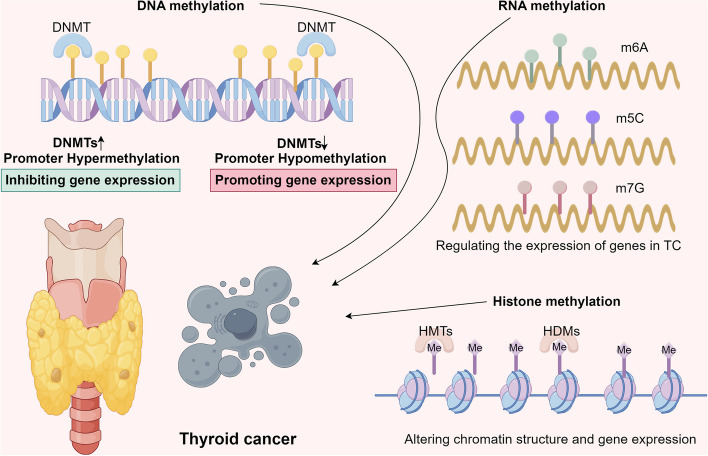
Abstract
Thyroid cancer (TC) represents one of the most prevalent endocrine malignancies, with a rising incidence worldwide. Epigenetic alterations, which modify gene expression without altering the underlying DNA sequence, have garnered significant attention in recent years. Increasing evidence underscores the pivotal role of epigenetic modifications, including DNA methylation, RNA methylation, and histone methylation, in the pathogenesis of TC. This review provides a comprehensive overview of these reversible and environmentally influenced epigenetic modifications, highlighting their molecular mechanisms and functional roles in TC. Additionally, the clinical implications, challenges associated with studying these epigenetic modifications, and potential future research directions are explored.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12957-024-03568-2.
Keywords: Thyroid cancer, Epigenetic methylations, DNA methylations, RNA methylations, Histone methylations
Introduction
The thyroid gland, a butterfly-shaped organ located at the base of the neck, secretes hormones crucial for regulating the body’s metabolism [1]. Thyroid cancer (TC) arises from the aberrant proliferation of thyroid cells [2]. The thyroid consists primarily of two cell types: follicular cells, which produce thyroid hormones, and C cells, which synthesize calcitonin [3]. TC is classified into five histological subtypes: papillary thyroid carcinoma (PTC), follicular thyroid carcinoma (FTC), medullary thyroid carcinoma (MTC), Hürthle cell carcinoma (HCC), and anaplastic thyroid carcinoma (ATC) [4]. These subtypes differ in prevalence, with a decreasing order of occurrence corresponding to increasing aggressiveness [5]. Advances in early diagnosis and personalized treatment strategies, including surgery, radioactive iodine therapy, chemotherapy, targeted therapy, and immunotherapy, have markedly improved survival rates in TC patients [6, 7]. However, the global rise in TC incidence has prompted increased scrutiny [8]. A deeper understanding of the molecular mechanisms underlying TC could enhance diagnostic precision, provide superior prognostic markers, offer targeted therapeutic approaches, and unveil novel treatment avenues.
Epigenetic modifications are defined as heritable changes in gene expression that do not involve changes to the DNA sequence itself [9]. Recent studies have demonstrated that these epigenetic alterations can influence the expression of genes and proteins involved in critical processes such as metastasis, apoptosis, and cell cycle regulation, contributing to the initiation and progression of TC [10]. Among these modifications, methylation plays a significant role, affecting the expression of genes and proteins crucial to cell cycle control, apoptosis, and metastasis, thereby driving TC pathogenesis [11]. For example, aberrant DNA methylation of specific genes may lead to gene silencing, promoting thyroid tumorigenesis or conferring resistance to apoptosis [12]. Similarly, RNA methylation, particularly N6-methyladenosine (m6A) modification, influences RNA molecule fate and function, regulating the expression of genes implicated in TC development, proliferation, and metastasis [13]. Furthermore, histone methylation alters chromatin structure and gene expression, while methylation of non-histone proteins modulates key signaling pathways involved in TC pathogenesis [14]. Given the reversible nature and external susceptibility of these epigenetic modifications, their study in TC opens new avenues for diagnostics, prognostics, and therapeutics. This review aims to provide a synthesis of the current understanding of epigenetic methylations at the DNA, RNA, and protein levels in TC, offering insights for future research and clinical application.
The role of DNA methylation in TC
DNA methyltransferases (DNMTs), including DNMT1, DNMT3A, and DNMT3B, catalyze the transfer of a methyl group to the 5-carbon position of the cytosine ring within cytosine-phosphate-guanine (CpG) dinucleotides [15]. CpG sites are often clustered in regions of the genome known as CpG islands, which are typically located within or near gene promoters [16]. Methylation of these CpG islands is commonly associated with transcriptional repression, playing a crucial role in processes such as genomic imprinting, X-chromosome inactivation, and the suppression of repetitive elements that might destabilize the genome [17]. In normal cells, DNA methylation is tightly regulated, ensuring appropriate gene expression for essential cellular functions [18]. However, aberrant DNA methylation patterns have been widely reported in TC. Table 1 and Fig. 1 summarize the effects of promoter methylation across different chromosomes in TC.
Table 1.
The role of DNA methylation in TC
| Methylation | Gene | Model | Role in TC | Reference |
|---|---|---|---|---|
| Hypermethylation | RASSF1A | Human TC cell lines and primary thyroid tumor samples from patients, including both benign and malignant tumors. | Regulating cell cycle and apoptosis | [19] |
| Hypermethylation | P16 | 74 PTC samples and 21 adjacent normal thyroid tissues | disrupting cell cycle control | [20] |
| Hypermethylation | TIMP3 |
Thyroid Tumor Samples: The cohort included 44 thyroid cancer samples and 44 benign thyroid lesions. Adjacent Normal Thyroid Tissue: Additionally, 15 samples of adjacent normal thyroid tissue were examined. |
Inhibiting invasion and metastasis by inhibiting MMPs | [21] |
| Hypermethylation | ATF3 |
Cell Lines: The study employed thyroid cancer cell lines to investigate the effects of ATF3 silencing and its subsequent impact on gene expression and pathway regulation. Clinical Samples: Tumor tissues from patients with thyroid cancer were used to analyze the methylation status of the ATF3 gene. These samples helped establish the correlation between ATF3 methylation and the progression of thyroid cancer. |
Inhibiting the proliferation and migration of TC cells | [22] |
| Hypermethylation | OGG1 | Malignant thyroid tissues from patients with PTC | Impairing the BER pathway | [23] |
| Hypermethylation | ATM |
Thyroid Tumor Samples: The cohort included 44 thyroid cancer samples and 44 benign thyroid lesions. Adjacent Normal Thyroid Tissue: Additionally, 15 samples of adjacent normal thyroid tissue were examined. |
Impairing DNA repair mechanisms | [21] |
| Hypermethylation | TSHR | Tissue samples from patients diagnosed with TC | Associated with the MAPK pathway | [24] |
| Hypomethylation | MAP17 | Various subtypes of thyroid tumors, including papillary, follicular, medullary, and anaplastic thyroid cancers | Promoting tumor growth | [25] |
| Hypomethylation | CTLA-4 | Peripheral blood samples from 200 patients with TC and 200 healthy controls. | Enhancing evasion of immune surveillance | [26] |
| Hypomethylation | MAP17 | Human PTC cell lines, Tumor tissues from patients with PTC and Xenograft models | Promoting EMT | [27] |
| Hypomethylation | TERT | Human PTC cell lines and Tumor tissues from patients with PTC | Correlating with poorer prognosis | [28] |
RASSF1A Ras association domain family 1 A, PTEN Phosphatase and tensin homolog, TIMP3 Tissue inhibitor of metalloproteinases 3, RARβ2 Retinoic acid receptor beta 2, ATF3 Activating transcription factor 3, MMPs Metalloproteinases, BER Base Excision Repair, OGG1 8-Oxoguanine DNA Glycosylase 1, MUTYH MutY Homolog, MLH1 MutL Homolog 1, ATM Ataxia telangiectasia mutated, NIS Sodium/Iodide Symporter, TSHR Thyroid stimulating hormone receptor, EMT Epithelial-mesenchymal transition, PFKFB2 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2, WDTC Well-differentiated thyroid carcinomas
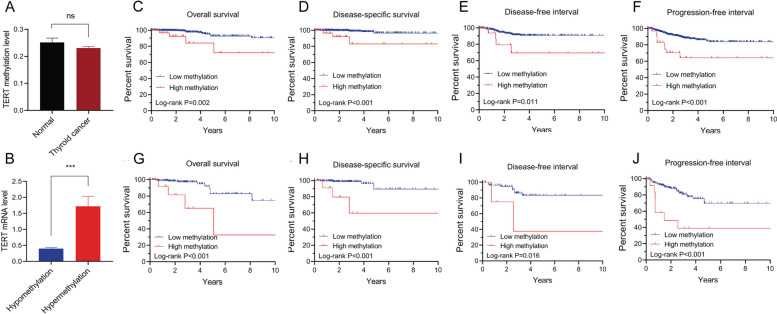
Fig. 1.
The Role of TERT promoter hypomethylation in TC. A TERT promoter methylation levels in 56 normal thyroid tissues vs. 571 TC tissues; (B) TERT mRNA expression in TERT hypermethylated (n = 33) and hypomethylated (n = 530) TC samples; (C-F). Kaplan-Meier analysis reveals the association of TERT methylation with all PTC clinical outcomes, including overall survival (C), disease-specific survival (D), disease-free interval (E), and progression-free interval (F); (G-J). Kaplan-Meier analysis reveals the association of TERT methylation with clinical outcomes of advanced PTC (stage III/IV), including overall survival (G), disease-specific survival (H), disease-free interval (I), and progression-free interval (J). Reproduced under the terms of the CC-BY license [28]. Copyright © 2024 The Authors, published by Frontiers
Promoter hypermethylation
Promoter hypermethylation is characterized by the increased methylation of specific gene promoter regions, leading to transcriptional silencing. This process can result in the inactivation of tumor suppressor genes (TSGs), the disruption of DNA repair pathways, and the epigenetic silencing of genes involved in iodine metabolism, all of which contribute to TC development.
Tumor suppressor gene silencing by promoter hypermethylation
Promoter hypermethylation-induced silencing of TSGs is a critical epigenetic mechanism that drives the initiation and progression of TC. Key TSGs implicated in TC include ras association domain family 1 A (RASSF1A), phosphatase and tensin homolog (PTEN), tissue inhibitor of metalloproteinases 3 (TIMP3), death-associated protein kinase (DAPK), retinoic acid receptor beta 2 (RARβ2), P16, and activating transcription factor 3 (ATF3). For instance, the CpG island in the promoter region of RASSF1A is fully methylated in nine TC cell lines, leading to its transcriptional silencing. RASSF1A, a critical tumor suppressor, regulates both the cell cycle and apoptosis [19]. In a study of 38 TC cases, 71% demonstrated hypermethylation of the RASSF1A CpG island, with higher methylation levels observed in more aggressive forms of TC [19]. Notably, RASSF1A methylation was found to be inversely correlated with BRAF mutations, the most common oncogenic mutation in TC, which activates the MAPK signaling pathway to promote tumor formation and progression [21]. Similarly, P16 encodes a protein that inhibits cyclin-dependent kinases, thereby regulating cell cycle progression [29]. In an analysis of 74 PTC cases, Wang et al. found that 27% exhibited significant promoter hypermethylation of P16 [20]. Excessive methylation of the P16 promoter leads to its transcriptional repression, disrupting cell cycle control and contributing to unchecked cellular proliferation [20].
DAPK is a key player in apoptosis (programmed cell death) and tumor suppression. Hypermethylation of the DAPK promoter leads to its silencing, which has been correlated with lymph node metastasis in TC patients, underscoring its critical role in TC pathogenesis [30]. The nuclear receptor RARβ2 mediates the growth-inhibitory effects of retinoic acid, a vital regulator of cell proliferation and differentiation [31]. Hypermethylation of the RARβ2 promoter, resulting in reduced RARβ2 expression, can disrupt cellular differentiation, thereby contributing to TC progression [31]. Additionally, a positive correlation has been observed between BRAF mutation and RARβ2 methylation [21]. The lipid phosphatase PTEN functions to inhibit the PI3K/AKT pathway, thereby regulating cell growth, survival, and proliferation [32]. Feng et al. reported elevated levels of PTEN promoter methylation in PTC [33]. Silencing of PTEN due to DNA hypermethylation results in uncontrolled activation of the PI3K/AKT pathway, promoting cell invasion, proliferation, and survival, which ultimately leads to TC [34].
TIMP3 is involved in inhibiting metalloproteinases (MMPs) from degrading the extracellular matrix, a critical step in tumor invasion and metastasis [35]. Promoter hypermethylation leading to the silencing of TIMP3 can result in increased MMP activity, thereby facilitating tumor metastasis [21]. ATF3 has the capacity to inhibit the proliferation and migration of TC cells [22]. Excessive DNA methylation within the ATF3 promoter impedes the binding of transcription factors SP1 and MYC-MAX, leading to gene silencing [22]. Maspin, a serine protease inhibitor, is involved in cell adhesion, migration, and apoptosis [36]. Ogasawara et al. found that undifferentiated TC samples exhibited higher levels of Maspin promoter methylation compared to normal thyroid tissue and differentiated TC [37]. Epigenetic silencing of Maspin may enhance the invasiveness and metastatic potential of TC [36].
Promoter hypermethylation-induced inactivation of DNA repair pathways
DNA repair pathways are essential for maintaining genomic integrity, correcting DNA damage caused by replication errors, environmental toxins, and radiation [38]. Promoter hypermethylation leading to the silencing of genes involved in these pathways can result in the accumulation of DNA mutations, genomic instability, and carcinogenesis [39]. DNA repair pathways affected by promoter hypermethylation include the Mismatch Repair (MMR) system, Base Excision Repair (BER) pathway, Nucleotide Excision Repair (NER) pathway, and both Homologous Recombination (HR) and Non-Homologous End Joining (NHEJ) [40]. The MMR system, for instance, corrects replication errors, ensuring the fidelity of base pairing [41]. MutL Homolog 1 (MLH1), a crucial component of the MMR system, is essential for preserving genomic integrity [42]. Guan et al. observed elevated MLH1 methylation in 8 out of 38 PTC samples [43].
The BER pathway repairs small base lesions caused by oxidative and alkylative damage [44]. 8-Oxyguanine DNA Glycosylase 1 (OGG1) is an enzyme that excises 8-oxoguanine (8-oxoG) and its ring-opened derivative, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG), from the DNA double helix [45]. 8-oxoG is a common marker of oxidative DNA damage induced by reactive oxygen and nitrogen species [45]. Guan et al. identified that 5% of PTC samples exhibited significant methylation of the OGG1 promoter [46]. This hypermethylation, leading to reduced OGG1 expression, impairs the BER pathway, thereby increasing the risk of mutations during DNA replication [23].
Homologous recombination (HR) repairs double-strand breaks using a homologous template, while non-homologous end joining (NHEJ) connects the ends of broken DNA without a template. Ataxia telangiectasia mutated (ATM) is a key gene in HR and NHEJ, responsible for detecting and repairing DNA double-strand breaks [47]. Hypermethylation-induced silencing of ATM impairs DNA repair mechanisms, leading to genomic instability and the accumulation of carcinogenic mutations [21]. Nucleotide excision repair (NER) corrects helix-distorting DNA lesions caused by UV radiation and chemical carcinogens [48]. Recent studies suggest that NER plays a significant role in the initiation and progression of TC [49]. In other tumors, such as bladder cancer, impaired NER capacity due to high methylation allows DNA damage to persist, leading to genomic instability and tumorigenesis [50]. Further research is necessary to fully understand the impact of elevated methylation on NER impairment in TC.
Promoter hypermethylation-induced epigenetic silencing of iodine metabolism genes
The thyroid’s ability to absorb and process iodide (I⁻) is crucial for its normal function and the production of thyroid hormones [51]. In TC, promoter hypermethylation-induced deregulation of iodine metabolism genes can alter thyroid hormone synthesis, contributing to tumorigenesis [52]. Moreover, this disruption may compromise the efficacy of treatments such as radioactive iodine therapy, which relies on the ability of TC cells to absorb iodide (I⁻). For example, hypermethylation-induced silencing of the SLC5A5 gene, which encodes the Sodium/Iodide Symporter (NIS), has been observed in TC [53]. This alteration is associated with reduced efficacy of radioactive iodine therapy in advanced TC patients, as NIS is essential for iodide (I⁻) uptake by thyroid cells [53]. The thyroid-stimulating hormone receptor (TSHR) gene is also frequently hypermethylated in TC. Studies have shown that PTC exhibits higher TSHR methylation rates, which may be linked to the pathophysiology of PTC [54]. Furthermore, TSHR gene methylation in PTC patients is strongly correlated with age, lymph node metastasis, clinical staging, and tumor size, suggesting that TSHR may serve as a marker for determining PTC severity [54]. Khan et al. found that among 60 TC tissues, 15 cases harbored the BRAF V600E mutation, and 73.3% of these cases exhibited TSHR promoter methylation, indicating a potential link between the TSHR pathway and the MAP kinase pathway [24].
Promoter hypermethylation-induced non-coding RNA silencing
Promoter hypermethylation also silences non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), adding another layer of complexity to the epigenetic regulation in TC. ncRNAs play pivotal roles in regulating gene expression at both the transcriptional and post-transcriptional levels [55]. They are involved in various cellular processes, including cell growth, differentiation, apoptosis, and stress responses [56]. In TC, ncRNAs have been shown to function as oncogenes or tumor suppressors [57, 58]. The silencing of ncRNAs through promoter hypermethylation disrupts normal cellular regulatory networks, contributing to thyroid carcinogenesis. For instance, miR-199a-3p is downregulated in PTC due to DNMT3a-mediated hypermethylation of its promoter region [59]. DNMT3a is upregulated in PTC, and inhibition of DNMT3a or demethylation treatment restores miR-199a-3p levels [59]. Functional studies have shown that miR-199a-3p suppresses cancer cell migration, invasion, and growth, and its overexpression in PTC cells delays tumor growth in xenografted mice [59]. Another study identified hypermethylation at 12 CpG sites in the promoter region of miR-204 in PTC tissues compared to normal tissues [60]. This hypermethylation was negatively correlated with the expression levels of miR-204 and its host gene, TRPM3. Moreover, downregulation of miR-204 was associated with PTC extrathyroidal extension, high T-stage, lymph node metastasis, the BRAF V600E mutation, and the aggressive tall cell variant [60]. Similarly, silencing lncRNAs through promoter hypermethylation can alter chromatin structure, affect gene expression, and promote tumorigenesis. For example, hypermethylation of the promoter region of lncRNA AB074169 is associated with decreased expression in PTC, suggesting a tumor-suppressive role for lncRNA AB074169 [59].
Promoter hypomethylation
Promoter hypomethylation occurs primarily in the regulatory regions of genes, resulting in the opening of chromatin structure and subsequent promotion of gene transcription [61]. In cancer, various environmental, genetic, and stochastic factors contribute to promoter hypomethylation by modulating the activity of DNA methyltransferases (DNMTs)—enzymes responsible for adding methyl groups to DNA—thus activating genes and promoting malignant transformation and tumor progression [62]. In TC, promoter hypomethylation significantly influences the pathophysiology and progression of the disease, primarily through the activation of oncogenes and alterations in genes involved in cell proliferation, migration, and invasion. Hypomethylation of genes such as FOXO3, ZEB2, and CDK6 has been associated with lymph node metastasis in PTC [63]. Unlike promoter hypermethylation, which typically involves the addition of methyl groups, promoter hypomethylation usually entails the loss of DNA methylation, leading to increased gene expression [64]. The consequences of promoter hypomethylation in TC include oncogene activation, genomic instability, altered gene expression, immune evasion, metastasis, and changes in the tumor microenvironment (TME).
Hypomethylation of oncogene promoters can result in their aberrant activation. For instance, the RAS family of oncogenes (HRAS, KRAS, and NRAS) frequently undergoes hypomethylation in TC [65]. Activated RAS signaling promotes cell proliferation, survival, and migration, contributing to tumor growth [66]. The RASSF1A-MST1-FoxO3 signaling pathway is regulated by the BRAF V600E mutation in PTC, leading to RASSF1A hypomethylation and influencing the degree of TC malignancy [46]. Hypomethylation can also lead to the overexpression of MAP17, which may further promote tumor growth in TC [25]. Additionally, in TC, the SYK gene is overexpressed due to hypomethylation in its promoter region, influencing various cellular processes and thereby contributing to the development of TC [67]. Non-coding genes are also implicated in promoter hypomethylation. For example, PTC tissue samples exhibit significantly higher expression levels of miR-21 and miR-146b compared to non-tumorous thyroid tissue, and this increase is associated with hypomethylation of specific loci within their genomic regions [68].
Promoter hypomethylation can also facilitate tumor immune evasion and metastasis. The CTLA-4 gene, an important immune checkpoint regulator, is overexpressed due to promoter hypomethylation [69]. This overexpression enhances the ability of TC cells to evade immune surveillance, allowing uncontrolled tumor growth and metastasis [26]. Wang et al. found that transmembrane 4 L six family member 1 (TM4SF1) was significantly overexpressed in PTC patients with lymph node metastasis, with these patients exhibiting lower levels of TM4SF1 promoter methylation [70]. Furthermore, hypomethylation of genes involved in epithelial-mesenchymal transition (EMT) promotes metastasis. EMT enables typically stationary epithelial cells to acquire a mesenchymal cell phenotype through biochemical alterations, facilitating metastasis [71]. Hypomethylation-induced overexpression of MAP17 can also promote EMT, further contributing to TC progression [27].
DNA hypomethylation, particularly in repetitive elements of the genome, contributes to genomic instability—a hallmark of cancer [72]. In thyroid cancer (TC), this instability can lead to chromosomal rearrangements, mutations, and other alterations, further driving tumor progression. Studies have shown that global Alu hypomethylation is increasingly prevalent in differentiated TC (DTC), poorly differentiated TC (PDTC), and ATC with distant metastases, suggesting that this epigenetic alteration may play a role in the progression and dedifferentiation of TC [73]. The TME may also be influenced by hypomethylation. For instance, the genetic and epigenetic landscape, including DNA methylation, affects the interaction between cancer stem cells in the thyroid tumor and cells within the TME [74]. In TC patients, hypomethylation has been associated with recurrence and prognosis. For example, a study by Li et al. found that methylation of the TERT promoter is associated with increased expression of TERT (telomerase reverse transcriptase), a key enzyme involved in maintaining telomere length and contributing to cellular immortality. High levels of TERT expression, driven by promoter methylation, were correlated with a worse prognosis in patients with PTC, including higher rates of tumor aggressiveness, recurrence, and reduced overall survival [28] (Fig. 1). Additionally, Camargo et al. identified that PFKFB2 methylation levels serve as an independent risk factor for recurrence in patients with well-differentiated thyroid carcinomas (WDTC), with 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2) promoter hypomethylation being linked to recurrence in WDTC [28, 74, 75].
The role of RNA methylation in TC
RNA modifications represent a form of epigenetic regulation that influences RNA structure and function, encompassing processes such as methylation, acetylation, lactylation, and glycosylation, with methylation and acetylation (e.g., ac4C-acetylation) being the most prevalent types [76]. RNA methylation, including modifications like N6-methyladenosine (m6A), 5-methylcytosine (m5C), N7-methylguanosine (m7G), and N1-methyladenosine (m1A), plays a critical role in splicing, translation, RNA stability, and localization [77]. These modifications introduce an additional layer of post-transcriptional regulation, significantly impacting TC pathogenesis and cellular function. Table 2 and Fig. 2 provide detailed insights into the effects of different RNA methylation modifications on TC.
Table 2.
The role of RNA and histone methylation in TC
| Modifications | Gene | Model | Role in TC | Reference |
|---|---|---|---|---|
| M6A | IGF2BP2 and YTHDF1 | Thirty pairs of thyroid cancer tissue samples (including PTC, FTC, ATC, and MTC) and adjacent normal thyroid tissues | Prognostic factors | [78, 79] |
| M6A | YTHDF3 | Thirty pairs of thyroid cancer tissue samples (including PTC, FTC, ATC, and MTC) and adjacent normal thyroid tissues | Prognostic biomarker | [78] |
| M6A | NF-κB | Mouse xenograft models and tumor tissues from PTC patients | METTL3-related modification inhibits tumor progression | [80] |
| M6A | STEAP2 | Mouse xenograft models | METTL3-related modification promoting proliferation and invasion of PTC cells | [81] |
| M6A | IGF2BP2 | Tissue samples from PTC patients | Stabilizing DPP4 mRNA, enhancing proliferation and invasion of PTC cells | [82] |
| M6A | IGF2BP2 | radioiodine refractory papillary thyroid cancer (RR-PTC) cell lines, specifically K1 and TPC1, and Tissue samples from RR-PTC patients | Stabilizing RUNX2 mRNA, inhibiting NIS expression, and hindering differentiation [83] | [83] |
| M6A | LINC01125 | Human PTC cell lines, specifically TPC-1 and IHH-4, and tumor tissues from patients with PTC | METTL3-related modification enhancing invasion, migration, and proliferation of TC cells | [84] |
| M6A | LINC00894 | Human PTC cell lines, including KTC-1 and BCPAP and tumor tissues from patients with TC | METTL3-related modification involved in the malignant phenotype of PTC | [85] |
| m5C | NSUN2 | Human ATC cell lines, mouse xenograft models, and tumor tissue samples from patients with ATC | Promoting proliferation, invasion, and migration of ATC cells | [86] |
| M7G | DOCK9-DT, DPP4-DT, TMEM105, SMG7-AS1 and HMGA2-AS1 | Transcriptome expression data from TCGA database, 567 samples were collected, including 509 THCA tissue samples and 58 normal paracancer tissue samples. | Prognostic model | [87] |
| Histone methylation | H3K4me3 | Tumor tissues from patients with different subtypes of TC | Linked with lymphatic vessel invasion | [88] |
METTL3 Methyltransferase-like 3, DPP4 Dipeptidyl peptidase-4, ATC Anaplastic thyroid carcinoma, H3K4me3 Histone H3 on lysine 4
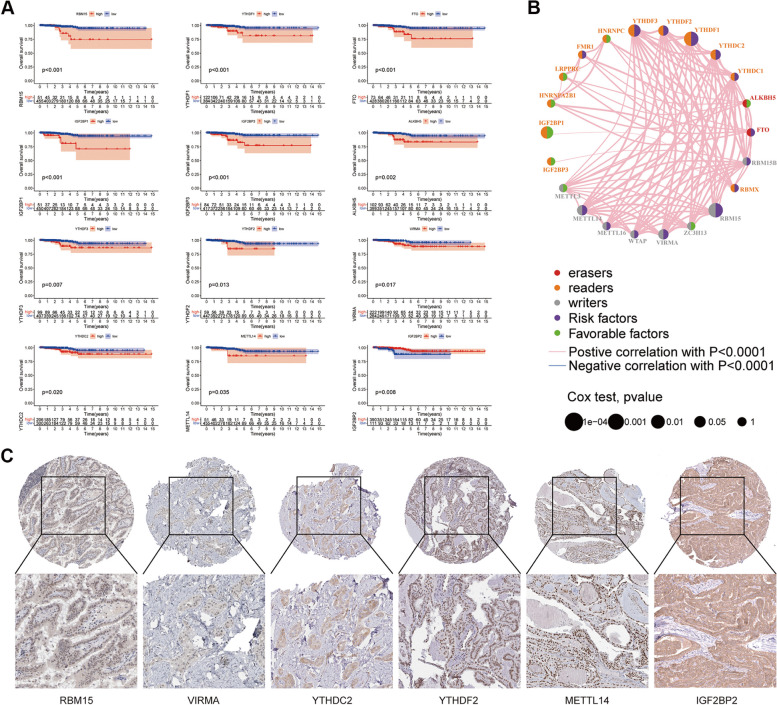
Fig. 2.
Relationship between m6A related genes and prognosis of TC. A Survival curves were used to reveal the relationship between survival time and survival of different m6A methylation-related genes; B The m6A prognostic network was used to demonstrate the expression and interactions of 23 m6A regulators in TC; C Protein expression of some m6A genes associated with prognosis in TC tissues. Reproduced under the terms of the CC-BY license [89]. Copyright © 2024 The Authors
m6A modification
m6A modification is dynamic and reversible, mediated by “writers” (methyltransferases), “erasers” (demethylases), and “readers” (binding proteins) [90, 91]. As the most common internal RNA modification in eukaryotic cells, m6A is involved in nearly all stages of the RNA lifecycle, including mRNA transcription, maturation, translation, degradation, and stability [92]. Initial research on m6A modification in TC has primarily focused on its prognostic significance. For instance, m6A methylation regulators such as IGF2BP2, YTHDF1, and YTHDF3 have been identified as potent independent prognostic factors in TC [78, 79]. Using comprehensive bioinformatics tools, Zhou et al. identified specific m6A regulators that are differentially expressed in TC tissues compared to normal tissues, which are associated with thyroid cancer progression and prognosis [89]. In addition, they developed a risk prediction model based on the expression levels of these m6A regulators, which was able to distinguish high-risk and low-risk thyroid cancer patients [89]. They also validated the results of the bioinformatics analysis using histologic samples, confirming that the identified m6A regulatory factors are indeed associated with thyroid cancer pathology. Notably, YTHDF3 is considered a potential prognostic biomarker for TC [78]. Furthermore, Yu et al. confirmed that FTO, RBM15, and KIAA1429 are independent prognostic biomarkers for TC patients, with this risk profile predicting outcomes more accurately in male patients [93]. Cell-based experiments have also shown that knocking down FTO, RBM15, and KIAA1429 inhibits the proliferation and migration of TC cells [93].
Recent studies have demonstrated that the aberrant expression of m6A regulatory factors is closely linked to the onset and progression of thyroid cancer (TC), playing a critical role in tumor drug resistance [94]. For instance, as a “writer” of m6A modification, Methyltransferase-like 3 (METTL3) inhibits tumor progression in a YTHDF2-dependent manner by modifying NF-κB mRNA, thereby reducing IL-8 production by PTC cells and limiting the recruitment of neutrophils [80]. Another study found that METTL3-associated STEAP2 m6A modification results in low STEAP2 expression, which is correlated with aggressive clinicopathological features and poor prognosis in PTC patients [81]. Mechanistic investigations revealed that STEAP2 overexpression reduces lung metastasis and tumorigenicity in vivo and inhibits PTC cell proliferation, migration, and invasion in vitro by blocking the Hedgehog signaling pathway and the epithelial-mesenchymal transition (EMT) [81]. IGF2BP2, an m6A “reader,” also plays a crucial role in TC. In PTC, upregulated IGF2BP2 stabilizes dipeptidyl peptidase-4 (DPP4) through an m6A-dependent mechanism, enhancing PTC cell proliferation, invasion, and migration, and ultimately promoting lymph node metastasis [82]. Moreover, in radioiodine-refractory PTC (RR-PTC), IGF2BP2 binds to m6A modification sites in the 3’-UTR of RUNX2 mRNA, stabilizing RUNX2 and inhibiting NIS expression, thereby hindering cellular differentiation [83]. Targeting IGF2BP2 has been shown to increase 125I uptake in RR-PTC cell lines and enhance NIS expression [83].
Several studies have also confirmed the relationship between m6A modification and ncRNAs, particularly long non-coding RNAs (lncRNAs). Huang et al., for example, investigated the role of m6A RNA methylation-related lncRNAs in PTC and developed a predictive model based on three lncRNAs: PSMG3-AS1, BHLHE40-AS1, and AC016747.3 [95]. The findings indicated that the risk score derived from this model correlates with molecular clusters, PD-L1 expression, immune cell infiltration, and immune checkpoint activity. In another study, He et al. discovered that METTL3 and its downstream target LINC01125 are downregulated in PTC, and that upregulating LINC01125 inhibits the invasion, migration, and proliferation of TC cells [84]. Additionally, research has shown that METTL3 and its downstream target LINC00894 are downregulated in PTC tissues, with METTL3 upregulating LINC00894 through an m6A-YTHDC2-dependent pathway, thereby inhibiting the malignant phenotype of PTC via the Hippo signaling pathway [85]. The association between m6A modification and microRNAs (miRNA) has also been established [96]. For example, in TC cells, METTL3 is overexpressed, while miR-222-3p is downregulated [97]. METTL3 enhances miR-222-3p expression by mediating m6A modification of pri-miR-222-3p [97]. miR-222-3p targets and inversely regulates serine/threonine stress kinase 4 (STK4) \, and knockdown of METTL3 increases STK4 expression by downregulating miR-222-3p, thereby suppressing TC malignancy and metastasis [97].
m5C modification
The m5C modification is one of several post-transcriptional modifications that occur in various RNA molecules, including mRNA, tRNA, and rRNA, influencing their stability, processing, and translation efficiency [98]. This modification is mediated by RNA methyltransferases, which catalyze the formation of m5C by transferring a methyl group from a donor molecule, typically S-adenosylmethionine (SAM), to the carbon-5 position of the cytosine base in RNA [99]. Among these RNA methyltransferases, NSUN2 (NOP2/Sun RNA methyltransferase 2) and DNMT2 (DNA methyltransferase 2) are the principal enzymes responsible for installing the m5C modification on a variety of RNAs, including mRNA, tRNA, and rRNA.
The first study investigating m5C modification in TC focused on its role in cell infiltration within the TME of PTC. The study found that a low m5C score correlates with activated immunity, indicating relatively favorable prognostic outcomes [100]. Additionally, ten distinct genes strongly correlated with the m5C score were used to construct a diagnostic model, which demonstrated high accuracy in diagnosing PTC [100]. Another study revealed that the tRNA m5C methyltransferase NSUN2 is upregulated in ATC, where it catalyzes tRNA structure-related m5C modification. This modification stabilizes tRNA, which is crucial for maintaining cellular homeostasis and efficiently transporting amino acids, particularly leucine [86]. The stabilized tRNA supports a pro-cancer translation program, promoting the synthesis of oncogenic proteins such as c-Myc, BCL2, RAB31, JUNB, and TRAF2 [86]. The functions of m5C writers and readers are believed to regulate gene expression at the post-transcriptional level and are involved in cellular metabolism and motility [101]. However, the mechanisms by which RNA m5C methylation influences cell mobility and metastasis in TC remain to be fully elucidated [101].
m7G modification
The m7G modification is found in various RNA types, including mRNA, tRNA, rRNA, miRNA, lncRNA, and at the 5’ cap end of eukaryotic mRNA [102]. The m7G cap is critical for RNA stability, nuclear export, splicing, and the initiation of translation [103]. This cap structure is essential for the efficient binding of mRNA to the ribosome, thereby promoting protein synthesis [103]. In tRNA, m7G modifications can affect tRNA folding, stability, and function, which in turn impacts the efficiency and fidelity of protein translation [104]. Increasing evidence suggests that aberrant m7G levels are closely associated with tumorigenesis and progression by regulating the expression of various oncogenes and tumor suppressor genes [105]. In mammals, the most well-studied regulator of m7G is methyltransferase-like 1 (METTL1), which, together with its cofactor WD repeat domain 4 (WDR4), installs m7G modifications in tRNA, miRNA, and mRNA [105]. RNA guanine-7 methyltransferase (RNMT) and its cofactor RNMT-activating miniprotein (RAM) are involved in the installation of m7G modifications at the 5’ caps of mRNA [105]. The role of m7G modifications in lncRNA has been partially elucidated in TC. Zhou et al. established a prognostic model for TC using five lncRNAs (DOCK9-DT, DPP4-DT, TMEM105, SMG7-AS1, and HMGA2-AS1) associated with m7G, demonstrating that this model has significant predictive capability, with patients scoring high on this model having a poorer prognosis [87]. Similar to m5C, research on the role of m7G in TC is still in its early stages, and further studies are needed to explore the biological mechanisms through which m7G modifications influence TC.
The role of histone methylation in TC
Histones are fundamental proteins that organize chromatin by forming nucleosomes, which consist of DNA segments, each with 147 base pairs, wrapped around an octamer of four core histone proteins (H3, H4, H2A, and H2B) [106]. The tails of these histones undergo extensive covalent posttranslational modifications (PTMs) that influence nucleosome dynamics, chromatin compaction, and transcriptional regulation [106]. These modifications can be induced by both internal and external stimuli [106]. Histone methylation, along with demethylation (catalyzed by histone demethylases [HDMs]), plays a crucial role in the regulation of gene activity by altering the accessibility of DNA to transcription factors. The functional outcome of histone methylation is determined by the specific amino acid residue and the number of methyl groups added (mono-, di-, or trimethylation).
Histone methylation can profoundly impact gene expression patterns in TC, thereby contributing to tumor initiation, progression, and metastasis. Specific histone methylation marks are associated with either gene activation or repression. For instance, lysine methyltransferases (HMTs) and demethylases (HDMs) modulate gene transcription by modifying lysine residues on histone proteins, and aberrant histone methylation has been implicated in cancer metastasis [107]. In TC, histone methylation modifications are often associated with gene repression. For example, trimethylation of histone H3 on lysine 4 (H3K4me3) is typically linked to gene activation [88], whereas overexpression of H3K27me3 has been correlated with increased lymph node metastasis and lymphatic vessel invasion [88].
In other cancers, abnormal histone methylation can lead to the epigenetic silencing of critical tumor suppressor genes. For instance, hypermethylation of H3K27, mediated by the polycomb repressive complex 2 (PRC2), can silence tumor suppressor genes such as RASSF1A and CDKN2A [108]. This repression facilitates uncontrolled cell growth and tumor development. Histone methylation also plays a vital role in maintaining chromatin structure and genome stability. In TC, alterations in histone methylation can disrupt chromatin organization, leading to genomic instability and an increased susceptibility to mutations [109]. This instability can further drive cancer progression and resistance to therapies.
Histone methylation often interacts with other epigenetic modifications, such as DNA methylation and histone acetylation, to coordinate gene expression regulation. In TC, these interactions contribute to complex epigenetic landscapes that influence cancer cell behavior. For example, histone methylation can recruit DNA methyltransferases to specific gene loci, leading to combined histone and DNA methylation, resulting in robust gene silencing. Moreover, histone methylation interacts with other genetic and epigenetic factors. For instance, the frequent BRAF gene mutation in TC is believed to regulate the RASSF1A-MST1-FoxO3 signaling pathway, which in turn influences TC malignancy through histone methylation modifications [46].
Clinical implications and challenges of methylation modifications
Clinical implications of DNA methylation
DNA methylation patterns have emerged as significant diagnostic and prognostic biomarkers in TC. These patterns can serve as robust indicators for the early detection of TC. Aberrant methylation of tumor suppressor genes, such as RASSF1A, PTEN, and CDKN2A, is frequently observed in thyroid malignancies. The hypermethylation of promoters in these genes can help distinguish malignant thyroid tissues from benign nodules, thereby aiding in early and accurate diagnosis. For instance, the hypermethylation of TIMP3, RARB2, SERPINB5, RASSF1, TPO, and TSHR has been reported as a common feature in papillary thyroid carcinoma (PTC) and can be used to differentiate it from benign thyroid conditions [110]. Advancements in liquid biopsy technologies have facilitated the detection of methylated DNA in circulating tumor DNA (ctDNA) from blood samples. This non-invasive approach allows for real-time monitoring of the methylation status of specific genes associated with TC. For example, the detection of methylated SLC5A8 and SLC26A4 genes in blood samples has been investigated as potential diagnostic markers for TC [111]. These non-invasive tools provide a less invasive and more patient-friendly method for early cancer detection and monitoring. Table 3 details the clinical implications of DNA methylation in TC.
Table 3.
The implication of different methylations in TC
| Modifications | Gene | Role in TC | Reference |
|---|---|---|---|
| DNA Methylation | TIMP3, RARB2, SERPINB5, RASSF1, TPO and TSHR | Differentiate PTC from benign thyroid conditions | [110] |
| DNA Methylation | SLC5A8 and SLC26A4 | Potential diagnostic markers | [111] |
| DNA Methylation | TIMP3 and SLC5A8 | Linked to higher tumor aggressiveness and worse prognosis | [112] |
| DNA Methylation | g10705422, cg17707274, and cg26849382 | Associated with higher risks of recurrent or persistent disease and distant metastasis in patients with DTC | [113] |
| DNA Methylation | BRAF | Hypomethylation of BRAF indicates a more favorable prognosis | [114] |
| DNA Methylation | PFKFB2 | PFKFB2 hypomethylation was associated with poor prognosis in PDTC/ATC and relapsed WDTC | [115] |
| RNA Methylation | YTHDF3 | Positively associated with the infiltration level of CD4 + T cells and macrophages | [78] |
| RNA Methylation | FTO, RBM15, and KIAA1429 | Independent prognostic biomarker in patients with thyroid carcinoma | [116] |
| Histone Methylation | H3K27me3 | Associated with extrathyroidal extension, lymphovascular invasion, lymph node metastasis, and higher risk of recurrence in DTC | [88] |
H3K4me3 Histone H3 on lysine 4, DTC Differentiated thyroid cancers, PDTC Poorly-differentiated thyroid carcinomas, ATC Anaplastic thyroid carcinomas, WDTC Well-differentiated thyroid carcinomas, PTC Papillary thyroid carcinoma
The DNA methylation status also offers valuable prognostic information, aiding in the stratification of TC patients based on their risk profiles. Certain methylation patterns are associated with more aggressive tumor behavior and poorer outcomes. For instance, hypermethylation of the TIMP3 and SLC5A8 genes has been linked to higher tumor aggressiveness and worse prognosis in TC patients [112]. Another study demonstrated that low DNA methylation levels at g10705422, cg17707274, and cg26849382 were significantly associated with higher risks of recurrent or persistent disease (odds ratio [OR] = 3.860) and distant metastasis (OR = 4.009) in patients with DTC [113]. Conversely, hypomethylation of oncogenes such as BRAF may indicate a more favorable prognosis [114]. Moreover, Camargo et al. revealed that PFKFB2 hypomethylation was associated with poor prognosis in PDTC/ATC and relapsed well-differentiated thyroid carcinomas (WDTC) compared to good-prognosis WDTC and non-malignant cases [115]. Lower PFKFB2 methylation levels were identified as an independent factor for high relapse risk in WDTC patients.
The pivotal role of DNA methylation in the onset and progression of TC provides a foundation for personalized treatment approaches. For example, tumors with methylation-induced DNA repair defects may exhibit specific vulnerabilities, such as increased sensitivity to DNA-damaging agents or PARP inhibitors, providing a rationale for targeted therapy strategies [117]. Reversing promoter hypermethylation with demethylating agents could restore the function of silenced DNA repair genes, potentially reversing the malignant phenotype [118]. However, studying the role of DNA methylation in TC presents a complex array of challenges. Methodological challenges include limitations in the sensitivity and specificity of DNA methylation detection methods (such as bisulfite sequencing and methylation-specific PCR) and the complexity of methylation patterns across different TC subtypes and stages [119]. Biological challenges include understanding the complex functional effects of specific methylation changes on TC gene expression and cellular behavior, the interaction of DNA methylation with other epigenetic modifications, and the heterogeneity of TCs, which can dilute aberrant methylation patterns. To address these challenges, future research must first establish a comprehensive atlas of DNA methylation patterns for different TC types, stages, and grades. Techniques such as methylation-specific PCR (MSP), bisulfite sequencing, and high-throughput methylation arrays can provide detailed DNA methylation profiles in tumor samples. Additionally, developing standardized protocols and guidelines for collecting, analyzing, and interpreting DNA methylation data will ensure consistent and reliable results across clinical settings.
Clinical implications of RNA methylation
RNA methylation marks, such as m6A, m5C, and m7G, have emerged as promising biomarkers for TC. Several m6A RNA methylation regulators have been identified as strong independent prognostic factors in TC. For instance, IGF2BP2, YTHDF1, and YTHDF3 have been highlighted as robust prognostic indicators in TC [78]. A risk score model was established to screen the predictors further [78]. A risk score model was subsequently developed to further refine these predictors The expression of YTHDF3 was found to be positively associated with the infiltration of CD4 + T cells and macrophages [78], and it exhibited strong correlations with various immune markers in TC. Another study identified a three-gene m6A RNA modification regulator-based risk signature (FTO, RBM15, and KIAA1429), which serves as an independent prognostic biomarker in patients with thyroid carcinoma [116]. Notably, this risk signature demonstrated better predictive accuracy in males than in females [116]. These findings underscore the significant impact of m6A methylation on TC progression, suggesting that m6A regulators could serve as potential prognostic markers and therapeutic targets. Table 3 details the clinical implications of RNA methylation in TC.
Exploring RNA methylation opens new avenues for understanding the complex epigenetic regulation of gene expression in cancers, including TC. However, deciphering the role of RNA methylation in TC presents numerous challenges, both technical and biological. Key challenges include the detection and quantification of RNA methylation modifications, the heterogeneity of modification patterns, the dynamic and reversible nature of RNA methylation, the complex effects on gene function and TC phenotypes, and the interactions of RNA methylation with other epigenetic and post-transcriptional mechanisms. To address these challenges, future research should not only map RNA methylation landscapes across different types and stages of TC but also investigate the functional impacts of specific RNA methylation modifications on the post-transcriptional regulation of TC gene expression. Targeting the enzymes responsible for adding, removing, and recognizing methylated RNA modifications (writers, erasers, and readers, respectively) could offer new therapeutic avenues for TC. The development of small molecule inhibitors or modulators of these proteins to manipulate RNA methylation states could also provide novel approaches to TC treatment [120].
Clinical implications of histone methylation
Specific histone methylation patterns can serve as valuable biomarkers for TC prognosis. For example, high levels of H3K27me3 expression have been significantly associated with extrathyroidal extension, lymphovascular invasion, lymph node metastasis, and a higher risk of recurrence in DTC, suggesting its involvement in the aggressiveness and dedifferentiation of TC [88]. EZH2, a methyltransferase responsible for H3K27me3, is frequently overexpressed in TC [121]. Inhibitors targeting EZH2 can reduce H3K27me3 levels, thereby reactivating tumor suppressor genes and inhibiting tumor growth [121]. Table 3 details the clinical implications of histone methylation in TC.
The dynamic and reversible nature of histone methylation plays a critical role in regulating gene expression in TC, influencing disease onset, progression, metastasis, and response to treatment [122]. Abnormal histone methylation patterns have the potential to serve as biomarkers for early detection, prognosis assessment, and prediction of treatment response, thereby facilitating the personalized management of TC patients [123]. However, the complexity of the enzyme networks and interactions involved in histone methylation presents a significant challenge in studying their role in TC. Future research aimed at elucidating how specific histone modifications influence transcription and epigenetic architecture will be crucial in clarifying their roles in TC pathogenesis, particularly concerning tumor suppressor genes, oncogenes, and genes involved in metastasis, angiogenesis, and drug resistance. The interaction between histone modifications and ncRNAs is another promising area for future investigation. Moreover, targeting histone modification mechanisms with small molecule inhibitors or epigenetic therapies offers a novel therapeutic approach. Drugs that regulate histone acetylation and methylation have already been tested in clinical trials for various cancers and hold significant promise for TC, particularly for patients in advanced stages or those who have developed resistance to standard treatments [124].
Supplementary Information
Acknowledgements
Not applicable.
Informed consent
Not applicable.
Institutional review board statement
Not applicable.
Authors’ contributions
Xiaojie Yu and Weiwei Zou: Conceptualization, Formal analysis, Investigation, Writing-original draft, Writing-review & editing; Xiaojie Yu, Weiwei Zou, Zhenlin Yang and Yong Han: Conceptualization, Formal analysis, Investigation, Writing - review & editing. Hao Zhang, Haojie Zhang, Changran Hou, Xiaohong Wang: Formal analysis and Investigation; Pengfei Gu, Xiaohong Wang and Weiwei Zou: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing - review & editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 81902702), Natural Science Foundation of Shandong Province (No.ZR2023MH115 and ZR2017LH072), National Key Research and Development Project (No. 2018YFC0114705).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yong Han, Email: hanyong2021@126.com.
Zhenlin Yang, Email: ikb0607@163.com.
Weiwei Zou, Email: sdbzzww@126.com.
References
- 1.Al-Suhaimi EA, Khan FA. Thyroid glands: physiology and structure. Emerging concepts in Endocrine structure and functions. Springer; 2022. pp. 133–60. 10.1007/978-981-16-9016-7_5.
- 2.Sloan LW. Of the origin, characteristics and behavior of thyroid cancer. J Clin Endocrinol Metabolism. 1954;14(11):1309–35. [DOI] [PubMed] [Google Scholar]
- 3.Al-Suhaimi EA, Al-Khater K. Functions of stem cells of thyroid glands in health and disease. Reviews Endocr Metabolic Disorders. 2019;20:187–95. [DOI] [PubMed] [Google Scholar]
- 4.Arrangoiz R, Cordera F, Caba D, Moreno E, Luque-de-Leon E, Muñ M. Thyroid cancer. Int J Otolaryngol Head Neck Surg. 2019;8(6):217–70. [Google Scholar]
- 5.Katoh H, Yamashita K, Enomoto T, Watanabe M. Classification and general considerations of thyroid cancer. Ann Clin Pathol. 2015;3(1):1045. [Google Scholar]
- 6.Campennì A, Barbaro D, Guzzo M, Capoccetti F, Giovanella L. Personalized management of differentiated thyroid cancer in real life–practical guidance from a multidisciplinary panel of experts. Endocrine. 2020;70:280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nabhan F, Dedhia PH, Ringel MD. Thyroid cancer, recent advances in diagnosis and therapy. Int J Cancer. 2021;149(5):984–92. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Gosnell JE, Roman SA. Geographic influences in the global rise of thyroid cancer. Nat Reviews Endocrinol. 2020;16(1):17–29. [DOI] [PubMed] [Google Scholar]
- 9.Wang N, Wang S, Li MY, Hu BG, Liu LP, Yang SL, et al. Cancer stem cells in hepatocellular carcinoma: an overview and promising therapeutic strategies. Ther Adv Med Oncol. 2018;10:1758835918816287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu X, Cheng S-y. Epigenetic modifications: novel therapeutic approach for thyroid cancer. Endocrinol Metabolism. 2017;32(3):326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iancu IV, Botezatu A, Plesa A, Huica I, Fudulu A, Albulescu A, et al. Alterations of regulatory factors and DNA methylation pattern in thyroid cancer. Cancer Biomarkers. 2020;28(2):255–68. [DOI] [PubMed] [Google Scholar]
- 12.Lv L, Cao L, Hu G, Shen Q, Wu J. Methylation-driven genes identified as novel prognostic indicators for thyroid carcinoma. Front Genet. 2020;11:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia M, Wang S, Ye Y, Tu Y, Huang T, Gao L. Effect of the m6ARNA gene on the prognosis of thyroid cancer, immune infiltration, and promising immunotherapy. Front Immunol. 2022;13:995645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Ruan X, Li Y, Zhi J, Hu L, Hou X, et al. KDM1A promotes thyroid cancer progression and maintains stemness through the Wnt/β-catenin signaling pathway. Theranostics. 2022;12(4):1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronner C, Alhosin M, Hamiche A, Mousli M. Coordinated dialogue between UHRF1 and DNMT1 to ensure faithful inheritance of methylated DNA patterns. Genes (Basel). 2019;10(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ioshikhes IP, Zhang MQ. Large-scale human promoter mapping using CpG islands. Nat Genet. 2000;26(1):61–3. [DOI] [PubMed] [Google Scholar]
- 17.Williams K, Christensen J, Helin K. DNA methylation: TET proteins—guardians of CpG islands? EMBO Rep. 2012;13(1):28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loaeza-Loaeza J, Beltran AS, Hernández-Sotelo D. DNMTs and impact of CpG content, transcription factors, consensus motifs, lncRNAs, and histone marks on DNA methylation. Genes (Basel). 2020;11(11):1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schagdarsurengin U, Gimm O, Hoang-Vu C, Dralle H, Pfeifer GP, Dammann R. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res. 2002;62(13):3698–701. [PubMed] [Google Scholar]
- 20.Wang P, Pei R, Lu Z, Rao X, Liu B. Methylation of p16 CpG islands correlated with metastasis and aggressiveness in papillary thyroid carcinoma. J Chin Med Assoc. 2013;76(3):135–9. [DOI] [PubMed] [Google Scholar]
- 21.Brait M, Loyo M, Rosenbaum E, Ostrow KL, Markova A, Papagerakis S, et al. Correlation between BRAF mutation and promoter methylation of TIMP3, RARβ2 and RASSF1A in thyroid cancer. Epigenetics. 2012;7(7):710–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao X, Chen M, Sang Y, Xue J, Jiang K, Chen Y, et al. Methylation-mediated silencing of ATF3 promotes thyroid cancer progression by regulating prognostic genes in the MAPK and PI3K/AKT pathways. Thyroid. 2023;33(12):1441–54. [DOI] [PubMed] [Google Scholar]
- 23.Eng ZH, Abdul Aziz A, Ng KL, Mat Junit S. Changes in antioxidant status and DNA repair capacity are corroborated with molecular alterations in malignant thyroid tissue of patients with papillary thyroid cancer. Front Mol Biosci. 2023;10:1237548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan MS, Pandith AA, Masoodi SR, Wani KA, Ul Hussain M, Mudassar S. Epigenetic silencing of TSHR gene in thyroid cancer patients in relation to their BRAF V600E mutation status. Endocrine. 2014;47(2):449–55. [DOI] [PubMed] [Google Scholar]
- 25.Rodríguez-Rodero S, Fernández AF, Fernández-Morera JL, Castro-Santos P, Bayon GF, Ferrero C, et al. DNA methylation signatures identify biologically distinct thyroid cancer subtypes. J Clin Endocrinol Metabolism. 2013;98(7):2811–21. [DOI] [PubMed] [Google Scholar]
- 26.Sun L, Niu T, Zhang Y. Association between thyroid cancer and CTLA-4 gene polymorphisms. Cell Mol Biol. 2023;69(4):31–6. [DOI] [PubMed] [Google Scholar]
- 27.Yu K, Lu H, Chen Y, Xin Y, Tan Z, Yang Q. 80MAP17 promotes the tumorigenesis of papillary thyroid carcinoma by reducing the stability of p53. Front Bioscience-Landmark. 2021;26(10):777–88. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Xue J, Jiang K, Chen Y, Zhu L, Liu R. TERT promoter methylation is associated with high expression of TERT and poor prognosis in papillary thyroid cancer. Front Oncol. 2024;14:1325345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding L, Cao J, Lin W, Chen H, Xiong X, Ao H, et al. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci. 2020;21(6):1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun D, Sun W, Zhou R, Dong A, Zhang H. Relationship between DAPK methylation and gene inactivation in papillary thyroid carcinoma. Eur J Inflamm. 2018;16:2058739218778710. [Google Scholar]
- 31.Sheikholeslami S, Zarif-Yeganeh M, Farashi S, Azizi F, Kia SK, Teimoori-Toolabi L, et al. Promoter Methylation of Tumor Suppressors in thyroid carcinoma: a systematic review. Iran J Public Health. 2021;50(12):2461–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nozhat Z, Hedayati M. PI3K/AKT pathway and its mediators in thyroid carcinomas. Mol Diagn Ther. 2016;20(1):13–26. [DOI] [PubMed] [Google Scholar]
- 33.Wei F, Wu Y, Wang Z, Li Y, Wang J, Shao G, et al. Diagnostic significance of DNA methylation of PTEN and DAPK in thyroid tumors. Clin Endocrinol (Oxf). 2020;93(2):187–95. [DOI] [PubMed] [Google Scholar]
- 34.Zhang K, Li C, Liu J, Tang X, Li Z. DNA methylation alterations as therapeutic prospects in thyroid cancer. J Endocrinol Invest. 2019;42(4):363–70. [DOI] [PubMed] [Google Scholar]
- 35.Bourboulia D, Stetler-Stevenson WG. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs): Positive and negative regulators in tumor cell adhesion. Semin Cancer Biol. 2010;20(3):161–8. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boltze C, Schneider-Stock R, Quednow C, Hinze R, Mawrin C, Hribaschek A, et al. Silencing of the maspin gene by promoter hypermethylation in thyroid cancer. Int J Mol Med. 2003;12(4):479–84. [PubMed] [Google Scholar]
- 37.Ogasawara S, Maesawa C, Yamamoto M, Akiyama Y, Wada K, Fujisawa K, et al. Disruption of cell-type-specific methylation at the maspin gene promoter is frequently involved in undifferentiated thyroid cancers. Oncogene. 2004;23(5):1117–24. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Bi K, Yang R, Li H, Nikitaki Z, Chang L. Role of DNA damage and repair in radiation cancer therapy: a current update and a look to the future. Int J Radiat Biol. 2020;96(11):1329–38. [DOI] [PubMed] [Google Scholar]
- 39.Casalino L, Verde P. Multifaceted roles of DNA methylation in neoplastic transformation, from tumor suppressors to EMT and metastasis. Genes (Basel). 2020;11(8):922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang M, Jia K, Wang L, Li W, Chen B, Liu Y, et al. Alterations of DNA damage repair in cancer: from mechanisms to applications. Annals Translational Med. 2020;8(24):1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patil SR, Hendre AS, Shelke SN, Kshirsagar MS, Salunke LS. DNA repair mechanisms insights into Cancer susceptibility and therapy. J ReAttach Ther Dev Diversities. 2023;6(7s):806–11. [Google Scholar]
- 42.Li G-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. [DOI] [PubMed] [Google Scholar]
- 43.Guan H, Ji M, Hou P, Liu Z, Wang C, Shan Z, et al. Hypermethylation of the DNA mismatch repair gene hMLH1 and its association with lymph node metastasis and T1799A BRAF mutation in patients with papillary thyroid cancer. Cancer. 2008;113(2):247–55. [DOI] [PubMed] [Google Scholar]
- 44.Gohil D, Sarker AH, Roy R. Base excision repair: mechanisms and impact in biology, disease, and medicine. Int J Mol Sci. 2023;24(18):14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ba X, Boldogh I. 8-Oxoguanine DNA glycosylase 1: beyond repair of the oxidatively modified base lesions. Redox Biol. 2018;14:669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaohong Z, Lijun X. Research progress of DNA methylation in thyroid cancer. DNA Methylation Mechanism. 2020;1:1. [Google Scholar]
- 47.Hu JL, Hu SS, Hou XX, Zhu X, Cao J, Jiang LH, et al. Abnormal expression of DNA double-strand breaks related genes, ATM and gammaH2AX, in thyroid carcinoma. Int J Endocrinolog. 2015;2015:136810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15(7):465–81. [DOI] [PubMed] [Google Scholar]
- 49.Cipollini M, Figlioli G, Maccari G, Garritano S, De Santi C, Melaiu O, et al. Polymorphisms within base and nucleotide excision repair pathways and risk of differentiated thyroid carcinoma. DNA Repair (Amst). 2016;41:27–31. [DOI] [PubMed] [Google Scholar]
- 50.Mijnes J, Veeck J, Gaisa NT, Burghardt E, de Ruijter TC, Gostek S, et al. Promoter methylation of DNA damage repair (DDR) genes in human tumor entities: RBBP8/CtIP is almost exclusively methylated in bladder cancer. Clin Epigenetics. 2018;10(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Köhrle J. Selenium, iodine and iron–essential trace elements for thyroid hormone synthesis and metabolism. Int J Mol Sci. 2023;24(4):3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galrao AL, Camargo RY, Friguglietti CU, Moraes L, Cerutti JM, Serrano-Nascimento C, et al. Hypermethylation of a New Distal Sodium/Iodide Symporter (NIS) enhancer (NDE) is associated with reduced NIS expression in thyroid tumors. J Clin Endocrinol Metabolism. 2014;99(6):E944–52. [DOI] [PubMed] [Google Scholar]
- 53.Zhang K, Li C, Liu J, Tang X, Li Z. DNA methylation alterations as therapeutic prospects in thyroid cancer. J Endocrinol Invest. 2019;42:363–70. [DOI] [PubMed] [Google Scholar]
- 54.Qu M, Wan S, Ren B, Wu H, Liu L, Shen H. Association between TSHR gene methylation and papillary thyroid cancer: a meta-analysis. Endocrine. 2020;69(3):508–15. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20(22):5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miguel V, Lamas S, Espinosa-Diez C. Role of non-coding-RNAs in response to environmental stressors and consequences on human health. Redox Biol. 2020;37:101580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pellecchia S, Sepe R, Decaussin-Petrucci M, Ivan C, Shimizu M, Coppola C, et al. The long non-coding RNA prader willi/angelman region RNA5 (PAR5) is downregulated in anaplastic thyroid carcinomas where it acts as a tumor suppressor by reducing EZH2 activity. Cancers (Basel). 2020;12(1):235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Gao H, Li Y. Inhibition of LncRNA FOXD3-AS1 suppresses the aggressive biological behaviors of thyroid cancer via elevating mir-296-5p and inactivating TGF-β1/Smads signaling pathway. Mol Cell Endocrinol. 2020;500:110634. [DOI] [PubMed] [Google Scholar]
- 59.Wu F, Lin X, Shan S-K, Li F, Xu F, Zhong J-Y, et al. The suppression of miR-199a-3p by promoter methylation contributes to papillary thyroid carcinoma aggressiveness by targeting RAP2a and DNMT3a. Front Cell Dev Biol. 2020;8:594528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia F, Wang W, Jiang B, Chen Y, Li X. DNA methylation-mediated silencing of miR-204 is a potential prognostic marker for papillary thyroid carcinoma. Cancer Manag Res. 2019;11:1249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Héberlé É, Bardet AF. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019;63(6):727–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011;12(2):206–22. [DOI] [PubMed] [Google Scholar]
- 63.Zhao F, Zhu S, Fang J, Dong H, Zhu C. Correlation of DNA methylation and lymph node metastasis in papillary thyroid carcinoma. Head Neck. 2023;45(7):1654–62. [DOI] [PubMed] [Google Scholar]
- 64.Smith J, Sen S, Weeks RJ, Eccles MR, Chatterjee A. Promoter DNA hypermethylation and paradoxical gene activation. Trends Cancer. 2020;6(5):392–406. [DOI] [PubMed] [Google Scholar]
- 65.Chai L, Li J, Lv Z. An integrated analysis of cancer genes in thyroid cancer. Oncol Rep. 2016;35(2):962–70. [DOI] [PubMed] [Google Scholar]
- 66.Kikuchi Y, Tsuji E, Yagi K, Matsusaka K, Tsuji S, Kurebayashi J, et al. Aberrantly methylated genes in human papillary thyroid cancer and their association with BRAF/RAS mutation. Front Genet. 2013;4:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zarkesh M, Zadeh-Vakili A, Azizi F, Foroughi F, Akhavan MM, Hedayati M. Altered epigenetic mechanisms in thyroid cancer subtypes. Mol Diagn Ther. 2018;22:41–56. [DOI] [PubMed] [Google Scholar]
- 68.Ortiz IMDP, Barros-Filho MC, Dos Reis MB, Beltrami CM, Marchi FA, Kuasne H, et al. Loss of DNA methylation is related to increased expression of miR-21 and miR-146b in papillary thyroid carcinoma. Clin Epigenetics. 2018;10(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vos Ld, Dietrich J, Strieth S, Bootz F, Dietrich D, Franzen A. PD-1, CTLA4, PD-L1 and PD-L2 DNA methylation in papillary thyroid carcinoma. Immunotherapy. 2020;12(12):903–20. [DOI] [PubMed] [Google Scholar]
- 70.Wang K, Zheng J, Wu Z, Fang J-G, Zhao J-Y, Yao J-M, et al. Hyper-expression and hypomethylation of TM4SF1 are associated with lymph node metastases in papillary thyroid carcinoma patients. Neoplasma. 2022;69(2):341–51. [DOI] [PubMed] [Google Scholar]
- 71.Seo J, Ha J, Kang E, Cho S. The role of epithelial–mesenchymal transition-regulating transcription factors in anti-cancer drug resistance. Arch Pharm Res. 2021;44:281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones PA, Gonzalgo ML. Altered DNA methylation and genome instability: a new pathway to cancer? Proceedings of the National Academy of Sciences. 1997;94(6):2103–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klein Hesselink EN, Zafon C, Villalmanzo N, Iglesias C, van Hemel BM, Klein Hesselink MS, et al. Increased global DNA hypomethylation in distant metastatic and dedifferentiated thyroid cancer. J Clin Endocrinol Metabolism. 2018;103(2):397–406. [DOI] [PubMed] [Google Scholar]
- 74.Veschi V, Verona F, Lo Iacono M, D’Accardo C, Porcelli G, Turdo A, et al. Cancer stem cells in thyroid tumors: from the origin to metastasis. Front Endocrinol (Lausanne). 2020;11:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Camargo Barros-Filho M, Menezes Barreto, de Lima L, Bisarro dos Reis M, Homem Bette, de Mello J, Moraes Beltrami C, Lopes Pinto CA, et al. Pfkfb2 promoter hypomethylation as recurrence predictive marker in well-differentiated thyroid carcinomas. Int J Mol Sci. 2019;20(6):1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karthiya R, Wasil SM, Khandelia P. Emerging role of N4-acetylcytidine modification of RNA in gene regulation and cellular functions. Mol Biol Rep. 2020;47(11):9189–99. [DOI] [PubMed] [Google Scholar]
- 77.Qiu L, Jing Q, Li Y, Han J. RNA modification: mechanisms and therapeutic targets. Mol Biomed. 2023;4(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, Chen Y, Chen R, Zhou H, Lin Y, Li B, et al. YTHDF3 as a prognostic predictive biomarker of thyroid cancer and its correlation with immune infiltration. BMC Cancer. 2023;23(1):882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X, Fu X, Zhang J, Xiong C, Zhang S, Lv Y. Identification and validation of m6A RNA methylation regulators with clinical prognostic value in papillary thyroid cancer. Cancer Cell Int. 2020;20:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He J, Zhou M, Yin J, Wan J, Chu J, Jia J, et al. METTL3 restrains papillary thyroid cancer progression via m6A/c-Rel/IL-8-mediated neutrophil infiltration. Mol Ther. 2021;29(5):1821–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu Y, Peng X, Zhou Q, Tan L, Zhang C, Lin S, et al. METTL3-mediated m6A modification of STEAP2 mRNA inhibits papillary thyroid cancer progress by blocking the hedgehog signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 2022;13(4):358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang W, Ding Y, Zhao Y, Li X. m6A reader IGF2BP2 promotes lymphatic metastasis by stabilizing DPP4 in papillary thyroid carcinoma. Cancer Gene Ther. 2024;31(2):285–99. [DOI] [PubMed] [Google Scholar]
- 83.Sa R, Liang R, Qiu X, He Z, Liu Z, Chen L. Targeting IGF2BP2 promotes differentiation of Radioiodine refractory papillary thyroid Cancer via Destabilizing RUNX2 mRNA. Cancers (Basel). 2022;14(5):1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He T, Xia H, Chen B, Duan Z, Huang C. m6A writer METTL3-Mediated lncRNA LINC01125 prevents the malignancy of papillary thyroid Cancer. Crit Rev Immunol. 2023;43(3):43–53. [DOI] [PubMed] [Google Scholar]
- 85.Zhou X, Chang L, Liang Q, Zhao R, Xiao Y, Xu Z, et al. The m6A methyltransferase METTL3 drives thyroid cancer progression and lymph node metastasis by targeting LINC00894. Cancer Cell Int. 2024;24(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li P, Wang W, Zhou R, Ding Y, Li X. The m5 C methyltransferase NSUN2 promotes codon-dependent oncogenic translation by stabilising tRNA in anaplastic thyroid cancer. Clin Transl Med. 2023;13(11):e1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou Y, Dai X, Lyu J, Li Y, Bao X, Deng F, et al. Construction and validation of a novel prognostic model for thyroid cancer based on N7-methylguanosine modification-related lncRNAs. Med (Baltim). 2022;101(42):e31075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai C-C, Chien M-N, Chang Y-C, Lee J-J, Dai S-H, Cheng S-P. Overexpression of histone H3 lysine 27 trimethylation is Associated with aggressiveness and dedifferentiation of thyroid Cancer. Endocr Pathol. 2019;30(4):305–11. [DOI] [PubMed] [Google Scholar]
- 89.Zhou W, Lin J, Liu J, Zhang R, Fan A, Xie Q, et al. Thyroid cancer risk prediction model using m6A RNA methylation regulators: integrated bioinformatics analysis and histological validation. Aging. 2023;15(3):846–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang S, Lv W, Li T, Zhang S, Wang H, Li X, et al. Dynamic regulation and functions of mRNA m6A modification. Cancer Cell Int. 2022;22(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karthiya R, Khandelia P. m6A RNA methylation: ramifications for gene expression and Human Health. Mol Biotechnol. 2020;62(10):467–84. [DOI] [PubMed] [Google Scholar]
- 92.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu Z-H, Feng S-T, Zhang D, Cao X-C, Yu Y, Wang X. The functions and prognostic values of m6A RNA methylation regulators in thyroid carcinoma. Cancer Cell Int. 2021;21(1):385. 10.1007/s12033-023-00921-w. [DOI] [PMC free article] [PubMed]
- 94.An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. 2022;21(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang Y, Li X, Chen W, He Y, Wu S, Li X, et al. Analysis of the prognostic significance and potential mechanisms of lncRNAs associated with m6A methylation in papillary thyroid carcinoma. Int Immunopharmacol. 2021;101(Pt B):108286. [DOI] [PubMed] [Google Scholar]
- 96.Jayasree PJ, Dutta S, Karemore P, Khandelia P. Crosstalk between m6A RNA methylation and miRNA biogenesis in cancer: An unholy nexus. Mol Biotechnol. 2023. [DOI] [PubMed]
- 97.Lin S, Zhu Y, Ji C, Yu W, Zhang C, Tan L, et al. METTL3-Induced Mir-222-3p Upregulation inhibits STK4 and promotes the malignant behaviors of thyroid carcinoma cells. J Clin Endocrinol Metab. 2022;107(2):474–90. [DOI] [PubMed] [Google Scholar]
- 98.Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m5C and its regulatory role in RNA processing. Wiley Interdisciplinary Reviews: RNA. 2021;12(4):e1639. [DOI] [PubMed] [Google Scholar]
- 99.Nombela P, Miguel-López B, Blanco S. The role of m6A, m5C and Ψ RNA modifications in cancer: novel therapeutic opportunities. Mol Cancer. 2021;20(1):1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li F, Deng Q, Pang X, Huang S, Zhang J, Zhu X, et al. m5C Regulator-mediated methylation modification patterns and Tumor Microenvironment Infiltration characterization in papillary thyroid carcinoma. Front Oncol. 2021;11:729887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Q, Liu F, Chen W, Miao H, Liang H, Liao Z, et al. The role of RNA m5C modification in cancer metastasis. Int J Biol Sci. 2021;17(13):3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tomikawa C. 7-Methylguanosine modifications in transfer RNA (tRNA). Int J Mol Sci. 2018;19(12):4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ramanathan A, Robb GB, Chan S-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44(16):7511–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol. 2021;22(6):375–92. [DOI] [PubMed] [Google Scholar]
- 105.Luo Y, Yao Y, Wu P, Zi X, Sun N, He J. The potential role of N 7-methylguanosine (m7G) in cancer. J Hematol Oncol. 2022;15(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nagarajan D, Nagaraja SS, Baisakhiya S. Role of histone methylation in Cancer: pathobiology and therapeutics. Handbook of oxidative stress in Cancer: therapeutic aspects. Springer; 2022. pp. 1411–28.
- 107.Gargalionis AN, Piperi C, Adamopoulos C, Papavassiliou AG. Histone modifications as a pathogenic mechanism of colorectal tumorigenesis. Int J Biochem Cell Biol. 2012;44(8):1276–89. [DOI] [PubMed] [Google Scholar]
- 108.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40(6):741–50. [DOI] [PubMed] [Google Scholar]
- 109.Norollahi SE, Vahidi S, Shams S, Keymoradzdeh A, Soleymanpour A, Solymanmanesh N, et al. Analytical and therapeutic profiles of DNA methylation alterations in cancer; an overview of changes in chromatin arrangement and alterations in histone surfaces. Horm Mol Biol Clin Investig. 2023;44(3):337–56. [DOI] [PubMed] [Google Scholar]
- 110.Stephen J, Chen K-M, Merritt J, Chitale D, Divine G, Worsham MJ. Methylation markers differentiate thyroid cancer from benign nodules. J Endocrinol Invest. 2018;41:163–70. [DOI] [PubMed] [Google Scholar]
- 111.Zane M, Agostini M, Enzo MV, Ide EC, Del Bianco P, Torresan F, et al. Circulating cell-free DNA, SLC5A8 and SLC26A4 hypermethylation, BRAFV600E: a non-invasive tool panel for early detection of thyroid cancer. Biomed Pharmacother. 2013;67(8):723–30. [DOI] [PubMed] [Google Scholar]
- 112.dos Bisarro M, Barros-Filho MC, Marchi FA, Beltrami CM, Kuasne H, Pinto CAL, et al. Prognostic classifier based on genome-wide DNA methylation profiling in well-differentiated thyroid tumors. J Clin Endocrinol Metabolism. 2017;102(11):4089–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Park J-L, Jeon S, Seo E-H, Bae DH, Jeong YM, Kim Y, et al. Comprehensive DNA methylation profiling identifies novel diagnostic biomarkers for thyroid cancer. Thyroid. 2020;30(2):192–203. [DOI] [PubMed] [Google Scholar]
- 114.Jasmine F, Aschebrook-Kilfoy B, Rahman MM, Zaagman G, Grogan RH, Kamal M, et al. Association of DNA promoter methylation and BRAF mutation in thyroid Cancer. Curr Oncol. 2023;30(3):2978–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Camargo Barros-Filho M, Barreto Menezes de Lima L, Bisarro dos Reis M, Bette Homem de Mello J, Moraes Beltrami C, Lopes Pinto CA, et al. Pfkfb2 promoter hypomethylation as recurrence predictive marker in well-differentiated thyroid carcinomas. Int J Mol Sci. 2019;20(6):1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu Z-H, Feng S-T, Zhang D, Cao X-C, Yu Y, Wang X. The functions and prognostic values of m6A RNA methylation regulators in thyroid carcinoma. Cancer Cell Int. 2021;21:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Srinivasan A, Gold B. Small-molecule inhibitors of DNA damage-repair pathways: an approach to overcome tumor resistance to alkylating anticancer drugs. Future Med Chem. 2012;4(9):1093–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hegi ME, Sciuscio D, Murat A, Levivier M, Stupp R. Epigenetic deregulation of DNA repair and its potential for therapy. Clin Cancer Res. 2009;15(16):5026–31. [DOI] [PubMed] [Google Scholar]
- 119.Papanicolau-Sengos A, Aldape K. DNA methylation profiling: an emerging paradigm for cancer diagnosis. Annu Rev Pathol. 2022;17:295–321. [DOI] [PubMed] [Google Scholar]
- 120.Biswas S, Rao CM. Epigenetic tools (the writers, the readers and the erasers) and their implications in cancer therapy. Eur J Pharmacol. 2018;837:8–24. [DOI] [PubMed] [Google Scholar]
- 121.de Mello DC, Saito KC, Cristovão MM, Kimura ET, Fuziwara CS. Modulation of EZH2 activity induces an antitumoral effect and cell redifferentiation in anaplastic thyroid cancer. Int J Mol Sci. 2023;24(9):7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gimeno-Valiente F, López-Rodas G, Castillo J, Franco L. Alternative splicing, epigenetic modifications and cancer: a dangerous triangle, or a hopeful one? Cancers (Basel). 2022;14(3):560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jia M, Liang J, Li Z, Qin Y, Lu X. Screening tumor stage-specific candidate neoantigens in thyroid adenocarcinoma using integrated exome and transcriptome sequencing. Front Immunol. 2023;14:1187160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.