Abstract
Background
Leaves are important sites for photosynthesis and can convert inorganic substances into organic matter. Photosynthetic performance is an important factor affecting crop yield. Leaf colour is closely related to photosynthesis, and leaf colour mutants are considered an ideal material for studying photosynthesis.
Results
We obtained a yellow-green leaf mutant jym165, using ethyl methane sulfonate (EMS) mutagenesis. Physiological and biochemical analyses indicated that the contents of chlorophyll a, chlorophyll b, carotenoids, and total chlorophyll in the jym165 mutant decreased significantly compared with those in Jiyu47 (JY47). The abnormal chloroplast development of jym165 led to a decrease in net photosynthetic rate and starch content compared with that of JY47. However, quality traits analysis showed that the sum of oil and protein contents in jym165 was higher than that in JY47. In addition, the regional yield (seed spacing: 5 cm) of jym165 increased by 2.42% compared with that of JY47 under high planting density. Comparative transcriptome analysis showed that the yellow-green leaf phenotype was closely related to photosynthesis and starch and sugar metabolism pathways. Genetic analysis suggests that the yellow-green leaf phenotype is controlled by a single recessive nuclear gene. Using Mutmap sequencing, the candidate regions related of leaf colour was narrowed to 3.44 Mb on Chr 10.
Conclusions
Abnormal chloroplast development in yellow-green mutants leads to a decrease in the photosynthetic pigment content and net photosynthetic rate, which affects the soybean photosynthesis pathway and starch and sugar metabolism pathways. Moreover, it has the potentiality to increase soybean yield under dense planting conditions. This study provides a useful reference for studying the molecular mechanisms underlying photosynthesis in soybean.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05740-y.
Keywords: Soybean, Yellow-green leaf mutant, Photosynthesis, Starch and sugar metabolism
Background
Leaves are important sites for photosynthesis and are crucial for plant growth and development [1, 2]. The photosynthetic pigments in the chloroplasts convert light energy into chemical energy, providing a material source for plant growth and development. Abnormal chloroplast development and chlorophyll synthesis metabolism can alter plant leaf colour [3–5]. Leaf colour mutants are closely related to chlorophyll content and photosynthesis; thus, the study of leaf colour mutants has important implications for a better understanding of the molecular mechanisms of light use efficiency. Over the past few decades, various leaf colour mutants have been discovered in different plants, such as albino [6], yellow [7], yellow-green/pale green [8], and purple [9]. Among these, yellow-green leaf mutants are considered an ideal material for studying light-use efficiency. In agricultural production, the full green crop variety decreases photosynthesis in the lower leaves by limiting the penetration of photosynthetically active radiation, which results in a decrease in overall light use efficiency [10–12]. However, pale green crops can make the distribution of effective photosynthetic radiation more uniform, resulting in the increasing of overall light use efficiency [5, 10, 13]. Therefore, yellow-green mutants have been extensively researched in various plants. For example, in rice, the mutation of OsYS83 gene, a yellow-green leaf gene, leads to leaf yellowing by decreasing photosynthetic pigment content and delaying chloroplast development [14]. In cotton, the mutant GaCHLH protein (glycine by a serine at the 517th amino acid) cannot bind to GaCHLD hindering chlorophyll synthesis and leading to a yellow leaf phenotype [15]. In Brassica napus, a mutation in a cytochrome P450-like gene (BnCDE1) inhibit chlorophyll synthesis, resulting in yellow leaf phenotype [16]. In wheat, the yellow-green mutant Ygm exhibits reduced chlorophyll contents and abnormal chloroplast development. Transcriptome analysis showed that CHLH and BCH are key enzymes for yellow-green leaf color phenotype [17].
Soybeans, an important grain crop, are used as a raw material for the production of high-quality protein and healthy plant oils [18, 19]. With improved quality of life, the demand for soybeans is increasing every day. In recent years, the annual soybeans consumption in China has exceeded 100 million tons. However, over 80% of China’s needs for soybean are imported (83.6% in 2020, 85.4% in 2021, and 81.8% in 2022), which significantly affects food security in China. Therefore, it is particularly important to improve China’s soybean production. In traditional breeding, breeders mainly focus on plant architecture, adversity resistance, and nutrient assimilation rather than on photosynthesis-related traits [10, 20, 21].
In this study, we obtained a yellow-green leaf mutant jym165, using ethyl methane sulfonate (EMS) mutagenesis. The photosynthetic pigment content, net photosynthetic rate, and starch content of jym165 decreased compared with Jiyu47 (JY47). Genetic analysis suggests that the yellow-green leaf phenotype is controlled by a single recessive nuclear gene. Furthermore, the differentially expressed genes between JY47 and jym165 were analyzed using transcriptome analysis. The photosynthetic pathways and starch and sugar metabolism pathways in jym165 were affected. The study could provide a useful reference for investigating the molecular mechanisms underlying soybean photosynthesis.
Materials and methods
Plant materials and ethyl methane sulfonate (EMS) mutagenesis
Three thousand high-quality seeds of JY47 were imbibed in 0.5% EMS in the dark for 6 h. The treated seeds were washed with 0.2 mol/L Na2S2O3 and then with running water for 2 h. Mutagenic seeds were sown in experimental fields at Jilin Agricultural University (125°24′25″N, 43°48′9″E). Approximately 10–20 M2 seeds were harvested from each individual M1 plant. Visual phenotypic variations in leaf colour, leaf morphology, plant height, branching, and seed size were recorded at M2 and M3.
The soybean stable genetic yellow-green mutant jym165 was identified from the ethyl methane sulfonate (EMS)-induced mutagenesis population of Jiyu47 (JY47). In order to quickly obtain M5 seeds, M4 seeds of jym165 were sowed in experimental fields of the School of Breeding and Multiplication, Hainan University (109°9′20″N, 18°22′56″E). Hybridization of jym165 (M5) and JY47 was used to construct a segregating population. The leaf colour separation ratio was counted for the F2 segregating population. The hybridization and phenotype identification experiments were completed in experimental fields of Jilin Agricultural University (125°24′25″N, 43°48′9″E).
Soybeans were planted in a mixture of vermiculite and soil (400 g) and cultivated in a climate-controlled room with a 16/8 h light/dark cycle at 26 °C and 50% relative humidity. Soybeans were subjected to drought treatment (no irrigation) at the two-week stage. Excessive watering (900 mL/pot) was performed which caused water-saturated soil before the drought treatment. After nine days without water, rehydration was implemented.
Measurement of photosynthetic pigment content and photosynthetic parameters
The leaves of JY47 and jym165 were collected to measure the photosynthetic pigment content at VC and R5 stages. At the VC stage, we collected 0.2 g shredded leaves from a real soybean leaf. At R5 stage, we collected 0.2 g shredded leaves from the third ternately compound leaf at the top of the soybean plant. The sample locations of JY47 and jym165 were consistent. Shredded leaves (0.2 g) was extracted in 20 ml of 80% (v/v) acetone in the dark for 36 h. The absorbances of extracting solution were measured using microplate photometer (Multiskan™ FC, Thermo Fisher Scientific, USA) at 470 nm, 646 nm, and 663 nm. The chlorophyll a (Chl a), chlorophyll b (Chl b), carotenoid (Car), and total chlorophyll contents were calculated according to the method described by Wellburn [22].
The net photosynthetic rate (A), intercellular CO2 concentration (Ci), stomatal conductance (gs), transpiration rate (E), and water use efficiency (WUE) of JY47 and jym165 were measured using a CIRAS-3 portable photosynthesis system (Hansatech, USA). Regarding parameters of CIRAS-3 portable photosynthesis system are as follows: CO2 reference, Ambient (remove chemicals); H2O reference, Fixed % of reference; Cuvette Flow, 250 cc/min; Analyser Flow, 100 cc/min; Light Source, 1500 µmol/m2/s LED; Temperature Sensor Measurement, Energy Balance; Temperature Control Type, Track cuvette to ambient. Photosynthesis-related parameters were measured on a sunny day (09:00–11:00, 23–28 °C) for 15 individual plants of JY47 and jym165.
Iodine potassium iodide (I2-KI) staining, starch content and soluble sugar content measurement
The mesophyll and leaf vein cell tissues of JY47 and jym165 were collected at R5 stages for tissue sectioning and I2-KI staining. We collected 0.4 cm × 0.5 cm sections from the third ternately compound leaf at the top of soybean plant for I2-KI staining (three biological replicates). For starch and sugar analyses, the third ternately compound leaf from the top of the plant was collected from three individual soybean plants. Simultaneously, the leaves were stained with I2-KI solution for 20 min under vacuum conditions. The starch content and soluble sugar contents were measured using a starch content assay kit (BC0705, Solarbio, Beijing) and a plant soluble sugar content assay kit (BC0035, Solarbio, Beijing), respectively.
Ultrastructural analysis of chloroplast
The fresh leaves of JY47 and jym165 at R5 stages were cut into 0.2 cm x 0.3 cm sections and fixed with 2.5% glutaraldehyde overnight at 4 °C (three biological replicates). The samples were washed with phosphate buffer three times and water one time, and then post-fixed with 1% osmium tetroxide for 3 h at 4 °C. The post-fixed samples were washed three times with phosphate buffe and dehydrated with 30%, 50%, 70%, 80%, 95%, 100%, and 100% ethanol. The dehydrated samples were immersed in a mixture of acetone and epoxy resin (3:1, 3 h; 1:1, 3 h; 1:3, 3 h; ), and embedded in epoxy resin. Sections were prepared using a Leica EM UC7 ultramicrotome (Leica, Germany), and the chloroplast ultrastructure was observed using a transmission electron microscope (HT7800, Hitachi, Japan).
Yield traits and quality traits analysis of the jym165
The grain yield per plant, grain number per plant, and hundred-grain weight of soybeans were analyzed using an intelligent test analyzer (TPKZ-3, TPYN, Zhejiang). The oil and protein contents of soybean seeds were detected using a Multifunctional near-infrared analyzer (NIRS DS2500, FOSS, Beijing). Ten biological replicates were used to assess the yield traits. Five biological replicates were used to assess the quality traits. To evaluate yield traits under different planting densities, the regional yield test (area: 4 m x 1.5 m) of high density (seed spacing: 5 cm) and low density (seed spacing: 10 cm) were conducted in experimental fields at Jilin Agricultural University (125°24′25″N, 43°48′9″E), with three replicates for each density.
Transcriptome analysis
The third ternately compound leaves at the top of the soybean from three individual plants were collected for RNA isolation (leaves were pooled). We performed sequence by Biomarker Technologies Co., Ltd (Beijing, China) (http://www.biomarker.com.cn/). The qualified library was sequenced using the PE150 mode (layout: paired; read length: 150 bp) on an Illumina NovaSeq6000 sequencing platform. High-quality clean data were obtained by filtering raw data, which were then mapped to the Zhonghuang13 reference genome (https://ngdc.cncb.ac.cn/gwh/Assembly/652/show) using HISAT2 [23]. Fragments Per Kilobase of transcript per million fragments mapped (FPKM) was used to quantify the expression level of a transcript. The genes with Fold Change (FC) ≥ 2 and False Discovery Rate (FDR) < 0.01 were regarded as differentially expressed genes (DEGs) between the two samples. We used DESeq2 [24] software for DEGs analysis The biological functions of DEGs were identified using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
Total RNA extraction and qRT-PCR analysis
The total RNA of JY47 and jym165 leaves were extracted using the RNAiso Plus (9108, Takara, DaLian, China). The cDNA was synthesized using M5 Sprint qPCR RT kit with gDNA remover (MF949-01, Mei5bio, Beijing, China). The qRT-PCR analysis with three biological replicates were completed using 2X M5 HiPer Realtime PCR mix (MF015-05, Mei5bio, Beijing, China). The GmEF1a gene [25] was used as the reference gene. Relative expression levels were calculated using the 2−ΔΔCt method [26]. The primers of qRT-PCR were list in Supplementary Table 1.
Genetic mapping of jym165 mutant
Leaves of 30 yellow-green phenotype in F2 segregating population and JY47 were collected for Mutmap analysis [27], which was implemented by Biomarker Technologies Company. High-quality clean data were mapped onto Zhonghuang13 reference genome to provide essential information for downstream variant analysis. SNP candidates were identified using GATK [28] and filtered to generate final SNP sets. Then, the SNP-index of all high-quality SNPs between parents and mixed pool were calculated based on reference genome, which was used for linkage analysis. The 99% of fitted SNP-index of all sites was taken as the linkage threshold.
Results
Phenotypic characterization of the yellow-green leaf mutant
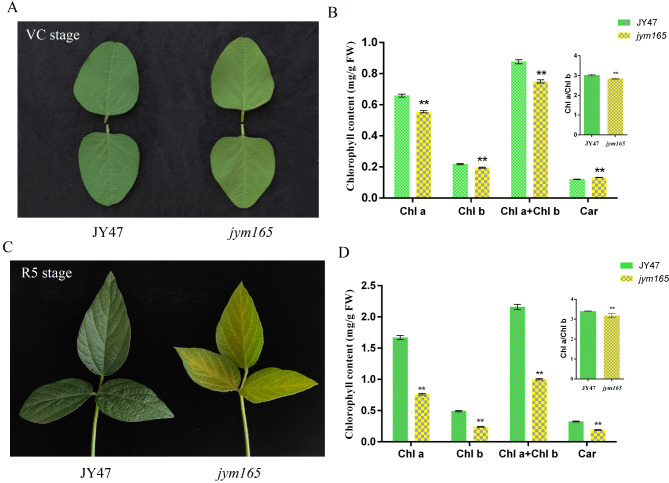
A yellow-green leaf mutant jym165 was identified in a population of EMS-mutagenized Jiyu47 (JY47). The mutant leaves were light-green at the VC stage and distinct yellow-green at the R5 stage (Fig. 1A, C). Chlorophyll content is a major determinant of leaf colour. The chlorophyll content of mutants was measured at the VC and R5 stages. The results showed that, compared with JY47, the chlorophyll content of jym165 was significantly lower, especially at the R5 stage (Fig. 1B, D). Compared with JY47, the total chlorophyll content of jym165 was significantly reduced by 54.30%, the Chl a content was significantly reduced by 51.13%, the Chl b content was significantly reduced by 53.57%, and the Car content was significantly reduced by 41.72% at the R5 stage (Fig. 1D). In addition, the chl a/chl b ratio of jym165 was lower than that of JY47 (Fig. 1B, D), which suggest that jym165 may adapt to low light condition by reducing the waste of light energy.
Fig. 1.
Plant phenotypes of JY47 and jym165 at the VC and R5 stages. A. Plant phenotypes at the VC stage. B. Photosynthetic pigment content of JY47 and jym165 at the VC stage. C. Plant phenotypes at the R5 stage. D. Photosynthetic pigment content of JY47 and jym165 at the R5 stage. These values represent means ± SDs of three biological replicates. Asterisks indicate significant differences according to the Student’s t-test (*P < 0.05, **P < 0.01)
To further investigate the influence of reduction in photosynthetic pigment content on photosynthesis, photosynthesis-related parameters were examined. The net photosynthetic rate (A) and water use efficiency (WUE) of jym165 were significantly lower than those of JY47 (Fig. 2A, E). The intercellular CO2 concentration (Ci) of jym165 was significantly higher than that of JY47 (Fig. 2B). And there was no significant difference in transpiration rate (E) and stomatal conductance (gs) between the mutant jym165 and the control JY47 (Fig. 2C, D). These results indicated that a reduction in photosynthetic pigment content may lead to a lower net photosynthetic rate by decreasing the utilization rate of CO2.
Fig. 2.
Photosynthesis-related parameters of JY47 and jym165 at R5 stages. A. Net photosynthetic rate (A). B. Intercellular CO2 concentration (Ci). C. Transpiration rate (E). D. Stomatal conductance (gs). E. Water use efficiency (WUE). These values represent means ± SDs of ten biological replicates. Asterisks indicate significant differences according to the Student’s t-test (*P < 0.05, **P < 0.01)
Ultrastructural analyses of chloroplast and starch content analyses
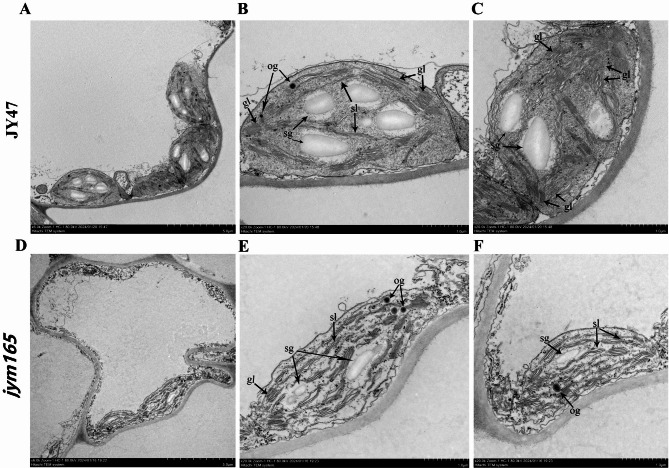
Plant leaf yellowing is typically caused by abnormal chloroplast development. Therefore, we examined the chloroplast ultrastructure of jym165 and JY47 using a transmission electron microscopy (TEM). The chloroplasts of JY47 are larger and plumper than jym165, and contain more starch granules than that in jym165 (Fig. 3A-C). In addition, a large number of grana lamellas were observed in the chloroplast of JY47; however, relatively few grana lamellae were observed in the chloroplast of jym165 (Fig. 3D-F). These results indicate that the chloroplast development of jym165 was abnormal.
Fig. 3.
Chloroplast ultrastructure of JY47 and jym165. A-C. Chloroplast ultrastructure of JY47. D-F. Chloroplast ultrastructure of jym165. sg, starch granules; og, osmiophilic granules; sl, stroma lamellae; gl, grana lamellae. A, D. Bar: 5.0 μm; B, C, E, F. Bar: 1.0 μm
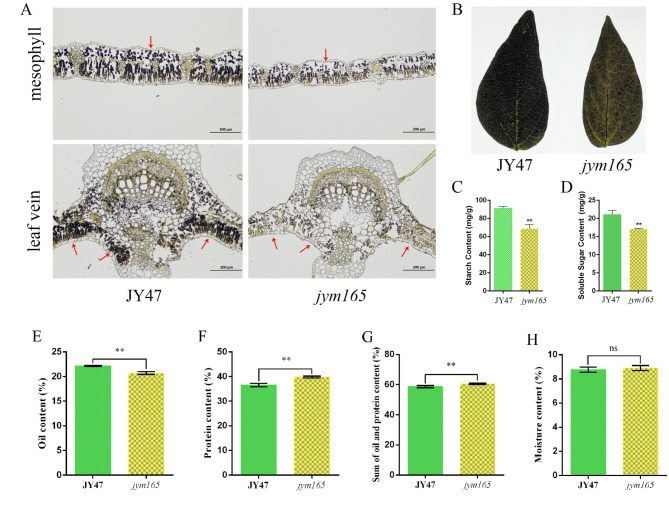
Based on these results, we speculate that the starch content of jym165 is lower than that of JY47. Therefore, iodine-potassium iodide staining was performed. The result of iodine-potassium iodide staining showed that the starch content in mesophyll and leaf vein cell of jym165 was obviously lower than that of JY47 (Fig. 4A, B). Further starch content determination showed that starch content of jym165 was significantly reduced by 24.63% compared with that of JY47 (Fig. 4C). In addition, the soluble sugar content of jym165 was significantly lower than that of JY47 (Fig. 4D). These results indicate that the abnormal chloroplast structure of jym165 leads to a decrease in chlorophyll content and net photosynthetic rate, which, in turn, results in a decrease in starch accumulation.
Fig. 4.
Leaves starch content and quality traits of seed analyses. A. Iodine-potassium iodide stain of mesophyll and leaf vein cell. The red arrows pointing out starch granules. B. Iodine-potassium iodide stain of the entire leaf. C. Starch content determination. D. Soluble sugar content determination. E-H. Quality traits of JY47 and jym165, E. Oil content; F. protein content; G. Sum of oil and protein content; H. Moisture content. The analyses of starch and soluble sugar content were three ten biological replicates, and the quality traits of seed analyses were ten biological replicates. These values represent means ± SDs. Asterisks indicate significant differences according to the Student’s t-test (*P < 0.05, **P < 0.01)
Yield and quality traits analysis
In recent years, breeding yellow-green crop which improves light use efficiency of high-density monocultures to enhance crop yield has become a topic of general interest. Quality traits, like yield traits, are important indicators for variety breeding. Quality trait analyses revealed that the oil content of jym165 was 1.47% lower than that of JY47, the protein content of jym165 was 3.33% higher than that of JY47, and the sum of oil and protein contents of jym165 was 60.54%, which was 1.86% higher than that of JY47 under the same moisture conditions (Fig. 4E–H). After confirming that the quality of jym165 mutant had not decreased, we evaluated its yield traits under different planting densities. Under low planting density, compared with JY47, the grain yield per plant and grain number per plant of jym165 were reduced by 17.93% and 22.52%, respectively (Table 1). Under high planting density, the grain yield per plant and grain number per plant of jym165 were slightly higher than those of JY47, but the difference was not significant. (Table 1). The results of regional yield test (area: 4 m x 1.5 m) showed that the yield of jym165 was 1.923 kg and 2.833 kg under low and high planting densities, respectively; whereas that of JY47 were 2.380 kg and 2.766 kg, respectively (Table 1). This indicates that the regional yield of jym165 increased by 2.42% compared with that of JY47 under high planting density. These results suggested that jym165 mutant can be used as an ideal material for improving grain yield under high-density cultivation.
Table 1.
The yield traits of jym165 mutant under different planting densities
| JY47 | jym165 | |||
|---|---|---|---|---|
| 10 cm | 5 cm | 10 cm | 5 cm | |
| Number of grains per plant | 103.9 ± 17.97 | 56.80 ± 11.08 | 80.50 ± 16.89 | 59.20 ± 5.41 |
| Grain yield per plant (g) | 23.19 ± 2.86 | 12.27 ± 2.89 | 19.03 ± 3.75 | 12.95 ± 1.30 |
| Hundred-grain weight (g) | 21.75 ± 0.97 | 21.46 ± 1.44 | 23.81 ± 2.37 | 21.95 ± 2.18 |
| Regional yield (kg/6m2) | 2.380 ± 0.25 | 2.766 ± 0.26 | 1.923 ± 0.23 | 2.833 ± 0.29 |
Transcriptome analysis
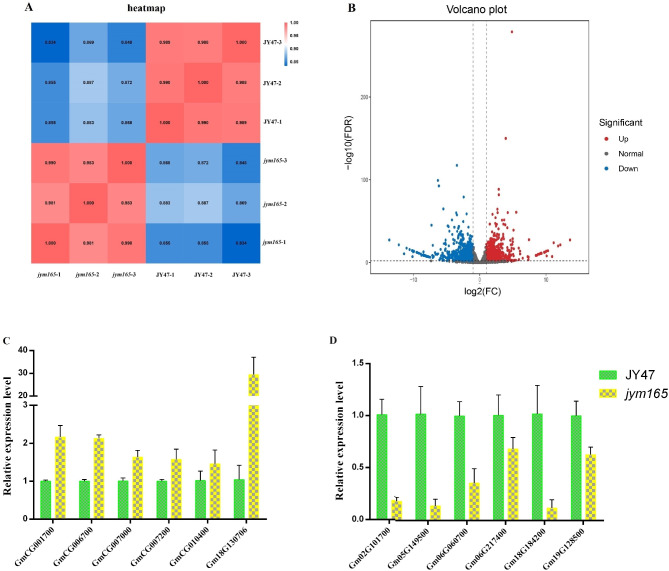
In order to better explain physiological phenotypes, comparative transcriptome analysis of JY47 and jym165 was performed. After trimming adapter contaminations and removing nucleotides with low-quality score, the clean data for each sample reaches 5.70 Gb, which map ratio ranged from 94.55 to 96.55%. The comparative transcriptome results showed a good correlation between repeated samples (Fig. 5A), and a total of 2261 differentially expressed genes (DEGs) were identified between jym165 and JY47, of which 952 genes were upregulated, and 1309 genes were downregulated in jym165 (Fig. 5B and Supplementary Table 2). To validate the reliability of DEGs expression, we selected 12 DEGs (six upregulated genes and six downregulated genes) for qRT-PCR analysis. The qRT-PCR and RNA-Seq results show similar expression patterns and an excellent Pearson correlation (r>0.92), indicating that the RNA-seq results were reliable (Fig. 5C, D and Supplementary Fig. 1).
Fig. 5.
Comparative transcriptome analysis between JY47 and jym165. A Correlation heatmap between samples. B Volcano plot of DEGs between JY47 and jym165. Red dots represent upregulated genes, blue dots represent downregulated genes (FC ≥ 2; FDR < 0.01). C-D. qRT-PCR analysis of DEGs. These values represent means ± SDs of three biological replicates
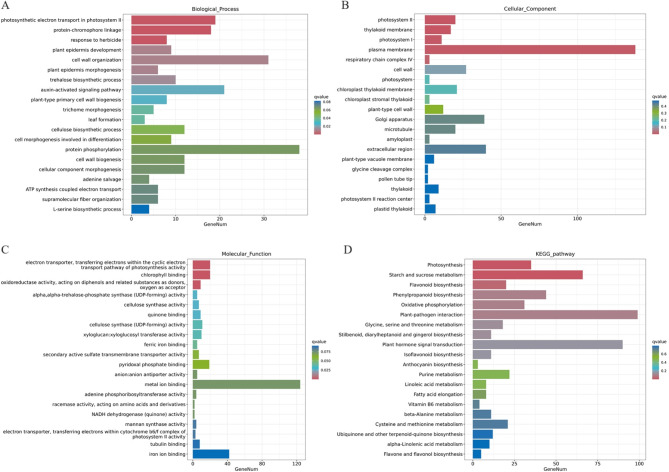
We performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on these DEGs (Fig. 6). In the biological process category (Fig. 6A), the DEGs were significantly enriched in photosynthetic electron transport in photosystem II (GO:0009772), protein-chromophore linkage (GO:0018298), and response to herbicide (GO:0009635). In the cellular component category (Fig. 6B), the DEGs were significantly enriched in photosystem II (GO:0009523), thylakoid membrane (GO:0042651), photosystem I (GO:0009522). In the molecular function category (Fig. 6C), the DEGs were significantly enriched in electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity (GO:0045156), chlorophyll binding (GO:0016168), oxidoreductase activity, acting on diphenols and related substances as donors, and oxygen as acceptor (GO:0016682). In general, all the significant enrichment terms were associated with photosynthesis. The top there enriched KEGG pathways were photosynthesis (ko00195), starch and sucrose metabolism (ko00500), flavonoid biosynthesis (ko00941) (Fig. 6D). These results suggest that the yellow-green leaf phenotype is closely related to photosynthesis, and that the decrease of starch content in yellow-green mutants is caused by abnormal starch and sugar metabolism pathways.
Fig. 6.
GO and KEGG analyses of DEGs between JY47 and jym165. A-C. GO enrichment analysis of the DEGs, A. Biological process; B. Cellular component; C. Molecular function. D. KEGG enrichment analysis of the DEGs. The P value is indicated by the color of the column
Identification of the candidate region for yellow-green leaf mutant
To investigate the inheritance pattern of yellow-green leaf mutant, the hybridization experiment (jym165 x JY47) was conducted. All F1 plants exhibit green leaf phenotype. Among 235 F2 plants, 185 plants showed a green leaf phenotype, and 50 plants showed a yellow-green leaf phenotype, which fitted a 3:1 segregation ratio by the Chi-square test (Xc2=1.5447 < X20.05,1=3.84) (Table 2). These results suggested that the yellow-green leaf phenotype is controlled by a single recessive nuclear gene.
Table 2.
Genetic inheritance analysis of yellow-green leaf mutant
| Observations of F2 (O) | Expectations of F2 (E) | O-E | (|O-E|-0.5)2/E | |
|---|---|---|---|---|
| Green leaf | 185 | 176.25 | 8.75 | 0.3862 |
| Yellow-green leaf | 50 | 58.75 | -8.75 | 1.1585 |
| Total | 235 | 235 | 0 | 1.5447 |
To quickly identify candidate genes for regulating leaf color, we conducted Mutmap sequencing. In this project, a total of 70.47Gbp clean data were obtained, and the average sequencing depth is 33.50x with a genome coverage of 97.78% (Min: 1X). Linkage analysis showed that eight trait-linked candidate regions with total length of 3.44 Mb on Chr 10 were related to leaf colour (Supplementary Fig. 2). These regions contain a total of 127 genes, 43 of which were non-synonymous mutated genes (Supplementary Table 3).
Discussion
The yellow-green mutant is closely related to photosynthesis
Leaves are the primary organs for plant photosynthesis, and their colour is mainly determined by chlorophyll content. Leaf colour mutants have been used as ideal materials for researching photosynthesis of plants over recent years. Researchers have found that the chlorophyll content and net photosynthetic rate of most leaf colour mutants declined [14–16, 29]. Similarly, in the present study, the chlorophyll content and net photosynthetic rate of the yellow-green mutant jym165 were significantly lower than those of the control JY47.
The light-harvesting chlorophyll a/b binding proteins family is a key protein family involved in photosynthetic reactions and is a mainly responsible for light energy capture and transfer [30, 31]. The transcriptome analysis showed that the expression level of the chlorophyll a/b binding protein (SoyZH13:08G070600) in jym165 mutant was significantly lower than that in the control JY47, which may be one of the reasons for decrease of photosynthesis. Previous studies have suggested that most photosynthesis-related genes in yellow-green mutant were downregulated [1, 17, 32]. However, in this study, all photosynthesis-related genes acquired by GO and KEGG enrichment analysis were upregulated in jym165 mutant, except for chlorophyll a/b binding protein (SoyZH13:08G070600). This was probably because the abnormal chloroplast development and low expression of chlorophyll a/b binding protein in jym165 led to a decrease in photosynthesis, and plant provide energy by improving the expression of photosynthesis-related genes to maintain growth. In addition, researchers have found that chlorophyll a/b binding protein can regulate the chlorophyll content and net photosynthetic rate of plants [31, 33–35]. For example, the chlorophyll a/b binding proteins of Actinidia chinensis, AcLhcb3.1/3.2, can increase the chlorophyll content of plants [34]. The chlorophyll a/b binding proteins of wheat, TaLhc2, can promote plant photosynthesis by increasing the chlorophyll content and net photosynthetic rate [35]. Thus, we speculate that the mutation of candidate gene results in a low expression of chlorophyll a/b-binding protein, which leads to the yellow-green phenotype.
In addition, transcriptome analysis revealed that multiple cytochrome P450 family member genes were regulated. Cytochrome P450 proteins belong to a superfamily and play important roles in the synthesis and metabolism of pigments, antioxidants, and phytohormones [16, 36]. The cytochrome P450 proteins CYP97A3 and CYP97C1 participate in lutein biosynthesis [37]. In addition, Yang et al. found that mutation of the cytochrome P450-like gene (BnCDE1) inhibits chlorophyll synthesis, resulting in a yellow-leaf phenotype. In our study, seven cytochrome P450 genes were down-regulated [16]. We speculate that the mutation of candidate genes results in the low expression of some cytochrome P450 proteins, which leads to a yellow-green phenotype.
There is a difference in drought tolerance between jym165 mutant and JY47
The WUE of plant leaves and soluble sugar content play important roles in plant stress resistance [38–42]. Many studies have shown that improving WUE and soluble sugar content can enhance drought tolerance. For example, under drought stress, the TaPYL1-1B gene, ABA receptor gene, can enhance drought tolerance of wheat by increasing WUE [43]. Pu-miR172d can enhances drought tolerance of poplar by targeting PuGTL1 to regulate water use efficiency [44]. The cotton DRE-binding transcription factor gene, GhDREB, can enhance drought tolerance of plants by increasing the content of soluble sugars [45]. The novel small signaling peptide of rice, OsDSSR1, promoted the accumulation of soluble sugars and free proline to enhance drought tolerance in plants [46]. In the present study, the photosynthetic parameters of field-grown soybeans showed that the WUE of jym165 was significantly lower than that of JY47 (Fig. 2E), and the soluble sugar content of jym165 was significantly lower than that of JY47 (Fig. 4D). Therefore, we speculate that the jym165 mutant is more sensitive to drought stress than the control JY47. However, we found that the jym165 has a stronger drought tolerance than JY47. After nine days without watering, the foliar damage of JY47 was more serious than that in jym165 (Supplementary Fig. 3). Similarly, barley breeders at the International Center for Agricultural Research in the Dry Areas (ICARDA, Aleppo, Syria) found that the barley variety Tadmor, with pale green leaves, is more adaptable to high-temperature and drought environments [47]. Tardy et al. [48] speculated that pale green leaves limit the heating effects of light by decreasing the light absorptance of the leaf under drought conditions, resulting in Tadmor being more adaptable to high-temperature and drought environments. In our study, we found that the E and gs of jym165 were lower than those of JY47. Thus, we speculate that the lower E and gs values of jym165 may reduce water loss, enhancing drought tolerance. In future studies, we will further investigate this molecular mechanism.
Yield traits and quality traits are difference between jym165 mutant and JY47
Photosynthesis is an important factor affecting crop yield. Breeders have proposed that improving canopy photosynthesis by reducing the leaf pigment content can increase the yield of high-density monocultures [10, 49]. In our study, the grain yield per plant and grain number per plant of jym165 were slightly higher than those of JY47 at high planting densities. This may be because the canopy leaves of JY47 limited the penetration of light, which resulted in a decrease in the overall light use efficiency; however, the jym165 mutant makes the distribution of effective photosynthetic radiation more uniform, resulting in an increase in the overall light use efficiency. In addition, the regional yield of jym165 increased by 2.42% compared to that of JY47 under high planting density. These results suggest that the jym165 mutant could be used as an ideal material for studying photosynthesis to improve grain yield under high-density cultivation.
Quality traits are important indicators in variety breeding, especially seed protein and oil content. Studies have shown a negative correlation between protein content and oil content [50, 51]. In our study, the oil content of jym165 was lower than that in JY47, and the protein content of jym165 was higher than that in JY47. Transcriptome analysis revealed 62 differentially expressed genes involved in lipid metabolism, among them, 41 genes were down-regulated and 21 genes were up-regulated (Supplementary Tables 2 and Supplementary Table 4). The change in lipid metabolism profile may be caused by the mutation of candidate gene causing yellow-green phenotype, or it may be caused by the mutation of a gene related to lipid metabolism affecting the lipid metabolism profile, which leads to a decrease in seed oil content and an increase in protein content.
RNA-binding protein CP33 may be the gene of regulating leaves color
Chloroplasts development is crucial for the growth and development of plants. When chloroplast development is abnormal, the leaf colour of plants will change, and in severe cases, it can lead to plant death. Chloroplast development is jointly regulated by nuclear and plastid genes. Among them, the nuclear-encoded chloroplast RNA binding protein plays an important role in chloroplast RNA metabolism and chloroplast biogenesis. In Arabidopsis thaliana, the chloroplast RNA binding protein mutants cp33a can only survive as albino seedlings on media containing sucrose [52]. The cp29a and cp31a mutants fail to develop normal chloroplasts and exhibit bleaching of newly emerging tissues under cold conditions [53, 54]. The chloroplast-targeted S1 RNA-binding domain protein mutant sdp displays dwarf and pale-green phenotypes [55]. Similarly, the tomato RNA binding protein SlRBP1 maintains chloroplast development by interacting with the eukaryotic translation initiation factor SleIF4A2 to promote its binding to RNA targets related to photosynthetic metabolic pathways. Loss-of-function of SlRBP1 results in dwarf tomato plants with yellow leaves [56]. In Nicotiana benthamiana, the chloroplast RNA-binding protein PRBP is involved in chloroplast RNA processing and chloroplast biogenesis. The PRBP-silenced plant displayed a pale-green to yellow phenotypes [57]. In our study, the candidate regions related of leaf color contain a chloroplast RNA-binding protein CP33 gene (SoyZH13_10G078400). Therefore, we speculate that the chloroplast RNA-binding protein CP33 may regulate leaf colour.
Conclusions
The abnormal chloroplast development in soybean yellow-green leaf mutants leads to a decrease in the photosynthetic pigment content and net photosynthetic rate, affecting soybean photosynthesis and starch and sugar metabolism pathways. Moreover, it has the potentiality to increase soybean yield under dense planting conditions. The study provides a useful reference for studying the molecular mechanisms underlying photosynthesis in soybeans.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 2: The primers of qRT-PCR
Supplementary Material 3: Differentially expressed genes between jym165 and JY47
Supplementary Material 4: Non synonymous mutation SNP sites within candidate regions
Supplementary Material 5: Differentially expressed genes related to lipid metabolism
Acknowledgements
We thank Dr. Qi Shan (Jilin Agricultural University) for helping us to collect samples.
Abbreviations
- EMS
Ethyl methane sulfonate
- Chl a
Chlorophyll a
- Chl b
Chlorophyll b
- Car
Carotenoid
- A
Photosynthetic rate
- Ci
Intercellular CO2 concentration
- Gs
Stomatal conductance
- E
Transpiration rate
- WUE
Water use efficiency
- FC
Fold Change
- FDR
False Discovery Rate
- FPKM
Fragments Per Kilobase of transcript per million fragments mapped
- DEGs
Differentially expressed genes
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- TEM
Transmission electron microscopy
Author contributions
YZ1 and KHX performed the experiments and wrote the manuscript. MXZ performed the RNA extraction. HTG performed the RT-qPCR experiment. YGZ analyzed the data. WBY and YZ2 performed the DNA extraction. WPZ, CF, YXL, and YJ assisted with the experiments. KHX directed the design of the experiments and revised the manuscript. All the authors have read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32301751), the Natural Science Foundation of Hainan Provincial (324RC451), and Postdoctoral Station Program of Hainan Province, China.
Data availability
Our data sets supporting the results of this study are included within the article and its additional files. Sequencing data used in this manuscript can be found in Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa) in National Genomics Data Center database [57] (GSA accession numbers: CRA018202 and CRA018205).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang T, Dong X, Yuan X, Hong Y, Zhang L, Zhang X, Chen S. Identification and characterization of CsSRP43, a major gene controlling leaf yellowing in cucumber. Hortic Res. 2022;9:uhac212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Chen N, Shen S. iTRAQ-based quantitative proteomics analysis reveals the mechanism of golden-yellow leaf mutant in hybrid paper mulberry. Int J Mol Sci. 2021;23(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Cui X, Wang Y, Wu J, Gu X, Lu T. The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol Plant. 2017;10(1):86–98. [DOI] [PubMed] [Google Scholar]
- 4.Wu Z, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L, et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007;145(1):29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakowska K, Alberti G, Genesio L, Peressotti A, Delle Vedove G, Gianelle D, Colombo R, Rodeghiero M, Panigada C, Juszczak R, et al. Leaf and canopy photosynthesis of a chlorophyll deficient soybean mutant. Plant Cell Environ. 2018;41(6):1427–37. [DOI] [PubMed] [Google Scholar]
- 6.Liu C, Zhu H, Xing Y, Tan J, Chen X, Zhang J, Peng H, Xie Q, Zhang Z. Albino Leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J Exp Bot. 2016;67(18):5339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandhu D, Coleman Z, Atkinson T, Rai KM, Mendu V. Genetics and physiology of the nuclearly inherited yellow foliar mutants in soybean. Front Plant Sci. 2018;9:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang S, Wang X, Yan W, Zhang Y, Song P, Guo Y, Xie K, Hu J, Hou J, Wu Y, et al. Melon yellow-green plant (cmygp) encodes a Golden2-like transcription factor regulating chlorophyll synthesis and chloroplast development. Theor Appl Genet. 2023;136(4):66. [DOI] [PubMed] [Google Scholar]
- 9.Lim GH, Kim SW, Ryu J, Kang SY, Kim JB, Kim SH. Upregulation of the MYB2 transcription factor is associated with increased accumulation of anthocyanin in the leaves of Dendrobium bigibbum. Int J Mol Sci. 2020, 21(16). [DOI] [PMC free article] [PubMed]
- 10.Cutolo EA, Guardini Z, Dall’Osto L, Bassi R. A paler shade of green: engineering cellular chlorophyll content to enhance photosynthesis in crowded environments. New Phytol. 2023;239(5):1567–83. [DOI] [PubMed] [Google Scholar]
- 11.Walker BJ, Drewry DT, Slattery RA, VanLoocke A, Cho YB, Ort DR. Chlorophyll can be reduced in crop canopies with little penalty to photosynthesis. Plant Physiol. 2018;176(2):1215–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slattery RA, VanLoocke A, Bernacchi CJ, Zhu XG, Ort DR. Photosynthesis, light use efficiency, and yield of reduced-chlorophyll soybean mutants in field conditions. Front Plant Sci. 2017;8:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Junfei G, Zhenxiang Z, Zhikang, Li Y, Chen, Zhiqin RWJFC. Rice (Oryza sativa L.) with reduced chlorophyll content exhibit higher photosynthetic rate and efficiency, improved canopy light distribution, and greater yields than normally pigmented plants. Field Crop Res. 2017;200:58–70. [Google Scholar]
- 14.Ma X, Sun X, Li C, Huan R, Sun C, Wang Y, Xiao F, Wang Q, Chen P, Ma F, et al. Map-based cloning and characterization of the novel yellow-green leaf gene ys83 in rice (Oryza sativa). Plant Physiol Biochem. 2017;111:1–9. [DOI] [PubMed] [Google Scholar]
- 15.Fan L, Hou Y, Zheng L, Shi H, Liu Z, Wang Y, Li S, Liu L, Guo M, Yang Z, et al. Characterization and fine mapping of a yellow leaf gene regulating chlorophyll biosynthesis and chloroplast development in cotton (Gossypium arboreum). Gene. 2023;885:147712. [DOI] [PubMed] [Google Scholar]
- 16.Yang M, Wan S, Chen J, Chen W, Wang Y, Li W, Wang M, Guan R. Mutation to a cytochrome P450 -like gene alters the leaf color by affecting the heme and chlorophyll biosynthesis pathways in Brassica napus. Plant J. 2023;116(2):432–45. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Shi N, An X, Liu C, Fu H, Cao L, Feng Y, Sun D, Zhang L. Candidate genes for yellow leaf color in common wheat (Triticum aestivum L.) and major related metabolic pathways according to transcriptome profiling. Int J Mol Sci. 2018, 19(6). [DOI] [PMC free article] [PubMed]
- 18.Ainsworth EA, Yendrek CR, Skoneczka JA, Long SP. Accelerating yield potential in soybean: potential targets for biotechnological improvement. Plant Cell Environ. 2012;35(1):38–52. [DOI] [PubMed] [Google Scholar]
- 19.Liu S, Zhang M, Feng F, Tian Z. Toward a Green Revolution for soybean. Mol Plant. 2020;13(5):688–97. [DOI] [PubMed] [Google Scholar]
- 20.Divi UK, Krishna P. Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. Nat Biotechnol. 2009;26(3–4):131–6. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, et al. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature. 2018;560(7720):595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wellburn AR, Lichtenthaler H. Formulae and program to determine total carotenoids and chlorophylls a and B of leaf extracts in different solvents. Springer Neth. 1984: 9–12.
- 23.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Deng Y, Zhou Y, Chen H, Dong Y, Wang N, Li X, Jameel A, Yang H, Zhang M, et al. Normalization for relative quantification of mRNA and microRNA in soybean exposed to various abiotic stresses. PLoS ONE. 2016;11(5):e0155606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol. 2012;30(2):174–8. [DOI] [PubMed] [Google Scholar]
- 28.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma YY, Shi JC, Wang DJ, Liang X, Wei F, Gong CM, Qiu LJ, Zhou HC, Folta KM, Wen YQ, et al. A point mutation in the gene encoding magnesium chelatase I subunit influences strawberry leaf color and metabolism. Plant Physiol. 2023;192(4):2737–55. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Yang W, Liu S, Meng Y, Zhu Z, Liang R, Cao K, Xie Y, Li X. Genome-wide analysis and identification of light-harvesting chlorophyll a/b binding (LHC) gene family and BSMV-VIGS silencing TaLHC86 reduced salt tolerance in wheat. Int J Biol Macromol. 2023;242(Pt 3):124930. [DOI] [PubMed] [Google Scholar]
- 31.Nick S, Meurer J, Soll J, Ankele E. Nucleus-encoded light-harvesting chlorophyll a/b proteins are imported normally into chlorophyll b-free chloroplasts of Arabidopsis. Mol Plant. 2013;6(3):860–71. [DOI] [PubMed] [Google Scholar]
- 32.Nie L, Zheng Y, Zhang L, Wu Y, Zhu S, Hou J, Chen G, Tang X, Wang C, Yuan L. Characterization and transcriptomic analysis of a novel yellow-green leaf wucai (Brassica campestris L.) germplasm. BMC Genomics. 2021;22(1):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietrzykowska M, Suorsa M, Semchonok DA, Tikkanen M, Boekema EJ, Aro EM, Jansson S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell. 2014;26(9):3646–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo J, Abid M, Tu J, Gao P, Wang Z, Huang H. Genome-wide identification of the LHC gene family in kiwifruit and regulatory role of AcLhcb3.1/3.2 for chlorophyll a content. Int J Mol Sci. 2022, 23(12). [DOI] [PMC free article] [PubMed]
- 35.Han X, Han S, Li Y, Li K, Yang L, Ma D, Fang Z, Yin J, Zhu Y, Gong S. Double roles of light-harvesting chlorophyll a/b binding protein TaLhc2 in wheat stress tolerance and photosynthesis. Int J Biol Macromol. 2023;253(Pt 5):127215. [DOI] [PubMed] [Google Scholar]
- 36.Renault H, Bassard JE, Hamberger B, Werck-Reichhart D. Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Curr Opin Plant Biol. 2014;19:27–34. [DOI] [PubMed] [Google Scholar]
- 37.Niu G, Guo Q, Wang J, Zhao S, He Y, Liu L. Structural basis for plant lutein biosynthesis from α-carotene. Proc Natl Acad Sci USA. 2020;117(25):14150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Driscoll AW, Bitter NQ, Sandquist DR, Ehleringer JR. Multidecadal records of intrinsic water-use efficiency in the desert shrub Encelia farinosa reveal strong responses to climate change. Proc Natl Acad Sci USA. 2020;117(31):18161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang H, Ma QJ, Zhong MS, Gao HN, Li YY, Hao YJ. The apple palmitoyltransferase MdPAT16 influences sugar content and salt tolerance via an MdCBL1-MdCIPK13-MdSUT2.2 pathway. Plant J. 2021;106(3):689–705. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Meng R, Liu Y, Chen S, Jiang J, Wang L, Zhao S, Wang Z, Fang W, Chen F, et al. Heterografted chrysanthemums enhance salt stress tolerance by integrating reactive oxygen species, soluble sugar, and proline. Hortic Res. 2022;9:uhac073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozturk M, Turkyilmaz Unal B, García-Caparrós P, Khursheed A, Gul A, Hasanuzzaman M. Osmoregulation and its actions during the drought stress in plants. Physiol Plant. 2021;172(2):1321–35. [DOI] [PubMed] [Google Scholar]
- 42.Wang A, Lam SK, Hao X, Li FY, Zong Y, Wang H, Li P. Elevated CO2 reduces the adverse effects of drought stress on a high-yielding soybean (Glycine max (L.) Merr.) Cultivar by increasing water use efficiency. Plant Physiol Biochem. 2018;132:660–5. [DOI] [PubMed] [Google Scholar]
- 43.Mao H, Jian C, Cheng X, Chen B, Mei F, Li F, Zhang Y, Li S, Du L, Li T, et al. The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use efficiency. Plant Biotechnol J. 2022;20(5):846–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Q, Wang Z, Yu S, Li W, Zhang M, Yang J, Li D, Yang J, Li C. Pu-miR172d regulates stomatal density and water-use efficiency via targeting PuGTL1 in poplar. J Exp Bot. 2021;72(4):1370–83. [DOI] [PubMed] [Google Scholar]
- 45.Gao SQ, Chen M, Xia LQ, Xiu HJ, Xu ZS, Li LC, Zhao CP, Cheng XG, Ma YZ. A cotton (Gossypium hirsutum) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep. 2009;28(2):301–11. [DOI] [PubMed] [Google Scholar]
- 46.Cui Y, Li M, Yin X, Song S, Xu G, Wang M, Li C, Peng C, Xia X. OsDSSR1, a novel small peptide, enhances drought tolerance in transgenic rice. Plant Sci. 2018;270:85–96. [DOI] [PubMed] [Google Scholar]
- 47.Oosterom EJV, Acevedo EJE. Adaptation of barley (Hordeum vulgare L.) to harsh mediterranean environments. Euphytica. 1992;62(1):29–38. [Google Scholar]
- 48.Tardy F, Créach A, Havaux M. Photosynthetic pigment concentration, organization and interconversions in a pale green Syrian landrace of barley (Hordeum vulgare L., Tadmor) adapted to harsh climatic conditions. Plant Cell Environ. 1998;21(5):479–89. [Google Scholar]
- 49.Croce R, Carmo-Silva E, Cho YB, Ermakova M, Harbinson J, Lawson T, McCormick AJ, Niyogi KK, Ort DR, Patel-Tupper D et al. Perspectives on improving photosynthesis to increase crop yield. Plant Cell. 2024, koae132. [DOI] [PMC free article] [PubMed]
- 50.Duan Z, Zhang M, Zhang Z, Liang S, Fan L, Yang X, Yuan Y, Pan Y, Zhou G, Liu S, et al. Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol J. 2022;20(9):1807–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teubner M, Fuß J, Kühn K, Krause K, Schmitz-Linneweber C. The RNA recognition motif protein CP33A is a global ligand of chloroplast mRNAs and is essential for plastid biogenesis and plant development. Plant J. 2017;89(3):472–85. [DOI] [PubMed] [Google Scholar]
- 52.Kupsch C, Ruwe H, Gusewski S, Tillich M, Small I, Schmitz-Linneweber C. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell. 2012;24(10):4266–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okuzaki A, Rühle T, Leister D, Schmitz-Linneweber C. The acidic domain of the chloroplast RNA-binding protein CP31A supports cold tolerance in Arabidopsis thaliana. J Exp Bot. 2021;72(13):4904–14. [DOI] [PubMed] [Google Scholar]
- 54.Han JH, Lee K, Lee KH, Jung S, Jeon Y, Pai HS, Kang H. A nuclear-encoded chloroplast-targeted S1 RNA-binding domain protein affects chloroplast rRNA processing and is crucial for the normal growth of Arabidopsis thaliana. Plant J. 2015;83(2):277–89. [DOI] [PubMed] [Google Scholar]
- 55.Ma L, Yang Y, Wang Y, Cheng K, Zhou X, Li J, Zhang J, Li R, Zhang L, Wang K, et al. SlRBP1 promotes translational efficiency via SleIF4A2 to maintain chloroplast function in tomato. Plant Cell. 2022;34(7):2747–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park YJ, Cho HK, Jung HJ, Ahn CS, Kang H, Pai HS. PRBP plays a role in plastid ribosomal RNA maturation and chloroplast biogenesis in Nicotiana Benthamiana. Planta. 2011;233(6):1073–85. [DOI] [PubMed] [Google Scholar]
- 57.Members CNCB-NGDC, Partners. Database Resources of the National Genomics Data Center. China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022;50(D1):D27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 2: The primers of qRT-PCR
Supplementary Material 3: Differentially expressed genes between jym165 and JY47
Supplementary Material 4: Non synonymous mutation SNP sites within candidate regions
Supplementary Material 5: Differentially expressed genes related to lipid metabolism
Data Availability Statement
Our data sets supporting the results of this study are included within the article and its additional files. Sequencing data used in this manuscript can be found in Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa) in National Genomics Data Center database [57] (GSA accession numbers: CRA018202 and CRA018205).