overview of the different analyses performed and their interrelationships.
Fig. 1.
Overview of the different analyses performed and their interrelationships
Functional enrichment analyses of AD/SID-related genes
We used IPA to perform functional enrichment analyses of the 219 input genes for the molecular landscape. In addition, we performed enrichment analyses of two subsets of input genes, namely, the 15 MAGMA genes that overlapped in the cross-trait analyses and the 19 overlapping genes obtained from the FUMA analyses. In addition, we screened the 492 genes from the auxiliary MAGMA cross-trait analyses with a 100kb annotation window, and the corresponding subset of 42 genes that overlapped between these cross-trait analyses. Supplementary Table 4 provides an overview of the results from all IPA analyses.
Within the set of all 219 input genes (see Supplementary Table 4), we found 5 enriched ‘canonical’ pathways related to immune signaling (e.g., ‘Antigen Presentation Pathway’). We found 9 enriched upstream regulators including the frontotemporal dementia-linked gene Progranulin (GRN – 8 target molecules in the set). A total of 12 protein interaction networks were enriched, with central molecules including Beta Estradiol (network #6) and APP (network #12). A large number of diseases and cellular functions were enriched, including ‘Abnormal morphology of synapse’, ‘Synaptic transmission of synapse’ and ‘Morphology of neurons’.
In the set of 15 input genes that emerged from all three MAGMA analyses (see Supplementary Table 4), we found no enriched pathways. We found 9 enriched upstream regulators, but – except for one – they all had only one target molecule in the set. Two networks were enriched with central molecules JUN and ESR1. After merging these two connected networks, ESR1 remained the central molecule of the merged network (Supplementary Fig. 1). A number of diseases and cellular functions were enriched, including ‘Synaptic transmission’.
In the set of 19 input genes that emerged from the FUMA analyses (see Supplementary Table 4), we found no enriched pathways. We found one enriched upstream regulator. Two networks were enriched, with central molecules APOE and APP. After merging these two connected networks, APP was the central molecule of the merged network (Supplementary Fig. 2). A number of diseases and cellular functions were enriched, including ‘Abnormal morphology of synapse’ and ‘Insulin resistance’.
In the set of 492 genes that emerged from the auxiliary MAGMA analyses (see Supplementary Table 4), we found 18 enriched pathways, mostly related to immune signaling (e.g. ‘Antigen Presentation Pathway’ and ‘Neuroinflammation Signaling Pathway’). We found GRN as the only enriched upstream regulator (10 target molecules in the set), and GRN itself was also part of the set. A total of 25 networks were enriched, with central molecules including BAG6 (network #5), SREBF1 (network #13), ESR1 (network #15) and APP (network #16). A large number of diseases and cellular functions were enriched, including ‘Development of neurons’ and ‘Abnormal morphology of neurons’.
In the set of 42 genes emerging from all three auxiliary MAGMA analyses (see Supplementary Table 4), we found one enriched pathway, ‘Neuroprotective Role of THOP1 in Alzheimer's Disease’. We found no enriched upstream regulators. Three networks were enriched, with central molecules Insulin (network #1 – Supplementary Fig. 3), TNF and MAPK3. A number of diseases and cellular functions were enriched including ‘Abnormal morphology of synapse’, ‘Synaptic transmission’ and ‘Development of neurons’.
In summary, enriched major functional themes that emerged from the IPA analyses were estrogen (ESR1) signaling, insulin signaling and synaptic functioning. GRN was identified as an enriched upstream regulator of interest.
Genetic sharing analyses for AD/SID-related genes and blood and CSF metabolite levels
After Bonferroni correction, we observed genetic sharing between AD and the levels of 10 blood metabolites (R2 of AD genetic risk explaining metabolic traits of up to 1.19%). Regarding SID, we found genetic sharing between DM2, MES and OBS, and the levels of 37, 66 and 16 blood metabolites, respectively (R2 of SID genetic risk explaining metabolic traits of up to 8.75%). We observed genetic sharing between AD and the levels of 11 CSF metabolites (R2 of AD genetic risk explaining metabolic traits of up to 14.26%). In the case of SID, we found genetic sharing between DM2, MES and OBS, and the levels of 18, 7 and 3 CSF metabolites, respectively (R2 of SID genetic risk explaining metabolic traits of up to 14.37%). See Supplementary Table 5 for all results.
From the aforementioned findings, the levels of 8 blood metabolites shared genetics with both AD and SID: DAG 34:2, LPE 18:0, TAG 50:4, TAG 52:4, TAG 52:5, TAG 54:5, TAG 54:6 and TAG 56:9. These are all lipids, mostly triacylglycerols (triglycerides, TAGs), one diacylglycerol (diglyceride, DAGs) and one lysophospholipid (LPE). For all these 8 lipid levels, the direction of genetic sharing was positive with AD. For 3 of these lipid levels (TAG 50:4, TAG 52:5 and TAG 54:6), the direction of genetic sharing was positive with DM2. For 7 of these lipid levels (all aforementioned except LPE 18:0), the direction of genetic sharing was positive with MES. For 2 of these lipid levels (LPE 18:0 and TAG 56:9), the direction of genetic sharing was negative with OBS. Supplementary Figs. 4–1 to 4–20 provide bar plots of the presence, extent and direction of genetic sharing for different PGS P-value thresholds for the 8 aforementioned blood metabolic traits (with AD and the SID mentioned). In addition to these 8 lipids, we observed other TAGs, DAGs, LPEs and other lipid species that also share genetics with AD and SID.
Regarding CSF metabolic traits, there were no overlapping metabolites between AD and SID, but also here we observed some lipid species (glycerophosphatidylcholines, GPCs; and fatty acids) that shared genetics with both AD and SID. A few CSF metabolic traits (pyruvate and isocitrate levels), together with a blood metabolic trait (fumarate, aggregated with maleate and α-ketoisovalerate levels) could be mapped to a common metabolic pathway, namely the Krebs cycle. The direction of genetic sharing was negative between AD and CSF pyruvate levels, and between MES and isocitrate levels. The direction of genetic sharing was positive between MES and blood fumarate, maleate and α-ketoisovalerate levels. Supplementary Figs. 5–1- to 5–3 provide bar plots of the presence, extent and direction of genetic sharing for different PGS P-value thresholds for the aforementioned 3 CSF and blood metabolic traits (with AD and the SID mentioned).
Taken together, our metabolome-wide genetic sharing analyses with AD/SID revealed lipid metabolism and energy metabolism (Krebs cycle) as potentially shared metabolic functional themes.
Molecular landscape of the genetic overlap between AD and SID
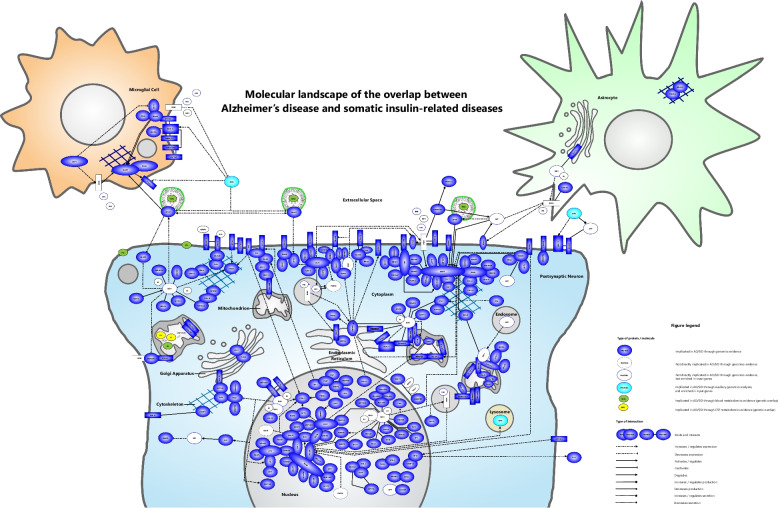
The landscape was built based on the literature-derived interactions between the proteins encoded by the 219 input genes shared by AD and at least one of the SID investigated. Subsequently, we added a number of blood and cerebrospinal fluid metabolites emerging from the shared genetic etiologies between AD and SID. In addition, the enriched functional themes estrogen (ESR1) signaling, insulin signaling, synaptic functioning and the upstream regulator GRN were integrated into the landscape. Given its important role in AD and its emergence in some of the enriched interacting protein networks, we also included APP and its interactions with other landscape proteins in the landscape. Furthermore, synaptic transmission was enriched in the landscape input genes/proteins and synaptic loss is a hallmark of AD that correlates with cognitive decline. Therefore and as most of the landscape proteins were located in the postsynaptic density/membrane, the landscape was mainly placed in a dendritic spine of a postsynaptic neuron. In addition, a microglial cell and astrocyte were added since certain landscape proteins are more highly expressed by these cell-types than by neurons.
The molecular landscape is shown in Fig. 2. In total, 147 of the proteins encoded by the 219 input genes (67%) could be placed into the landscape. In the Supplementary Information, we provide extensive detail about how the landscape was built and about all the protein–protein interactions in the landscape – organized by cellular compartment – and the blood and CSF metabolites that emerged from the shared genetic etiology analyses.
Fig. 2.
Molecular landscape of the overlap between Alzheimer’s disease (AD) and somatic insulin-related diseases (SID)
Differentially expressed landscape genes in AD-related brain regions (AD patients versus controls)
To corroborate our landscape, we investigated for all genes encoding proteins in the landscape whether they are significantly differentially expressed in one or more of 7 AD-related brain regions, comparing AD patients to control individuals [82]. In Supplementary Table 6, all landscape genes are listed, with their respective fold changes (at FDR P < 0.05) in one or more AD brain regions. In total, 110 of the landscape genes were also found to be differentially expressed in at least one AD-related brain region.
Identification of putative novel drug targets for AD
After building the molecular landscape, we considered the five broad aspects of drug target specificity that are described in the Methods to identify putative novel drug targets for AD among the landscape proteins. We identified 10 such putative targets and in Supplementary Table 7, we have indicated how each target adheres to the five aspects of specificity. In the Discussion, we will elaborate on the five most fitting examples of landscape proteins that we consider to be potentially interesting novel drug targets for AD: MARK4, TMEM219, FKBP5, NDUFS3 and IL34.
Discussion
We built a molecular landscape of the overlap between AD and SID by analyzing and integrating the available GWAS data for these diseases and assessing the functions and interactions of the proteins encoded by the genes found to be relevant for both AD and SID. In addition, we investigated genetic sharing of AD/SID with the levels of 237 blood and 338 CSF metabolites. In total, 147 of the proteins encoded by the 219 input genes (67%) could be placed into the landscape. A number of functional themes play a role in the molecular landscape of the overlap between AD and SID. First, we found enrichment of estrogen (ESR1) signaling, insulin signaling and synaptic functioning in the input genes/proteins for the landscape. Furthermore, from the genetic sharing analyses, we observed energy metabolism (oxidative phosphorylation and mitochondrial functioning), lipid metabolism and tau signaling as possibly shared metabolic pathways. From the proteins interact in the molecular landscape, we further discuss five potential (novel) drug targets for AD, some of which could also be of relevance for SID given their molecular overlap with AD.
Estrogen receptor 1 (ESR1) signaling was identified as a major functional theme in the AD/SID landscape. ESR1 is a nuclear hormone receptor and transcription factor regulating many genes in different target tissues, including the insulin receptor [104]. Furthermore, estradiol (E2) treatment increases insulin sensitivity through FOXO1 inhibition (via the PI3K/AKT pathway) [103]. Estradiol and other estrogens binds to the estrogen receptors ESR1 and ESR2, which are mainly located in the nucleus and cytoplasm, after which the ESRs can dimerize [9, 26]. In the nucleus, ESR1 and ESR2 can bind to estrogen response elements at the promoter region of their target genes and exert different effects on transcription. In addition to directly binding to target DNA, there are also indirect transcriptional effects of ESR1 and ESR2 [9, 26]. With regard to the brain, estrogen signaling is important for normal brain functioning and reduced estrogen signaling has been thought to be involved in the etiology of AD [92]. Similarly, we observed reduced expression of ESR1 in AD versus controls for two AD-related brain areas (temporal cortex and parahippocampal gyrus). Furthermore, estrogen signaling regulates the expression of glucose transporter type 4 (GLUT4) [29] and possibly also of GLUT3 [18]. Important landscape proteins that are regulated by ESR1 include CKB (involved in energy metabolism), MAPT, the MHC class 1 complex, PHB1 (involved in mitochondrial functioning and transcription) and RTN2. In addition, ESR1 binds and interacts with other mitochondrial landscape proteins (such as MTCH2, TOMM22 and TOMM40), and the important lipogenic transcription factor SREBF1. Therefore, ESR1 also connects different functional themes in the landscape, including estrogen signaling, insulin signaling, lipid metabolism, tau signaling and mitochondrial functioning/energy metabolism.
Another important functional landscape theme is insulin signaling. Brain insulin resistance has been thought to be a key mechanism in AD [38, 47]. Insulin binds to the insulin receptor (INSR) and the insulin-INSR complex translocates from the cell membrane to the cytoplasm and nucleus. It is then either recycled back to the plasma membrane or sent to lysosomes for degradation [28, 39]. Insulin increases glucose uptake by muscle and adipose tissue through the insertion of GLUT4 in the cell membrane [27]. Furthermore, insulin inhibits hepatic gluconeogenesis and glycogenolysis, affects fatty acid and protein synthesis, and has effects on growth and development [7, 99]. As can also be seen from the landscape, insulin signaling is linked to both APP and MAPT, and, indeed, has been implicated in the formation of both amyloid plaques and neurofibrillary tangles, the two key pathological characteristics of AD [38]. Furthermore, brain insulin signaling plays roles in multiple processes including growth and maturation of neurons, synaptic functioning, and regulating overall glucose homeostasis through hypothalamic feedback on hepatic glucose release [10, 28],Martina, Ribeiro and Antunes, 2018; [78]. Modulation of synaptic plasticity by insulin may be related to its effects on dopamine, norepinephrine and serotonin signaling [10, 28]. Furthermore, while GLUT4 plays a key role in peripheral insulin signaling, in the brain, GLUT1 and GLUT3 play major roles in glucose uptake across the blood–brain-barrier and by neurons and glia cells [32, 45, 56]. Although GLUT1 and -3 have been considered as insulin-insensitive, evidence suggests that there might be (indirect) mechanisms through which insulin signaling may affect GLUT1/3-mediated glucose uptake in the brain [57].
In the landscape, insulin signaling plays an important role. The landscape protein CELF1, strongly associated with both AD and SID in the latest available GWASs, is a splicing regulator that causes skipping of exon 11 of the INSR, resulting in the neuronal, A-isoform [72, 75]. On the contrary, peripheral INSRs are mainly the B-isoform, which includes exon 11 [28, 56]. The INSR A-isoform can bind both insulin and IGF2, and also plays a role in fetal growth [75, 99] and cancer [73]. The INSR regulates the expression of numerous important landscape proteins, including SREBF1 [85], PEMT [51] (converts phosphatidylethanolamine to phosphatidylcholine, of which blood/CSF-levels shows genetic sharing with AD/SID), the transcription factor POU5F1 (OCT4)(Kee Keong [37]) and RTN2 [11] (regulated by ESR1 and regulates APP, GLUT4 and MAPT [74]). In addition, GRN – an important upstream regulator in the landscape with downstream targets APOC1, APOE, APP and the MHC (class 1 and 2) complex and known for its link with frontotemporal dementia [80] – is also regulated by the INSR. Therefore, in summary, insulin signaling plays a central role and links different themes in the molecular landscape, including estrogen signaling, lipid metabolism, synaptic functioning and tau signaling.
In addition to estrogen and insulin signaling, energy metabolism emerged as a functional theme from the landscape and genetic sharing analyses. In the mitochondria, ATP – as a cellular energy source – is generated through oxidative phosphorylation. To this end, the mitochondrial electron transport chain consists of a series of protein complexes that are located in the mitochondrial membrane. Electrons are passing through this chain of protein complexes through a number of redox reactions. This electron transport creates a proton gradient across the mitochondrial membrane, which subsequently drives the synthesis of ATP from ADP by the final complex of the chain (ATP synthase). The Krebs, tricarboxylic acid or citric acid cycle is a critical step in cellular respiration, by providing the electron carriers NADH and FADH2 to the electron transport chain. NADH and FADH2 are converted back to their oxidized forms, NAD + and FAD—by the electron transport chain—that are then used again in the Krebs cycle and in glycolysis [25, 64]. Interestingly, in the genetic sharing analyses, we found that genetic variants/SNPs that increase AD risk also contribute to lower CSF levels of pyruvate, whereas SNPs that increase MES risk also contribute to lower CSF-levels of isocitrate and increased blood levels of fumarate. These metabolites can be mapped to the Krebs cycle as a potentially shared metabolic pathway between AD and SID (MES). In addition, a number of mitochondrial proteins from the landscape – such as CKB, DNAJC11, LACTB, NDUFS2, NDUFS3, PHB1, TOMM22 and TOMM40 – are involved in energy metabolism or other mitochondrial functions such as membrane organization, mitochondrial dynamics and protein import. Further, different interactions of CELF1, ESR1, MAPT and SREBF1 with (the aforementioned) mitochondrial landscape proteins point to the link of mitochondrial energy production and functioning with other functional themes in the landscape, including insulin signaling, estrogen signaling, lipid metabolism and tau signaling.
We found that there is genetic sharing between AD as well as SID and the blood levels of eight specific lipids (mainly triglycerides (TAGs), a diglyceride (DAG 34:2) and a lysophospholipid (LPE 18:0)). The direction of genetic sharing for AD and SID (DM2 and MES) was positive with regard to these lipid species, whereas for OBS the direction of genetic sharing was negative. In addition, we identified genetic sharing between AD/SID and the blood levels of several other TAGs, DAGs and some lysophospholipids. DAGs may play a role in (peripheral) insulin resistance through disrupted fatty acid processing and consequent activation of certain protein kinase C isoforms [21, 71]. TAGs are strongly related to and serve as biomarkers for insulin resistance, both in peripheral tissues and in the brain [6, 86]. In addition, there is genetic sharing between AD/SID and the blood and CSF levels of different other lipid species such as lysophosholipids and phosphatidylcholines (PCs). PCs are important for normal lipid metabolism [49] and increased breakdown of PCs has been linked to AD [12]. Thus, lipid metabolism emerges as an important functional theme from the molecular landscape, and is closely connected with (brain) insulin signaling. In addition, the expression of genes related to cholesterol synthesis has been linked to tau pathology [91]. Lastly, as can also be seen in the landscape, the lipid-transport protein encoded by APOE, and in particular by the AD risk factor APOE4, increases the phosphorylation of MAPT and is (consequently) involved in tau hyperphosphorylation [31, 88]. Importantly, APOE4 has also been shown to impair brain insulin signaling through trapping of the INSR in endosomes, which, in turn adversely affects insulin-dependent energy metabolism in the mitochondria [106]. These effects can be accelerated through increased dietary intake of fatty acids [106].
Lastly, tau (or MAPT) is thought to play a key role in AD. Hyperphosphorylation of tau decreases its binding to microtubules and its aggregation results in neurofibrillary tangles inside affected neurons in AD and other ‘tauopathies’ [15, 35]. Tau signaling also emerges as an important functional theme in the landscape. Specifically, tau has many interactions with other landscape proteins such as APOE(4), CRHR1, FKBP5, MARK4 and NDUFS3, and it is connected to insulin signaling, estrogen signaling and APP. Moreover, knockout of the neuronal INSR increases tau phosphorylation, thereby (indeed) pointing to a role for brain insulin resistance in AD [74]. Therefore, Tau signaling plays an important role in the overlap between AD and SID, both through its link with insulin signaling and other functional themes in the landscape. In addition, tau interacts with a number of potential novel drug targets for AD (see below).
Putative (novel) drug targets emerging from the molecular landscape of the overlap between AD and SID
MAP/microtubule affinity-regulating kinase 4 (MARK4, localized to the cytoplasm) is a serine/threonine-protein kinase that phosphorylates MAPT [87]. MARK4 shows regional specificity for AD, as it is differentially expressed (downregulated) in the hippocampus and parahippocampal gyrus from AD patients versus controls (Supplementary Table 6). Furthermore, MARK4 is associated with early tau phosphorylation in AD granulovacuolar degeneration bodies [52], pointing to its temporal specificity for AD. Dl-3-n-butylphthalide inhibits MARK4 and reduces cognitive deficits, synaptic loss and tau phosphorylation (in tau transgenic mice) [17]. Inactivation of MARK4 leads to increased insulin sensitivity (in mice) [84]. This shows that MARK4 also has symptomatic specificity, in that it is involved in different clinical signs/symptoms of AD (and SID). In the molecular landscape of AD-SID overlap, MARK4 interacts with multiple other important landscape proteins, including tau, constituting its molecular specificity. Importantly, inhibition of MARK4 could be beneficial for AD and SID through possible effects on tau phosphorylation, neuroinflammation, and insulin resistance [76, 77, 84], reflecting its modulatory specificity. MARK4 is also inhibited by acetylcholinesterase (AChE) inhibitors (e.g. donepezil, rivastigmine) – that are among the only approved drugs for AD [67] – antidiabetics (e.g. metformin, linagliptin), and a number of other compounds (including serotonin, irisin). Taken together, as it adheres to all five aspects of target specificity, we would submit that MARK4 would be an excellent drug target for AD.
Another putative drug target from the landscape of AD-SID overlap is insulin-like growth factor-binding protein 3 receptor (IGFBP3R, other name:TMEM219), a cell membrane receptor specific for IGFBP3. TMEM219 is highly expressed in the brain, including in the hippocampus [89]. In addition, binding of IGFBP3 to TMEM219 leads to pancreatic beta cell loss and dysfunction [20]. These findings show the regional specificity of TMEM219 for the brain and pancreas. In addition, decreased blood levels of the TMEM219-ligand IGFBP3 are linked with increased age and decreased cognitive skills in AD patients [34], and higher Aβ42 CSF levels (a robust biomarker of AD) [36], constituting the temporal and symptomatic specificity of TMEM219 in AD. Nevertheless, the decrease in IGFBP3 blood levels with age may be a confounding factor here [34] and increased IGFBP3 blood levels have also been linked to AD [36]. Furthermore, TMEM219 interacts with multiple other (important) landscape proteins such as the MHC class II complex and the acetylcholine receptor, suggesting its molecular specificity for AD. Moreover, inhibition of TMEM219/IGFBP3 signaling has been suggested to be beneficial for both AD and SID (DM), by decreasing the loss/dysfunction of TMEM219-expressing cells affected in these diseases [63]. However, TMEM219 has been found to be suppressed in different types of cancers, which may be a contra-indication for TMEM219-antagonism as a treatment for AD [63]. Summarizing, TMEM219 could be a potential drug target for AD that warrants further investigation.
Further, peptidyl-prolyl cis–trans isomerase (FKBP5, localized to the cytoplasm) is a peptidyl prolyl isomerase chaperone that is involved in multiple functions, including protein folding, activation and degradation [87]. FKBP5 has regional specificity for AD, since it is differentially expressed (upregulated) in AD cases versus controls in the hippocampus (Supplementary Table 6). Furthermore, FKBP5 is a biomarker of metabolic dysfunction [81] and is linked with Aβ-induced memory impairment [3] and insulin resistance [83], pointing to its temporal and symptomatic specificity for AD and SID. In the molecular landscape, FKBP5 interacts with many other (important) landscape proteins, such as tau and AKT, reflecting its molecular specificity. In addition, Apelin-13 inhibits FKBP5 and protects against Aβ-induced memory impairment [3], while increased expression of FKBP5 is linked to insulin resistance [79]. This suggests that inhibition of FKBP5 could be beneficial for AD and SID, constituting its modulatory specificity. In this respect, existing FKBP5-inhibitors include Rapamycin, Apelin-13 and selective serotonin reuptake inhibitors class antidepressants (e.g. fluoxetine, citalopram, sertraline). To summarize, FKBP5 is a potential novel drug target for AD – with already existing modulating compounds – that could be further studied/developed.
NADH dehydrogenase (ubiquinone) iron-sulfur protein 3 (NDUFS3, localized to the mitochondrion), is a core subunit of mitochondrial electron transport chain Complex 1 (NADH dehydrogenase) [87]. NDUFS3 expression is downregulated in the hippocampus and parahippocampal gyrus from AD patients versus controls (Supplementary Table 6), pointing to its regional specificity. In addition, plasma neuroexosomal NDUFS3 is increased at the early clinical stages of AD in people with SID (DM2) and may serve as an early prognostic and diagnostic biomarker of AD (onset) [19], thereby constituting its temporal specificity. Furthermore, inhibition of Complex 1 improves cognitive function, synaptic plasticity, phosphorylated tau (p-tau) and Aβ levels, neuroinflammation, insulin resistance, and neurodegeneration [90], reflecting the symptomatic specificity of NDUFS3/Complex 1 and suggesting that inhibition of Complex 1 could be beneficial for AD. Furthermore, NDUFS3 is regulated by tau and interacts with other mitochondrial landscape protein (such as prohibitin), pointing to its molecular specificity in AD/SID. Moreover, different Complex 1 inhibitors already exist, and especially a small molecule tricyclic pyrone compound (CP2) appears to be promising. CP2 showed good pharmacological properties, low toxicity and good efficacy in animal studies, including improvement in brain and peripheral energy homeostasis (including insulin resistance), synaptic activity and long-term potentiation, dendritic spine maturation, cognitive function, as well as reducing p-tau and Aβ levels and brain and peripheral inflammation, reflecting the modulatory specificity for NDUFS3/Complex 1 [90]. Therefore, we consider NDUFS3 (and mitochondrial Complex 1) as a putative novel drug target for AD that should be further investigated.
Lastly, interleukin-34 (IL34, localized extracellular) is a pro-inflammatory cytokine that promotes proliferation and differentiation of monocytes and macrophages [87]. IL34 expression is downregulated in the hippocampus from AD patients versus controls (Supplementary Table 6), constituting its regional specificity. Furthermore, by promoting microglial proliferation and thereby possibly neurodegeneration [65], IL34 may contribute to AD disease progression over time, and it has been associated with cognitive decline in vascular dementia [96]. In addition, IL34 impairs the ability of macrophages to ‘clear’ pathological amyloid beta [107] and plasma IL34 levels correlate positively with insulin resistance [60]. All these findings point to the temporal and symptomatic specificity of IL34 in AD. Further, IL34 interacts with one other landscape protein (HEY2) and is involved in neuroprotective signaling in neurodegeneration [53], constituting its molecular specificity. As for its modulatory specificity, inhibition of IL34 reduces microglial proliferation (and hence conveys a protective effect in AD) [65], although at the same time, IL34 was found to enhance the neuroprotective effects of microglia to attenuate amyloid beta neurotoxicity [58]. Therefore, with regard to AD, both IL34 inhibition and activation could have beneficial effects, so maintaining an optimal IL34 level may be recommended. With regard to SID (DM), it appears that IL34 inhibition may be beneficial, given the positive association of IL34 levels with insulin resistance, obesity [16], and beta cell dysfunction and apoptosis [68]. Summarizing, IL34 may be a putative novel drug target for AD, but this needs to be further investigated.
Strengths and limitations
In the current study, we explored the hypothesis that there is molecular overlap between AD and SID through altered insulin signaling from an unbiased perspective. By analyzing and integrating data from the largest and latest GWASs of AD and SID, complemented with metabolome-wide GWAS data and an extensive literature search on interactions between the AD/SID-relevant proteins and metabolites, we built a molecular landscape of the AD-SID overlap. This molecular landscape provides insights into the shared molecular processes underlying AD and SID, and, importantly, it allows for the identification of biologically meaningful and potentially novel drug target (and biomarker) candidates that could be further investigated and developed in future studies. A limitation of the genome-wide screen for genetic sharing between AD/SID and blood and CSF metabolite levels is that we considered only the genome-wide component of genetic sharing in the PGS-based-analyses. More specifically, we considered the aggregated SNP effects across the genome rather than genetic signals originating from specific loci. In addition, while the sample sizes for the GWAS data of AD/SID were sufficiently large, the sample sizes of some of the metabolite GWASs – in particular the ones from cerebrospinal fluid – were (relatively) small. Therefore, it may have been the most sound approach to study genome-wide shared genetic effects, rather than locus-specific effects. Furthermore, the accessed literature and databases may overrepresent certain diseases or topics (e.g., oncology) and their investigated substrates and pathways, and are (unavoidably) still incomplete with regard to protein–protein and protein-metabolite interactions. Therefore, future studies regarding the protein interactome in general and with regard to the described functional themes and proposed drug target leads for AD are needed.
Conclusions
In conclusion, we have analyzed the latest available GWAS data for AD and SID, as well as an extensive range of blood and CSF metabolites, to identify genes/proteins relevant to both AD and SID, and to investigate genetic overlap between AD/SID and blood/CSF metabolite levels. We investigated functional enrichment in the identified genes/proteins and performed an extensive literature search on the interactions between the proteins encoded by the genes relevant to AD/SID, and the metabolites of which the blood/CSF levels show genetic overlap with AD/SID. We integrated all data and results into a molecular landscape of the overlap between AD and SID, and we identified a number of functional biological themes including estrogen signaling, insulin signaling, synaptic functioning, energy metabolism and tau signaling. Based on the landscape, we identified 5 interesting leads for potential (novel) drug targets for AD. That being said, future in-depth studies are necessary to further investigate the identified molecular themes. In addition, in silico, in vitro, in vivo and ultimately human clinical studies are needed to (possibly) ‘develop’ the proposed drug target leads into new drugs to treat AD.
Supplementary Information
Acknowledgements
This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative (Grant no. EINF1824). Part of the results in this study are based on data obtained from the AMP-AD Knowledge Portal (10.7303/syn2580853). The data available in the AD Knowledge Portal would not be possible without the participation of research volunteers and the contribution of data by collaborating researchers. Study data were provided by the following sources: The Mayo Clinic Alzheimers Disease Genetic Studies, led by Dr. Nilufer Taner and Dr. Steven G. Younkin, Mayo Clinic, Jacksonville, FL using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimers Disease Research Center, and the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216, R01 AG003949, National Institute of Neurological Disorders and Stroke (NINDS) grant R01 NS080820, CurePSP Foundation, and support from Mayo Foundation.
Authors’ contributions
Conceptualization, I.H.R. and G.P.; Methodology, I.H.R., J.W., W.D.W., and G.P.; Investigation, I.H.R., J.W., W.D.W., and G.P.; Data curation, I.H.R.; Writing—original draft manuscript preparation, I.H.R. and G.P.; Writing – review and editing, I.H.R., J.W., W.D.W., N.R.M., G.F., V.V.G., W.J.J., S.J.B.V., A.F., C.B., S.B., K.A.A., A.M., J.H., A.O.L., D.S., M.S., J.G., J.K.B., J.B., B.F. and G.P.; Visualization, I.H.R. and G.P; All authors have read, contributed and agreed to the published version of the manuscript.
Funding
This study received funding from the European Community’s Horizon 2020 Programme (H2020/2014 – 2020) under grant agreement no. 847879 (PRIME). In addition, S.J.B.V. receives research support from EPND. This project has received funding from the IMI 2 Joint Undertaking (JU) under grant agreement no. 101034344. The IMI JU receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA. B.F.’s contribution was supported by funding from the European Community’s Horizon 2020 Programme (H2020/2014 – 2020) under grant agreement no. 847879 (PRIME) and from the PRISM2 project (www.prism-project.eu; grant agreement no. 101034377) through the Innovative Medicines Initiative 2 Joint Undertaking. She also received relevant funding from the Dutch Ministry of Education, Culture and Science of the government of The Netherlands for the NWO Gravitation programme GUTS (grant 024.005.011). Research reported in this publication was also supported by the National Institute Of Mental Health of the National Institutes of Health under Award Number R01MH124851 and the Research Council of Norway (Project no. 331725). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding organizations.
Data availability
All key data that support the findings from this study are available within the manuscript or supplementary information and materials.
Declarations
Ethics approval and consent to participate
Not applicable (ethical approval and consent to participate was obtained by each individual study, of which the published results and/or publicly available data were used as input for the current study).
Consent for publication
Not applicable.
Competing interests
G.P. is director and I.H.R., J.W. and W.D.W. are employees of Drug Target ID, Ltd., but their activities at this company do not constitute competing interests with regard to this paper. J.K.B. has been in the past 3 years a consultant to / member of advisory board of / and/or speaker for Takeda, Roche, Medice, Angelini, Neuraxpharm, and Servier, all unrelated to this manuscript. He is not an employee of any of these companies, and not a stock shareholder of any of these companies. He has no other financial or material support, including expert testimony, patents, royalties. B.F. has received educational speaking fees from Medice GmbH. N.R.M., G.F., V.V.G., W.J.J., S.J.B.V., A.F., C.B., S.B., K.A.A., A.M., J.H., A.O.L., D.S., M.S., J.G. and J.B. do not report any conflicts of interest. In addition, the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alberini CM. IGF2 in memory, neurodevelopmental disorders, and neurodegenerative diseases. Trends Neurosci. 2023;46:488–502. Available at: 10.1016/j.tins.2023.03.007. Elsevier Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer’s association. 2021 Alzheimer’s disease facts and figures. Alzheimer’s Dementia. 2021;17(3):327–406. Available at: 10.1002/alz.12328. [DOI] [PubMed] [Google Scholar]
- 3.Aminyavari S, et al. Anxiolytic impact of Apelin-13 in a rat model of Alzheimer’s disease: Involvement of glucocorticoid receptor and FKBP5. Peptides. 2019;118:170102. Available at: 10.1016/j.peptides.2019.170102. [DOI] [PubMed] [Google Scholar]
- 4.Auton, A. et al. (2015) ‘A global reference for human genetic variation’, Nature. Nature Publishing Group, pp. 68–74. Available at: 10.1038/nature15393. [DOI] [PMC free article] [PubMed]
- 5.Ballard C, et al. Alzheimer’s disease. Lancet. 2011;377(9770):1019–31. Available at: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 6.Banks WA, et al. Triglycerides cross the blood-brain barrier and induce central leptin and insulin receptor resistance. Int J Obes. 2018;42(3):391–7. Available at: 10.1038/ijo.2017.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedinger D, Adams S. Metabolic, anabolic, and mitogenic insulin responses: A tissue-specific perspective for insulin receptor activators. Mol Cell Endocrinol. 2015;415:143–56. [DOI] [PubMed] [Google Scholar]
- 8.Bellenguez C, et al. ‘New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Gen. 2022;54(4):412–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–42. [DOI] [PubMed] [Google Scholar]
- 10.Blázquez, E. et al. (2014) ‘Insulin in the Brain: Its Pathophysiological Implications for States Related with Central Insulin Resistance, Type 2 Diabetes and Alzheimer’s Disease’. Front Endocrinol. 5(OCT). Available at: 10.3389/FENDO.2014.00161 [DOI] [PMC free article] [PubMed]
- 11.Bluher M, et al. Intrinsic heterogeneity in adipose tissue of fat-specific insulin receptor knock-out mice is associated with differences in patterns of gene expression. J Biol Chem. 2004;279(30):31891–901. [DOI] [PubMed] [Google Scholar]
- 12.Blusztajn JK, Slack BE. Accelerated Breakdown of Phosphatidylcholine and Phosphatidylethanolamine Is a Predominant Brain Metabolic Defect in Alzheimer’s Disease. J Alzheimer’s Dis: JAD. 2023;93(4):1285–9. Available at: 10.3233/JAD-230061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braak H, et al. Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J Neuropathol Exp Neurol. 2011;70(11):960–9. 10.1097/NEN.0b013e318232a379. [DOI] [PubMed]
- 14.Bradburn, S., Murgatroyd, C. and Ray, N. (2019) ‘Neuroinflammation in mild cognitive impairment and Alzheimer’s disease: A meta-analysis’. Ageing Res Rev. Elsevier Ireland Ltd, pp. 1–8. Available at: 10.1016/j.arr.2019.01.002. [DOI] [PubMed]
- 15.Butner KA, Kirschner MW. Tau protein binds to microtubules through a flexible array of distributed weak sites. J Cell Biol. 1991;115(3):717–30. Available at: 10.1083/JCB.115.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang, E.J. et al. (2014) ‘IL-34 is associated with obesity, chronic inflammation, and insulin resistance’, J Clin Endocrinol Metab. 99(7). Available at: 10.1210/jc.2013-4409. [DOI] [PubMed]
- 17.Chang, Y. et al. (2021) ‘Dl-3-n-Butylphthalide Reduces Cognitive Deficits and Alleviates Neuropathology in P301S Tau Transgenic Mice’, Front Neurosci. 15. Available at: 10.3389/fnins.2021.620176 [DOI] [PMC free article] [PubMed]
- 18.Cheng C, et al. Estrogen augments glucose transporter and IGF1 expression in primate cerebral cortex. FASEB J. 2001;15(6):907–15. [DOI] [PubMed] [Google Scholar]
- 19.Chi H, et al. Blood Neuroexosomal Mitochondrial Proteins Predict Alzheimer Disease in Diabetes. Diabetes. 2022;71(6):1313–23. Available at: 10.2337/DB21-0969. [DOI] [PubMed] [Google Scholar]
- 20.D’Addio, F. et al. (2022) ‘The IGFBP3/TMEM219 pathway regulates beta cell homeostasis’. Nat Commun. 13(1). Available at: 10.1038/s41467-022-28360-2 [DOI] [PMC free article] [PubMed]
- 21.Erion DM, Shulman GI. Diacylglycerol-mediated insulin resistance. Nat Med. 2010;16(4):400–2. Available at: 10.1038/NM0410-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466–8. Available at: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fanelli G, et al. Insulinopathies of the brain? Genetic overlap between somatic insulin-related and neuropsychiatric disorders. Transl Psychiatry. 2022;12(1):59. Available at: 10.1038/s41398-022-01817-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fanelli, G. et al. (2024) ‘Local patterns of genetic sharing challenge the boundaries between neuropsychiatric and insulin resistance-related conditions.’, MedRxiv preprint [Preprint]. Available at: 10.1101/2024.03.07.24303921.
- 25.Fernie AR, Carrari F, Sweetlove LJ. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opinion Plant Biol. 2004;7(3):254–61. Available at: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Fuentes N, Silveyra P. Estrogen receptor signaling mechanisms. Adv Protein Chem Struct Biol. 2019;116:135–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Funaki M, Randhawa P, Janmey P. Separation of insulin signaling into distinct GLUT4 translocation and activation steps. Mol Cell Biol. 2004;24(17):7567–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gralle M. The neuronal insulin receptor in its environment. J Neurochem. 2017;140(3):359–67. [DOI] [PubMed] [Google Scholar]
- 29.Gregorio K, Laurindo C, Machado U. Estrogen and Glycemic Homeostasis: The Fundamental Role of Nuclear Estrogen Receptors ESR1/ESR2 in Glucose Transporter GLUT4 Regulation. Cells. 2021;10(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hane, F.T., Lee, B.Y. and Leonenko, Z. (2017) ‘Recent Progress in Alzheimer’s Disease Research, Part 1: Pathology’. J Alzheimer’s Dis. IOS Press, pp. 1–28. Available at: 10.3233/JAD-160882. [DOI] [PubMed]
- 31.Harris F, et al. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: modulation by zinc. J Biol Chem. 2004;279(43):44795–801. [DOI] [PubMed] [Google Scholar]
- 32.He C, et al. Recurrent moderate hypoglycemia accelerates the progression of Alzheimer’s disease through impairment of the TRPC6/GLUT3 pathway. JCI Insight. 2022;7(5):e154595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Heide, L.P., Ramakers, G.M.J. and Smidt, M.P. (2006) ‘Insulin signaling in the central nervous system: Learning to survive’, Progress Neurobiol. pp. 205–221. Available at: 10.1016/j.pneurobio.2006.06.003. [DOI] [PubMed]
- 34.Hu X, Yang Y, Gong D. Circulating insulin-like growth factor 1 and insulin-like growth factor binding protein-3 level in Alzheimer’s disease: a meta-analysis. Neurol Sci. 2016;37(10):1671–7. Available at: 10.1007/s10072-016-2655-1. [DOI] [PubMed] [Google Scholar]
- 35.Hwang, K. et al. (2022) ‘Tauopathy and Epilepsy Comorbidities and Underlying Mechanisms’, Front Aging Neurosci. 14. Available at: 10.3389/FNAGI.2022.903973 [DOI] [PMC free article] [PubMed]
- 36.Johansson P, et al. Serum but not cerebrospinal fluid levels of insulin-like growth factor-I (IGF-I) and IGF-binding protein-3 (IGFBP-3) are increased in alzheimer’s disease. Psychoneuroendocrinol. 2013;38(9):1729–37. Available at: 10.1016/j.psyneuen.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Kee Keong Teo, A. et al. (2021) ‘Defective insulin receptor signaling in hPSCs skews pluripotency and negatively perturbs neural differentiation’, J Biol Chem, Jan-Jun(296), p. 100495. [DOI] [PMC free article] [PubMed]
- 38.Kellar, D. and Craft, S. (2020) ‘Brain insulin resistance in Alzheimer’s disease and related disorders: mechanisms and therapeutic approaches’. Lancet Neurol. Lancet Publishing Group, pp. 758–766. Available at: 10.1016/S1474-4422(20)30231-3. [DOI] [PMC free article] [PubMed]
- 39.Kesten D, et al. (2018) ‘Insulin-induced translocation of IR to the nucleus in insulin responsive cells requires a nuclear translocation sequence.’ Biohim Biophys Acta Mol Cell Res. 1865;4:551–9. [DOI] [PubMed] [Google Scholar]
- 40.Kishi, T. et al. (2017) ‘Memantine for Alzheimer’s Disease: An Updated Systematic Review and Meta-analysis’, J Alzheimer’s Dis. IOS Press, pp. 401–425. Available at: 10.3233/JAD-170424. [DOI] [PubMed]
- 41.Klein M, et al. Genetic markers of ADHD-related variations in intracranial volume. Am J Psychiatry. 2019;176(3):228–38. Available at: 10.1176/appi.ajp.2018.18020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klemann, C.J.H.M. et al. (2017) ‘Integrated molecular landscape of Parkinson’s disease’, npj Parkinson’s Dis. 3(1). Available at: 10.1038/s41531-017-0015-3 [DOI] [PMC free article] [PubMed]
- 43.Klemann CJHM, et al. Integrated molecular landscape of amyotrophic lateral sclerosis provides insights into disease etiology. Brain Pathol. 2018;28(2):203–11. Available at: 10.1111/bpa.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knopman, D.S. et al. (2021) ‘Alzheimer disease’. Nat Rev Dis Primers, 7(1). Available at: 10.1038/s41572-021-00269-y [DOI] [PMC free article] [PubMed]
- 45.Koepsell H. Glucose transporters in brain in health and disease. Pflugers Arch. 2020;472(9):1299–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krämer A, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics (Oxford, England). 2014;30(4):523–30. Available at: 10.1093/BIOINFORMATICS/BTT703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de la Monte SM. The Full Spectrum of Alzheimer’s Disease Is Rooted in Metabolic Derangements That Drive Type 3 Diabetes. Adv Experiment Med Biol. 2019;1128:45–83. Available at: 10.1007/978-981-13-3540-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Leeuw CA, et al. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput Biol. 2015;11(4):1–19. Available at: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Vance DE. Phosphatidylcholine and choline homeostasis. J Lipid Res. 2008;49(6):1187–94. Available at: 10.1194/JLR.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- 50.Lind L. Genome-Wide Association Study of the Metabolic Syndrome in UK Biobank. Metab Syndrome Related Disord. 2019;17(10):505–11. Available at: 10.1089/met.2019.0070. [DOI] [PubMed] [Google Scholar]
- 51.Ling A, et al. FoxO1 Is Required for Most of the Metabolic and Hormonal Perturbations Produced by Hepatic Insulin Receptor Deletion in Male Mice. Endocrinology. 2018;159(3):1253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lund H, et al. MARK4 and MARK3 associate with early tau phosphorylation in Alzheimer’s disease granulovacuolar degeneration bodies. Acta Neuropathol Commun. 2014;2:22. 10.1186/2051-5960-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo J, et al. Colony-stimulating factor 1 receptor (CSF1R) signaling in injured neurons facilitates protection and survival. J Experiment Med. 2013;210(1):157–72. Available at: 10.1084/jem.20120412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–13. Available at: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martina I, Ribeiro R, Antunes VR. The role of insulin at brain-liver axis in the control of glucose production. Am J Physiol Gastrointest Liver Physiol. 2018;315:538–43. Available at: 10.1152/ajpgi.00290.2017.-Glucose. [DOI] [PubMed] [Google Scholar]
- 56.McNay E, Pearson-Leary J. GluT4: A central player in hippocampal memory and brain insulin resistance. Exp Neurol. 2020;323:113076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Milstein, J.L. and Ferris, H.A. (2021) ‘The brain as an insulin-sensitive metabolic organ’, Mole Metab. Elsevier GmbH. Available at: 10.1016/j.molmet.2021.101234 [DOI] [PMC free article] [PubMed]
- 58.Mizuno T, et al. Interleukin-34 selectively enhances the neuroprotective effects of microglia to attenuate oligomeric amyloid-β neurotoxicity. Am J Pathol. 2011;179(4):2016–27. Available at: 10.1016/j.ajpath.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moss, D.E. (2020) ‘Improving anti-neurodegenerative benefits of acetylcholinesterase inhibitors in alzheimer’s disease: Are irreversible inhibitors the future?’. Int J Mole Sci. MDPI AG. Available at: 10.3390/ijms21103438 [DOI] [PMC free article] [PubMed]
- 60.Mostafa TM, El-Gharbawy NM, Werida RH. Circulating IRAPe, Irisin, and IL-34 in Relation to Insulin Resistance in Patients With Type 2 Diabetes. Clin Ther. 2021;43(7):e230–40. Available at: 10.1016/j.clinthera.2021.05.003. [DOI] [PubMed] [Google Scholar]
- 61.Mota NR, et al. Cross-disorder genetic analyses implicate dopaminergic signaling as a biological link between Attention-Deficit/Hyperactivity Disorder and obesity measures. Neuropsychopharmacol. 2020;45(7):1188–95. Available at: 10.1038/s41386-019-0592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moyaert P, et al. Effect of Acetylcholinesterase Inhibitors on Cerebral Perfusion and Cognition: A Systematic Review. J Alzheimer’s Dis. 2023;93(4):1211–21. Available at: 10.3233/jad-221125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naseri N, Mirian M, Mofid MR. Expression of recombinant insulin-like growth factor-binding protein-3 receptor in mammalian cell line and prokaryotic (Escherichia coli) expression systems. Adv Biomed Res. 2022;11(1):19. Available at: 10.4103/abr.abr_197_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nolfi-Donegan, D., Braganza, A. and Shiva, S. (2020) ‘Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement’, Redox Biol. 37. Available at: 10.1016/J.REDOX.2020.101674 [DOI] [PMC free article] [PubMed]
- 65.Obst, J. et al. (2020) ‘Inhibition of IL-34 Unveils Tissue-Selectivity and Is Sufficient to Reduce Microglial Proliferation in a Model of Chronic Neurodegeneration’, Front Immunol. 11. Available at: 10.3389/fimmu.2020.579000 [DOI] [PMC free article] [PubMed]
- 66.Panyard DJ, et al. Cerebrospinal fluid metabolomics identifies 19 brain-related phenotype associations. Commun Biol. 2021;4(1):63. Available at: 10.1038/s42003-020-01583-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Passeri, E. et al. (2022) ‘Alzheimer’s Disease: Treatment Strategies and Their Limitations’. Int J Mole Sci. MDPI. Available at: 10.3390/ijms232213954 [DOI] [PMC free article] [PubMed]
- 68.Piao C, et al. IL-34 causes inflammation and beta cell apoptosis and dysfunction in gestational diabetes mellitus. Endocr Connect. 2019;8(11):1503–12. Available at: 10.1530/EC-19-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rhee EP, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18(1):130–43. Available at: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ribe, E.M. and Lovestone, S. (2016) ‘Insulin signalling in Alzheimer′s disease and diabetes: from epidemiology to molecular links’. J Intern Med. Blackwell Publishing Ltd, pp. 430–442. Available at: 10.1111/joim.12534. [DOI] [PubMed]
- 71.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet (London, England). 2010;375(9733):2267–77. Available at: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Savkur R, Philips A, Cooper T. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29(1):40–7. [DOI] [PubMed] [Google Scholar]
- 73.Scalia P, Giordano A, Williams SJ. The IGF-II–insulin receptor isoform-a autocrine signal in cancer: Actionable perspectives. Cancers. 2020;12(2):366. Available at: 10.3390/cancers12020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schubert M, et al. Role for neuronal insulin resistance in neurodegenerative diseases. Proc Natl Acad Sci. 2004;101(9):3100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sen S, Talukdar I, Webster N. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol Cell Biol. 2009;29(3):871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shamsi A, et al. MARK4 inhibited by AChE inhibitors, donepezil and rivastigmine tartrate: Insights into alzheimer’s disease therapy. Biomolecules. 2020;10(5):789. Available at: 10.3390/biom10050789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shamsi A, et al. Inhibition of MARK4 by serotonin as an attractive therapeutic approach to combat Alzheimer’s disease and neuroinflammation. RSC Med Chem. 2022;13(6):737–45. Available at: 10.1039/d2md00053a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shaughness, M. et al. (2020) ‘Role of Insulin in Neurotrauma and Neurodegeneration: A Review’. Front Neurosci. Frontiers Media S.A. Available at: 10.3389/fnins.2020.547175. [DOI] [PMC free article] [PubMed]
- 79.Sidibeh CO, et al. FKBP5 expression in human adipose tissue: potential role in glucose and lipid metabolism, adipogenesis and type 2 diabetes. Endocrine. 2018;62(1):116–28. Available at: 10.1007/s12020-018-1674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simon, M.J. et al. (2023) ‘Lysosomal functions of progranulin and implications for treatment of frontotemporal dementia’, Trends Cell Biol. Elsevier Ltd, pp. 324–339. Available at: 10.1016/j.tcb.2022.09.006. [DOI] [PubMed]
- 81.Smedlund, K.B., Sanchez, E.R. and Hinds, T.D. (2021) ‘FKBP51 and the molecular chaperoning of metabolism’. Trends Endocrinol Metab. Elsevier Inc., pp. 862–874. Available at: 10.1016/j.tem.2021.08.003. [DOI] [PMC free article] [PubMed]
- 82.Soheili-Nezhad S, et al. Long genes are more frequently affected by somatic mutations and show reduced expression in Alzheimer’s disease: Implications for disease etiology. Alzheimer’s Dementia. 2021;17(3):489–99. Available at: 10.1002/alz.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strączkowski M, et al. Relation of adipose tissue and skeletal muscle FKBP5 expression with insulin sensitivity and the regulation of FKBP5 by insulin and free fatty acids. Endocrine. 2022;76(3):536–42. Available at: 10.1007/s12020-022-03018-7. [DOI] [PubMed] [Google Scholar]
- 84.Sun C, et al. Inactivation of MARK4, an AMP-activated protein kinase (AMPK)-related kinase, leads to insulin Hypersensitivity and resistance to diet-induced obesity. J Biol Chem. 2012;287(45):38305–15. Available at: 10.1074/jbc.M112.388934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun X, et al. Insulin Dissociates the Effects of Liver X Receptor on Lipogenesis, Endoplasmic Reticulum Stress, and Inflammation. J Biol Chem. 2016;291(3):1115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tahapary, D.L. et al. (2022) ‘Challenges in the diagnosis of insulin resistance: Focusing on the role of HOMA-IR and Tryglyceride/glucose index’. Diabetes Metab Syndrome: Clin Res Rev. Elsevier Ltd. Available at: 10.1016/j.dsx.2022.102581. [DOI] [PubMed]
- 87.The UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023;51(D1):D523–31. Available at: 10.1093/NAR/GKAC1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Theendakara V, et al. Neuroprotective Sirtuin ratio reversed by ApoE4. Proc Natl Acad Sci U S A. 2013;110(45):18303–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thul PJ, et al. A subcellular map of the human proteome. Science (New York, NY). 2017;356(6340):eaal3321. Available at: 10.1126/SCIENCE.AAL3321. [DOI] [PubMed] [Google Scholar]
- 90.Trushina, E., Trushin, S. and Hasan, M.F. (2022) ‘Mitochondrial complex I as a therapeutic target for Alzheimer’s disease’. Acta Pharma Sinica B. Chinese Acad Med Sci. pp. 483–495. Available at: 10.1016/j.apsb.2021.11.003. [DOI] [PMC free article] [PubMed]
- 91.Tsumagari, K. et al. (2022) ‘Co-expression network analysis of human tau-transgenic mice reveals protein modules associated with tau-induced pathologies’. iScience. 25(8). Available at: 10.1016/J.ISCI.2022.104832 [DOI] [PMC free article] [PubMed]
- 92.Uddin, M.S. et al. (2020) ‘Estrogen Signaling in Alzheimer’s Disease: Molecular Insights and Therapeutic Targets for Alzheimer’s Dementia’. Mole Neurobiol. Springer, pp. 2654–2670. Available at: 10.1007/s12035-020-01911-8. [DOI] [PubMed]
- 93.Veyrieras, J.B. et al. (2008) ‘High-resolution mapping of expression-QTLs yields insight into human gene regulation’, PLoS Genet. 4(10). Available at: 10.1371/JOURNAL.PGEN.1000214 [DOI] [PMC free article] [PubMed]
- 94.Villemagne VL, et al. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: A prospective cohort study. Lancet Neurol. 2013;12(4):357–67. Available at: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- 95.van de Vondervoort I, et al. An integrated molecular landscape implicates the regulation of dendritic spine formation through insulin-related signalling in obsessive–compulsive disorder. J Psychiatr Neurosci. 2016;41(4):280–5. Available at: 10.1503/jpn.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang, Y. et al. (2021) ‘Decreased Levels of Serum IL-34 Associated with Cognitive Impairment in Vascular Dementia’. BioMed Res Int. 2021. Available at: 10.1155/2021/6793860 [DOI] [PMC free article] [PubMed]
- 97.Watanabe, K. et al. (2017) ‘Functional mapping and annotation of genetic associations with FUMA’. Nat Commun. 8(1). Available at: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed]
- 98.Watanabe K, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51(9):1339–48. Available at: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 99.White V, et al. IGF2 stimulates fetal growth in a sex- and organ-dependent manner. Pediatr Res. 2018;83(1–1):183–9. [DOI] [PubMed] [Google Scholar]
- 100.Widomska, J. et al. (2023) ‘Molecular Landscape of Tourette’s Disorder’. Int J Mole Sci. 24(2). Available at: 10.3390/ijms24021428 [DOI] [PMC free article] [PubMed]
- 101.Wightman DP, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat Genet. 2021;53(9):1276–82. Available at: 10.1038/s41588-021-00921-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu, L. et al. (2012) ‘Early-onset familial alzheimer’s disease (EOFAD)’. Canadian J Neurol Sci. pp. 436–445. Available at: 10.1017/S0317167100013949. [DOI] [PubMed]
- 103.Yan H, et al. Estrogen improves insulin sensitivity and suppresses gluconeogenesis via the transcription factor Foxo1. Diabetes. 2019;68(2):291–304. Available at: 10.2337/db18-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yasrebi A, et al. Activation of Estrogen Response Element-Independent ERα Signaling Protects Female Mice From Diet-Induced Obesity. Endocrinology. 2017;158(2):319–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao N, et al. Apolipoprotein E4 Impairs Neuronal Insulin Signaling by Trapping Insulin Receptor in the Endosomes. Neuron. 2017a;96(1):115-129.e5. Available at: 10.1016/J.NEURON.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao N, et al. Apolipoprotein E4 Impairs Neuronal Insulin Signaling by Trapping Insulin Receptor in the Endosomes. Neuron. 2017b;96(1):115-129.e5. Available at: 10.1016/j.neuron.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zuroff LR, et al. Effects of IL-34 on Macrophage Immunological Profile in Response to Alzheimer’s-Related Aβ42 Assemblies. Front Immunol. 2020;11:1449. Available at: 10.3389/fimmu.2020.01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All key data that support the findings from this study are available within the manuscript or supplementary information and materials.