Abstract
Hepadnaviruses are enveloped viruses, each with a DNA genome packaged in an icosahedral nucleocapsid, which is the site of viral DNA synthesis. In the presence of envelope proteins, DNA-containing nucleocapsids are assembled into virions and secreted, but in the absence of these proteins, nucleocapsids deliver viral DNA into the cell nucleus. Presumably, this step is identical to the delivery of viral DNA during the initiation of an infection. Unfortunately, the mechanisms triggering the disintegration of subviral core particles and delivery of viral DNA into the nucleus are not yet understood. We now report the identification of a sequence motif resembling a serine- or threonine-proline kinase recognition site in the core protein at a location that is required for the assembly of core polypeptides into capsids. Using duck hepatitis B virus, we demonstrated that mutations at this sequence motif can have profound consequences for RNA packaging, DNA replication, and core protein stability. Furthermore, we found a mutant with a conditional phenotype that depended on the cell type used for virus replication. Our results support the hypothesis predicting that this motif plays a role in assembly and disassembly of viral capsids.
An apparent paradox of virus replication is that infected cells must be permissive for both assembly and disassembly of viral nucleocapsids. Since both pathways cannot act on nucleocapsids at the same time, an important question concerns the mechanisms responsible for the switch from the uncoating to the assembly mode. To solve this problem, viruses have adopted different strategies, among which the best-known strategy relies on the use of different cellular compartments for assembly and disintegration (21). For example, in adenoviruses and orthomyxoviruses, viral disintegration occurs in acidic compartments, and assembly occurs in the cytosol of infected cells (6).
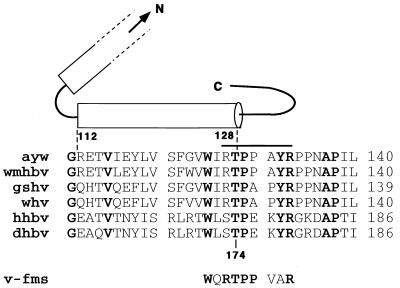
The mechanism controlling assembly and disintegration of the icosahedral core particles of hepadnaviruses is not well understood. Assembly occurs in the cytosol in a two-step process where core proteins form dimers and 120-dimer subunits build capsids (23, 29). Disintegration of viral particles occurs after DNA synthesis and may be activated by a switch that is created on the surface of core particles in response to DNA synthesis. If correct, such a model would predict the presence of regulatory sequence motifs on the surface of viral capsids effecting assembly and disassembly. During a search for known consensus motifs that signify recognition sites for posttranslational processing, we found a threonine-proline kinase recognition site that is conserved among core polypeptides of all known ortho-and avihepadnaviruses. Based on structural and biochemical data, this motif is located at the end of an α-helix that is required for the multimerization of the core dimers to form the icosahedral core shells (1, 2, 5, 16, 23) (Fig. 1). Notably, the motif is located close to the fivefold and twofold icosahedral symmetry axes of the capsid, where it forms a junction between the interior of the capsid and a surface-exposed loop (23). Hence, this motif is in an ideal location to relay a signal, possibly created by phosphorylation and dephosphorylation reactions that could produce changes in the local conformation leading to the destabilization of the critical dimer-dimer interaction and, consequently, to the disintegration of capsids. Such a model predicts that mutations at this site might affect RNA packaging, DNA replication, and the stability of core particles. The genetic approach described in this report yielded variants with such defects and, thus, supported a model predicting that this site plays a pivotal role in viral DNA replication.
FIG. 1.
Conservation of the threonine-proline kinase motif on hepadnavirus capsid proteins. The figure shows the predicted structure of the HBV capsid protein as described by Bottcher et al. (2). The cylinders represent the complete carboxy-terminal α-helix and a portion of the penultimate α-helix. The bar depicts the segment which is exposed on the surface of core particles, as determined by Pushko et al. (16). The numbering of the amino acids refers to the core sequences of HBV (top) and DHBV (bottom). Aligned were the sequences of HBV (ayw), woolly monkey HBV (wmhbv), ground squirrel and woodchuck hepatitis viruses (gshv and whv), heron virus (hhbv), and DHBV (dhbv). The proline kinase recognition site present on the oncogene v-fms is also shown. Conserved residues are shown in boldface. C, carboxy terminal; N, amino terminal.
Mutations at Thr 174 prevent capsid formation or interfere with DNA replication.
To examine whether mutations at the cdc2 kinase motif can destabilize cores and interfere with RNA packaging or DNA synthesis, we replaced Thr 174 with aspartic acid (T174D) or alanine (T174A) to mimic phosphorylated or unphosphorylated Thr, respectively. We then tested the ability of the mutants to produce virus in transfected LMH cells (7). Five days after transfection, viral DNA was isolated as described by Summers et al. (19) and Yang et al. (26) and analyzed by Southern blot hybridization.
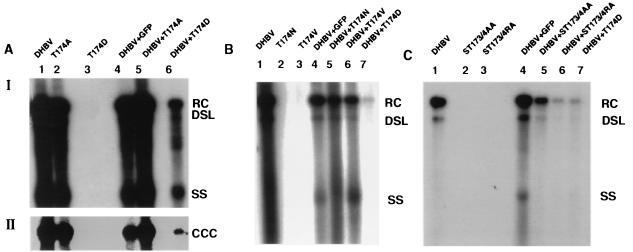
The results showed that variant T174D was defective for viral DNA synthesis and RNA packaging and exhibited a dominant negative phenotype (Table 1; Fig. 2A, lanes 1, 3, and 6; Fig. 3, lane 5). Furthermore, accumulation of core polypeptides expressed with this variant was reduced compared to that in wild-type duck hepatitis B virus (DHBV) (Fig. 3, lane 5). In contrast, mutant T174A produced the same viral DNAs as wild-type DHBV, including covalently closed circular DNA (cccDNA) (Fig. 2A, lanes 1 and 2). The levels of viral DNA intermediates that accumulated in cells transfected with this variant exhibited a slight, twofold reduction compared to the wild type. As expected, cotransfection of the wild-type construct with variant T174A or with a plasmid expressing green fluorescent protein did not interfere with the production of viral DNA intermediates (Fig. 2A through C). Under the assumption that aspartic acid mimics phosphorylated Thr, the results obtained with T174D were consistent with the hypothesis predicting that phosphorylation at Thr 174 could trigger the disintegration of viral capsids. The observed dominant negative effect of T174D suggested that the mutant is competent for the formation of multimeric complexes and, possibly, that only a fraction of core proteins in a nucleocapsid need to be phosphorylated to signal capsid disintegration. As predicted, mutant T174A was competent for viral DNA synthesis, although the presence of cccDNA suggested either that capsids can disintegrate without a requirement for phosphorylation at Thr 174 or that the mutant activated an alternate site, such as serine 173 (Fig. 1), as a substrate for a cdc2-like kinase.
TABLE 1.
Summary of mutants
| Mutant | Effect on
|
Dominant negative phenotype | ||
|---|---|---|---|---|
| DNA synthesis | RNA packaging | Core polypeptides | ||
| T174A | Yesa | Yes | Yes | N/A |
| T174D | No | No | Reduced | Yes |
| T174V | No | Yes | Yes | No |
| T174N | No | No | Reduced | No |
| ST173/4AA | No | No | Yes | No |
| ST173/4RA | No | Yesb | Yes | Yes |
In LMH cells but not in PDHs.
Truncated RNA.
FIG. 2.
Viral DNA replication in transfected LMH cells. The figure shows Southern blots of viral core DNA (panels A I, B, and C) and cccDNA (panel AII) extracted from LMH cells (7) transfected with equal amounts of the indicated plasmids. The cells were maintained in Dulbecco's modified Eagle medium–F-12 medium supplemented with 10% fetal bovine serum, kanamycin (100 μg/ml), penicillin (50 U/ml), and streptomycin (50 μg/ml) and transfected with plasmid DNA using a calcium phosphate cell transfection kit (CalPhos mammalian transfection kit; Clontech, Palo Alto, Calif.). Plasmid DHBV directs expression of the DHBV pg from the cytomegalovirus immediate-early promoter (22). Plasmid T174A contains a dA-to-dG substitution at nucleotide 145 (the nomenclature is according to Mandart et al. [12]). Plasmid T174D contains a dAC-to-dGA substitution at nucleotides 145 and 146. Plasmid T174V contains a dAC-to-dGT substitution at nucleotides 145 and 146. Plasmid T174N contains a dC-to-dA substitution at nucleotide 146. Plasmid ST173/4AA contains a dT-to-dG substitution at nucleotide 142 and a dA-to-G substitution at nucleotide 145. Plasmid ST173/4RA contains a dTC-to-dCG substitution at nucleotides 142 and 143 and a dA-to-dG substitution at nucleotide 145. All mutations were verified by nucleotide sequence analysis. RC, relaxed circular, DSL, double-stranded linear; SS; single stranded; CCC, covalently closed circular.
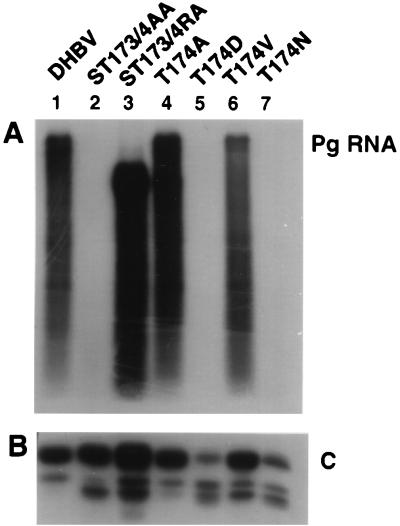
FIG. 3.
Accumulation of packaged pg RNA and core protein in transfected LMH cells. (A) Northern blot analysis of packaged RNA from LMH cells transfected with the indicated plasmids. Encapsidated RNA was extracted as described by Schultz et al. (17). The blot was hybridized with radiolabeled DHBV RNA. (B) Western blot analysis of a total extract from LMH cells transfected with the indicated plasmids. Cells (35-mm-diameter dishes) were lysed in 250 μl of lysis buffer (50 mM Tris-HCl [pH 8], 1% sodium dodecyl sulfate, 1 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin per ml, and 0.7 μg of pepstatin per ml). Three microliters of extract was analyzed by Western blot using antibodies obtained from a rabbit immunized with DHBV core protein. C, core protein; pg RNA, pregenomic RNA.
To examine whether the phenotypes of the two mutants were specific to the selected amino acid changes, we produced two additional mutants with large and small side chains, changing Thr 174 to asparagine (T174N) and valine (T174V), respectively. Because the DHBV core contains a serine residue at position 173 that could have served as an alternate phosphorylation site in the absence of Thr 174, we created two additional variants, ST173/4AA and ST173/4RA. The Ser 173-to-arginine mutation was selected based on the corresponding amino acid sequence motif on the hepatitis B virus (HBV) core protein (Fig. 1).
In contrast to the observations made with T174A, none of the other four variants was competent for viral DNA synthesis (Table 1; Fig 2B [lanes 2 and 3] and C [lanes 2 and 3]). However, two variants, T174V and ST173/4RA, were competent for particle assembly and pregenome (pg) packaging (Fig. 3). Mutant ST173/4RA appeared to accumulate an RNA species with a reduced size compared to the wild type, which could signify replication of a truncated minus-strand DNA species. The other two variants, T174N and ST173/4AA, did not appear to package pg RNA. As with T174D, reduced packaging of pg RNA with T174N could be explained with the results obtained from the Western blot showing that the core polypeptides expressed with this variant accumulated approximately 20-fold less than wild-type core protein (Fig. 3B). In contrast, ST173/4AA displayed normal levels of core protein, suggesting that this variant lost the capacity to form capsids.
Cotransfection of the wild type with ST173/4RA reduced the levels of viral DNA synthesis approximately 10-fold (Fig. 2A, lane 6, and C, lane 6). This result suggested that this variant, like T174D, expressed core polypeptides that were competent to interact with wild-type core subunits and prevent the formation of functional capsids in a dominant negative fashion. In contrast, T174N, T174V, and ST173/4AA did not inhibit wild-type replication, suggesting either that the core polypeptides expressed with these plasmids were incompetent for assembly of capsids or that hybrid capsids supported DNA replication (Fig. 2B, lanes 5 and 6, and C, lane 5). The latter possibility may be relevant in the case of T174V, which is competent for the formation of RNA-containing capsids (Fig. 3A).
These results indicated that replacement of Thr 174 with amino acids carrying bulky side chains interferes with assembly of capsids as well as with the accumulation of core protein in transfected cells. However, the dominant negative inhibition appears to be a specific property of phosphorylated Thr as simulated with T174D. In contrast, replacement of Thr 174 with amino acids containing small side chains did not block capsid formation. In the case of T174V, it blocked DNA synthesis, possibly because it prevented proper arrangement of pg RNA in capsids or induced a steric inhibition of the polymerase to synthesize DNA. Finally, the results showed that serine 173 is required for capsid formation presumably as a component of the helix motif and that even conservative changes, such as in ST173/4RA, can interfere with viral DNA synthesis and, presumably, with the packaging of pg RNA.
The phenotype of DHBV mutant T174A can vary depending on the host cell.
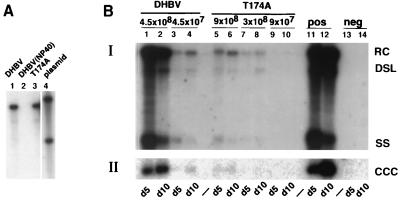
The results obtained so far seemed to suggest that the replacement of Thr 174 with alanine interfered neither with the assembly nor with the subsequent disintegration of mature core particles. However, it is not known whether the mechanisms for the disintegration of cores during the intracellular amplification of cccDNA as measured in LMH cells and following de novo infection are the same. Therefore, we used primary duck hepatocytes (PDHs) to determine whether the virus produced from T174A can initiate an infection. PDHs were prepared as previously described (15, 20). Virus was obtained from the culture supernatants of LMH cells transfected with wild-type DHBV or with T174A. The virus was precipitated with 10% polyethylene glycol 8000, and the pellets were resuspended in culture medium (19). The titer of the concentrated virus suspension was determined by Southern blot analysis using plasmid DNA as a standard and was found to be approximately 3 × 108 virus particles per ml for both the wild type and the mutant (Fig. 4A). Infection of PDHs with the two samples revealed an approximately 40-fold difference in viral DNA replication between the wild-type and the mutant viruses (Fig. 4B). This difference did not depend on the time point used for DNA analysis, because the results obtained with samples taken either 5 or 10 days postinfection (p.i.) yielded comparable results. The levels of nuclear cccDNA expressed from the mutant virus were also approximately 40-fold lower than observed with wild-type virus at both time points (Fig. 4B, panel II, lanes 1 through 6). Thus, the formation of cccDNA did not appear to be inhibited.
FIG. 4.
Infection of PDHs. (A) Southern blot of DNA extracted from enveloped virions present in samples of transfected LMH cells used for the infection of PDHs. A plasmid standard (lane 4) was used to determine the amount of DNA present in lanes 1 and 3. The concentrated culture supernatants were protease treated and incubated with DNase I to remove contaminating core particles, as described by Yu and Summers (27). A sample containing wild-type virus was incubated with NP-40 as a control to demonstrate the efficacy of the protease to digest nonenveloped core particles (lane 2). (B) Southern blot from core (BI) and cccDNA (BII) extracted from PDHs infected with the indicated virus samples. Cells were harvested 5 (d5) and 10 (d10) days postinfection. RC, relaxed circular; DSL, double-stranded linear; SS, single stranded; CCC, covalently closed circular.
To determine whether the reduction in viral DNA synthesis was due to a block in the initiation of infection, we next asked whether the mutant virus could convert its relaxed circular genome into cccDNA. The levels of cccDNA derived directly from the infecting virus in the absence of DNA replication were determined with PDHs that were infected in the presence of phosphonoformic acid (PFA), a known inhibitor of the viral reverse transcriptase (13). Under these conditions, cccDNA was synthesized in PDHs infected with the mutant, albeit at approximately twofold reduced levels compared to wild-type virus (Fig. 5A). Based on Northern blot analysis, the cccDNA derived from T174A was competent for the transcription of the three known viral mRNAs, pg RNA, pre-S-RNA, and S-RNA (Fig. 5B). These results showed that the mutant virus was competent to infect PDHs, enter the uncoating pathway, convert the DNA genome into cccDNA, and produce all three viral RNAs.
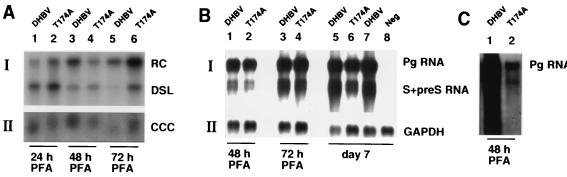
FIG. 5.
Formation of cccDNA from virion DNA, viral RNA expression, and packaged RNA in PDHs. (A) Southern blot of core DNA (input virus) (panel AI) and cccDNA (panel AII) extracted from PDHs infected with the indicated virus samples in the presence of PFA. (B) Northern blot analysis of poly(A)-enriched RNA isolated from PDHs infected with the indicated virus samples. The blot was first hybridized with radiolabeled DHBV DNA (panel BI) and subsequently with a probe corresponding to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (panel BII). Plates were maintained in the presence of PFA and collected 48 and 72 h p.i. or on day 7 p.i. PDHs were infected with 5 × 107 virions per 60-mm culture dish for DHBV and 4 × 107 virions for T174A (panels A and B). (C) Northern blot analysis of packaged RNA from PDHs infected with the indicated virus samples at 3.5 × 108 virions per dish. Encapsidated RNA was extracted as described by Schultz et al. (17). The blot was hybridized with radiolabeled DHBV RNA.
Because the 40-fold reduced level of viral DNA synthesis did not result from a deficiency in nucleocapsid breakdown and cccDNA formation during initiation of infection, we examined whether RNA-containing core particles accumulated in the PDH infected with the mutant. The results showed that RNA packaging occurred at an approximately 100-fold reduced rate compared to that in wild-type virus (Fig. 5C). As noted above, packaging of the same mutant occurred at normal levels in LMH cells, indicating that the defect is host cell specific.
Implications for viral replication.
Our results showed that mutations at a conserved cdc2 kinase-like motif on the hepadnavirus core protein can have profound effects on DNA replication, RNA packaging, and the stability of core particles and, hence, can support a model predicting that this site might play a role in assembly and disintegration of capsids. These experimental observations are in agreement with structural data obtained with the HBV core protein revealing the presence of hydrogen bonds between the hydroxyl of Thr 128 (corresponding to Thr 174 in DHBV) and the backbone of valine 124 and tryptophan 125, respectively (Fig. 1). These interactions stabilize the carboxyl end of the α-helix and thus dimer-dimer formation of core subunits. Hence, changes in the local conformation around Thr 128 are predicted to destabilize the core structure, essentially as demonstrated by our results. The proposal that this motif could relay a signal, created as a consequence of viral DNA synthesis, from the interior of capsids to the outside is supported by the results showing that different mutations in this motif can interfere with DNA replication (ST173/4RA and T174V) and RNA packaging (ST173/4AA), as well as with the stability of core polypeptides (T174D and T174N) (Table 1). Additional support for a functional role of this motif in the viral life cycle has been provided by the observation showing that the phenotype of the variant T174A depends on the cell line selected for virus replication. One possible explanation is that the mutant core protein expressed with T174A is less stable in PDHs than in LMH cells. However, the fact that cccDNA formation was not affected by the T174A mutant indicates that phosphorylation at this site does not play a role in virus disassembly. Nevertheless, independent support for a role of this region in the regulation of viral replication also comes from a recent report by Yuan and Shih (28). These authors found that proline 130 of HBV (Fig. 1) is involved in relaying a signal from the interior of capsids to the outside, as a consequence of DNA synthesis, which induces the interaction of cores with envelope components.
Although the oncogene v-fms is phosphorylated at a Thr residue within a cdc2 sequence motif similar to the conserved motif in hepadnaviruses (Fig. 1) (18), we so far have not found any evidence to suggest that cores expressed in LMH cells are phosphorylated at this site. However, because our results with variant T174D, mimicking phosphorylated Thr, indicated that phosphorylation of the Thr residue could induce rapid degradation of the core protein, our results do not exclude a role for phosphorylation in capsid disassembly. Phosphorylation is known to regulate assembly and disintegration of several multisubunit structures in eukaryotic cells. For example, phosphorylation triggers the breakdown of the nuclear membrane and the fragmentation of the Golgi complex during mitosis (8, 14). The role of phosphorylation in disassembly has found major support through the identification of the proline isomerase PIN1, which induces isomerization of prolines adjacent to phosphorylated serine or Thr residues (9, 25). Proline isomerization may also play a major role during the assembly and disintegration of human immunodeficiency virus type 1, which depends on the presence of cyclophilin in nucleocapsids (3, 10, 11). In addition, the life cycle of the yeast retrotransposon Ty1 appears to be regulated by the kinase Fus3. Fus3 could suppress transposition by phosphorylation of Ty1 capsids, which, in turn, could trigger the degradation of the viruslike particles (4, 24). We are in the process of examining the possible role of core phosphorylation in capsid disintegration with the help of a recently developed in vitro system permissive for viral DNA synthesis in vitro.
Acknowledgments
We acknowledge services provided by the Fox Chase Cancer Center nucleotide sequencing facility. We thank Mike Sauder for help with structural analyses.
This work was supported by grants from the National Institutes of Health and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Beames B, Lanford R E. Insertions within the hepatitis B virus capsid protein influence capsid formation and RNA encapsidation. J Virol. 1995;69:6833–6838. doi: 10.1128/jvi.69.11.6833-6838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 3.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conte D, Jr, Barber E, Banerjee M, Garfinkel D J, Curcio M J. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol Cell Biol. 1998;18:2502–2513. doi: 10.1128/mcb.18.5.2502. . (Erratum, 18:5620.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 6.Greber U F, Singh I, Helenius A. Mechanisms of virus uncoating. Trends Microbiol. 1994;2:52–56. doi: 10.1016/0966-842x(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 7.Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 1987;47:4460–4464. [PubMed] [Google Scholar]
- 8.Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin D J, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- 9.Lu K P, Hanes S D, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 10.Luban J. Absconding with the chaperone: essential cyclophilin-Gag interaction in HIV-1 virions. Cell. 1996;87:1157–1159. doi: 10.1016/s0092-8674(00)81811-5. [DOI] [PubMed] [Google Scholar]
- 11.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 12.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mason W S, Lien J, Petcu D J, Coates L, London W T, O'Connell A, Aldrich C, Custer R P. In vivo and in vitro studies on duck hepatitis B virus replication. In: Robinson W S, Koike K, Will H, editors. Hepadna viruses. A. R. New York, N.Y: Liss; 1987. pp. 3–16. [Google Scholar]
- 14.Moir R D, Goldman R D. Lamin dynamics. Curr Opin Cell Biol. 1993;5:408–411. doi: 10.1016/0955-0674(93)90004-a. [DOI] [PubMed] [Google Scholar]
- 15.Pugh J C, Yaginuma K, Koike K, Summers J. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J Virol. 1988;62:3513–3516. doi: 10.1128/jvi.62.9.3513-3516.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pushko P, Sallberg M, Borisova G, Ruden U, Bichko V, Wahren B, Pumpens P, Magnius L. Identification of hepatitis B virus core protein regions exposed or internalized at the surface of HBcAg particles by scanning with monoclonal antibodies. Virology. 1994;202:912–920. doi: 10.1006/viro.1994.1413. [DOI] [PubMed] [Google Scholar]
- 17.Schultz U, Summers J, Staeheli P, Chisari F V. Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. J Virol. 1999;73:5459–5465. doi: 10.1128/jvi.73.7.5459-5465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smola U, Hennig D, Hadwiger-Fangmeier A, Schutz B, Pfaff E, Niemann H, Tamura T. Reassessment of the v-fms sequence: threonine phosphorylation of the COOH-terminal domain. J Virol. 1991;65:6181–6187. doi: 10.1128/jvi.65.11.6181-6187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summers J, Smith P M, Huang M J, Yu M S. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuttleman J, Pugh J, Summers J. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J Virol. 1986;58:17–25. doi: 10.1128/jvi.58.1.17-25.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittaker G R, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 22.Wu T-T, Condreay L D, Coates L, Aldrich C, Mason W S. Evidence that less-than-full-length pol gene products are functional in hepadnavirus DNA synthesis. J Virol. 1991;65:2155–2163. doi: 10.1128/jvi.65.5.2155-2163.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wynne S A, Crowther R A, Leslie A G. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, Boeke J D. Inhibition of Ty1 transposition by mating pheromones in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2736–2743. doi: 10.1128/mcb.11.5.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaffe M B, Schutkowski M, Shen M, Zhou X Z, Stukenberg P T, Rahfeld J U, Xu J, Kuang J, Kirschner M W, Fischer G, Cantley L C, Lu K P. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 26.Yang W, Mason W S, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu M, Summers J. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol. 1994;68:4341–4348. doi: 10.1128/jvi.68.7.4341-4348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan T T, Shih C. A frequent, naturally occurring mutation (P130T) of human hepatitis B virus core antigen is compensatory for immature secretion phenotype of another frequent variant (I97L) J Virol. 2000;74:4929–4932. doi: 10.1128/jvi.74.10.4929-4932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou S, Standring D N. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]