Abstract
Background
Nicotinamide and tryptophan metabolism play important roles in regulating tumor synthesis metabolism and signal transduction functions. However, their comprehensive impact on the prognosis and the tumor immune microenvironment of glioma is still unclear. The purpose of this study was to investigate the association of nicotinamide and tryptophan metabolism with prognosis and immune status of gliomas and to develop relevant models for predicting prognosis and sensitivity to immunotherapy in gliomas.
Methods
Bulk and single-cell transcriptome data from TCGA, CGGA and GSE159416 were obtained for this study. Gliomas were classified based on nicotinamide and tryptophan metabolism, and PPI network associated with differentially expressed genes was established. The core genes were identified and the risk model was established by machine learning techniques, including univariate Cox regression and LASSO regression. Then the risk model was validated with data from the CGGA. Finally, the effects of genes in the risk model on the biological behavior of gliomas were verified by in vitro experiments.
Results
The high nicotinamide and tryptophan metabolism is associated with poor prognosis and high levels of immune cell infiltration in glioma. Seven of the core genes related to nicotinamide and tryptophan metabolism were used to construct a risk model, and the model has good predictive ability for prognosis, immune microenvironment, and response to immune checkpoint therapy of glioma. We also confirmed that high expression of TGFBI can lead to an increased level of migration, invasion, and EMT of glioma cells, and the aforementioned effect of TGFBI can be reduced by FAK inhibitor PF-573,228.
Conclusions
Our study evaluated the effects of nicotinamide and tryptophan metabolism on the prognosis and tumor immune microenvironment of glioma, which can help predict the prognosis and sensitivity to immunotherapy of glioma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12883-024-03924-5.
Keywords: Glioma, Nicotinamide metabolism, Tryptophan metabolism, Prognosis, Immunotherapy sensitivity, Core genes
Introduction
Glioma is the most prevalent primary malignant tumor that originates from neuroglia cells in the central nervous system. Its malignant characteristics, such as extensive drug resistance, rapid postoperative recurrence, and extensive infiltration of surrounding tissues, contribute to its status as the most malignant tumor with the worst prognosis in the central nervous system [1]. The main treatment methods for glioblastoma currently include surgical resection of the primary lesion, combined treatment with various methods such as temozolomide chemotherapy, radiotherapy, targeted therapy, and electric field therapy [2]. Immunotherapy, such as immune checkpoint therapy, chimeric antigen receptor T cell therapy, dendritic cell vaccine, and tumor-associated macrophage therapy, have demonstrated certain anti-tumor effects in many tumors and can be considered as promising approaches. However, the application of immunotherapy to glioblastoma presents serious challenges due to the immunosuppressive property and drug resistance caused by the blood-brain barrier preventing entry of most antitumor drugs or cells into the brain [3, 4]. Therefore, it is essential to explore the pathogenesis of glioma and discover potential therapeutic targets to inhibiting glioma progression and improving patient prognosis.
Niacinamide is synthesized through biocatalytic events to produce the crucial coenzyme nicotinamide adenine dinucleotide (NAD+). NAD+ is involved in several critical biological activities in the human body, including immune cell activation, cell proliferation, DNA repair, and aging [5, 6]. Nicotinamide exhibits significant potential in the prevention and treatment of tumors. It can inhibit the growth and migration of triple-negative breast cancer by accelerating tumor cell death, activating ROS, and modulating lipid metabolism [7]. Nicotinamide phosphoribosyltransferase, the rate-limiting enzyme for NAD+ synthesis from nicotinamide, controls tumor immune escape by promoting the expression of programmed cell death 1 ligand 1 (PD-L1) and regulating CD8+ T cells activity [8]. Moreover, three clinical trials have demonstrated that oral nicotinamide substantially reduces the incidence rate of actinic keratosis and non-melanoma skin cancer [9]. However, the involvement of nicotinamide in the malignancy of glioma and its potential contribution to glioma treatment require further elucidation.
Tryptophan, an essential amino acid, regulates immunity, neuronal homeostasis and the intestinal microenvironment through the kynurenine metabolic pathway [10]. There are three rate-limiting enzymes involved in tryptophan metabolism: isoleamine 2,3-dioxygenase (IDO) 1, IDO2, and tryptophan 2,3-dioxygenase (TDO2). These enzymes catalyze the conversion of tryptophan to kynurenine and modulate the immune response against cancer tissues by producing metabolites that can activate the aryl hydrocarbon receptor [11, 12]. Previous research has demonstrated that tryptophan metabolism is associated with cancer progression through several pathways. For example, TDO2 is associated with poor prognosis in hepatocellular carcinoma patients and can promote the migration and invasion of liver cancer cells through Wnt signaling pathway [13]. In addition, the novel oral IDO1 inhibitor eos200271/pf-06840003, either alone or in combination with PD-L1-blocking antibodies, can reverse the anergy of T cells against tumor cells and effectively suppressing tumor progression [14]. However, the combination therapy of the IDO1 inhibitor Navoximod with the PD-L1 inhibitor Atezolizumab applied to multiple advanced cancers, has not demonstrated definitive therapeutic effects [15]. Thus, further studies are needed to determine the specific role of tryptophan metabolism in tumor immune escape.
Considering the close relationship between nicotinamide and tryptophan metabolism with tumor immune status, we performed a study to correlate nicotinamide and tryptophan metabolism with glioma immune cell infiltration status and immunotherapy sensitivity. 691 glioma patients in the Cancer Genome Atlas (TCGA) were included in this study. We used cluster analysis to categorize glioma patients, and the specific metabolic status of each group of gliomas as well as the relationship between gliomas and tumor immune infiltration in each group was analyzed. Subsequently, we developed a risk model using least absolute shrinkage and selection operator (LASSO) regression, and the data in CGGA verified the accuracy of the model for prognostic prediction of glioma patients. Additionally, we explored the predictive value of the model for the sensitivity of glioma patients to immune checkpoint inhibitor (ICI) therapy. Epithelial-mesenchymal transition (EMT) refers to the process by which epithelial cells acquire mesenchymal characteristics. In cancer, EMT is associated with tumor initiation, invasion, metastasis, and treatment resistance [16]. We also explored the effects and potential mechanisms of the gene involved in modelling on migratory, invasive and EMT of glioma cells. In summary, our study highlights the potential of nicotinamide and tryptophan metabolism in predicting prognosis, immune microenvironment status, and the response to immune checkpoint inhibitor therapy in glioma patients. These findings provide new research directions and potential targets for immunotherapy of glioma.
Materials and methods
Data sources and processing
This study included a total of 691 patients with glioblastoma, grade II and grade III glioma from TCGA database. The above classification of gliomas followed the 2016 World Health Organization Classification of Tumors of the Central Nervous System [17]. Supplementary Table 1 provides the specific details from TCGA. Additionally, we obtained gene expression profile data and corresponding clinical features from CGGA datasets, namely CGGA mRNAarra301, CGGA mRNAseq693, and CGGA mRNAseq325, to validate the risk prediction model. To enhance the accuracy of data analysis, we standardized the data in CGGA mRNAseq325 and CGGA mRNAseq693 using log2(x + 1) transformation. Supplementary Tables 2, 3, and 4 present the relevant clinical information obtained from CGGA. We curated 42 genes related to nicotinamide metabolism from Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway hsa00760 and REACTOME pathway R-HSA-196,807. Similarly, we identified 40 genes associated with tryptophan metabolism from KEGG pathway hsa00380.
Cluster analysis and principal component analysis (PCA)
We first employed consensus clustering to examine the differences in gene expression levels among different subgroups of gliomas. This involved applying agglomerative pam clustering with a 1-pearson correlation distance, supplemented by resampling 80% of the samples for 10 repetitions using the ConsensuClusterPlus package in R. The optimal number of clusters was determined by analyzing the empirical cumulative distribution function plot. To validate the results of the cluster analysis, we conducted PCA using the stats package in R. Specifically, we first standardized the expression spectrum using Z-scores, and then performed dimensionality reduction analysis using the prcomp function, resulting in a reduced matrix.
Identification of the differently expressed genes (DEGs) and functional enrichment analysis
The Limma package was utilized for identifying genes displaying differential expression levels among subgroups obtained through cluster analysis. Genes with an absolute log2 fold change (|Log2FC|) greater than or equal to 1 and a false discovery rate (FDR) lower than 0.05 were considered as differentially expressed between the groups. To elucidate the cellular pathways associated with these differentially expressed genes among the subgroups, the clusterprofiler package in R was employed for Gene Ontology (GO) and KEGG pathway enrichment analysis. Data visualization was conducted using the ggplot2 package in R. Results were deemed statistically significant if the p-value was below 0.05 and the FDR was below 0.1.
Immune microenvironment analysis and ICI therapy response
ESTIMATE (Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data) calculates the abundance of immune cells, stromal cells, and tumor cells associated with the tumor microenvironment based on transcriptional profiles of cancer samples. Stromal Score shows the level of stroma in the tumor tissue. Immune Score represents the level of immune cell infiltration in the tumor tissue. ESTIMATE Score is a composite score calculated from the Stroma Score and the Immune Score. The higher the ESTIMATE Score, the higher the level of stromal and immune cells and the lower the level of tumor cells in the tumor microenvironment. The gene expression profile data of glioma samples was analyzed using the IOBR package in R to estimate the Stromal Score, Immune Score, and ESTIMATE Score for each sample. The gene expression characteristics of 28 immune infiltrating cells in tumors were obtained from TISIDB. The degree of enrichment of immune infiltrating cells in tumor samples was evaluated using single sample gene set enrichment analysis (ssGSEA) with the GSVA package in R software. Additionally, the CIBERSORT tool was utilized to determine the scores of 22 immune cells in each sample. To assess the sensitivity of each sample to immunotherapy, the Tumor Immune Dysfunction and Exclusion (TIDE) approach was employed.
Protein-protein interaction (PPI) analysis
In this study, our objective was to identify key genes associated with nicotinamide and tryptophan metabolism. To achieve this, we analyzed the intersecting genes between the differential genes in nicotinamide metabolism and tryptophan metabolism. The protein-protein interaction network was then determined using the String database. Next, the PPI network was further examined using the MCC algorithm of Cytohubba plugins in Cyotscape, allowing us to identify the top 20 Hub genes with the highest linkage degrees. Finally, we visualized the PPI network formed by these 20 Hub genes using the String database.
Construction of machine learning risk model
In this experiment, we utilized the gene expression data of the aforementioned 20 hub genes and the survival information of relevant patients to construct a prediction model. To assess the accuracy of our model, we extracted gene expression data related to patient survival from CGGA as a test cohort. Firstly, we employed univariate Cox regression in the survival package of R to evaluate the correlation between the 20 hub genes and the prognosis of glioma patients. Then, we selected the genes that showed a significant association with prognosis of patients. And the glmnet package in R was used to perform LASSO multivariate Cox regression algorithm with 1000 cross-validations, enabling us to identify a robust prognostic model from the selected genes. The genes exhibiting positive results were screened based on optimal penalty parameters, and risk score characteristics were constructed using the corresponding coefficients. Lastly, we utilized Kaplan Meier curves to depict the survival curves of both the training and test sets, while employing 1-year, 3-year, and 5-year receiver operating curve (ROC) analysis to assess the prediction accuracy of the developed prognostic model.
Analysis of single-cell RNAseq Data
Single-cell transcriptome data from 18 glioblastoma samples in GSE159416 were used for single-cell analysis in this study. Seurat package in R was used for expression matrix analysis. The PercentageFeatureSet function was used to calculate the percentage of mitochondrial genes. And the genes were filtered by the number of genes > 50 and mitochondrial percentage < 5. Next, the data were normalized and the 1500 genes with the largest coefficients of variation were extracted for PCA downscaling. Twenty PCA groupings were selected for subsequent TSNE cluster analysis. The critical distance was calculated using the FindNeighbors function, then the FindClusters function grouped the cells, and the RunTSNE function performed TSNE clustering for the cells. Differential genes and characteristic genes for each cluster were obtained by analysis of variance with logFC > 1 and adjpvalue < 0.05. HumanPrimaryCellAtlasData in Celldex package in R was used to provide cellular annotation information. SingleR package in R is used to annotate clustered cells, and Cellchat package in R is used to analyze cellular receptor-ligand pairs and to calculate cellular communication relationships. In addition, transcriptomic data from the Ivy Glioma Atlas were used to analyze the expression patterns of genes in risk models at the anatomical level in glioblastoma [18].
Cell culture
The human glioblastoma cell lines LN229 and U87 were obtained from the China Infrastructure of Cell Line Resource (National Science & Technology Infrastructure, NSTI). In vitro culture and experimentation of the cell lines were conducted using Dulbecco’s Modified Eagle’s Medium (DMEM, Biosharp) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution. The cells were maintained at a temperature of 37℃ and a CO2 concentration of 5%. To inhibit focal adhesion kinase (FAK), the FAK inhibitor (PF-573228) was purchased from MCE and applied at a concentration of 10µM during the in vitro experiment [19].
Transfection
The transforming growth factor beta induced (TGFBI) overexpression plasmid and the negative control plasmid were fabricated by RiboBio. To induce TGFBI overexpression, cells were transfected with the pEXP-RB-Mam-TGFBI plasmid. The cDNA sequence encoding the entire length of TGFBI was cloned into the pEXP-RB-Mom vector, and its accuracy was verified by DNA sequencing. Cell transfection was conducted using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s instructions. The protein levels of the cells were assessed by Western blotting 72 h post-transfection.
Western blot
Cellular proteins were lysed using Ripa lysate (Beyotime), supplemented with protease inhibitors. The lysates were then separated on SDS-PAGE gels using a voltage of 100 V for 1.5 h. Subsequently, equal amounts of protein were electrotransferred to PVDF membranes. The membranes were blocked with TBST in 5% nonfat milk for 2 h and subsequently incubated with the primary antibody overnight at 4℃. Following three washes with TBST, the membranes were incubated with secondary antibodies and visualized using enhanced chemiluminescence (ECL) reagent (Meilunbio). The primary antibodies used were TGFBI (Abclonal, 1:1000), N-cadherin (Affinity, 1:1000), Vimentin (Affinity, 1:500), and GAPDH (Abclonal, 1:10000).
Invasion and migration assays
Glioma cell invasion was observed using transwell chambers containing Matrigel (8 μm pore size, LABSELECT). In the upper chamber, 1000 glioma cells were suspended in 250 µL of serum-free DMEM culture medium, while the lower chamber contained 750 µL of complete medium with 10% FBS. Following a 16-hour incubation, the cells on the upper surface of the chamber were removed, and the cells on the lower surface were fixed with a 4% paraformaldehyde solution (PFA) for 15 min. Subsequently, they were stained with 1% crystal violet for 10 min and then counted. For the glioma cell migration experiment, the same type and concentration of culture medium were used in both the upper and lower chambers of the transwell chamber as in the invasion experiment described above. The experimental procedures for glioma cell migration were similar to those for invasion, except that matrigel-free transwell chambers were used and the incubation time was reduced to 12 h. Each experiment was conducted three times.
Immunofluorescence staining
The cultured LN229 and U87 cells were incubated with 4% paraformaldehyde and subsequently sealed with 5% BSA. Following this, specific antibodies were applied to the cells overnight at 4℃, and secondary antibodies were applied for 1 h. The cells were then restained with DAPI for 10 min and visualized using a fluorescence microscope.
Statistical analysis
The data collected in this study were subjected to statistical analysis using R (version 4.3.3) and visualized using the online network tools Sangerbox [20]. The differences in ssGSEA scores among subgroups were analyzed using the chi-square test. Student’s t-test was employed to compare the two groups with normally distributed continuous indicators, while a nonparametric test was used for skewed distribution continuous indicators. Survival status differences between groups were assessed using Kaplan-Meier (KM) curves analyzed with the survival package in R. The prediction performance of the risk model was evaluated using the pROC package in R, which generated time-dependent ROC curves. A value of area under curve (AUC) > 0.7 indicated the accuracy of the prediction model. Pearson’s coefficient was utilized to analyze the correlation between two continuous variables. A significance level of P < 0.05 was applied.
Results
The specific process of this study is shown in Fig. 1.
Fig. 1.
Flow chart of this study
Establishment of glioma nicotinamide metabolic subtypes
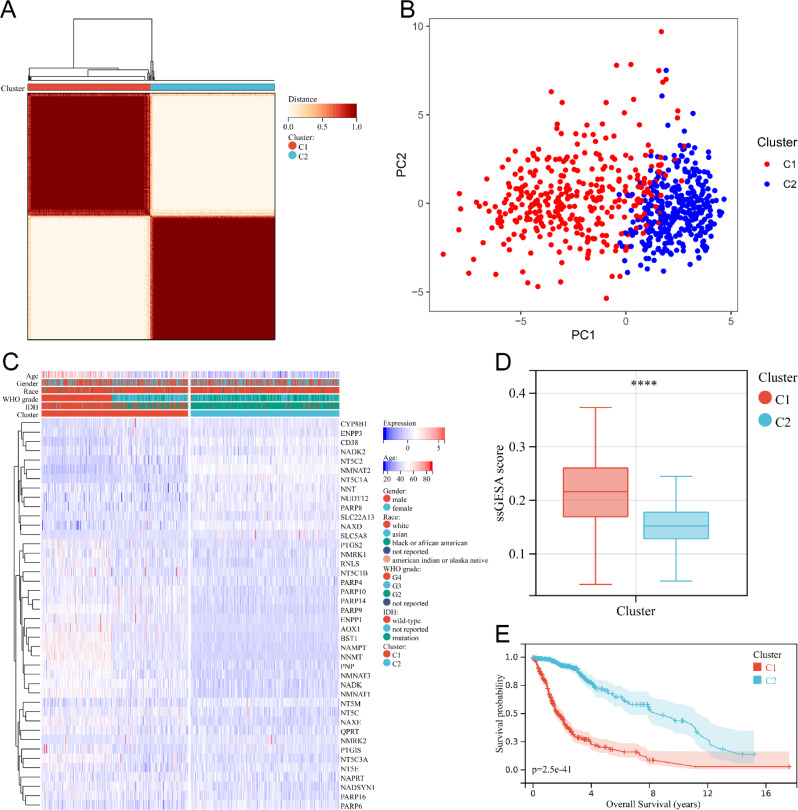
To explore the potential impact of nicotinamide metabolism on the biological behavior of glioma, our study used cluster analysis to divide glioma patients into two clusters. Consensus clustering, based on 42 genes related to nicotinamide metabolism, was employed to achieve the most stable grouping, with k set to 2 (Fig. 2A). Cluster 1 consisted of 343 glioma patients, while Cluster 2 included 348 glioma patients. PCA confirmed the clear distinction between Cluster 1 and Cluster 2 in a two-dimensional space (Fig. 2B). The heatmap exhibited substantial disparities in gene expression levels between the two subgroups (Fig. 2C). The heatmap showed that Cluster 1 contained the majority of WHO grade IV gliomas, more IDH wild-type gliomas and more elderly patients (Fig. 2C). Differences in race and gender did not show significant differences between the two groups (Fig. 2C). And ssGSEA analysis showed that the scores of nicotinamide metabolism genes were significantly higher in Cluster 1 than in Cluster 2 (Fig. 2D). Moreover, Kaplan Meier curves demonstrated that patients in Cluster 1 had a significantly worse prognosis compared to Cluster 2 (Fig. 2E). Collectively, these findings illustrate that glioma patients can be categorized into two distinct molecular subtypes based on the expression level of nicotinamide metabolism.
Fig. 2.
Cluster analysis of gene expression profiles related to nicotinamide metabolism in glioma patients. (A) Consensus clustering matrix for two subgroups. (B) PCA of gene expression profiles for two subgroups. (C) Heat map of the expression levels of nicotinamide metabolism related genes in two subgroups. (D) Box plot of nicotinamide metabolism related scores for both groups. **** P < 0.0001. (E) Kaplan Meier survival curves of glioma patients in two subgroups
Functional analysis of nicotinamide metabolism-related genes
To conduct a detailed comparison of the biological characteristics of the two subtypes of glioma, we initially examined the variations in gene expression between the two clusters. Cluster 1 had 510 up-regulated differentially expressed genes and 616 down-regulated differentially expressed genes compared with Cluster 2 (Supplementary Fig. 1A). KEGG pathway enrichment analysis showed that signaling pathways such as ECM-receptor interaction, cell adhesion molecules, and Th1 and Th2 cell differentiation were significantly enriched in cluster1 (Supplementary Fig. 1B), and signaling pathways such as cAMP signaling pathway, Calcium signaling pathway, and Wnt signaling pathway were significantly enriched in cluster2 (Supplementary Fig. 1C). In addition, GO enrichment analysis showed that defense response, external encapsulating structure and signaling receptor binding were significantly enriched in cluster1 (Supplementary Fig. 1D-F), and cell cell signaling, synapse and cation transmembrane transporter activity were significantly enriched in cluster2 (Supplementary Fig. 1G-I). In conclusion, our study shows that the two subgroups of gliomas have different biological behaviours and metabolic profiles.
The relationship between nicotinamide metabolism and immune infiltration in glioma
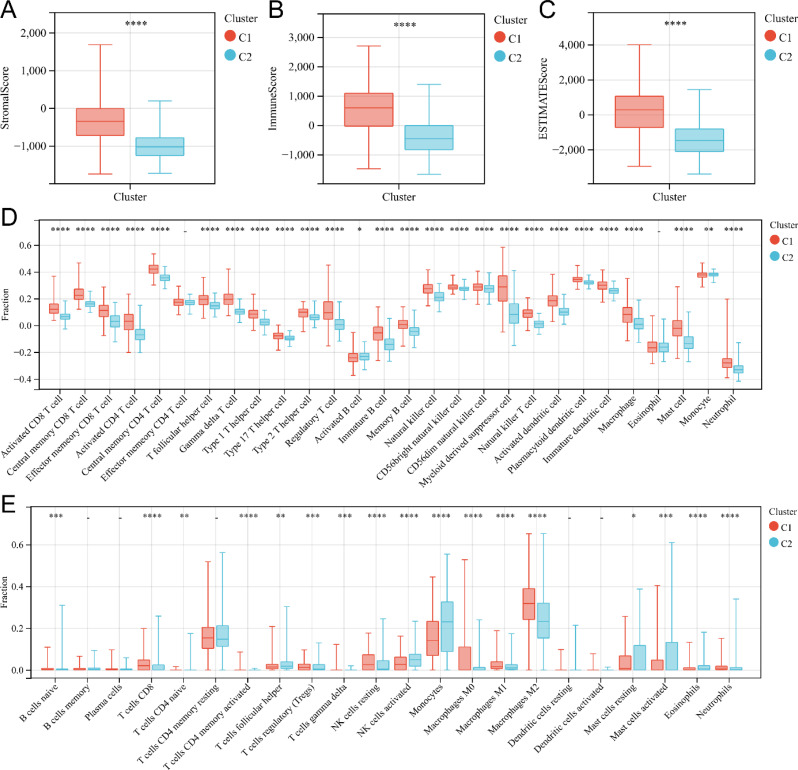
To investigate the relationship between nicotinamide metabolism and immune status in gliomas, we examined immune infiltration in two subtypes. Firstly, we used ESTIMATE algorithm to assess the immune-related scores in two subgroups of glioma patients. Our study found that Stromal Score (Fig. 3A), Immune Score (Fig. 3B) and ESTIMATED Score (Fig. 3C) of glioma patients in the high-risk subtype (Cluster1) were significantly higher than those of glioma patients in the low-risk subtype (Cluster2) (P < 0.0001). Secondly, we employed the ssGSEA algorithm to evaluate the enrichment degree of 28 immune cells in glioma. The analysis revealed significant differences in the enrichment of immune cells, excluding effector memory CD4+ T cells and eosinophils, between the two subtypes (Fig. 3D) (P < 0.01). And CIBERSORT algorithm also demonstrated significant differences in the infiltration levels of multiple immune cells between the two subtypes (Fig. 3E) (P < 0.05). In conclusion, our study highlights the significant differences in the immune microenvironment status of glioma between the two molecular subtypes associated with nicotinamide metabolism.
Fig. 3.
Immunocyte infiltration analysis of two subgroups related to nicotinamide metabolism. (A-C) Box plot of interstitial score(A), immune score (B), and ESTIMATE score (C) for patients with high and low nicotinamide metabolism subgroups. (D) Box plot of immune cell infiltration levels in high nicotinamide metabolism and low nicotinamide metabolism subgroups analyzed by ssGSEA. (E) Box plot of immune cell infiltration levels in high nicotinamide metabolism and low nicotinamide metabolism subgroups calculated by CIBERSORT. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001; -, not significant
Establishment of glioma tryptophan metabolic subtypes
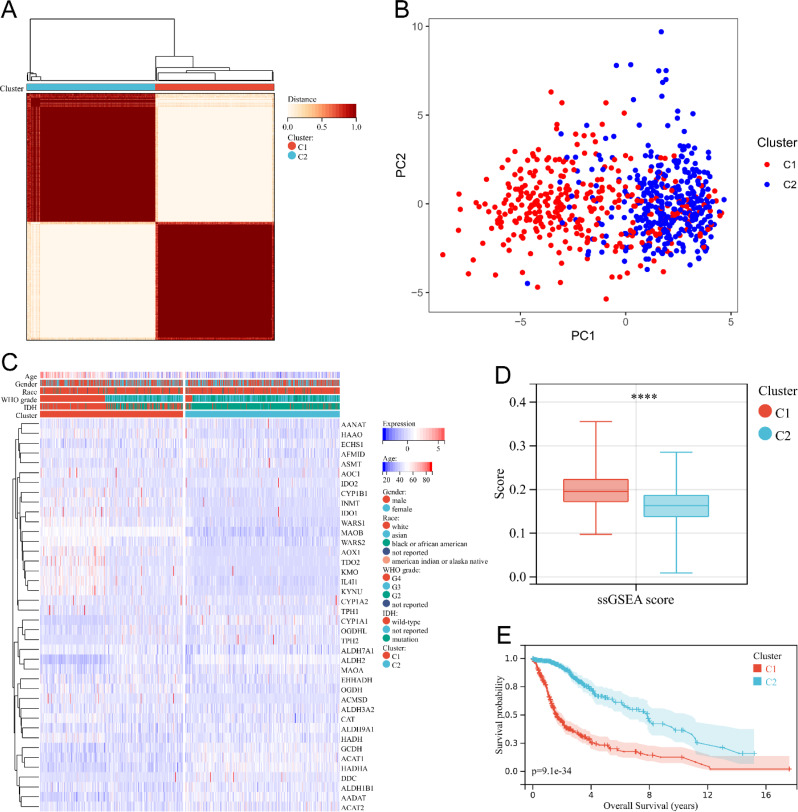
In light of the fact that nicotinamide metabolism and tryptophan metabolism can both modulate the sensitivity of tumor immunotherapy through the regulation of T cell activity within the tumor microenvironment, we sought to investigate whether the two metabolic pathways have a synergistic impact on the infiltration of immune cells in glioma. To accomplish this, we divided glioma patients into two subgroups according to genes related to tryptophan metabolism using a consensus clustering approach (Fig. 4A), resulting in 332 patients in cluster 1 and 359 patients in cluster 2. PCA showed the difference in the distribution of gliomas in two-dimensional space between the two subgroups (Fig. 4B). The heatmap showed the difference in gene expression levels between the two subgroups (Fig. 4C). We found that Cluster1 contained the majority of WHO grade IV gliomas, more IDH wild-type gliomas, and more elderly glioma patients (Fig. 4C). Differences in race and gender did not show significant differences between the two subgroups (Fig. 4C). And ssGSEA analysis showed that the scores of tryptophan metabolism genes were significantly higher in Cluster 1 than in Cluster 2 (Fig. 4D). In addition, patients in cluster1 had a worse prognosis compared to patients in cluster2 (Fig. 4E).
Fig. 4.
Cluster analysis of gene expression profiles related to tryptophan metabolism in glioma patients. (A) Consensus clustering matrix for two subgroups. (B) PCA of gene expression profiles for two subgroups. (C) Heat map of the expression levels of tryptophan metabolism related genes in two subgroups. (D) Box plot of tryptophan metabolism related scores for both groups**** P < 0.0001. (E) Kaplan Meier survival curves of glioma patients in two subgroups
The interrelationship between nicotinamide metabolism and tryptophan metabolism
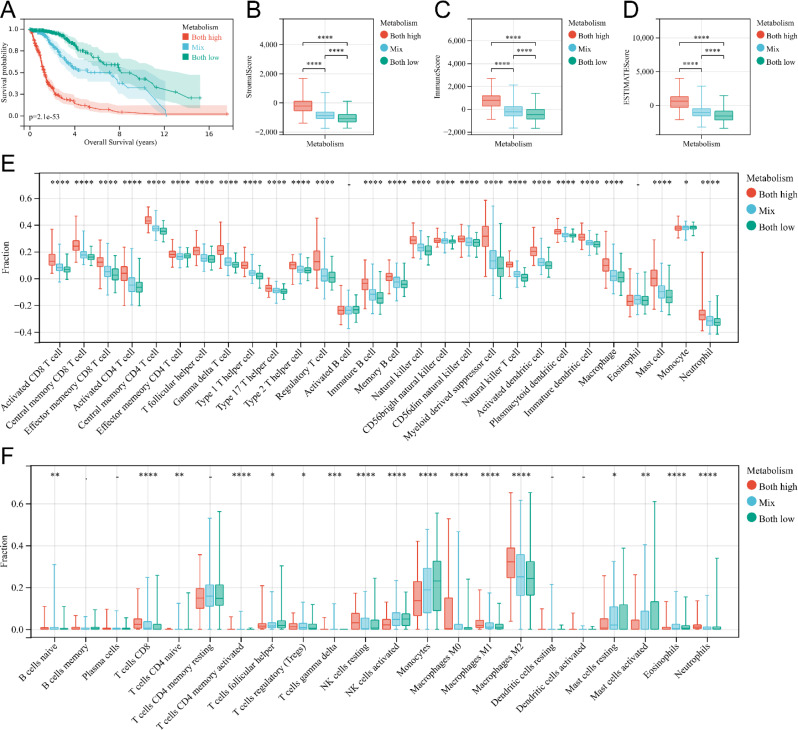
Then, glioma patients were classified into three groups based on the status of nicotinamide metabolism and tryptophan metabolism: the nicotinamide metabolism high / tryptophan metabolism high group, the mixed group, and the nicotinamide metabolism low / tryptophan metabolism low group. The results showed that KM curve demonstrated that patients in the nicotinamide metabolism high / tryptophan metabolism high group had the worst prognosis, whereas patients in the nicotinamide metabolism low / tryptophan metabolism low group had the best prognosis (Fig. 5A). Additionally, ESTIMATE algorithm indicated that Stromal score (Fig. 5B), Immune score (Fig. 5C), and ESTIMATE score (Fig. 5D) were highest in the high nicotinamide metabolism / high tryptophan metabolism group out of the three groups. And ssGSEA algorithm also revealed significantly highest scores for various immune infiltrating cells in the tumor samples of the high nicotinamide metabolism / high tryptophan metabolism group compared to the other two groups (Fig. 5E). MCPcounter analysis showed different cell infiltration in different subgroups (Fig. 5F). For example, the level of infiltration of resting NK cells was higher in the nicotinamide metabolism high / tryptophan metabolism high group, whereas the level of infiltration of activated NK cells was lower in the nicotinamide metabolism low / tryptophan metabolism low group (Fig. 5F). In addition, the infiltration level of M2-type macrophages was higher in the nicotinamide metabolism high / tryptophan metabolism high group (Fig. 5F). In conclusion, our study confirms that there is a strong correlation between nicotinamide metabolism, tryptophan metabolism and the degree of immune infiltration in gliomas and that the higher the degree of immune cell infiltration, the worse the prognosis of gliomas. The discovery may facilitate the establishment of reliable prognostic models for evaluating the prognosis of glioma patients.
Fig. 5.
Immunocyte infiltration analysis of nicotinamide-tryptophan metabolism related subgroups. (A) Kaplan Meier survival curves of glioma patients of high nicotinamide-tryptophan metabolism group, mixed group, and low nicotinamide-tryptophan metabolism group. (B-D) Box plot of stromal score (B), immune score (C), and ESTIMATE score (D) for the three groups of patients mentioned above. (E) Box plot of immune cell infiltration levels in high nicotinamide-tryptophan metabolism group, mixed group, and low nicotinamide-tryptophan metabolism group analyzed by ssGSEA. (F) Box plot of immune cell infiltration levels in high nicotinamide-tryptophan metabolism group, mixed group, and low nicotinamide-tryptophan metabolism group analyzed by CIBERSORT
Identification of nicotinamide and tryptophan metabolism-related core genes
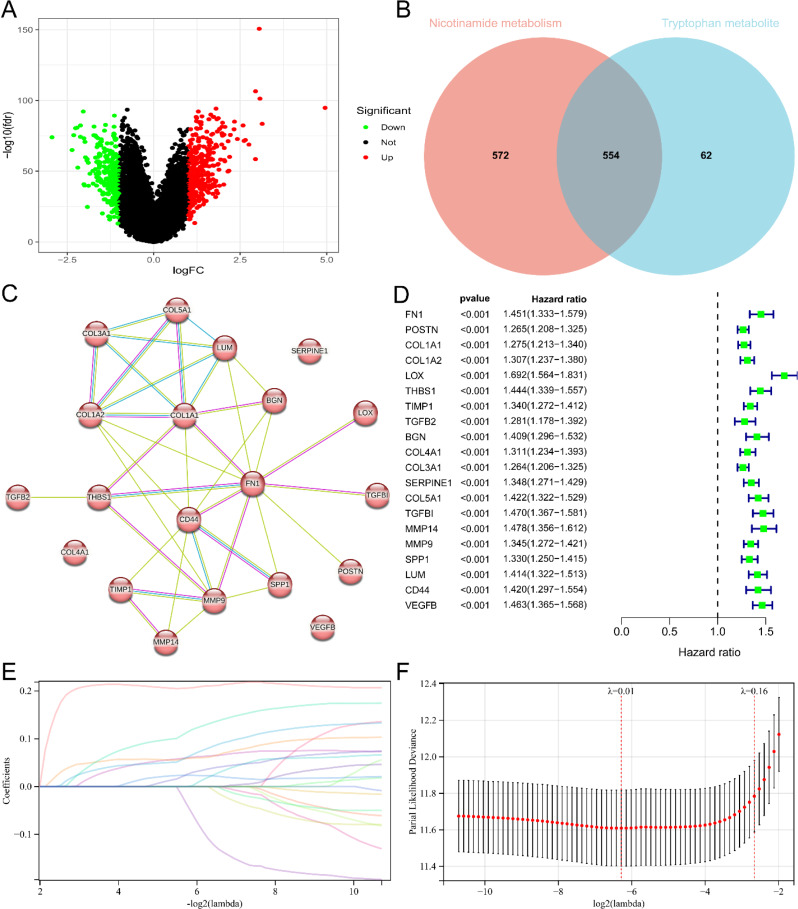
To establish a risk model concerning nicotinamide and tryptophan metabolism, we screened the core genes associated with these metabolic processes in glioma. Limma analysis revealed 616 differentially expressed genes within the two subgroups of tryptophan metabolism, with 362 genes up-regulated and 254 genes down-regulated (Fig. 6A). By intersecting the 616 differentially expressed genes of tryptophan metabolism with the 1126 differentially expressed genes of nicotinamide metabolism, we obtained a set of 554 shared genes (Fig. 6B). Subsequently, PPI networks were constructed using the STRING database. The MCC algorithm in Cytoscape was employed to identify 20 hub genes in the PPI network (Fig. 6C). The univariate Cox regression analysis also highlighted a significant correlation between the prognosis of glioma patients and the 20 hub genes (Fig. 6D).
Fig. 6.
Identification of core genes related to nicotinamide and tryptophan metabolism and establishment of prognostic risk score signatures. (A) Volcano map of differentially expressed genes in two subgroups related to tryptophan metabolism. Red and green represent differentially expressed genes with upregulation and downregulation of gene expression. (B) Venn plot showing the overlap relationship between differentially expressed genes in two subgroups of nicotinamide metabolism and differentially expressed genes in two subgroups of tryptophan metabolism. Numbers represent the number of genes. (C) PPI network formed by 20 core genes. The network nodes are proteins. The edges represent the predicted functional associations. Purple line - experimental evidence. Yellow line - textmining evidence. Light blue line - database evidence. (D) Single factor Cox regression analysis of 20 core genes related to nicotinamide and tryptophan metabolism. (E) Lasso coefficient spectra of 20 core genes. The positive coefficient means that the algorithm believes that high expression of the gene predicts a poorer prognosis for glioma. The negative coefficient means that the algorithm believes that high expression of the gene predicts a better prognosis for gliomas. (F) Cross validation fitting curve calculated by lasso regression method. The range between the two dashed lines shows the range of optimal λ values considered by the algorithm
Construction and validation of machine learning risk model
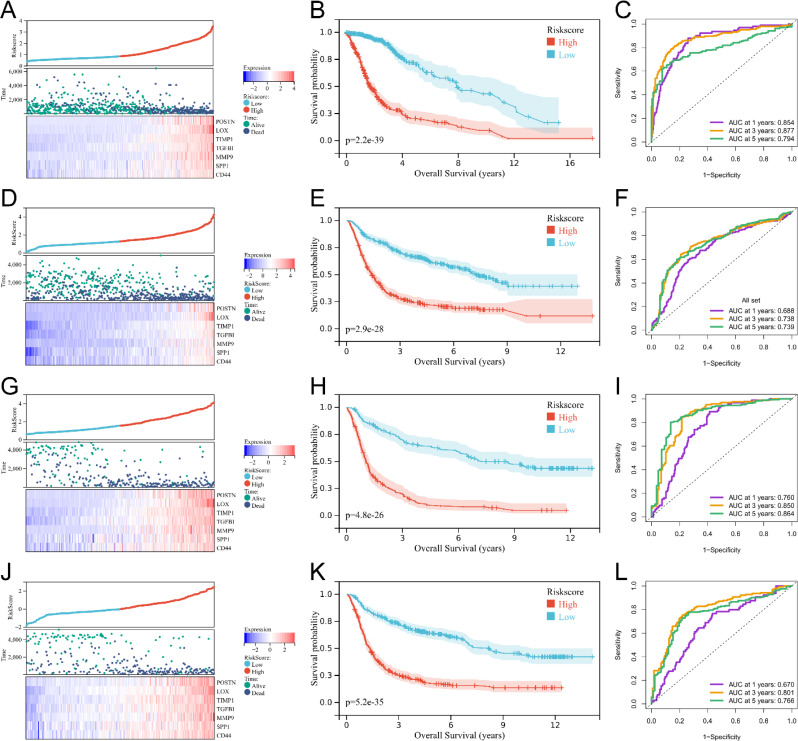
Based on the 20 core genes mentioned above, we developed a risk model focusing on nicotinamide and tryptophan metabolism for assessing the prognosis of glioma patients. We determined the risk formula by lasso analysis (Fig. 6E and F). The risk score is computed as follows: Risk score = (0.057POSTN exp) + (0.206LOX exp) + (0.056TIMP1 exp) + (0.099TGFBI exp) + (0.051MMP9 exp) + (0.022SPP1 exp) + (0.014*CD44 exp). Figure 7A, D and G, and 7J showed the sample distribution of gliomas in each dataset sorted according to the risk score. The Kaplan-Meier curve for the TCGA dataset showed a correlation between higher risk score and poorer prognosis among glioma patients (Fig. 7B). In addition, the area under the curve for 1-year survival, 3-year survival, and 5-year survival exceeded 0.7, indicating the superior predictive ability of the model for the prognosis of glioma patients (Fig. 7C). To validate the accuracy of the risk model, we examined three glioma transcriptome datasets: CGGA mRNAseq693, CGGA mRNAseq325, and CGGA mRNAarray. The Kaplan-Meier curve of these datasets consistently showed a correlation between higher risk scores and poorer prognosis in patients with gliomas (Fig. 7E, H and K). In addition, the area under the curve for 3-year survival and 5-year survival exceeded 0.7 for all three datasets, and the area under the curve for 1-year survival exceeded 0.7 for the CGGA mrnaseq325 dataset (Fig. 7F, I and L). In summary, our findings establish the accuracy of the risk model, which is based on genes associated with tryptophan and nicotinamide metabolism, in predicting the prognosis of glioma patients, specifically in the long term.
Fig. 7.
Verification of the signature of the prognostic risk score for nicotinamide-tryptophan metabolism. (A) Heat map of glioma patients in TCGA based on the distribution of the risk score. (B) Kaplan Meier survival curves for glioma patients in TCGA based on the risk score. (C) ROC curves for predicting 1-, 3-, and 5-year survival of glioma patients by the risk score in TCGA. (D) Heatmap of glioma patients in CGGA mRNAseq693 based on the distribution of the risk score. (E) Kaplan Meier survival curves for glioma patients in CGGA mRNAseq693 based on the risk score. (F) ROC curves for predicting 1-, 3-, and 5-year survival of glioma patients by the risk score in CGGA mRNAseq693. (G) Heatmap of glioma patients in CGGA mRNAseq325 based on the distribution of the risk score. (H) Kaplan Meier survival curves for glioma patients in CGGA mRNAseq325 based on the risk score. (I) ROC curves for predicting 1-, 3-, and 5-year survival of glioma patients by the risk score in CGGA mRNAseq325. (J) Heatmap of glioma patients in CGGA mRNAarray based on the distribution of the risk score. (K) Kaplan Meier survival curves for glioma patients in CGGA mRNAarray based on the risk score. (L) ROC curves for predicting 1-, 3-, and 5-year survival of glioma patients by the risk score in CGGA mRNAarray
The relationship between risk score and immune microenvironment of glioma
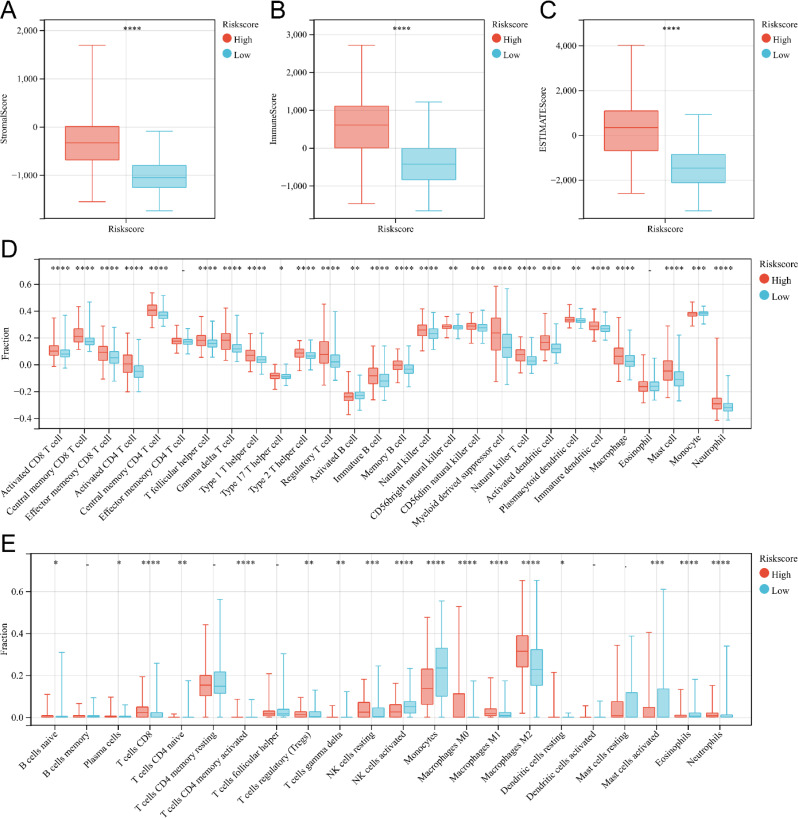
We further explored the relationship between risk scores and immune cell infiltration in gliomas. The glioma patients were categorized into high-risk score group and low-risk score group based on the median risk score. The application of the ESTIMATE algorithm revealed significantly higher Immune score, Stromal score, and ESTIMATE score values in the high-risk score group compared to the low-risk score group, indicating a greater presence of immune cells and stroma in the gliomas of the high-risk score group (Fig. 8A, B and C). To further analyze the immune microenvironment of gliomas, we utilized TISIDE and CIBERSORT to assess the degree of infiltration by different immune cells. The ssGSEA score derived from immune cell markers in TISIDE demonstrated significantly higher infiltration levels of almost all immune cell types in the high-risk score group compared to the low-risk score group (Fig. 8D), suggesting distinct immune states in these two groups. Additionally, the CIBERSORT algorithm showed higher infiltration levels of CD8+ T cells, resting NK cells, M0 macrophages, M1 macrophages, and M2 macrophages in the high-risk score group, whereas activated NK cells exhibited lower infiltration levels (Fig. 8E). We also achieved similar findings when analyzing CGGA mRNAseq693, CGGA mRNAseq325, and CGGA mRNAarray data (Supplementary Fig. 2, Supplementary Fig. 4, Supplementary Fig. 6). In conclusion, our study demonstrates that the risk model can effectively assess the immune microenvironment of glioma.
Fig. 8.
Analysis of immune cell infiltration in two subgroups related to the risk score of TCGA. (A-C) Box plot of stromal score (A), immune score (B), and ESTIMATE score (C) for patients in the high and low-risk score groups. (D)Box plot of immune cell infiltration levels high-risk score and low-risk score groups analyzed by ssGSEA. (E)Box plot of immune cell infiltration levels in high-risk score and low-risk score groups analyzed by CIBERSORT. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001; -, not significant
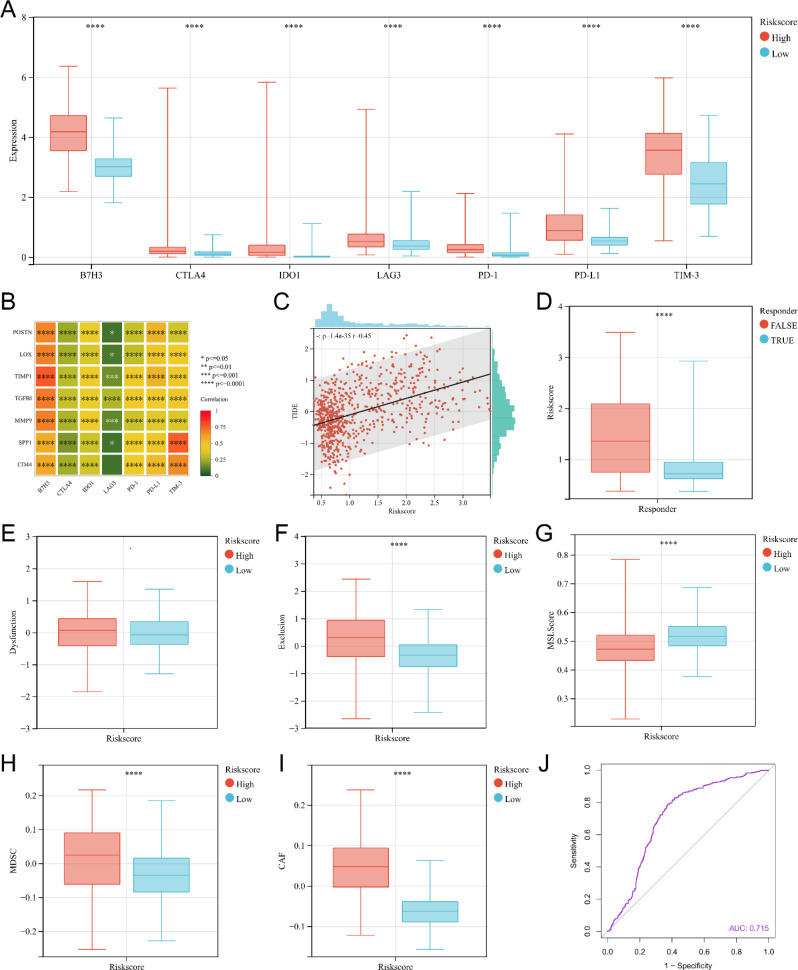
Risk score may be used as an indicator to predict ICI therapy
To further assess the predictive ability of the risk score for ICI treatment sensitivity in gliomas, our study investigated the relationship between the risk score and the expression of immune checkpoints in tumors, as well as the TIDE score. TIDE algorithm consists of three main scores: Dysfunction score, Exclusion score and TIDE score. The higher Dysfunction score usually means that the T cells in the tumor are functioning poorly. The higher score usually means that immune cells have difficulty engaging with tumor cells through the tumor microenvironment and thus exerting a killing effect. The higher TIDE score usually implies a poorer response to ICI treatment and a poorer prognosis. We found that the high-risk score group in TCGA exhibited significantly elevated expression levels of seven immunological checkpoints (Fig. 9A). There was also a positive correlation between the expression of immunological checkpoints and seven genes included in the risk model (Fig. 9B). And we obtained similar results when analyzing CGGA mRNAseq693, CGGA mRNAseq325, and CGGA mRNA array (Supplementary Fig. 3A, B, Supplementary Fig. 5A, B, Supplementary Fig. 7A, B). Then, our study showed a significant positive correlation between TIDE score and the risk score (Fig. 9C, Supplementary Fig. 3C, Supplementary Fig. 5C, Supplementary Fig. 7C). And the risk score for gliomas were significantly higher in the ICI treatment false responder group predicted by TIDE (Fig. 9D, Supplementary Fig. 3D, Supplementary Fig. 5D, Supplementary Fig. 7D). And there was no statistically difference in immune dysfunction scores for gliomas between the high-risk score group and the low-risk score group (Fig. 9E, Supplementary Fig. 3E, Supplementary Fig. 5E, Supplementary Fig. 7E). The immune exclusion score, MDSC and CAF were significantly higher for gliomas in the high-risk score group than in the low-risk score group (Fig. 9F, H, I, Supplementary Fig. 3F, H, I, Supplementary Fig. 5F, H, I, Supplementary Fig. 7F, H, I). The microsatellite instability (MSI) score for gliomas in the high-risk scoring group were significantly lower than that in the low-risk scoring group (Fig. 9G, Supplementary Fig. 3G, Supplementary Fig. 5G, Supplementary Fig. 7G). Finally, ROC curve analysis showed that risk scores had high accuracy in predicting the sensitivity of gliomas to ICI treatment in all four datasets (AUC = 0.715 for TCGA, AUC = 0.719 for CGGA mRNAseq693, AUC = 0.796 for CGGA mRNAseq325, and AUC = 0.718 for CGGA mRNAarray) (Fig. 9J, Supplementary Fig. 3J, Supplementary Fig. 5J, Supplementary Fig. 7J). In conclusion, our findings provide evidence that the risk model effectively predicts the susceptibility of glioma to ICI treatment.
Fig. 9.
The predictive role of risk model of TCGA for ICI treatment response. (A) The box plot of immune checkpoint expression in the high-risk score group and low-risk score group. (B) Correlation analysis between the expression levels of risk model-related genes and the expression levels of immune checkpoints. (C) Correlation analysis between the risk score and TIDE score. (D) The box plot of risk score for predicting ICI treatment true responder and false responder gliomas using the TIDE algorithm. (E) The box plot of Dysfunction score for high-risk score group and low-risk score group. (F) The box plot of Dysfunction score for high-risk score group and low-risk score group. (G) The box plot of MSI score for high-risk score group and low-risk score group. (H) The box plot of MDSC for high-risk score group and low-risk score group. (I) The box plots of CAF for high-risk score group and low-risk score group. (J) The ROC curve of the risk score predicting response to ICI treatment
Analysis of single-cell sequencing
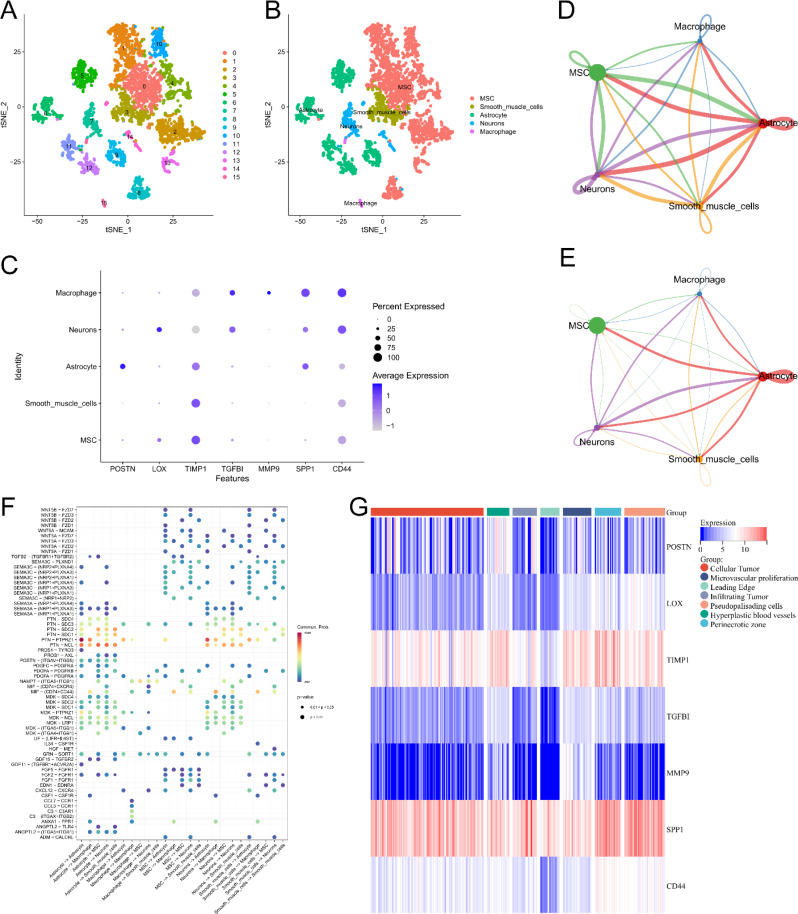
To obtain a more complete picture of the specific distribution of risk model-related genes in glioma tissues, single-cell RNA-seq analysis of GSE159416 was performed. UMAP was clustered for the cells in the dataset to form 15 different clusters (Fig. 10A). Cell type annotation was performed for the above 15 clusters, yielding five known cell types (Fig. 10B). LOX was highly expressed in neurons. MMP9 was highly expressed in macrophage. TGFBI was highly expressed in macrophage and neurons. SPP1 was highly expressed in macrophage, neurons, and astrocyte. CD44 and TIMP1 were expressed in all five cell types (Fig. 10C). We also explored different cellular interactions in gliomas. The intercellular communication network showed a strong communication relationship between astrocyte and the other four cells (Fig. 10D-E). Notably, PTN-PTPRZ1 and PTN-NCL are involved in ligand receptor interactions in multiple cells (Fig. 10F). In addition, transcriptomic data from the Ivy Glioblastoma Atlas Project showed that POSTN was highly expressed in Hyperplastic blood vessels. LOX was highly expressed in pseudopalisading cells. TIMP1 was highly expressed in the perinecrotic zone. TGFBI was highly expressed in the TGFBI was highly expressed in perinecrotic zone, hyperplastic blood vessels and microvascular proliferation. MMP9 was highly expressed in microvascular proliferation. SPP1 and CD44 were highly expressed in Perinecrotic zone (Fig. 10G). In conclusion, our results demonstrated the distribution of model-associated genes in different cells of glioma, laying the foundation for subsequent investigation of the mechanism of action of different genes and the signaling of intercellular signals.
Fig. 10.
Results of data analysis for single-cell sequencing. (A-B) Cluster and cell type annotation of tumor microenvironment cells. (C) Expression characteristics of risk model related genes in different cells. (D) Circle interaction plots showing the number of cell-to-cell communication. The size of the circle represents the number of cells. The thickness of the lines between the cells represents the number of times the two cells communicate. The color of the lines represents the type of cell that serves as a ligand. (E) Circle interaction plots showing the strength of cell-to-cell communication. The size of the circle represents the number of cells. The thickness of the lines between the cells represents the strength of communication between the two cells. The color of the lines represents the type of cell that serves as a ligand. (F) The ligand-receptor interactions between cells. X-axis shows the type of cells that interact. The cells in the front are the cells where the ligand is located and the cells in the back are the cells where the receptor is located. Y-axis shows the names of the ligand-receptors. The colors of the circles in the picture represent the average expression levels of these two genes in cells. A redder color indicates a higher level of expression. The size of the circle represents the size of the p-value. (G) Heat map of risk model related genes in anatomical locations of tumor samples according to Ivy Glioblastoma Atlas. Leading Edge is the tumor cells at the boundary of the tumor as determined by histological methods. Recurrence of gliomas often begins at this site. Infiltrating Tumor is the portion of tumor tissue that invades normal tissue. Cellular Tumor is the core portion of tumor tissue as determined by histological method
TGFBI mediates glioma cell migration, invasion, and EMT through FAK
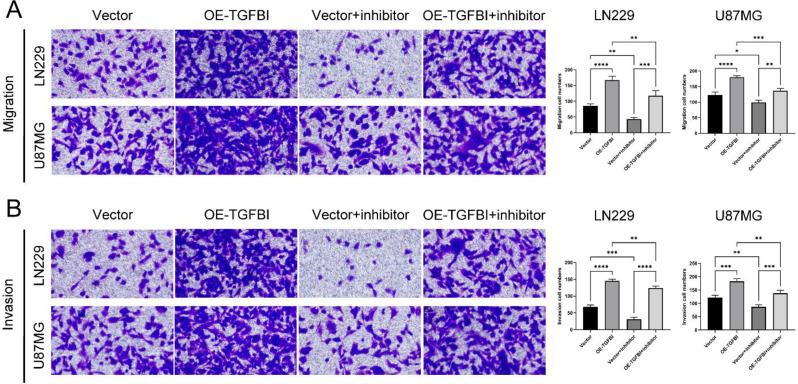
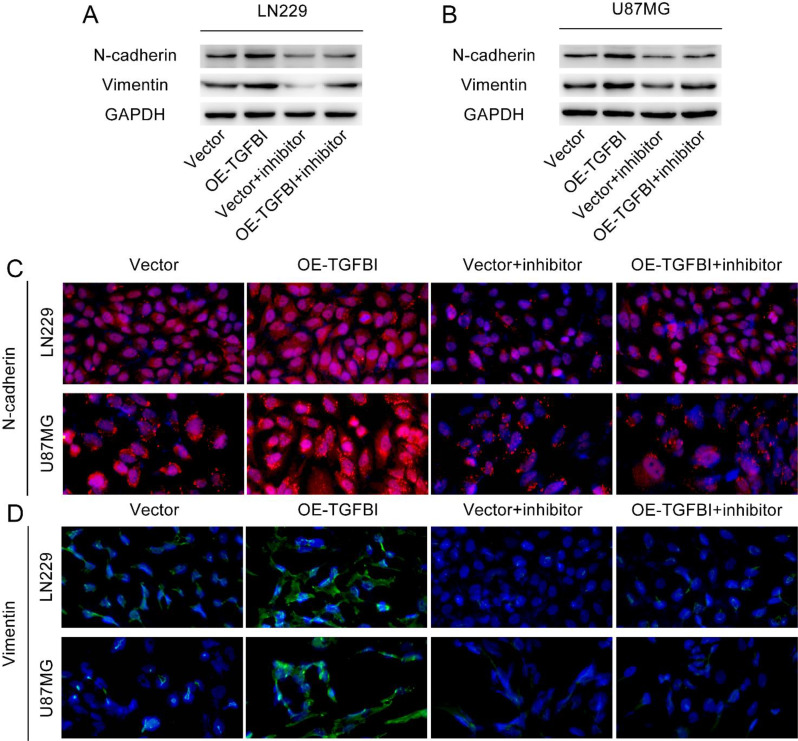
Higher coefficient of a gene in the risk model indicates that the expression level of the gene has a greater influence on the risk score. The higher coefficient of TGFBI indicate that the expression level of TGFBI has a greater effect on the risk score. Therefore, we conducted a further investigation into the impact of TGFBI on the specific biological behavior of glioma cells. The Supplementary Fig. 8 illustrates the scenario of TGFBI overexpression. Our study revealed a significant enhancement in the migration and invasion capabilities of ln229 cells and U87 cells due to the overexpression of TGFBI (Fig. 11A, B). Western blot analysis of EMT-related markers demonstrated a notable increase in the expression levels of mesenchymal markers, such as N-cadherin and Vimentin, in ln229 cells and U87 cells with TGFBI overexpression (Fig. 12A, B). Immunofluorescence examination also confirmed these alterations in EMT-related proteins resulting from TGFBI overexpression (Fig. 12C, D). FAK is a non-receptor tyrosine kinase that promotes tumor progression, metastasis and the formation of an immunosuppressive microenvironment in tumors [21, 22]. Notably, pretreatment with the FAK inhibitor PF-573,228 effectively mitigated TGFBI-mediated migration and invasion of ln229 cells and U87 cells (Fig. 11A, B), thereby reversing the augmented expression levels of N-cadherin and Vimentin caused by TGFBI overexpression (Fig. 12A-D). To summarize, our findings strongly suggest that TGFBI promotes glioma cell migration, invasion, and EMT through FAK.
Fig. 11.
Migration and invasion experiments on LN229 cells and U87-MG cells after overexpression of TGFBI and pretreatment with FAK inhibitors. (A) Representative images of migration experiments and the number of transmembrane cells. (**P < 0.01, ***P < 0.001, ****P < 0.0001, n = 3, Student t test). (B) Representative images of invasion experiments and the number of transmembrane cells. (**P < 0.01, ***P < 0.001, ****P < 0.0001, n = 3, Student t test). Vector: transfect empty plasmid. OE-TGFBI: transfect TGFBI overexpression plasmid. Vector + inhibitor: transfect empty plasmid and pretreatment with PF-573,228 for 1 h before the experiment. OE-TGFBI + inhibitor: transfect TGFBI overexpression plasmid and pretreatment with PF-573,228 for 1 h before the experiment. The magnification of the above image is 200×
Fig. 12.
The expression of EMT-related proteins in LN229 cells and U87-MG cells after overexpression of TGFBI and pretreatment with FAK inhibitors. (A-B) Western blot was used to detect the expression levels of N-cadherin and Vimentin in LN229 cells and U87-MG cells. (C-D) immunofluorescence analysis showed the expression levels of N-cadherin and Vimentin in LN229 cells and U87-MG cells. The magnification of the image is 200×. Vector: transfect empty plasmid. OE-TGFBI: transfect TGFBI overexpression plasmid. Vector + inhibitor: transfect empty plasmid and pretreatment with PF-573,228 for 1 h before the experiment. OE-TGFBI + inhibitor: transfect TGFBI overexpression plasmid and pretreatment with PF-573,228 for 1 h before the experiment
Discussion
Since the discovery of aerobic glycolysis, which is a distinct metabolic mode in tumors as compared to normal human cells, researchers have gradually started exploring and studying its implications. In tumors, not only do the metabolic processes related to sugars and lipids differ from that of normal human tissues, but also substances like niacinamide and various amino acids, including tryptophan, play significant roles in cell structure and metabolism, thus influencing the occurrence and progression of tumors [23]. Previous studies have indicated that nicotinamide induces apoptosis by reducing mitochondrial membrane potential and ATP production while increasing the oxidation of fatty acids in triple-negative breast cancer [7]. Tryptophan metabolism, serving as a crucial intermediate link, has been found to be involved in the development of liver cancer in mice due to changes in intestinal flora [24]. Furthermore, nicotinamide and tryptophan have been shown to regulate tumor immune escape and the response to immunotherapy by influencing the activity of immune cells in the tumor immune microenvironment [25–28]. Considering that the effectiveness of immunotherapy in treating glioma still requires improvement, our study aims to enhance the accuracy of predicting prognosis and immunotherapy sensitivity in patients with gliomas.
To investigate the role of nicotinamide and tryptophan metabolism in glioma, we conducted cluster analysis to categorize glioma patients into high and low-metabolism groups. The rationality of the grouping was confirmed using the PCA method and ssGSEA score. Considering the impact of IDH mutation status on glioma metabolic status as well as prognosis, the heatmap showed the distribution of glioma IDH mutation status with respect to nicotinamide metabolism and tryptophan metabolism in subgroups. Our results showed a higher distribution of IDH wild-type gliomas in the high nicotinamide metabolism and high tryptophan metabolism groups. Since IDH mutations lead to increased consumption of NADPH in the tricarboxylic acid cycle of the cell [29] and nicotinamide is an important precursor for NADPH synthesis, this seems to predict that the status of IDH may influence the level of nicotinamide metabolism in gliomas. However, the specific relationship between IDH mutation status and nicotinamide and tryptophan metabolism in gliomas may still require further experimental studies. Next, we examined the effects of nicotinamide metabolism, tryptophan metabolism, and their combined effects on the prognosis of glioma patients. Our results revealed that patients with high levels of both nicotinamide and tryptophan metabolism had the poorest overall prognosis, suggesting that these two metabolic pathways may act together to cause poor prognosis in glioma patients. Previous studies have investigated the impact of nicotinamide and tryptophan metabolism on the biological behavior and prognosis of various tumors. High expression of nicotinamide N-methyltransferase is associated with tumor progression, metastasis and worse clinical outcomes [30]. Tryptophan metabolites, such as 5-hydroxytryptamine and 3-hydroxybenzoic acid, can promote tumor growth by suppressing ferroptosis in tumor cells [31]. In short, our study reveals the correlation between increased nicotinamide and tryptophan metabolism and poor prognosis in glioma patients. The research on nicotinamide and tryptophan metabolism in gliomas may provide new directions for glioma treatment and improvement of patient prognosis.
To investigate the role of nicotinamide metabolism in the pathophysiology of gliomas, we conducted KEGG enrichment analysis analysis to identify potential signaling pathways associated with this metabolism. Our findings elucidated the close relationship between nicotinamide metabolism and multiple signaling pathways, including cell adhesion molecules (CAMs), Th1 and Th2 cell differentiation, and Th17 cell differentiation. Previous studies have demonstrated the significance of these signaling pathways in the biological behavior of different tumors. Alterations in cell adhesion molecules in the tumor microenvironment are not only directly involved in tumor invasion and distant metastasis, but also contribute to malignant transformation and immune evasion of tumors [32, 33]. Th1 and Th2 cells, regulated by cytokines produced by themselves as well as other immune cells, have a crucial role in regulating specific immune responses within tumors and are thus a focal point of current immunotherapy research [34]. The observed association between nicotinamide metabolism and signaling pathways implicated in different malignancies further suggests the involvement of this metabolism in the development and immune escape of gliomas.
Afterwards, we classified gliomas into the nicotinamide metabolism high / tryptophan metabolism high group, the mixed group, and the nicotinamide metabolism low / tryptophan metabolism low group based on nicotinamide and tryptophan metabolism status and analyzed the prognostic status and immune cell infiltration status of the three groups of glioma patients. Our results showed that the high nicotinamide tryptophan metabolism group exhibited more infiltrating immune cells and poorer prognosis. And the group with more immune cell infiltration had a worse prognosis. This is because the killing effect of immune cells on tumor cells in the tumor microenvironment is influenced by multiple factors. The cells in glioma tissue mainly include tumor cells, stromal cells and immune cells. A portion of immune cells play tumor-killing roles, such as CD8 + T cells and NK cells [35, 36], and some cells play immunosuppressive roles, such as Treg cells and Th17 cells [37, 38]. And the immunosuppressive microenvironment of tumor tissues can inhibit the killing effect of immune cells. In addition, stromal cells in tumor tissues can also inhibit the killing effect of immune cells by hindering the contact between immune cells and tumor cells [39]. Tumor cells, immune cells and the tumor immune microenvironment together determine the efficacy of the immune system in tumor clearance. It has also been shown that high levels of immune cell infiltration in gliomas are associated with poor prognosis [40], which is consistent with our results. In conclusion, our study reveals a close relationship between high nicotinamide tryptophan metabolism and poor prognosis and high immune cell infiltration in gliomas.
In a subsequent study, we aimed to validate a prognostic risk model by identifying core genes associated with tryptophan and nicotinamide metabolism. Our results showed that the expression of the seven genes involved in modelling are all highly elevated factors for gliomas. And the data in the CGGA also showed that the model can more accurately predict the prognosis of patients with gliomas. It has also been shown that seven genes associated with model establishment are closely associated with poor prognosis of various malignancies and are involved in the development and progression of different tumors. For instance, POSTN activates the PI3K/Akt pathway through interaction with integrins αVβ3, promoting EMT and facilitating migration and invasion of ovarian cancer cells [41]. LOX-1 inhibits autophagy in esophageal cancer by binding to RACK1 and activating the MAPK-ERK signaling pathway [42]. TIMP1 stimulates the FAK-AKT signaling pathway, promoting the proliferation and invasion of colon cancer cells, which can adversely affect patients’ prognoses [43]. MMP9, a well-known secretory endopeptidase, has been extensively studied due to its involvement in critical biological processes such as cell migration, neovascularization, and immune response [44]. The inclusion of these genes in our model indirectly supports its accuracy, as they play a crucial role in controlling different aspects of tumor biology. Moreover, these genes may serve as potential targets for understanding glioma incidence, progression, and immune evasion.
Previous research has shown that the abundance of immune cells in tumors affects the response to immunotherapy in cancer patients [45]. Our study also explored the correlation between risk scores and the degree of immune cell infiltration in gliomas in the hope of finding potential predictive targets for glioma immunotherapy. The results of the ESTIMATE algorithm suggest that tumor stroma and immune cell infiltration are also higher in gliomas with higher glioma risk scores. Additionally, using the ssGSEA score based on immune cell markers obtained from TISIDB, we observed that gliomas in the high-risk score group exhibited higher levels of infiltration from immune cells such as regulatory T (Treg) cells and Th17 cells. It is worth noting that Treg cells [37] and Th17 cells [38] have been shown to have immunosuppressive effects on various malignancies. Thus, high-risk scores may indicate greater immunosuppression and immune evasion in glioma. However, CD8+ T cells, NK cells and dendritic cells were also more abundant in the high-risk scoring group. The three cells are thought to promote killing effect in immune responses against cancer cells [46–48]. This may be explained by the immunosuppressive microenvironment created by tumor cells, which hampers the function of these cells in antigen presentation and tumor killing by reducing self-antigens or changing the antigen type [35, 49], making it difficult for immune cells to exert their tumor-killing effects. Moreover, CIBERSORT analysis revealed an increased abundance of resting NK cells and a decreased abundance of active NK cells in the high-risk score group. The high-risk score group also showed a significantly higher abundance of M2 macrophages, which are known to promote tumor growth, angiogenesis, and immunosuppression [50]. NK cells release cytotoxic particles, such as granzyme and perforin, that directly lyse tumor cells [51], thus suggesting a correlation between high-risk score and enhanced immunosuppressive effects of the glioma microenvironment. To summarize, our results indicate a correlation between risk score and immune cell infiltration in glioma. These findings suggest that tryptophan and nicotinamide metabolism may regulate the tumor microenvironment of glioma and could be potential targets for future glioma immunotherapy research.
The immune checkpoint plays a crucial role in regulating the immune system, maintaining autoimmune tolerance, and controlling the duration and extent of immune responses in peripheral tissues. However, cancer cells can evade immune surveillance and induce immune tolerance through immune checkpoints. Immune checkpoint inhibitor therapy is used to block the immunosuppressive effects of immune checkpoints, thereby regulating the activity of normal immune cells to exert an anti-tumor effect [52]. Nevertheless, the low response rate of this therapy remains a major obstacle to achieving its desired effects [53]. Thus, it is imperative to identify appropriate markers that can predict individual responses to immune checkpoint therapy. Considering that immune checkpoint molecules are major targets for ICI therapy [54], our study explored the correlation between immune checkpoint expression and risk scores to assess the predictive value of risk scores for ICI therapy sensitivity. Our study showed that the high-risk score group exhibited higher levels of immune checkpoint molecule expression. This suggests that the higher risk score predicts a better response to ICI therapy in glioma patients. But our TIDE algorithm showed the higher risk score in patients predicted to be non-responders to ICI treatment, and Pearson coefficient also showed a significant positive correlation between TIDE and risk score. Thus, the results of TIDE algorithm showed poor sensitivity to ICI treatment in the high-risk score group, which contradicts the conclusions of the analysis of the expression of immune checkpoint molecules. The antitumor immune response is not only influenced by immune checkpoints, but also by other factors involved in regulating the dynamic process of tumor immunity and the response of the immune checkpoint blockade. Tumor-intrinsic factors such as tumor mutation burden, MSI and extracellular vesicles, stromal and immune cell status in the tumor microenvironment, metabolic status of the tumor microenvironment, and host systemic factors such as systemic immune response, gut microbiota, and hormone levels also have an impact on the level of response to ICI therapy [39, 55]. Because multiple factors influence the sensitivity of ICI therapy, it is not rigorous enough to judge the sensitivity of ICI therapy solely by the expression level of immune checkpoint molecules. Then, the dysfunction score did not show significant differences between the high-risk score and low-risk score group, suggesting that the functional status of T cells was similar in both groups. The exclusion score of the high-risk score group were significantly higher than that of the low-risk score group, suggesting that the tumor microenvironment of the high-risk score group had a higher degree of rejection of immune cells, and that it was difficult for immune cells to play their killing role in the tumor microenvironment. And MSI score of the high-risk score group was significantly lower than that of the low-risk score group, suggesting that gliomas in the high-risk score group were less likely to be recognized by the immune system and responded poorly to ICI. Myeloid-derived suppressor cells (MDSCs) and cancer-associated fibroblasts (CAF) can prevent T-cell infiltration in tumor tissues by secreting extracellular matrix and suppressor factors [56, 57]. MDSC scores and CAF scores were higher in the high-risk score group suggesting that the immunosuppressive state of the tumor microenvironment of gliomas in the high-risk score group was stronger, which was not conducive to the anti-tumor effects of the immune system. In conclusion, our study suggests that the risk score might serve as a valuable predictor of ICI treatment sensitivity in gliomas.
Single-cell sequencing analysis is able to analyze the interior of a tumor at the individual cell level, allowing for a more profound study of tumor heterogeneity, immune microenvironmental status, and other factors that influence the degree of tumor malignancy. In this study, single-cell RNA analysis was performed for seven genes involved in risk modeling. Our study shows that five of the seven genes are expressed at high levels in macrophages. And TIMP1 and CD44 were expressed at high levels in mesenchymal stem cells (MSCs). In the immune microenvironment of glioblastomas, tumor-associated macrophages and microglia are the most cellular immune cells in the tumor [58]. Macrophages in the glioblastoma immune microenvironment play an important role in promoting tumor progression and inducing tumor immune escape [59]. Blocking TAM infiltration and inhibiting the immunosuppressive polarization state of TAM by chemical agents and activating TAM-mediated phagocytosis against tumor cells have emerged as therapeutic strategies for glioblastomas targeting macrophages in the tumor microenvironment [60]. The role of MSCs in glioma development and progression is more complex. Some studies have shown that MSCs can suppress the migration and invasion of U251 cells by inhibiting EMT [61]. However, it has also been shown that MSCs inhibit apoptosis in glioblastoma cells by promoting chemokine expression [62]. In addition, MSCs are being explored as drug carriers in the treatment of IDH wild-type and mutant gliomas due to their advantage of crossing the blood-brain barrier to enter the interior of the tumor [63].
Given the high-risk coefficient of TGFBI in our risk model, we investigated its unique function in glioma cells and its potential involvement of FAK in regulating the biological behavior of TGFBI. TGFBI is an extracellular matrix protein that binds to various ECM proteins, including fibronectin, laminin, glass connexin, and several collagens, thereby contributing to the formation of the tumor microenvironment [64]. Epithelial-mesenchymal transition refers to the process by which epithelial cells acquire mesenchymal characteristics. It gives cells the ability to metastasize and invade and is involved in the processes of cancer development, metastasis, drug resistance and immunosuppression [16]. FAK, a non-receptor tyrosine kinase, is involved in a variety of cellular activities, including growth factor signaling, cell motility, and cell cycle progression [21]. FAK can promote tumor progression, metastasis, and the formation of a tumor-immunosuppressive microenvironment by affecting the interaction of cancer cells with cells in the tumor microenvironment [21, 22]. Our research revealed that high levels of TGFBI expression significantly enhance the migratory, invasive, and EMT abilities of glioma cells. Importantly, the addition of an FAK inhibitor counteracted the beneficial effects of TGFBI, indicating that TGFBI facilitates these processes by phosphorylating FAK. Numerous studies have demonstrated the involvement of TGFBI in various biological activities associated with tumors. For example, the RNA binding protein LIN28B promotes the migration and epithelial-mesenchymal transition (EMT) changes of cholangiocarcinoma cells through TGFBI [65]. The interaction between DKK3 and TGFBI in the extracellular matrix controls cell adhesion and motility via signaling through focal adhesion kinase [66]. In breast cancer, TGFBI affects blood perfusion and tumor tissue hypoxia, leading to its participation in tumor stemness and metastasis [67]. Moreover, the role of TGFBI in various cancers through the FAK pathway has been extensively studied. TGFBI has been identified as a promoter molecule in gastrointestinal cancer and is involved in the regulation of survival and proliferation of liver cancer cells via activation of the FAK-AKT pathway [68]. Furthermore, TGFBI induces microtubule stability and sensitizes ovarian tumors to paclitaxel through FAK and Rho-dependent microtubule stabilization [69]. Given the high-risk coefficient of TGFBI in our risk model, we investigated its unique function in glioma cells and its potential involvement of FAK in regulating the biological behavior of TGFBI. Our research revealed that high levels of TGFBI expression significantly enhance the migratory, invasive, and EMT abilities of glioma cells. Importantly, the addition of an FAK inhibitor counteracted the beneficial effects of TGFBI, indicating that TGFBI facilitates these processes by phosphorylating FAK. These findings validate the dependability of our risk model and suggest that TGFBI plays a role in the malignant progression of gliomas.
In this study, a risk model related to nicotinamide and tryptophan metabolism in glioma was developed based on bioinformatics. The model has good predictive ability for the prognosis of glioma patients, the immune microenvironmental status of glioma, and the effect of ICI treatment. However, this study still has some limitations. First, in the analysis of the seven genes involved in modelling with scRNA dataset, we used only the scRNA dataset for IDH wild-type gliomas for the analysis due to the lack of single-cell sequencing data for IDH mutant gliomas in the GEO database. This led to a lack of rigor in this study in determining the distribution of model-building-related genes in glioma cells. Second, there is a lack of further in vitro and in vivo experiments to validate the effects and potential mechanisms of the modelling related genes in the metabolism of nicotinamide and tryptophan of glioma cells and in the clearance of glioma cells by immune cells. Therefore, the next step of this study aims to collect tumour samples from IDH wild-type and mutant gliomas for single-cell sequencing analysis, supplemented by in vivo and in vitro experiments to explore the specific relationship between glioma metabolism and immunotherapy, as well as the underlying mechanisms.
Conclusion
In summary, our study unveiled the notable elevation of nicotinamide and tryptophan metabolism was associated with increased malignancy and increased immune infiltration in gliomas. And we developed and validated prognostic models based on these metabolic pathways. These findings offer a novel and targeted approach to predict the prognosis of glioma patients and their responsiveness to immunotherapy. Moreover, they open up potential avenues for identifying immunotherapy targets in glioma patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge TCGA, CGGA and GEO databases for providing their platform and the contributors for uploading their meaningful datasets.
Author contributions
XZ contributed to the project design. YJ, and BX participated in collecting samples and clinical analysis. XF, HZ, MQ and KZ participated in the implementation of the experiments. SW, SG and SL contributed to drafting the manuscript. XZ contributed to revising the manuscript critically. All authors read and approved the final manuscript.
Funding
Not applicable.
Data availability
The datasets TCGA-GBM and TCGA-LGG analyzed in the current study can be found in the Cancer Genome Atlas database (https://portal.gdc.cancer.gov/projects). Datasets CGGA mRNAseq_693, CGGA mRNAseq_125, and CGGA mRNA array_101 were collected in Chinese Glioma Genome Atlas (http://www.cgga.org.cn/). GSE159416 were downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159416).
Declarations
Ethics approval and consent to participate
Extra informed consent is not essential because the data were all obtained from public databases.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sen Wang, Shen Gao and Shaochong Lin contributed equally to this work.
References
- 1.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23(8):1231–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40(1):1–14. [DOI] [PubMed] [Google Scholar]
- 3.Wainwright DA, Chang AL, Dey M, Balyasnikova IV, Kim CK, Tobias A, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin cancer Research: Official J Am Association Cancer Res. 2014;20(20):5290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ladomersky E, Zhai L, Lenzen A, Lauing KL, Qian J, Scholtens DM, et al. IDO1 inhibition synergizes with Radiation and PD-1 blockade to Durably Increase Survival against Advanced Glioblastoma. Clin cancer Research: Official J Am Association Cancer Res. 2018;24(11):2559–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navas LE, Carnero A. NAD(+) metabolism, stemness, the immune response, and cancer. Signal Transduct Target Therapy. 2021;6(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasperi V, Sibilano M, Savini I, Catani MV. Niacin in the Central Nervous System: an update of Biological aspects and clinical applications. Int J Mol Sci. 2019;20(4). [DOI] [PMC free article] [PubMed]
- 7.Jung M, Lee KM, Im Y, Seok SH, Chung H, Kim DY, et al. Nicotinamide (niacin) supplement increases lipid metabolism and ROS-induced energy disruption in triple-negative breast cancer: potential for drug repositioning as an anti-tumor agent. Mol Oncol. 2022;16(9):1795–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv H, Lv G, Chen C, Zong Q, Jiang G, Ye D, et al. NAD(+) metabolism maintains inducible PD-L1 expression to Drive Tumor Immune Evasion. Cell Metabol. 2021;33(1):110–e275. [DOI] [PubMed] [Google Scholar]
- 9.Chen AC, Martin AJ, Choy B, Fernández-Peñas P, Dalziell RA, McKenzie CA, et al. A phase 3 Randomized Trial of Nicotinamide for skin-Cancer chemoprevention. N Engl J Med. 2015;373(17):1618–26. [DOI] [PubMed] [Google Scholar]
- 10.Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discovery. 2019;18(5):379–401. [DOI] [PubMed] [Google Scholar]
- 11.Cheong JE, Sun L. Targeting the IDO1/TDO2-KYN-AhR pathway for Cancer Immunotherapy - challenges and opportunities. Trends Pharmacol Sci. 2018;39(3):307–25. [DOI] [PubMed] [Google Scholar]
- 12.Peyraud F, Guegan JP, Bodet D, Cousin S, Bessede A, Italiano A. Targeting Tryptophan Catabolism in Cancer Immunotherapy Era: challenges and perspectives. Front Immunol. 2022;13:807271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Xiang Y, Zong QB, Dai ZT, Wu H, Zhang HM et al. TDO2 modulates liver cancer cell migration and invasion via the Wnt5a pathway. Int J Oncol. 2022;60(6). [DOI] [PubMed]
- 14.Gomes B, Driessens G, Bartlett D, Cai D, Cauwenberghs S, Crosignani S, et al. Characterization of the selective indoleamine 2,3-Dioxygenase-1 (IDO1) catalytic inhibitor EOS200271/PF-06840003 supports IDO1 as a critical resistance mechanism to PD-(L)1 blockade therapy. Mol Cancer Ther. 2018;17(12):2530–42. [DOI] [PubMed] [Google Scholar]
- 15.Jung KH, LoRusso P, Burris H, Gordon M, Bang YJ, Hellmann MD, et al. Phase I study of the indoleamine 2,3-Dioxygenase 1 (IDO1) inhibitor Navoximod (GDC-0919) administered with PD-L1 inhibitor (atezolizumab) in Advanced Solid tumors. Clin cancer Research: Official J Am Association Cancer Res. 2019;25(11):3220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to Overcome Drug Resistance in Cancer. Molecules. 2016;21(7). [DOI] [PMC free article] [PubMed]
- 17.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–20. [DOI] [PubMed] [Google Scholar]
- 18.Puchalski RB, Shah N, Miller J, Dalley R, Nomura SR, Yoon JG, et al. An anatomic transcriptional atlas of human glioblastoma. Sci (New York NY). 2018;360(6389):660–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Mejia IC, Pijuan J, Navaridas R, Santacana M, Gatius S, Velasco A, et al. Oxidative stress-induced FAK activation contributes to uterine serous carcinoma aggressiveness. Mol Oncol. 2023;17(1):98–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen W, Song Z, Zhong X, Huang M, Shen D, Gao P et al. Sangerbox: a comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 2022;1(3). [DOI] [PMC free article] [PubMed]
- 21.Chuang HH, Zhen YY, Tsai YC, Chuang CH, Hsiao M, Huang MS et al. FAK in Cancer: from mechanisms to therapeutic strategies. Int J Mol Sci. 2022;23(3). [DOI] [PMC free article] [PubMed]
- 22.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14(9):598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21(10):669–80. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Wen L, Bao Y, Tang Z, Zhao J, Zhang X, et al. Gut flora disequilibrium promotes the initiation of liver cancer by modulating tryptophan metabolism and up-regulating SREBP2. Proc Natl Acad Sci USA. 2022;119(52):e2203894119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhai L, Spranger S, Binder DC, Gritsina G, Lauing KL, Giles FJ, et al. Molecular pathways: Targeting IDO1 and other Tryptophan dioxygenases for Cancer Immunotherapy. Clin cancer Research: Official J Am Association Cancer Res. 2015;21(24):5427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhai L, Bell A, Ladomersky E, Lauing KL, Bollu L, Nguyen B, et al. Tumor Cell IDO enhances Immune suppression and decreases Survival Independent of Tryptophan Metabolism in Glioblastoma. Clin cancer Research: Official J Am Association Cancer Res. 2021;27(23):6514–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro MN, Gómez de Las Heras MM, Mittelbrunn M. Nicotinamide adenine dinucleotide metabolism in the immune response, autoimmunity and inflammageing. Br J Pharmacol. 2022;179(9):1839–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo SK, Kwon B. Immune regulation through tryptophan metabolism. Exp Mol Med. 2023;55(7):1371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74(12):3317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberti A, Fernández AF, Fraga MF. Nicotinamide N-methyltransferase: at the crossroads between cellular metabolism and epigenetic regulation. Mol Metabolism. 2021;45:101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu D, Liang CH, Huang B, Zhuang X, Cui W, Yang L, et al. Tryptophan Metabolism acts as a New Anti-ferroptotic Pathway to Mediate Tumor Growth. Adv Sci (Weinheim Baden-Wurttemberg Germany). 2023;10(6):e2204006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janiszewska M, Primi MC, Izard T. Cell adhesion in cancer: beyond the migration of single cells. J Biol Chem. 2020;295(8):2495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Läubli H, Borsig L. Altered cell adhesion and glycosylation promote Cancer Immune suppression and metastasis. Front Immunol. 2019;10:2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basu A, Ramamoorthi G, Albert G, Gallen C, Beyer A, Snyder C, et al. Differentiation and regulation of T(H) cells: a Balancing Act for Cancer Immunotherapy. Front Immunol. 2021;12:669474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philip M, Schietinger A. CD8(+) T cell differentiation and dysfunction in cancer. Nat Rev Immunol. 2022;22(4):209–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers JA, Miller JS. Exploring the NK cell platform for cancer immunotherapy. Nat Reviews Clin Oncol. 2021;18(2):85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tay C, Tanaka A, Sakaguchi S. Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell. 2023;41(3):450–65. [DOI] [PubMed] [Google Scholar]
- 38.Knochelmann HM, Dwyer CJ, Bailey SR, Amaya SM, Elston DM, Mazza-McCrann JM, et al. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell Mol Immunol. 2018;15(5):458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morad G, Helmink BA, Sharma P, Wargo JA. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell. 2021;184(21):5309–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C, Cheng W, Ren X, Wang Z, Liu X, Li G, et al. Tumor Purity as an underlying key factor in Glioma. Clin cancer Research: Official J Am Association Cancer Res. 2017;23(20):6279–91. [DOI] [PubMed] [Google Scholar]
- 41.Yue H, Li W, Chen R, Wang J, Lu X, Li J. Stromal POSTN induced by TGF-β1 facilitates the migration and invasion of ovarian cancer. Gynecol Oncol. 2021;160(2):530–8. [DOI] [PubMed] [Google Scholar]
- 42.Li C, Wang L. TFEB-dependent autophagy is involved in scavenger receptor OLR1/LOX-1-mediated tumor progression. Autophagy. 2022;18(2):462–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma B, Ueda H, Okamoto K, Bando M, Fujimoto S, Okada Y, et al. TIMP1 promotes cell proliferation and invasion capability of right-sided colon cancers via the FAK/Akt signaling pathway. Cancer Sci. 2022;113(12):4244–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Augoff K, Hryniewicz-Jankowska A, Tabola R, Stach K. MMP9: a tough target for targeted therapy for Cancer. Cancers. 2022;14(7). [DOI] [PMC free article] [PubMed]
- 45.Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17(8):807–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y, Jia A, Wang Y, Liu G. CD8(+) T cell exhaustion in anti-tumour immunity: the new insights for cancer immunotherapy. Immunology. 2023;168(1):30–48. [DOI] [PubMed] [Google Scholar]
- 47.Chu J, Gao F, Yan M, Zhao S, Yan Z, Shi B, et al. Natural killer cells: a promising immunotherapy for cancer. J Translational Med. 2022;20(1):240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. 2020;20(1):7–24. [DOI] [PubMed] [Google Scholar]
- 49.Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer. 2021;21(5):298–312. [DOI] [PubMed] [Google Scholar]
- 50.Boutilier AJ, Elsawa SF. Macrophage polarization States in the Tumor Microenvironment. Int J Mol Sci. 2021;22(13). [DOI] [PMC free article] [PubMed]
- 51.Liu S, Galat V, Galat Y, Lee YKA, Wainwright D, Wu J. NK cell-based cancer immunotherapy: from basic biology to clinical development. J Hematol Oncol. 2021;14(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bagchi S, Yuan R, Engleman EG. Immune Checkpoint inhibitors for the treatment of Cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–49. [DOI] [PubMed] [Google Scholar]
- 53.He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020;30(8):660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pérez-Ruiz E, Melero I, Kopecka J, Sarmento-Ribeiro AB, García-Aranda M, De Las Rivas J. Cancer immunotherapy resistance based on immune checkpoints inhibitors: targets, biomarkers, and remedies. Drug Resist Updates: Reviews Commentaries Antimicrob Anticancer Chemother. 2020;53:100718. [DOI] [PubMed] [Google Scholar]
- 55.Ladomersky E, Zhai L, Lauing KL, Bell A, Xu J, Kocherginsky M, et al. Advanced Age increases Immunosuppression in the brain and decreases immunotherapeutic efficacy in subjects with Glioblastoma. Clin cancer Research: Official J Am Association Cancer Res. 2020;26(19):5232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Law AMK, Valdes-Mora F, Gallego-Ortega D. Myeloid-derived suppressor cells as a therapeutic target for Cancer. Cells. 2020;9(3). [DOI] [PMC free article] [PubMed]
- 57.Chen Y, McAndrews KM, Kalluri R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Reviews Clin Oncol. 2021;18(12):792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xuan W, Lesniak MS, James CD, Heimberger AB, Chen P. Context-dependent Glioblastoma-Macrophage/Microglia symbiosis and Associated mechanisms. Trends Immunol. 2021;42(4):280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simonds EF, Lu ED, Badillo O, Karimi S, Liu EV, Tamaki W et al. Deep immune profiling reveals targetable mechanisms of immune evasion in immune checkpoint inhibitor-refractory glioblastoma. J Immunother Cancer. 2021;9(6). [DOI] [PMC free article] [PubMed]
- 60.Khan F, Pang L, Dunterman M, Lesniak MS, Heimberger AB, Chen P. Macrophages and microglia in glioblastoma: heterogeneity, plasticity, and therapy. J Clin Investig. 2023;133(1). [DOI] [PMC free article] [PubMed]
- 61.Lu L, Chen G, Yang J, Ma Z, Yang Y, Hu Y, et al. Bone marrow mesenchymal stem cells suppress growth and promote the apoptosis of glioma U251 cells through downregulation of the PI3K/AKT signaling pathway. Volume 112. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie; 2019. p. 108625. [DOI] [PubMed]
- 62.Akimoto K, Kimura K, Nagano M, Takano S, To’a Salazar G, Yamashita T, et al. Umbilical cord blood-derived mesenchymal stem cells inhibit, but adipose tissue-derived mesenchymal stem cells promote, glioblastoma multiforme proliferation. Stem Cells Dev. 2013;22(9):1370–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oishi T, Koizumi S, Kurozumi K. Mesenchymal stem cells as therapeutic vehicles for glioma. Cancer Gene Ther. 2024;31(9):1306–14. [DOI] [PubMed]
- 64.Corona A, Blobe GC. The role of the extracellular matrix protein TGFBI in cancer. Cell Signal. 2021;84:110028. [DOI] [PubMed] [Google Scholar]
- 65.Puthdee N, Sriswasdi S, Pisitkun T, Ratanasirintrawoot S, Israsena N, Tangkijvanich P. The LIN28B/TGF-β/TGFBI feedback loop promotes cell migration and tumour initiation potential in cholangiocarcinoma. Cancer Gene Ther. 2022;29(5):445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kano J, Wang H, Zhang H, Noguchi M. Roles of DKK3 in cellular adhesion, motility, and invasion through extracellular interaction with TGFBI. FEBS J. 2022;289(20):6385–99. [DOI] [PubMed] [Google Scholar]
- 67.Fico F, Santamaria-Martínez A. TGFBI modulates tumour hypoxia and promotes breast cancer metastasis. Mol Oncol. 2020;14(12):3198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han B, Cai H, Chen Y, Hu B, Luo H, Wu Y, et al. The role of TGFBI (βig-H3) in gastrointestinal tract tumorigenesis. Mol Cancer. 2015;14:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, et al. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12(6):514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets TCGA-GBM and TCGA-LGG analyzed in the current study can be found in the Cancer Genome Atlas database (https://portal.gdc.cancer.gov/projects). Datasets CGGA mRNAseq_693, CGGA mRNAseq_125, and CGGA mRNA array_101 were collected in Chinese Glioma Genome Atlas (http://www.cgga.org.cn/). GSE159416 were downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159416).