Abstract
Background
Skin cutaneous melanoma (SKCM) poses a significant public health challenge due to its aggressive nature and limited treatment options. To address this, the study introduces the Tumor Mutational Burden-Derived Immune lncRNA Prognostic Index (TILPI) as a potential prognostic tool for SKCM.
Methods
TILPI was developed using a combination of gene set variation analysis, differential expression analysis, and COX regression analysis. Additionally, functional experiments were conducted to validate the findings, focusing on the effects of STARD4-AS1 knockdown on SKCM tumor cell behavior. These experiments encompassed assessments of tumor cell proliferation, gene and protein expression, migration, invasion, and in vivo tumor growth.
Results
The results demonstrated that knockdown of STARD4-AS1 led to a significant reduction in tumor cell proliferation and impaired migration and invasion abilities. Moreover, it resulted in the downregulation of ADCY4, PRKACA, and SOX10 gene expression, as well as decreased protein expression of ADCY4, PRKACA, and SOX10. In vivo experiments further confirmed the efficacy of STARD4-AS1 knockdown in reducing tumor growth.
Conclusions
This study elucidates the mechanistic role of STARD4-AS1 and its downstream targets in SKCM progression, highlighting the importance of the ADCY4/PRKACA/SOX10 pathway. The integration of computational analysis with experimental validation enhances the understanding of TILPI and its clinical implications. Overall, the findings underscore the potential of novel computational frameworks like TILPI in predicting and managing SKCM, particularly through targeting the ADCY4/PRKACA/SOX10 pathway.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05732-4.
Keywords: Skin cutaneous melanoma, Tumor mutational burden, STARD4-AS1 knockdown, ADCY4/PRKACA/SOX10 pathway, Immune landscape
Introduction
The incidence of skin cutaneous melanoma (SKCM) continues to rise, but the mortality from advanced SKCM has declined over the past decade [1]. Immunotherapy and targeted therapy are the preferred clinical approaches for treating SKCM [2]. Biomarkers such as tumor mutation burden (TMB), lactate dehydrogenase (LDH), and neoantigen load (NAL) have been used to predict the response of ICI to cutaneous melanoma. Among them, TMB is a predictive biomarker for ICI treatment of SKCM.
TMB indicates the mutation counts per million bases. FDA had approved the PD-1 antibody called pembrolizumab for the treatment of high TMB (≥ 10 mutations/megabase) solid tumors (https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/125514s068lbl.pdf). Several researchers have demonstrated that high TMB patients can achieve better effects from immunotherapy [3–6]. This suggests that TMB is a novel and independent biomarker for assessing immunotherapy efficacy [7, 8]. Furthermore, the predictive effect of high TMB extends to SKCM [9].
Long non-coding RNA (lncRNA) is over 200 bp and disabled to encode protein. Several pieces of evidence suggest that lncRNA regulates the expression of nearby genes. For example, lncRNAs mediate resistance to BRAF inhibitors in human SKCM cells and regulate nearby genes [10]. CDR1as depletion is connected to the epigenetic silencing of LINC00632, which is the original lncRNA of CDR1as. Moreover, the level of CDR1 as indicates cellular states connected with distinct effects of immunotherapy [11]. Therefore, the effect of lncRNA on the therapy of SKCM cannot be ignored.
However, the further development of TMB has encountered some problems in recent years. The accuracy of the TMB measurement method has been questioned, and incorporating patients' TMB into predictive models to guide clinical decisions has proven to be challenging [12]. Despite an extensive literature review, we found no existing TMB-based computational framework for predicting patient prognosis or the potential benefits of immunotherapy combined with targeted therapy. Therefore, we hoped to contribute to the clinical application of TMB (Fig. 1).
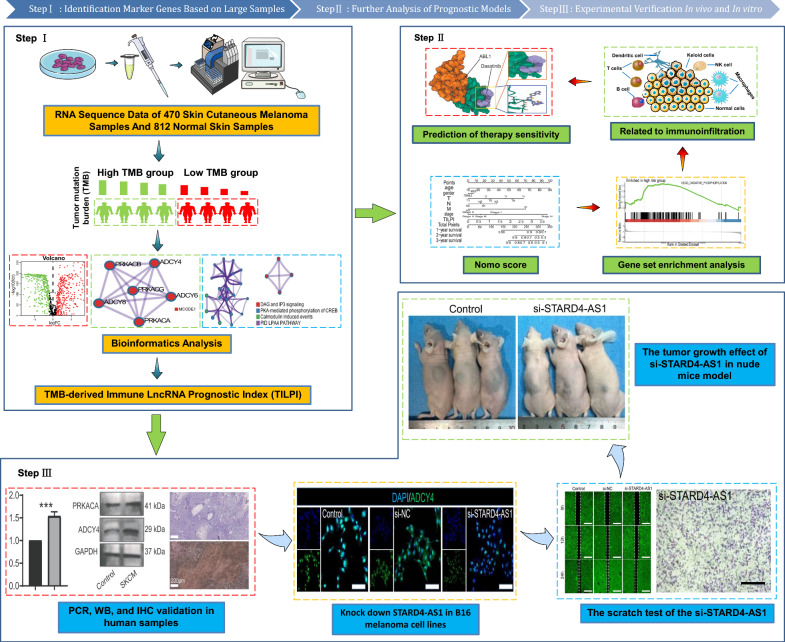
Fig. 1.
The experiment flowchart and analysis flowchart. Step I: RNA sequencing data from 470 SKCM and 812 normal skin samples were analyzed. Tumor Mutation Burden (TMB) was calculated to classify samples into high and low TMB groups. Bioinformatics analysis identified immune-related lncRNAs. Step II: Gene set enrichment analysis explored immunoinfiltration and therapy sensitivity. TILPI was established and validated. Various immune cells were studied, and STARD4-AS1 knockdown in B16 melanoma cell lines was performed. Scratch test and tumor growth analysis in nude mice were conducted. Step III: PCR, Western Blot (WB), and Immunohistochemistry (IHC) validations were performed on human samples. In vivo experiments included tumor growth and gene expression analyses in a nude mice model
Methods
The datasets source
In this study, we obtained RNA sequence data of 470 SKCM samples from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and 812 normal skin samples from the Genotype-Tissue Expression (GTEx) (https://gtexportal.org/). We also compiled clinical information, including overall survival (OS) time, survival status, age, gender, pathological TNM, and pathological stage (Table S1). The RNA sequencing data we collected was in transcripts per kilobase of exon model per million mapped reads (TPM) format.
Identify the TMB-derived immune lncRNA set (TILncSet)
In the initial step, we utilized the TCGAmutations package of R [13] to obtain the TMB value of 470 SKCM samples. However, only 468 SKCM samples had available TMB values. We then categorized these samples into high and low TMB groups based on the median TMB value. To identify different gene sets between these two groups, we utilized gene set variation analysis (GSVA) [14], a non-parametric, unsupervised method for estimating gene-set enriched variation from expression data sets. We also obtained 2524 immune-related mRNAs from the ImmPort database (https://www.immport.org/) and the InnateDB database (http://www.innatedb.com/).
Furthermore, we identified differentially expressed lncRNAs between 812 normal skin samples and 470 SKCM samples based on the limma package of R [15]. We also used the normalizeBetweenArrays function from the limma package to remove batch effects between samples. We screened qualified lncRNAs using two criteria: (1) and (2) False discovery rate (FDR)-adjusted P value < 0.05 to identify oncogenic lncRNAs as candidate lncRNAs. We then conducted Pearson analysis between candidate lncRNAs and candidate mRNAs, with high correlation coefficients and statistical significance (cor > 0.4, P < 0.05) as qualifications.
Functional enrichment analysis
We employed the Metascape database to explore the possible functions of candidate genes. (https://metascape.org/). Firstly, we used the Molecular Complex Detection (MCODE) algorithm to establish a protein–protein interaction (PPI) network. Then, we conducted enrichment analysis using the Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRUST) to identify potential transcription factors associated with TIgeneSet. Additionally, enrichment analysis was performed in transcription factor targets. We also conducted functional enrichment analysis based on the GO (http://geneontology.org/) and KEGG (https://www.kegg.jp/) databases. We applied a filter of P value < 0.05 to the analysis. For KEGG and GO analyses, we utilized the following R packages: clusterProfiler for functional classification and enrichment analysis, org.Hs.eg.db for mapping gene IDs to biological information specific to Homo sapiens, enrichplot for visualizing enrichment results, and ggplot2 for creating detailed and customizable plots.
Construction and validation of a computational framework for the prediction of prognosis
The TCGA database contains 470 skin melanoma samples, of which only 468 have TMB values. A single patient may have multiple sequencing samples, but each patient has only one survival time. Therefore, we retained samples with a Vial sequence number of 01A and removed five duplicate samples. As a result, only 463 samples were eventually. We randomly divided 463 SKCM samples with OS into two groups: the training group (n = 232) and the testing group (n = 231). The TCGA group (n = 463) was formed by combining the training and testing groups. We identified TILncSet as more likely to participate in SKCM progression than other lncRNAs. Therefore, we performed univariate and multivariate Cox proportional risk regression analyses to evaluate the TMB-derived immune lncRNA signature (TILncSig) in the training group. In multivariate Cox, we used the following parameters: Dependent Variable (Survival Time and Status): The response variable in the model is the patient's survival time and status (alive or deceased). Independent Variables (Covariates): The predictors include the expression levels of the identified lncRNAs, adjusted for potential confounders such as age, gender, and clinical stage. Coefficients (β): These represent the estimated log hazard ratios for each covariate, indicating the strength and direction of the association with patient survival. Baseline Hazard (h₀(t)): The baseline hazard function represents the hazard when all covariates are zero. The Cox proportional risk regression analysis formulas used were:
Using following formula, the value of h0(t) can be calculated. h0(t) is the baseline hazard function. The Risk Score (TILncSig) is the hazard function associated with the covariate X at time t. After modeling with multivariate Cox regression by R package survival, the risk score calculated using the predict () function is actually the Risk Score (TILncSig). By subtracting the product of the expression level of lncRNA and its coefficient from the risk score, lnh0(t) can be obtained. Taking the exponential of lnh0(t) gives h0(t).
Risk Score (TILncSig) represents the prognostic index of each SKCM patient. We also referred to risk score (TILncSig) as TMB-derived immune lncRNA prognostic index (TILPI). The value of n is the number of lncRNA signatures. The stands for the coefficient of obtained by multivariate Cox regression analysis. Expr () refers to the expression level of . In addition, we used the scatterplot3d package to draw a 3D PCA graph. The data was then subjected to PCA analysis using the prcomp function with scaling. The PCA results were predicted using the predict function. Next, a 3D scatter plot of the PCA results was created using the scatterplot3d function, where the color of the points was determined by the sample groups. Furthermore, four lncRNA prognostic models of SKCM reported in the past two years were collected, including Ma et al. [16], Zhang et al. [17], Liu et al. [18], and Tian et al. [19].
Additionally, we conducted gene set enrichment analysis (GSEA) to identify the pathways enriched in different risk subgroups [20]. The gene sets of GSEA analysis, including 10,647 pathways of GO and KEGG databases, were collected from Molecular Signatures Database (MSigDB, https://www.gsea-msigdb.org/gsea/msigdb/). We utilized four R packages for Gene Set Enrichment Analysis (GSEA): clusterProfiler, org.Hs.eg.db, limma, and enrichplot. The clusterProfiler package was employed to perform GSEA on custom gene sets derived from the MSigDB database. The org.Hs.eg.db package facilitated gene ID conversion necessary for GSEA. The limma package was used to generate gene rankings, which were crucial for subsequent GSEA. Finally, the enrichplot package was utilized to visualize the results of the enrichment analyses. Finally, we constructed a novel model called a nomogram to predict individual survival probability, which included TILPI and eight types of clinical characteristics (age, gender, smoking, race, pathological stage, and pathological TNM) [21].
Cell culture and plasmid transfection
Mouse melanoma cell line B16-F10 was used in this experiment. All cells were cultured in 1640 medium containing 10% fetal bovine serum. GC cells were transfected with si-NC (negative Control) and si-STARD4-AS1 (small-interfering RNAs against STARD4-AS1). Si-STARD4-AS1 was used to knockdown STARD4-AS1. The siRNA sequence was listed in Table S2. All plasmids were purchased from GenePharma (Shanghai, China).
Gene expression of LINC00189, LINC00861 and STARD4-AS1 in human tissue
Total RNA was extracted from plasma by the QIAamp RNA Kit (Qiagen). After the concentration, purity and integrity of the total RNA were evaluated, and the total RNA was reversely tran‐scribed into complementary DNA using PrimeScript™ RT reagent Kit (Takara). Second, quantitative polymerase chain reaction (qPCR) was performed by using KODSYBR®qPCR Mix (Toyobo). GAPDH was used as an internal reference. Finally, the results were calculated by the 2−ΔΔCt formula. Primers used in the qPCR were listed in Table S3.
CCK8 assay
The procedure for the CCK-8 test was carried out in the following manner: After cultivating in 12-well plates filled with CCK-8 solution for a duration of three days, the proliferation rate of cells from each group was assessed. Into each well of the 12-well plates, a volume of 100 mL of CCK-8 solution was introduced for every distinct cell cluster. Post a two-hour interval, the optical density (OD) readings of the supernatant were determined using a BioTech microplate reader.
Cell migration test
To assess the migratory capabilities of B16-F10 cells within each batch, longitudinal incisions were made on the cellular surface once cell fusion within each group approximated 90%, followed by a cultivation period of 12 and 24 h. Subsequently, cells that were isolated received a wash using PBS. In the concluding step, staining with Calcein AM (procured from Invitrogen, USA) was executed, employing a confocal laser scanning microscope (Zeiss LSM 980, located in Germany) for observation.
Transwell cell migration assay
Cell migration was analyzed using transwell apparatuses. Groups of B16-F10 cells categorized into Control, si-NC, and si-STARD4-AS1 were subjected to a 24-h fasting period. Post-digestion and centrifugation, cells in a serum-deprived medium were tallied with the aid of a microscope. For each well in a 24-well plate, a 100 ml cell mixture was deposited at the bottom. The cell density was regulated to attain a count of 1 × 105 cells/ml. Into every well, 600 ml of 30% FBS medium was introduced, followed by the addition of the cell mixture to a 96-well MTS plate. The optical density at OD570 was gauged to assess metastatic activity by incorporating 5000 cells into each well. After a 24-h incubation at 37 °C, an inner section was cleansed using a cotton swab. Cells that migrated were stained using a 1% crystal violet solution after fixation in methanol. A light microscope facilitated the enumeration and imagery capture of cells from three arbitrarily chosen fields.
Transwell cell invasion chamber experiments
To assess cell invasion levels, transwell invasion chambers were utilized. B16-F10 cell populations were categorized into Control, si-NC, and si-STARD4-AS1 groups. These cells were seeded onto fresh 24-well plates, with both upper and lower chambers receiving 500 ml of serum-free medium, and the bottom chamber an additional 100 ml of the same medium. Following a 2-h incubation at 37 °C, the cell count was performed under a microscope, and a cell suspension devoid of serum was adjusted to a density of 1 × 105 cells/ml. Preparing a new plate, cells were transferred, the medium from the upper chamber was discarded, and 500 ml of the cell suspension was introduced, with the lower chamber being supplemented with 750 ml of 30% FBS medium. To serve as a metastasis reference, the optical density at OD570 was determined after distributing the cell suspension across a 96-well MTS plate, with an allocation of 5000 cells for each well. Post a 22-h incubation at 37 °C, the inner cell chamber remained untouched by a cotton swab. The invasive cells were then stained using a crystal violet solution. For analyzing cell invasiveness variances among the groups, three fields were randomly chosen for enumeration and imaging via a light microscope.
Western bolt
A protein extraction kit was used to extract the proteins, and cells were lysed in protease inhibitors and phosphatase inhibitors. The BCA protein Kit was used to determine the protein concentrations. The proteins were loaded onto SDS gels, separated by electrophoresis for 35 min, and then transferred onto PVDF membranes for 30 min. Finally, they were blocked with milk and incubated with primary antibodies overnight. After washing with TBST thrice, incubation with the secondary antibodies was conducted for 60 min at room temperature, and the protein was added to an ECL chromogenic solution. The results were analyzed with Image J software, and the gray values of serum protein bands were calculated.
Subcutaneous tumor in nude mice
In this study, we investigated the role of STARD4-AS1 in the development of cutaneous melanoma (SKCM) by establishing a subcutaneous tumor formation model in nude mice. Healthy male nude mice, aged 6–8 weeks and weighing about 20–22 g, were fed and watered freely in a specific pathogen free (SPF) environment. B16-F10 skin melanoma cell lines and si-STARD4-AS1 cell lines were cultured in DMEM medium containing 10% fetal bovine serum until logarithmic growth, then washed with PBS and counted. The cells were adjusted to the concentration of 1 × 107 cells /mL, suspended with serum-free DMEM medium, and each nude mouse was injected subcutaneously with 1 × 106 cells (100 μL). The injection site was disinfected with 70% alcohol and injected subcutaneously using a 1 mL syringe with a 26G needle. The health status and tumor growth of the nude mice were monitored regularly after injection, and the nude mice were euthanized with carbon dioxide after 28 days.
Histochemical staining
The fresh tissue was fixed with 4% paraformaldehyde and then embedded with paraffin after gradient dehydration. The sections with a thickness of 5 μm were prepared by paraffin sectioning mechanism, and the corresponding antibodies were used for histochemical staining of the pathological sections.
Mapping of the immune landscape
We analyzed the tumor immune microenvironment (TIME) using eight different quantification algorithms to provide a detailed description of the infiltration landscape of immune cells. These algorithms included CIBERSORT [22], CIBERSORT-absolute mode [22, 23], EPIC [24], MCPCOUNTER [25], QUANTISEQ [26], TIMER [23, 27], TISIDB [28], and XCELL [29]. Through intersection analyses, we identified types of immune cells that were more prevalent in two TILPI subgroups.
In addition to immune cells, we also examined the stromal components in the TIME. We collected the TIME score and stroma score from XCELL and calculated the cytotoxicity score of MCPCOUNTER. We also obtained the stromal score and tumor purity using ESTIMATE [30]. Furthermore, we aimed to determine the immune subtype of samples using 6 types of immune subtypes, which consisted of wound healing (immune C1), IFN-γ dominant (immune C2), inflammatory (immune C3), lymphocyte depleted (immune C4), immunologically quiet (immune C5), and TGF-β dominant (immune C6) [31].
Prediction of immunotherapy sensitivity
To assess the potential response to immunotherapy, we calculated the expression levels of five significant immune checkpoint-associated proteins, including CTLA4, PD-L1, PD-1, TIGIT, and LAG-3. We explored the differential expression of these immune checkpoint proteins between different TILPI subgroups to determine which population may be more sensitive to immunotherapy. The tumor inflammation signature (TIS) model has been shown to retrospectively predict the clinical benefit of anti-PD-1 treatment in clinical trials [32]. We connected TILPI with the TIS model to predict the response to immunotherapy.
Prediction of sensitive drugs
We employed a selection strategy to identify drugs that were sensitive to different TILPI subgroups based on the R package called pRRophetic [33] and oncopredict [34]. We used the semi-inhibitory concentration (IC50) as the boundary to identify sensitive drugs, where lower IC50 values indicated that the drug was sensitive to the TILPI subgroup. To ensure statistical significance, we applied a filter where the P value of the Wilcoxon test had to be less than 0.05. Additionally, we utilized the connectivity map (CMap) (https://clue.io/) to identify drugs that inhibited pathways more present in the low TMB subgroup. We only considered drugs with an absolute normalized CMap score greater than 1. Finally, we intersected the three derived drug lists to identify candidate drugs.
In addition, we conducted an analysis of the modes of interaction between the candidate drugs and their targets using Autodock Vina 1.2.2 [35]. We obtained the molecular structures of candidate drugs from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and the 3D coordinates of their targets from the PDB (http://www.rcsb.org/). To analyze the interaction between the drugs and their targets, we constructed molecular docking models using Autodock Vina 1.2.2 (http://autodock.scripps.edu/).
Statistical analysis
The statistical analysis software utilized in our study was R version 4.2.2, which aided in analyzing data and generating Figure and tables (https://www.r-project.org). Statistical evaluations utilized SPSS Statistics 23.0 (IBM, USA) and GraphPad Prism 8.0 (GraphPad Software, USA). For comparing two distinct groups, unpaired t-tests were applied, whereas one-way ANOVA, accompanied by Bonferroni's correction, was used when assessing three or more groups. The data from experiments were expressed as the mean ± standard deviation (Mean ± SD). A P-value less than 0.05 indicated statistical significance.
Results
Identify the TMB-derived immune lncRNA set (TILncSet)
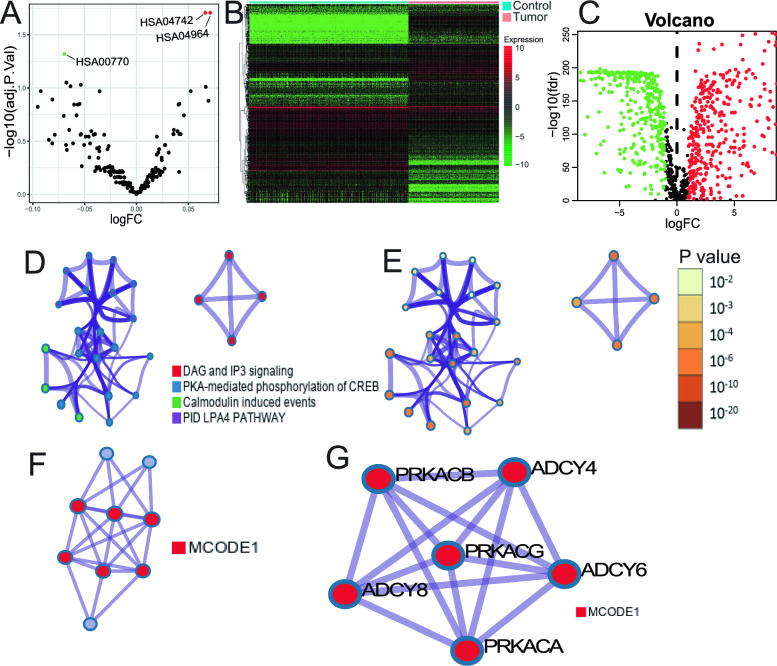
Based on GSVA analysis, we found three gene sets that were differentially expressed between different TMB groups, namely HSA04742, HSA04964, and HSA00770 (Fig. 2A). These gene sets were composed of 91 mRNAs. We then obtained 2524 immune-related mRNAs from the ImmPort and InnateDB databases and intersected them with the 91 mRNAs from GSVA analysis to identify TMB-derived immune mRNAs.
Fig. 2.
Comprehensive Analysis of TMB Groups, Differential Expression, Functional Pathways, and Network Interactions. A Gene Set Variation Analysis (GSVA) results comparing high and low TMB groups, highlighting pathway activity differences. B, C Volcano plots illustrating differential expression of lncRNAs between normal skin tissues and Skin Cutaneous Melanoma (SKCM), with log fold change (logFC) and adjusted p-values represented. D, E Network diagrams showing relationships among four functional pathways identified through pathway and process enrichment analysis, with nodes color-coded based on pathways. and statistical significance. F, G Protein–Protein Interaction (PPI) network visualized with MCODE components identified within the TIgeneSet, highlighting significant molecular interactions and gene clusters
Furthermore, we conducted a differential expression analysis of lncRNAs between 812 normal skin samples and 470 SKCM samples using the limma package and identified 793 differentially expressed lncRNAs (Fig. 2B, C) (Table S4). We then performed a Pearson correlation analysis to investigate the relationship between differentially expressed lncRNAs and TMB-derived immune mRNAs, resulting in the identification of 12 immune TMB-derived oncogenic lncRNAs, collectively known as TMB-derived immune lncRNA set (TILncSet). These lncRNAs include AC090559.1, AC096921.2, BACH1-IT1, C1RL-AS1, CADM3-AS1, CARD8-AS1, FAM66C, LINC00189, LINC00861, LINC00996, LINC01197, and STARD4-AS1. We also identified a set of 10 mRNAs after Pearson correlation analysis, known as TMB-derived immune mRNA set (TImSet), which included ADCY4, ADCY6, ADCY8, DDOST, ITPR3, PDE1A, PRKACA, PRKACB, PRKACG, and PRKX. The TMB-derived immune gene set (TIgeneSet) consisted of TILncSet and TImSet.
Explore the biological functional pathways of TIgeneSet
We utilized Metascape to explore the potential functions of TIgeneSet in SKCM. We set three conditions to screen for enriched pathways: P value < 0.01, a minimum count of 3, and an enrichment factor > 1.5. Eligible pathways were grouped based on their similarity, and we used Kappa scores to measure the similarity. Pathways with a similarity > 0.3 were considered a cluster. Based on the pathway clusters, a network of enriched terms was constructed, consisting of four pathway clusters with the smallest P value (Fig. 2D, E). These clusters included DAG and IP3 signaling, PKA-mediated phosphorylation of CREB, calmodulin-induced events, and PID LPA4 pathway, which were associated with metabolic processes, multicellular organismal processes, and signal transmission.
We also conducted a PPI network based on three pathways with the smallest P value, which included DAG and IP3 signaling, PLC beta-mediated events, and G-protein-mediated events. Using the MCODE algorithm, we identified the density of the network when a subset contained between 3 and 500 proteins. The PPI network showed the relationship between nine protein subsets consisting of the three pathways. The six high-density protein subsets were represented by MCODE1 (Fig. 2F), which included ADCY4, ADCY6, ADCY8, PRKACA, PRKACB, and PRKACG. The proteins of MCODE1 had functional interactions between them (Fig. 2G).
Enrichment analysis in DisGeNET showed that leukocyte adhesion deficiency type 1 was the most significant (Fig. S1A). Leukocyte adhesion deficiency has been linked to cancer metastasis, which corresponds to the fact that most sample types of SKCM are metastatic rather than primary. In addition, we identified eight interconnected targets by the enrichment analysis of transcription factor targets (Fig. S1B). Among them, SOX10 was the most significant and was shown to promote angiogenesis in melanoma and metastatic progress.
To further explore the potential functions of TIgeneSet in SKCM, we conducted GO and KEGG functional enrichment analysis. We identified 40 significant pathways in the GO analysis, including 27 pathways for biological process (BP), two for cell component (CC), and 11 for molecular function (MF). We showed 10 pathways to draw intuitive diagrams (Fig. S1C-D). The BP pathways were related to the cAMP biosynthetic process, cyclic nucleotide metabolic process, protein kinase A signaling, negative regulation of posttranscriptional gene silencing, and adenylate cyclase-modulating G protein-coupled receptor signaling pathway. The CC pathway was associated with the cAMP-dependent protein kinase complex, while the MF pathways were related to protein kinase A regulatory subunit binding, protein serine/threonine kinase activity, protein kinase C binding, and pre-mRNA binding. We also obtained 92 functional pathways in the KEGG analysis and showed 16 significant pathways (Fig. S1E, F). These pathways were divided into the cell cycle, signaling pathway, and protein production. The cell cycle part included apoptosis, the Ras signaling pathway, cellular senescence, and so on. The signaling pathway consisted of the cAMP signaling pathway, cGMP-PKG signaling pathway, phospholipase D signaling pathway, Wnt signaling pathway, Rap1 signaling pathway, and MAPK signaling pathway. Ribosome biogenesis in eukaryotes and protein processing in the endoplasmic reticulum belonged to protein production. Overall, the potential functions of TIgeneSet were mostly based on the ADCY and PRKA protein families, which were related to cAMP-dependent protein kinase and adenylate cyclase.
Construct the TILPI computational framework in the training group
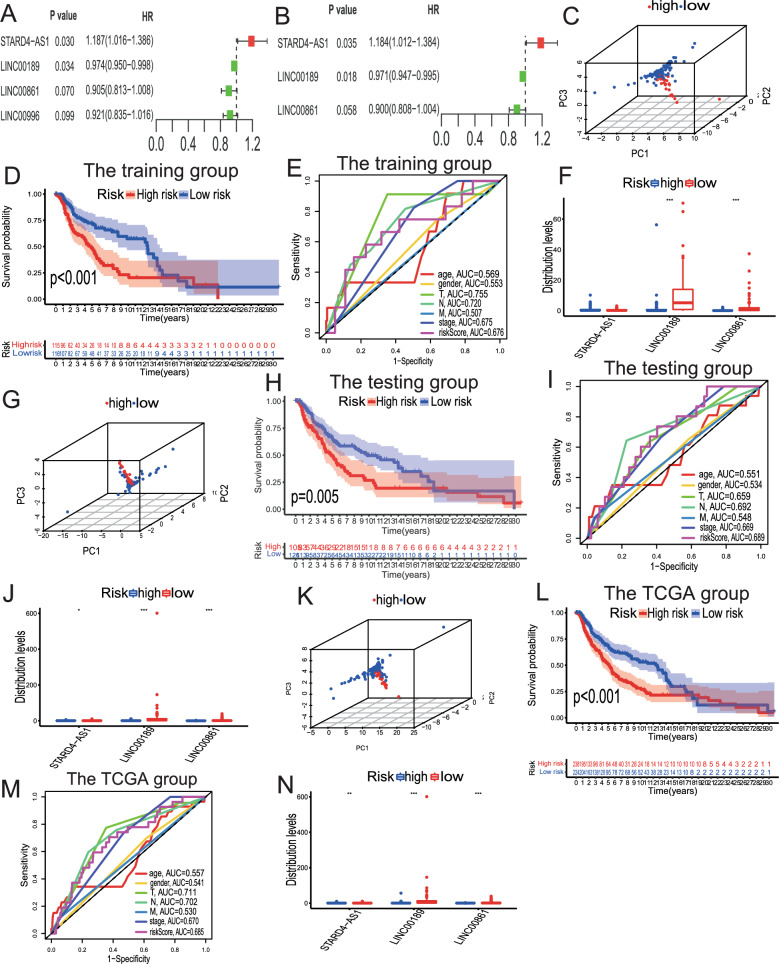
In this section, we present the TILPI computational framework that we constructed in the training group to predict the prognoses of SKCM patients. Firstly, we randomly divided all samples (n = 463) from the TCGA group into the training group (n = 232) and the testing group (n = 231). We performed statistical analysis to ensure that the clinical characteristics of patients in each group were evenly distributed (Table S1). We then conducted univariate Cox proportional risk regression analysis in the training group and identified four lncRNAs that were closely associated with SKCM patients’ OS (STARD4-AS1, LINC00189, LINC00861, LINC00996) (Fig. 3A). Additionally, we accurately obtained three TMB-derived immune lncRNA signatures (TILncSig) by multivariate Cox proportional risk regression analysis (STARD4 − AS1, LINC00189, LINC00861) (Fig. 3B). Among these lncRNAs, two coefficients were negative (protective factors), and one coefficient was positive (risk factor). We then constructed the computational framework to assess the risk score resulting from the expression of TILncSig in each patient. The framework was as follows:
Fig. 3.
Identification and Validation of TILncSig and TILPI in Training, Testing, and TCGA Groups. A, B In the training group, TILncSig was identified by univariate and multivariate Cox proportional hazards regression analyses. C–E 3D PCA was conducted to visualize the separation of samples based on the TILPI, demonstrating its discriminative power. Survival analysis was performed to evaluate the prognostic significance of TILPI, showing a clear distinction in survival outcomes between different risk groups. The ROC curve was used to assess the predictive accuracy of TILPI, confirming its reliability and robustness as a prognostic tool. F The different expression of TILncSig in the high-risk group and the low-risk group (***P < 0.001). G–J We verified the reliability of TILPI in the testing group. (K-N) We verified the reliability of TILPI in the TCGA group
For a certain patient, their TILPI was calculated as expression (0.1842 + 0.1686 * expression (STARD4-AS1)—0.0295 * expression (LINC00189)—0.1050 * expression (LINC00861)). We used the median TILPI of the training group as the boundary to divide all patients into high-risk (TILPI > 1.1338) and low-risk (TILPI ≤ 1.1338) subgroups. We used 3D PCA to verify the reliability of the grouping, and the result showed that our grouping had high reliability (Fig. 3C). In addition, the OS of the low-risk subgroup was signally higher than the high-risk subgroup (P = 0.001) (Fig. 3D). The 5-year survival rate in the low-risk subgroup (41.7%) was significantly better than the high-risk subgroup (22.2%). The area under curve (AUC) value of TILPI in the training group was 0.676 (Fig. 3E). We also evaluated the predicted ability of other clinical characteristics such as age (AUC = 0.569), gender (AUC = 0.553), pathological stage (AUC = 0.675), pathological T (AUC = 0.755), pathological N (AUC = 0.702), and pathological M (AUC = 0.507) (Fig. 3E). Furthermore, we found that the expression of LIN00189 and LIN00861 in the low-risk subgroup was significantly higher than that in the high-risk subgroup (P < 0.001) (Fig. 3F). However, the expression of STARD4-AS1 was not statistically significant between high-risk and low-risk subgroups (P > 0.05) (Fig. 3F). Therefore, LIN00189 and LIN00861 may be beneficial for patients.
Verify the reliability of the TILPI computational framework in the testing group and the TCGA group
In our previous study, we demonstrated the potential of TILPI in predicting the prognoses of SKCM patients in the training group. However, it was necessary to conduct rigorous experiments to further validate our findings in the testing groups. We also used 3D PCA to verify the reliability of our grouping, which showed high reliability (Fig. 3G). Furthermore, we found a statistically significant difference in OS between the low-risk subgroup and high-risk subgroup (P = 0.005) (Fig. 3H). The AUC value of TILPI in the testing group was also satisfactory, and it performed better than other clinical features, including age, gender, pathological T, N, M, and stage (Fig. 3I). Additionally, we found that STARD4-AS1, LINC00189, and LINC00861 were differentially expressed in different risk subgroups (P < 0.05) (Fig. 3J).
In the TCGA group, we also found that the TILPI computational framework grouping was reliable based on 3D PCA (Fig. 3K). Furthermore, we found that the OS of patients in the low-risk subgroup was significantly higher than that of patients in the high-risk subgroup (P < 0.001) (Fig. 3L). The AUC value of TILPI in the TCGA group was also satisfactory, and it outperformed other clinical features (Fig. 3M). Compared with prognostic lncRNA markers of SKCM reported by several other researchers in the past two years, including Ma et al. (AUC = 0.615, 2024), Zhang et al. (AUC = 0.64, 2023), Liu et al. (AUC = 0.674, 2023), and Tian et al. (AUC = 0.677, 2023), the average AUC value of TILPI was higher (0.685). Similarly, we found that STARD4-AS1, LINC00189, and LINC00861 in the TCGA group were differentially expressed in different risk subgroups (P < 0.05) (Fig. 3N).
To evaluate the independence of TILPI on patients' prognoses, we conducted an independence analysis based on univariate Cox analysis. We found that TILPI was an independent factor of prognosis (P = 0.003) (Fig. S2A). Age and pathological T were also independent factors (Fig. S2A). And the further multivariate Cox analysis also confirmed the independence of TILPI on prognosis (P = 0.007) (Fig. S2B). Age, pathological T, N, and stage were also independent factors in prognosis (Fig. S2B). Through GSEA analysis, we found patients of the high-risk group were more enriched in oxidative phosphorylation, cell adhesion, and keratinocyte differentiation (Fig. S2C), indicating that oxidative phosphorylation and tumor cell migration may be the conditions required for cancer cells to adapt to changes in the metabolic environment. Therefore, inhibiting the specific oxidative phosphorylation and migration antigens of cancer cells may be a potential treatment for SKCM patients. In contrast, the low-risk group was more related to the immunity (Fig. S2C), suggesting that activation of immune pathways may be a protective factor in the low-risk group. Additionally, we combined TILPI with several clinical feares (age, gender, pathological TNM, and stage) to construct a new computational model that connects prognosis with clinical characteristics (Fig. S2E). The predicted ability of the new model in the prediction of OS looked well-matched with the actual 1, 2, and 3-year OS (Fig. S2F-H). We believe that the new model is more convenient for clinicians to evaluate SKCM patients' prognoses based on clinical characteristics.
Genes and proteins are expressed in human samples
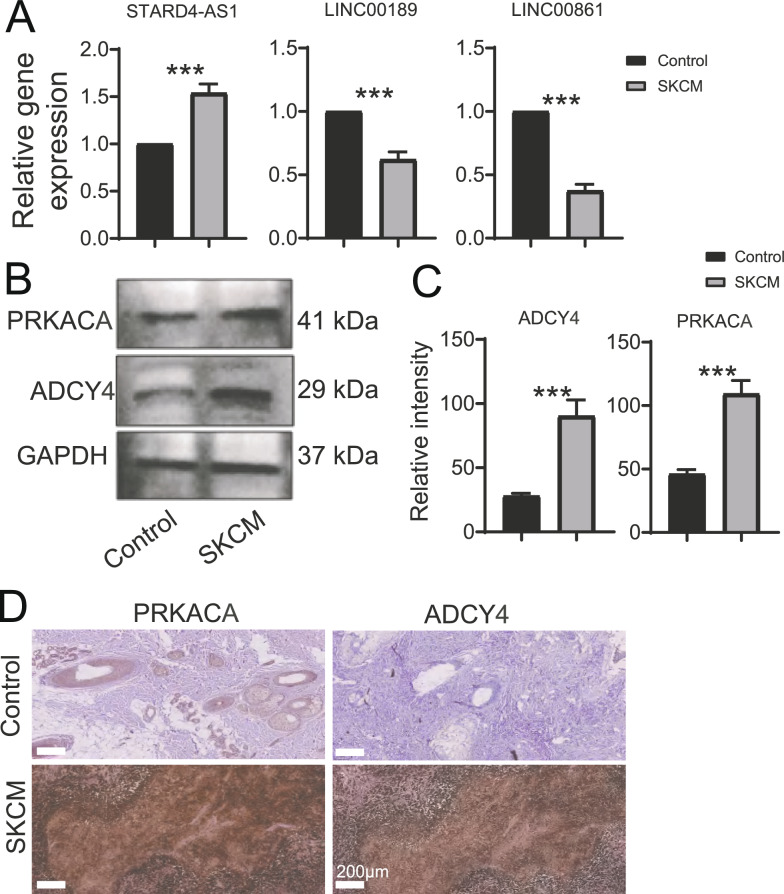
The above enrichment analysis suggests that TIgeneSet was highly enriched in the ADCY-PRKA pathway, and this biological correlation may extend to TILPI. Based on the aforementioned PPI network analysis, we reviewed the literature and found that there were few studies on the role of ADCY4 and PRKACA in SKCM, so we decided to further explore their mechanisms of action. In this study, PCR detection found that STARD4-AS1 expression was higher in human melanoma samples than in normal skin tissues. The expression of LINC00189 and LINC00861 in human melanoma samples was lower than that in normal skin tissue (Fig. 4A). In addition, the expression of ADCY4 and PRKACA in human melanoma samples was higher than that in normal skin tissues (Fig. 4B, C). Furthermore, ADCY4 and PRKACA showed similar trends to western bolt in histochemical staining (Fig. 4D).
Fig. 4.
Analysis of Gene and Protein Expressions in Human Samples. A Column graph displaying the PCR results for the expression of three lncRNAs, STARD4-AS1, LINC00189, and LINC00861, in human samples (n = 3). B Western Bolt analysis detected the protein expressions of PRKACA and ADYC4 in human samples. C Quantitative analysis of Western Bolt (mean ± SD, n = 3). D Immunohistochemistry analysis detected the protein expressions of PRKACA and ADYC4 in human samples. For all panels, unpaired two-tailed Student’s t test was used for betweengroup comparisons. Data are expressed as mean ± SD. (***P < 0.001)
Effects of STARD4-AS1 Knockdown on Tumor Cell Behavior
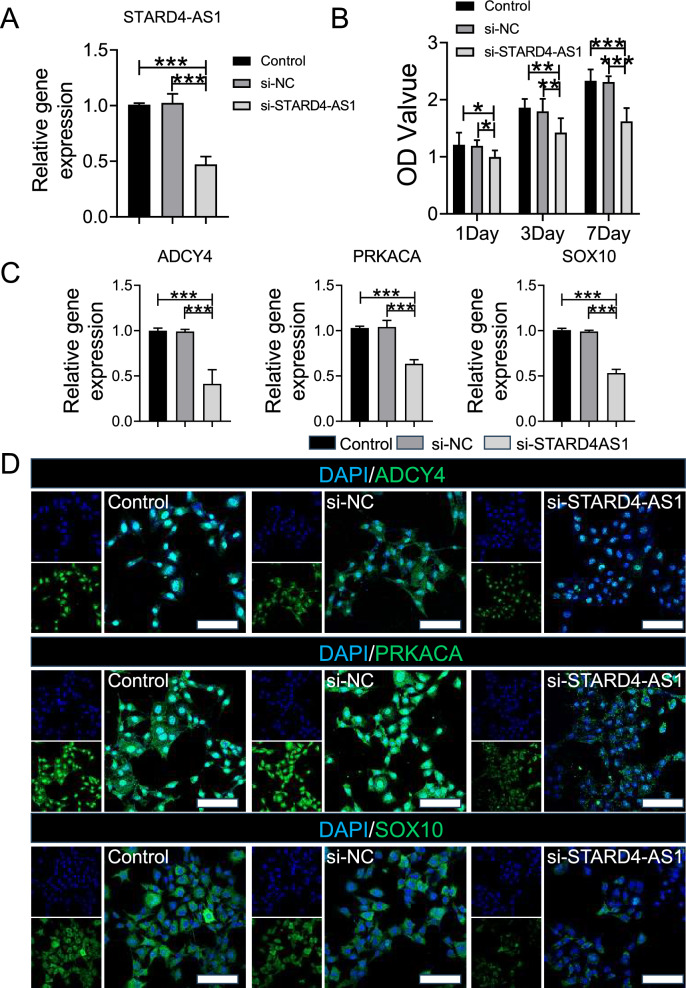
In this study, we used siRNA to knockdown STARD4-AS1 expression in B16 melanoma cell lines (Fig. 5A). Subsequently, CCK8 results showed no significant difference between the Control group and the si-NC group after 1, 3, and 7 days, while the proliferation of the si- STARD4-AS1 group decreased significantly compared with the Control group and the si-NC group (Fig. 5B). PCR results showed that the gene expressions of ADYC4, PRKACA and SOX10 in the si-STARD4-AS1 group were significantly lower than those in the Control and si-NC groups (Fig. 5C). Similarly, immunofluorescence results showed significantly reduced protein expression of ADYC4, PRKACA, and SOX10 in the si-STARD4-AS1 group compared to the Control and si-NC groups (Fig. 5D). Further, the results of the scratch test showed that the tumor proliferation ability of the si-STARD4-AS1 group was lower than that of the Control and si-NC groups in 12 h and 24 h (Fig. S3A, B). The tumor migration (Fig. S3C) and invasion (Fig. S3D) experiments in each group showed that the migration and invasion ability of tumor cells in si-STARD4-AS1 group and si-NC group decreased significantly compared with Control group and si-NC group.
Fig. 5.
Analysis of Gene Expression and Tumor Cell Proliferation. A The expression of STARD4-AS1 gene was detected by PCR. B CCK8 was used to detect the proliferation of tumor cells in each group. C The expression of ADCY4, PRKACA and SOX10 genes were detected by PCR. D The expression of ADCY4, PRKACA and SOX10 in tumor cells were detected by immunofluorescence (Scale bar represents 50 μm). Statistical differences were determined by utilizing One-way ANOVA with Bonferroni's multiple comparison tests when comparing three groups. When comparing two groups, the unpaired t-test was utilized. Data are expressed as mean ± SD (*P < 0.05, **P < 0.01, and ***P < 0.001)
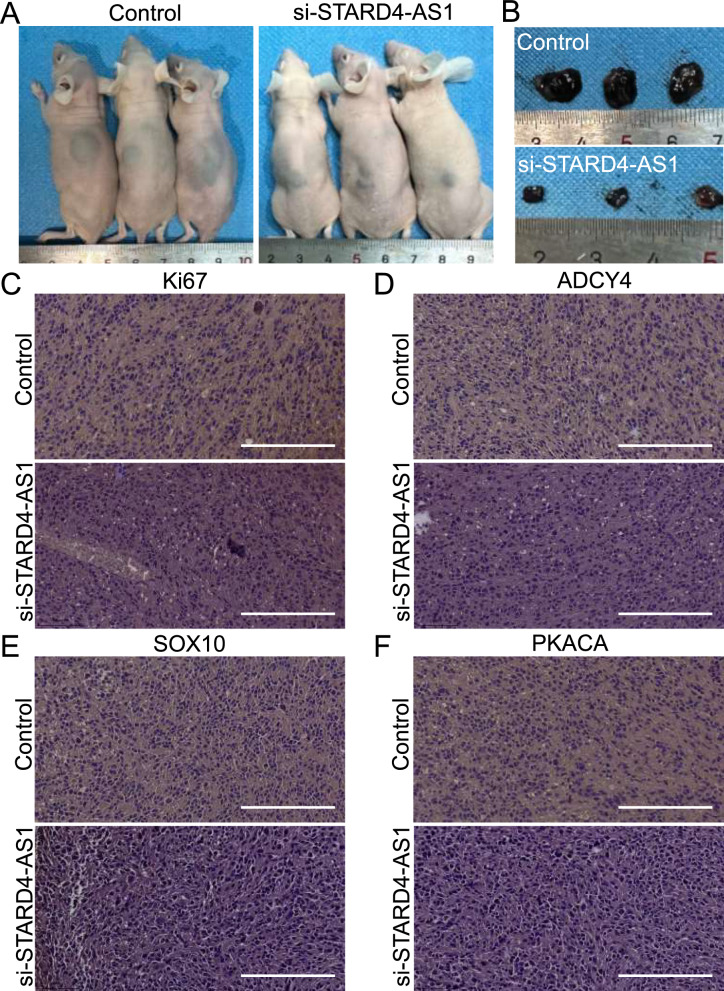
In addition, tumor cells in the Control group and the si-STARD4-AS1 group were injected into the subcutaneous skin of nude mice for 28 days to observe the tumor growth of each group. The results showed that the tumor size in the si-STARD4-AS1 group was significantly smaller than that in the Control group (Fig. 6A, B). Immunohistochemical results showed that Ki67, ADCY4, PKACA and SOX10 in si-ST group were significantly reduced compared with those in Control group (Fig. 6C–F).
Fig. 6.
Tumor Analysis in Nude Mice with Control and si-STARD4-AS1 Treatments. A Tumor status of nude mice in control group and si-STARD4-AS1 group after 28 days of injection. B The size of tumor after removal in two groups of nude mice. C Immunohistochemical analysis of Ki67 expression in tumor tissue of two groups of nude mice (Scale bar represents 200 μm). D Immunohistochemical analysis of ADCY4 expression in tumor tissue of two groups of nude mice. E Immunohistochemical analysis of SOX10 expression in tumor tissue of two groups of nude mice (Scale bar represents 200 μm). F Immunohistochemical analysis of PRKACA expression in tumor tissue of two groups of nude mice (Scale bar represents 200 μm)
Mapping of immune landscape based on the computational framework
In this study, we collected immune cells from 8 algorithms and mapped the immune landscape of SKCM. The CIBERSORT algorithm showed that more immune cells existed in the low-risk subgroup (Fig. S4A-D) (Table S5). Similarly, 7 other algorithms, more immune cells existed in the low-risk subgroup (Fig. S4E-F) (Fig. S5A-E) (Table S5). We then identified the types of immune cells that were more distributed in different TILPI subgroups through intersection analyses. CD8 T cells, memory B cells, and M1 macrophages were more distributed in the low TILPI subgroup, while uncharacterized cells were more distributed in the high TILPI subgroup (Table S5). Furthermore, we explored the correlation between TILPI and the immune subtype and found that TILPI was correlated with the immune subtype (Fig. S5F). The population with low TILPI was classified more often as immune C2, while the high TILPI subgroup was classified more often as immune C1 (Fig. S5F).
As TIME includes not only immune cells but also stromal components, we planned to connect TILPI with other stromal components. We found that the cytotoxicity score of MCPcounter was negatively correlated with TILPI (R = − 0.41, p < 2.2e − 16) (Fig. S6A), which was also supported by the result of grouping (P < 0.001) (Fig. S6B). We obtained the stroma score, immune score, and TIME score based on XCELL, and found that the immune score and TILPI had a negative correlation (R = − 0.56, P < 0.05) (Fig. S6C), while the correlation between TIME and TILPI was not statistically significant (R = − 0.018, p = 0.7) (Fig. S6D). Furthermore, we found a negative correlation between TIME and TILPI (R = − 0.54, P < 0.05) (Fig. S6E). We then performed correlation analyses to verify the above results by grouping. The TIME score and immune score were higher in the low-risk group, while the stroma score was significantly higher in the high-risk group (P < 0.001) (Fig. S6F). The stroma score, immune score, and estimate score were all negatively correlated with TILPI (R = − 0.24, P = 0.05) (R = − 0.46, P < 0.05) (R = − 0.4, p < 0.05) (Fig. S6G-I). Moreover, we found a positive correlation between tumor purity and TILPI (R = 0.42, P < 0.05) (Fig. S6J), which was consistent with the result of grouping (Fig. S6L-K). In summary, our findings suggest that more immune cells are present in the immune landscape of the low TILPI subgroup, which may correspond to the good prognosis of SKCM. Further exploration of this finding is warranted.
Prediction of immunotherapy sensitivity
We aimed to evaluate whether the TILPI computational framework could predict the immune landscape of TIME and its potential role in predicting immunotherapy efficacy. Therefore, we measured the expression levels of five well-known immune checkpoint-associated proteins in our SKCM samples. Our findings showed that TILPI was negatively correlated with PD-L1, PD-1, TIGIT, and LAG-3 (R = − 0.19, P = 5.2e−05) (R = − 0.35, P = 3.7e − 15) (R = − 0.46, P < 2.2e − 16) (R = − 0.27, P = 6e − 09) (Fig. S7A–D). Although CTLA4 was not associated with TILPI, it was highly expressed in the low TILPI subgroup (R = − 0.073, P = 0.11) (P < 0.001) (Fig. S7E-F). Similarly, other immune checkpoint-associated proteins, including PD-L1, PD-1, TIGIT, and LAG-3, were overexpressed in the low TILPI subgroup (P < 0.001) (Fig. S7F). Furthermore, TILPI was negatively correlated with TMB (R = − 0.11, P = 0.015) (Fig. S7G), and TMB was higher in the low TILPI subgroup (P = 0.017) (Fig. S7H). We also investigated the correlation between TILPI and TIS, which is a biomarker that predicts the clinical benefit of PD-1 targeted therapy based on 18 genes. Our results showed a negative correlation between TIS and TILPI (R = − 0.33, P = 3.2e − 13) (Fig. S7I), and TIS was higher in the low TILPI subgroup (P < 0.001) (Fig. S7J). Overall, our results suggest that the low TILPI subgroup may respond better to immunotherapy. This finding warrants further investigation.
Prediction of sensitive drugs
We performed drug sensitivity analyses using the pRRophetic package and identified 73 drugs that were more sensitive in the low-TILPI subgroup and only 12 drugs for the high-TILPI subgroup. We further conducted drug sensitivity analyses using the oncopredict package. We found 41 drugs that were more sensitive in the low-TILPI subgroup and 27 drugs for the high-TILPI subgroup. These findings suggest that the low-TILPI subgroup may be more responsive to certain drugs, and further studies are needed to validate these results. Additionally, we used the CMap database to identify drugs that inhibited pathways more prevalent in the low TMB subgroup, with a positive connectivity score greater than 1. In addition, we intersected the results of drug sensitivity analyses based on pRRophetic, oncopredict, and CMap for the population with a good prognosis, specifically the low-TILPI subgroup. We identified three drugs that were sensitive in this population, including dasatinib, gemcitabine, and ruxolitinib. These findings provide potential treatment options for SKCM patients with a good prognosis, but further research is needed to validate these results.
Furthermore, we used molecular docking models to evaluate the affinity of these candidate drugs to their targets. The best binding energy of gemcitabine to RRM1 was − 66.741 kcal/mol (Fig. S7K), indicating low affinity. In contrast, the best binding energy of dasatinib to ABL14 was − 1.921 kcal/mol (Fig. S7L), and the best binding energy of ruxolitinib to JAK1 was − 8.173 kcal/mol (Fig. S7M). These findings suggest that dasatinib and ruxolitinib have excellent binding energy for their targets, indicating high stability and potential. Overall, these three sensitive drugs for the low-TILPI subgroup warrant further exploration.
Discussion
Skin cutaneous melanoma (SKCM) remains a significant cause of mortality. and understanding its molecular mechanisms is crucial for developing effective treatments. Long non-coding RNAs (lncRNAs) play an important role in various biological processes [36, 37]. In this study, we aimed to identify better signatures to reduce the mortality rate of SKCM. We utilized Gene Set Variation Analysis (GSVA) to identify different pathways between high and low tumor mutation burden (TMB) groups and intersected TMB-derived mRNAs of different pathways and immune-related mRNAs to determine candidate mRNAs. By comparing oncogenic lncRNAs between 812 normal skin samples and 470 SKCM samples and identified candidate mRNAs and oncogenic lncRNAs through Pearson analysis, we then obtained TILncSet. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses showed that TIgeneSet was involved in SKCM primarily through the cAMP-dependent protein kinase and adenylate cyclase pathways. DisGeNET analysis also revealed a relationship between TIgeneSet and leukocyte adhesion deficiency type 1, a condition linked to cancer metastasis. This finding aligns with the observation that most SKCM samples are metastatic rather than primary. Notably, SOX10 emerged as a significant transcription factor target of TIgeneSet, promoting melanoma angiogenesis and metastatic progression. These findings provide new insights into the molecular mechanisms of SKCM, but further research is needed to validate our findings [38].
Based on Kaplan–Meier analysis, univariate Cox proportional risk regression, and multivariate Cox proportional risk regression, we identified TILncSet (STARD4-AS1, LINC00189, LINC00861) that closely correlated with patients' overall survival (OS). STARD4-AS1 is a risk factor, whereas LINC00189 and LINC00861 are protective factors. Our comprehensive in vitro and in vivo experiments demonstrated that STARD4-AS1 knockdown significantly inhibited tumor cell proliferation, migration, and growth, reinforcing its role as a key regulator of SKCM development and a potential therapeutic target. These results are consistent with existing literature on the role of lncRNAs in regulating tumor cell behavior [39]. In particularly, our results suggest that STARD4-AS1 may play an important role in SKCM tumor cell proliferation and migration by influencing the activity of ADCY and PRKA protein families, thereby regulating CAMP-dependent signaling pathways. In vivo experiments, we further confirmed that STARD4-AS1 knockdown significantly inhibited SKCM tumor growth. This finding not only confirms the biological relevance of the results of the in vitro experiment, but also provides direct in vivo evidence for the function of STARD4-AS1 in the development of SKCM. This suggests that STARD4-AS1 may promote the proliferation and spread of SKCM by activating a specific signaling pathway, and its knockdown therefore inhibits tumor growth. Some researchers have found that TILncSet has known functions related to skin diseases [40, 41]. In addition to identifying prognostic markers for SKCM, we also developed a new computational framework called TILPI based on TILncSet. The framework effectively divided SKCM patients into good and bad prognostic groups, with higher TILPI associated with worse prognosis. The reliability of TILPI was verified through survival analyses and time-dependent ROC curves, and correlation analyses showed that age, pathological stage, and pathological T were correlated with TILPI. However, the specific relationship and mechanism between TILPI and clinical characteristics require further investigation. The construction of the prognostic model is of great significance to clinical decision-making and medical care, but an unverified or adjusted model cannot be applied to clinical practice [42]. Moreover, we identified 31 significantly enriched pathways through GSEA, with 13 immune pathways correlating with prolonged OS.There was reported that immune escape was one of the most significant problems leading to the progression and therapeutic resistance of SKCM [43, 44] and immune activation may be protecting these patients [45]. We used 8 algorithms to describe the immune infiltration landscape of SKCM and found that CD8 T cells, memory B cells, and M1 Macrophages were more prevalent in the population with good prognoses. In contrast, uncharacterized cells were more prevalent in the bad prognosis population.
Immunotherapy has become an important adjuvant therapy for SKCM [46–48], and selecting sensitive groups before treatment can maximize its efficacy. Failure of SKCM targeted therapy has become common in recent years, and these patients often choose to receive immunotherapy as an alternative [49]. Based on our computational framework, we proposed identifying the SKCM population sensitive to immunotherapy. We found that a low TILPI population expressed more immune checkpoint-associated proteins and had higher TIS scores, indicating it is likely more sensitive to immunotherapy. Targeted therapy and chemotherapy are also important treatment options for SKCM. Additionally, we identified dasatinib targeting Kit mutation and ruxolitinib inhibiting JAK as potential targeted therapies, and gemcitabine as a potential cytotoxic agent. There have been clinical trials of dasatinib in treating melanoma [50]. Nevertheless, ruxolitinib has been reported to induce skin cancers [51, 52]. Chemotherapy is the secondary therapy to SKCM, and we found that gemcitabine may be an effective cytotoxic agent. It was reported that gemcitabine promotes antitumor immunity against melanoma [53]. However, these findings need to be confirmed in subsequent clinical trials before being applied in clinical practice.
Overall, our study provides new insights into the molecular mechanisms and potential treatment options for SKCM, but further research is needed to validate our findings.
Conclusion
We developed a computational framework called TILPI based on TILncSig, which we identified using TMB-derived immune genes and oncogenes in SKCM. TILPI successfully divided SKCM patients into two populations with different prognoses. CD8 T cells, memory B cells, and M1 Macrophages were more prevalent in the population with a good prognosis. We also found that the low TILPI population with a good prognosis was sensitive to a combination immunotherapy and targeted therapy.
To validate the findings, the researchers conducted functional experiments. The results demonstrated that knockdown of STARD4-AS1 reduced tumor cell proliferation and impaired migration and invasion abilities. These experimental findings provided mechanistic insights into the role of STARD4-AS1 and its downstream targets in SKCM progression, emphasizing the significance of the ADCY4/PRKACA/SOX10 pathway. The combination of computational analysis and experimental validation enhanced the understanding of TILPI and its clinical implications. This study reveals underlying biology and validates the prognostic and predictive capabilities of novel computational frameworks like TILPI, emphasizing the significant changes observed in the ADCY4/PRKACA/SOX10 pathway following STARD4-AS1 knockdown. These treatment options provide potential treatment options for SKCM, but further research is needed to validate them.
Supplementary Information
Abbreviations
- BP
Biological process
- CC
Cell component
- CIBERSORT
Cell type identification by estimating relative subsets of RNA transcripts
- CIBERSORT-ABS
CIBERSORT-absolute mode
- CMap
Connectivity map
- XCELL
Digitally portraying the tissue cellular heterogeneity landscape
- EPIC
Estimating the proportions of immune and cancer cells
- ESTIMATE
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data
- FDR
False discovery rate
- GO
Gene Ontology
- GSEA
Gene set enrichment analysis
- GSVA
Gene set variation analysis
- GTEx
Genotype-tissue expression
- ICI
Immune checkpoint inhibitor
- ImmPort
Immunology database and analysis portal
- InnateDB
Innate immune response
- KM
Kaplan–Meier
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LDH
Lactate dehydrogenase
- LncRNA
Long non-coding RNA
- MCPCOUNTER
Microenvironment cell populations-counter
- MCODE
Molecular complex detection
- MF
Molecular function
- NAL
Neoantigen load
- OS
Overall survival
- PPI
Protein–protein interaction
- QUANTISEQ
Quantifying immune contexture of human tumors
- SKCM
Skin cutaneous melanoma
- TCGA
The Cancer Genome Atlas
- TIgeneSet
TMB-derived immune gene set
- TILPI
TMB-derived immune lncRNA prognostic index
- TILncSet
TMB-derived immune lncRNA set
- TILncSig
TMB-derived immune lncRNA signature
- TImSet
TMB-derived immune mRNA set
- TRRUST
Transcriptional regulatory relationships unraveled by sentence-based text mining
- TPM
Transcripts per kilobase of exon model per million mapped reads
- TISIDB
Tumor and Immune System Interaction Database
- TIMER
Tumor Immune Estimation Resource
- TIME
Tumor immune microenvironment
- TIS
Tumor inflammation signature
- TMB
Tumor mutational burden
Author contributions
J. L., Y. W., G. L., and L. F. conceived and designed the work. G. L., C. W., H. L., and P. G. conducted the experiments and gathered the data. X. H., C. W., and Z. L. analyzed and interpreted the data. G. L. and C. W. drafted the manuscript. J. L., Y. W., G. L., and L. F. provided critical revisions and intellectual input to the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by grants from the Guangdong Provincial Key Laboratory of Precision Medicine for Gastrointestinal Cancer (2020B121201004), the Guangdong Provincial Major Talents Project (No. 2019JC05Y361), the Guangzhou Science and Technology Planning Project (202206010135), the Natural Science Foundation of Guangdong Province of China (2018A030313537).
Availability of data and materials
The raw data and code are available in the figshare (https://www.jianguoyun.com/p/Dd0uFBAQgOC2CxjGxPcEIAA).
Declarations
Ethics approval and consent to participate
Frozen fresh and paraffin-embedded skin cutaneous melanoma and non-cancerous tissues were collected from patients at the Nanfang Hospital of Southern Medical University. All samples were collected with informed consent from patients, and all procedures were performed after the internal review and approval of the Ethics Committees of Southern Medical University and the Nanfang Hospital.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gaohua Li, Tingting Wu, Heping Li and Chuzhong Wei have contributed equally to this work and shares first authorship.
Contributor Information
Jianwei Li, Email: ovenli1979@smu.edu.cn.
Yanan Wang, Email: wyn8116@163.com.
Guoxin Li, Email: gzliguoxin@163.com.
Lei Fan, Email: fanl1006@163.com.
References
- 1.Siegel RL, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Curti BD, Faries MB. Recent advances in the treatment of melanoma. N Engl J Med. 2021;384(23):2229–40. 10.1056/NEJMra2034861. [DOI] [PubMed] [Google Scholar]
- 3.Chan TA, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fusco MJ, West HJ, Walko CM. Tumor mutation burden and cancer treatment. JAMA Oncol. 2021;7(2):316. 10.1001/jamaoncol.2020.6371. [DOI] [PubMed] [Google Scholar]
- 5.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–99. 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–6. 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–13. 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–20. 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forschner A, et al. Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma - results of a prospective biomarker study. J Immunother Cancer. 2019;7(1):180. 10.1186/s40425-019-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joung J, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548(7667):343–6. 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanniford D, et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37(1):55-70 e15. 10.1016/j.ccell.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anagnostou V, et al. The status of tumor mutational burden and immunotherapy. Nat Cancer. 2022;3(6):652–6. 10.1038/s43018-022-00382-1. [DOI] [PubMed] [Google Scholar]
- 13.Ellrott K, et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6(3):271-281. e7. 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7): e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma A, et al. GRASLND regulates melanoma cell progression by targeting the miR-218–5p/STAM2 axis. J Transl Med. 2024;22(1):684. 10.1186/s12967-024-05397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, et al. Comprehensive prediction of immune microenvironment and hot and cold tumor differentiation in cutaneous melanoma based on necroptosis-related lncRNA. Sci Rep. 2023;13(1):7299. 10.1038/s41598-023-34238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, et al. Necroptosis-related LncRNAs in skin cutaneous melanoma: evaluating prognosis, predicting immunity, and guiding therapy. BMC Cancer. 2023;23(1):752. 10.1186/s12885-023-11246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian C, Liu S, Huo R. Identification of the ageing-related prognostic gene signature, and the associated regulation axis in skin cutaneous melanoma. Sci Rep. 2023;13(1):24. 10.1038/s41598-022-22259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimand J, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14(2):482–517. 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balachandran VP, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16(4):e173-80. 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7. 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sturm G, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–45. 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Racle J, et al. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017. 10.7554/eLife.26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becht E, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finotello F, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11(1):34. 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ru B, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–2. 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 29.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsson V, et al. The immune landscape of cancer. Immunity. 2018;48(4):812-830. e14. 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayers M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–40. 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9): e107468. 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021. 10.1093/bib/bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris GM, Huey R, Olson AJ. Using AUTODOCK for ligand-receptor docking. Curr Protoc Bioinformatics. 2008. 10.1002/0471250953.bi0814s24. [DOI] [PubMed] [Google Scholar]
- 36.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36. 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 37.Wei C, et al. Phase separation: “the master key” to deciphering the physiological and pathological functions of cells. Adv Biol (Weinh). 2022;6(7): e2200006. 10.1002/adbi.202200006. [DOI] [PubMed] [Google Scholar]
- 38.Hu F, et al. DEPDC1B promotes melanoma angiogenesis and metastasis through sequestration of ubiquitin ligase CDC16 to stabilize secreted SCUBE3. Adv Sci (Weinh). 2022;9(10): e2105226. 10.1002/advs.202105226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez Calle A, et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109(7):2093–100. 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosche S, et al. Rare variant analysis in eczema identifies exonic variants in DUSP1, NOTCH4 and SLC9A4. Nat Commun. 2021;12(1):6618. 10.1038/s41467-021-26783-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L, et al. Excavating novel diagnostic and prognostic long non-coding RNAs (lncRNAs) for head and neck squamous cell carcinoma: an integrated bioinformatics analysis of competing endogenous RNAs (ceRNAs) and gene co-expression networks. Bioengineered. 2021;12(2):12821–38. 10.1080/21655979.2021.2003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moons KG, et al. Prognosis and prognostic research: application and impact of prognostic models in clinical practice. BMJ. 2009;338: b606. 10.1136/bmj.b606. [DOI] [PubMed] [Google Scholar]
- 43.Wei CY, et al. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370–3p/CXCL12 axis in melanoma. Mol Cancer. 2020;19(1):84. 10.1186/s12943-020-01191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerezo M, et al. Translational control of tumor immune escape via the eIF4F-STAT1-PD-L1 axis in melanoma. Nat Med. 2018;24(12):1877–86. 10.1038/s41591-018-0217-1. [DOI] [PubMed] [Google Scholar]
- 45.Kalaora S, et al. Mechanisms of immune activation and regulation: lessons from melanoma. Nat Rev Cancer. 2022;22(4):195–207. 10.1038/s41568-022-00442-9. [DOI] [PubMed] [Google Scholar]
- 46.Dimitriou F, et al. Double trouble: immunotherapy doublets in melanoma-approved and novel combinations to optimize treatment in advanced melanoma. Am Soc Clin Oncol Educ Book. 2022;42:1–22. 10.1200/EDBK_351123. [DOI] [PubMed] [Google Scholar]
- 47.Reijers ILM, et al. Personalized response-directed surgery and adjuvant therapy after neoadjuvant ipilimumab and nivolumab in high-risk stage III melanoma: the PRADO trial. Nat Med. 2022;28(6):1178–88. 10.1038/s41591-022-01851-x. [DOI] [PubMed] [Google Scholar]
- 48.Bhave P, et al. Efficacy of anti-PD-1 and ipilimumab alone or in combination in acral melanoma. J Immunother Cancer. 2022. 10.1136/jitc-2022-004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pires da Silva I, et al. Efficacy and safety of anti-PD1 monotherapy or in combination with ipilimumab after BRAF/MEK inhibitors in patients with BRAF mutant metastatic melanoma. J Immunother Cancer. 2022. 10.1136/jitc-2022-004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalinsky K, et al. A phase 2 trial of dasatinib in patients with locally advanced or stage IV mucosal, acral, or vulvovaginal melanoma: A trial of the ECOG-ACRIN Cancer Research Group (E2607). Cancer. 2017;123(14):2688–97. 10.1002/cncr.30663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blechman AB, et al. Aggressive skin cancers occurring in patients treated with the janus kinase inhibitor ruxolitinib. J Drugs Dermatol. 2017;16(5):508–11. [PubMed] [Google Scholar]
- 52.Demirel Ogut N, Mizrak B. Possible ruxolitinib-induced acquired epidermodysplasia verruciformis and multiple non-melanoma skin cancers in a patient with polycythemia vera. J Cosmet Dermatol. 2022;21(11):6502–3. 10.1111/jocd.15253. [DOI] [PubMed] [Google Scholar]
- 53.Qu X, et al. Antitumor effects of anti-CD40/CpG immunotherapy combined with gemcitabine or 5-fluorouracil chemotherapy in the B16 melanoma model. Int Immunopharmacol. 2013;17(4):1141–7. 10.1016/j.intimp.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and code are available in the figshare (https://www.jianguoyun.com/p/Dd0uFBAQgOC2CxjGxPcEIAA).