Abstract
Background
Micrococcus antarcticus (M. antarcticus) is an aerobic Gram-positive spherical actinobacterium that was initially isolated from Chinese Great-Wall station in Antarctica in 2000. M. antarcticus was considered to be of low pathogenicity, no previous cases of human infection by this organism have been reported. Here we describe the first report with community-acquired pneumonia (CAP) caused by M. antarcticus.
Case presentation
An 87-year-old female was presented to the Central Hospital of Wuhan in November 2023 with a chief complaint of cough, sputum production, and chest tightness for 2 weeks. Microbial culture of the patient’s bronchoalveolar lavage fluid (BALF) and identification of the isolates using Matrix-assisted laser desorption ionization/time of flight mass spectrometry (MALDI-TOF MS) and 16S rRNA gene sequencing revealed M. antarcticus infection. Combined with clinical symptoms, laboratory and imaging examination, the patient was diagnosed with CAP. Then cefoperazone/sulbactam and levofloxacin was administrated, the patient’s condition was improved and she was discharged after a week after admission, no abnormalities were detected during a 5-month follow-up.
Conclusions
This case highlights that M. antarcticus, first identified from a patient with CAP, is an extremely rare pathogenic microorganism. Clinicians should be aware of its potential as a pathogen in the diagnosis and treatment of CAP.
Keywords: Micrococcus antarcticus, 16S rRNA gene sequencing, MALDI-TOF MS, Community-acquired pneumonia, Case report
Background
The genus Micrococcus is a Gram-positive, aerobic, non-motile, and non-sporeforming actinomycetes that are generally isolated from various habitats, such as activated sludge [1], feces of black pig [2], aquatic plant [3–6], oreochromis niloticus in Egypt [7], human skin [8], soil [9], air [10], and dairy industry waste [11]. The typical perception of Micrococcus species is that they are non-pathogenic, yet there have been occasional instances of Micrococcus infections among individuals with compromised immune systems [12]. M. antarcticus, a member of the genus Micrococcus, was originally isolated from Chinese Great-Wall station in Antarctica in 2000 [13]. Literature review revealed a series of researches on the discovery and identification of β-glucosidase BglU, which is related to the cold-adapted mechanism of M. antarcticus, and its relevant molecular structure basis [14, 15]. However, the role and pathogenicity of this bacterium in the field of medicine remains unknown. Currently, no reports of human infection caused by M. antarcticus have been found. Hence, we present the first case of community-acquired pneumonia case caused by M. antarcticus in China.
Case presentation
On November 21, 2023, an 87-year-old female patient, known for a medical history comprising coronary atherosclerotic heart disease, hypertension, renal insufficiency, colon polyps, and previous treatment for endometrial cancer, was admitted to our hospital with a chief complaint of “cough, sputum production, and chest tightness for 2 weeks”. The patient presented with symptoms including a runny nose, nasal congestion, mild cough, fatigue, poor appetite, without fever, headache, or palpitations, but accompanied by nausea after catching a cold 2 weeks ago. Despite self-administering of “Chinese herbal medicine” in an attempt to alleviate symptoms, the cough exhibited a progressive aggravation with no relief. The physical examination revealed the following: body temperature of 36.5 °C, pulse rate of 87 beats per minute, blood pressure of 113/81mmHg, blood oxygen saturation of 97%, coarse breath sounds in both lungs, and moist rales audible in the lower left lung. The laboratory test results are as follows: white blood cell count of 5.51 × 109/L (75.2% neutrophils), platelet count of 100 × 109/L (normal 125–350 × 109/L), erythrocyte sedimentation rate of 51 mm/h (normal 0–20 mm/h), hypersensitive C-reactive protein (whole blood) of 8.82 mg/dL (normal 0-0.8 mg/dL), interleukin-6 of 8.54 pg/mL (normal < 7 pg/mL), creatinine of 108.9 umol/L (normal 41–81 umol/L), uric acid of 437 umol/L (normal 155–357 umol/L), and estimated glomerular filtration rate of 39.46 mL/min/1.73m2 (normal > 90 mL/min/1.73m2). In addition, no significant abnormalities were identified in the tests for cardiac troponin, SARS-CoV-2, Mycobacterium tuberculosis, Chlamydia pneumoniae, Mycoplasma pneumoniae, influenza A, and influenza B. Furthermore, the chest computed tomography scan showed enhanced texture in both lungs, and a patchy clouding opacity can be seen in the left lower lung, which suggested a newly developed lung infection in the patient (Fig. 1A). Based on the results mentioned above, the patient was diagnosed with CAP and was prescribed empirical antimicrobial treatment with intravenous (i.v.) cefoperazone/sulbactam 2 × 1 g/day (q 12 h) and levofloxacin 1 g/day (qd). This empirical treatment was selected to provide broad-spectrum coverage against potential Gram-positive and Gram-negative pathogens.
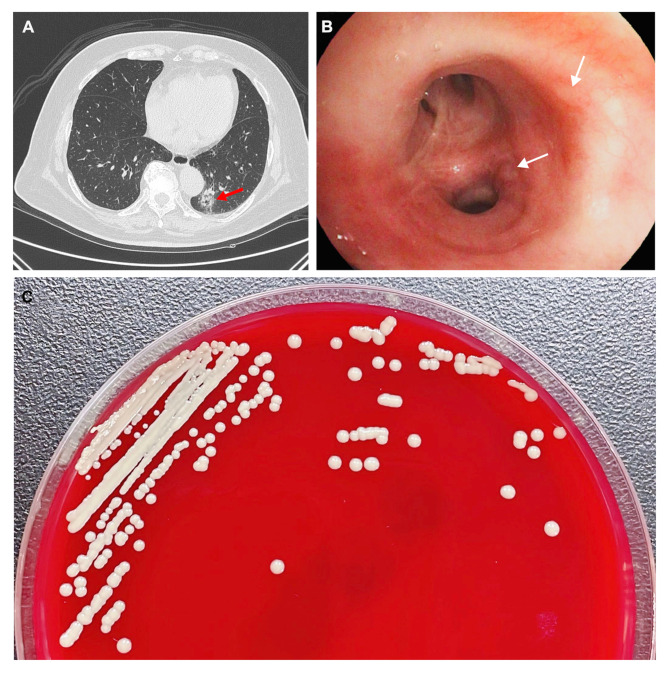
Fig. 1.
Results of computed tomography (CT) and electronic bronchoscope, and the morphology of the Micrococcus antarcticus strain CAP44B11. (A) Chest CT demonstrating infectious lesions in the lower left lung (red arrow). (B) The electronic bronchoscope shows bronchial mucosa inflammation with redness and purulent secretion as indicated by white arrows. (C) Bacterial colonies on Columbia blood agar after being cultured at 37 °C in the presence of 5% CO2 for 24 h.
Following that, a bronchoscopy was performed on the patient under general anesthesia, which showed inflammation of the bronchial mucosa bilaterally (Fig. 1B). BALF was then collected from the affected area, gram stained and inoculated onto Columbia blood agar plates, MacConkey agar plates, chocolate agar plates, and Sabouraud agar plates (Guangzhou Dijing Microbial Technology Co., Ltd., Guangzhou, China). The plates were incubated at 35 °C with 5% CO2 for 18–24 h.
Actually, a large number of gram-positive cocci were observed under the microscope by Gram staining of BALF, some of which were distributed in and around the leukocytes. Moreover, in the BALF culture of the patient, three bacterial species have been isolated and classified by MALDI-TOF MS (Bruker Daltonik GmbH, Germany), including Micrococcus sp., Streptococcus oralis, and Streptococcus parasanguinis, among which Micrococcus sp. was the dominant bacterium. Since Streptococcus oralis and Streptococcus parasanguinis are part of common normal flora, and their colony counts were significantly lower than that of Micrococcus sp., the isolated Micrococcus sp. named CAP44B11 was considered to be the causative agent in this case. The strain CAP44B11 is yellow, mucoid, smooth-edged and circular colony on the Columbia blood agar plate which was confirmed to be Gram-positive cocci through Gram staining (Fig. 1C).
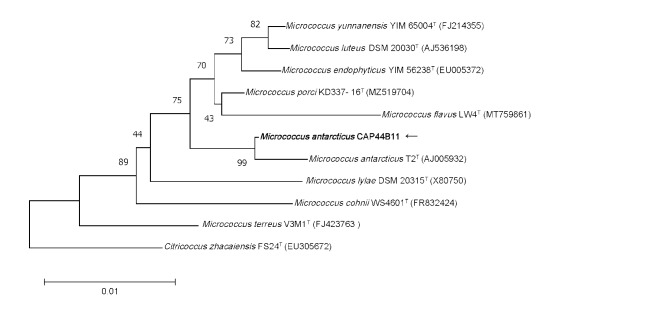
Subsequently, 16S rRNA gene sequencing was employed for further identification. Genomic DNA was extracted from purified bacteria using the TIANamp Bacteria DNA Kit (TIANGEN Biotech, Co, Ltd, Beijing, China) in accordance with the instructions provided. The 16S rRNA gene was amplified using universal primers (forward, 5’-AGTTTGATCCTGGCTCAG-3’; reverse, 5’-GTATTGCCGCGGCTGCTG-3’) and sequenced. A total of 1,413 contiguous nucleotides were obtained. The complete 16S rRNA sequence of the strain CAP44B11 was analyzed with the EzBioCloud Database [16]. The strain CAP44B11 exhibited the highest (99.58%) 16S rRNA gene sequence similarity with the type strain of M. antarcticus T2T (accession no. AJ005932). Among the 1,413 bases, there was only a six-base difference between the two strains. The CLUSTALW algorithm was used to calculate the multiple alignments and sequence similarity levels of the most closely related Micrococcus species [17]. The phylogenetic tree was constructed using the neighbor-joining method in MEGA software version 11 [18]. Bootstrap resampling method of Felsenstein (1985) was employed to repetitively evaluate the topology of the phylogenetic tree through 1000 replicates [19]. The above results demonstrated that strain CAP44B11 was clustered with the type strain M. antarcticus T2T, with a bootstrap value of 99% (Fig. 2), which indicated that the isolated strain CAP44B11 belongs to the M. antarcticus species. The 16S rRNA sequence of M. antarcticus CAP44B11 has been deposited into GenBank under accession number of PP535566.
Fig. 2.
The phylogenetic tree based on the 16S rRNA gene sequences showing the relationship of isolated strain CAP44B11 (black arrow) and members within genus Micrococcus. The tree was reconstructed by the neighbor-joining method, and Citricoccus zhacaiensis FS24T (EU305672) was used as an outgroup. Bootstrap values (> 50%) based on 1,000 replicates are shown at branch nodes. T, type strain.
AutoMic-i600 platform (Autobio Co., Ltd., Zhenzhou, China) was used to test the antimicrobial susceptibility of the strain M. antarcticus CAP44B11, and Staphylococcus aureus ATCC 25,923 and Escherichia coli ATCC 25,922 were used as the quality control strains. The drug susceptibility was executed in accordance with the EUCAST 2024 standard V 14.0 [20]. According to the results, the M. antarcticus demonstrates sensitivity to a majority of antibiotics, including ceflorin, ciprofloxacin, and moxifloxacin, etc. (Table 1). Although the M. antarcticus was sensitive to ceflorin and cefoxitin, cefoperazone/sulbactam offers a broader antimicrobial spectrum. The use of levofloxacin was based on the patient’s sensitivity to ciprofloxacin and moxifloxacin, indicating a good response to fluoroquinolones. Combining cefoperazone/sulbactam with levofloxacin can provide a synergistic effect, enhancing overall treatment efficacy and reducing the risk of resistance to a single drug. After a comprehensive evaluation of the patient, blood cultures were not performed before initial antibiotics. However, after 5 days of treatment with cefoperazone/sulbactam combined with levofloxacin, the patient’s symptoms showed significant improvement, indicating that the treatment is feasible and effective. Then the patient was discharged after a week, no abnormalities were detected during a 5-month follow-up.
Table 1.
Drug susceptibility results of M. Antarcticus CAP44B11
| Antibiotics | MIC (ng/µL)* | Antibiotics | MIC (ng/µL) |
|---|---|---|---|
| Penicillin | ≤ 0.12(S) | Ampicillin | ≤ 0.5(S) |
| Ceflorin | ≤ 0.12(S) | Cefoxitin | ≤1(S) |
| Oritavancin | ≤ 0.015(S) | Teicoplanin | ≤1(S) |
| Ciprofloxacin | ≤ 0.12(S) | Vancomycin | ≤0.5(S) |
| Tetracycline | ≤ 1(S) | Moxifloxacin | ≤0.12(S) |
| Gentamicin | ≤ 2(S) | Tigecycline | ≤0.03(S) |
| Co-trimoxazole | ≤ 0.5/9.5(S) | Linezolid | ≤1(S) |
| Rifampicin | ≤ 0.015(S) | Daptomycin | ≤0.12(S) |
| Clindamycin | ≤ 0.25(S) | Erythromycin | ≤0.25(S) |
* Drug sensitivity was judged according to EUCAST 2024 standard V 14.0. S: sensitive; I: intermediate; R: resistant
Discussion and conclusions
The genus Micrococcus, classified within the family Micrococcaceae, order Micrococcales, and phylum Actinomycetota, was originally proposed by Cohn (1872) and later revised by Stackebrandt et al. [21] and Wieser et al. [22] based on the utilization of glucose, analysis of the G + C content of the genomic DNA and phylogenetic analysis of the 16S rRNA gene. Up to date, eight species have been identified with validly published and correct names under the List of Prokaryotic Names with Standing in Nomenclature (LPSN) [23]: Micrococcus antarcticus, Micrococcus cohnii, Micrococcus endophyticus, Micrococcus flavus, Micrococcus luteus, Micrococcus lylae, Micrococcus porci, and Micrococcus terreus. Members of the genus Micrococcus are Gram-positive, aerobic, non-motile, and non-sporeforming actinomycetes. Among Micrococcus spp., only a few species have been reported to be associated with human infectious diseases. Notably, Micrococcus luteus have been implicated in various conditions, such as brain abscess [24], bloodstream infection [25], infecting endocarditis [26], bacteremia in immunocompromised patients [27]. Additionally, peritoneal dialysis-associated peritonitis has been attributed to Micrococcus aloeverae [28], while community-acquired pneumonia has been linked to Micrococcus yunnanensis [12]. Nevertheless, there are no accounts of human infection with M. antarcticus, so its pathogenicity remains largely unexplored. In this instance, we reported the debut case of CAP caused by M. antarcticus, aspiring to offer further insights into this rare infection.
CAP presents as a significant public health challenge worldwide, characterized by high mortality and morbidity rates along with economic burdens. Despite its impact, research on CAP in China has been relatively limited, with increasing attention being drawn to this issue only in recent years. The research conducted by Academician Xu’s team at the Chinese Center for Disease Control and Prevention has revealed that the incidence of CAP in China is higher than previously understood [29]. Moreover, studies indicate that bacteria still hold a dominant position in adult pneumonia cases [30]. Therefore, the prompt and accurate identification of causative pathogens is of paramount importance for the early diagnosis and management of CAP. There have been previous reports about CAP caused by Micrococcus yunnanensis [12], and in this case, we have documented a case of CAP caused by M. antarcticus, highlighting the importance of considering the potential for Micrococcus spp. infections in the diagnosis of CAP in the future.
M. antarcticus was initially discovered from Chinese Great-Wall station in Antarctica, with studies indicating its optimal temperature to be 15–17 °C [13]. However, it is noteworthy that M. antarcticus was detected under the conditions of overnight cultivation in a 5% CO2, 35 °C incubator, which diverges from its typical cold adapted habitat. This finding in a clinical patient raises intriguing questions about its environmental adaptability and potential implications. As demonstrated in Chen’s study [31], Aeromonas salmonicida is a commonly known cold-water pathogenic bacterium, and its most notable and characteristic virulence factors are aerolysin and hemolysin. Interestingly, the genes aerA and hlyA, which encode these toxins, were found to be significantly upregulated at both 28 °C and 37 °C. This indicates that the virulence and environmental adaptation-related genes of certain bacteria are significantly influenced by the environmental temperature. Although the optimal growth temperature for M. antarcticus is relatively low, it may have acquired the ability to survive and grow under non-optimal conditions through genetic mutations or other evolutionary mechanisms. This could enable it to briefly survive within the human body and potentially cause infection. Nevertheless, further investigations are required to substantiate this hypothesis.
Previous research has revealed that individuals with immunocompromised function are more susceptible to be infected by Micrococcus spp. [24, 25, 32], and age is a well-known risk factor for CAP, especially among the older population [33]. In this case, the patient is an 87-year-old elderly woman with a previous medical history of surgical intervention for endometrial cancer in 2022 as well as diagnoses of coronary artery atherosclerosis heart disease, hypertension, and renal dysfunction. Hence, she is considered to be part of the immunocompromised group, which could be the underlying reason for her CAP caused by the infection of M. antarcticus. However, the patient has denied having been to Antarctica recently, so the exact source of the isolated bacteria remains unknown. Nonetheless, the elevated inflammatory markers in the patient and the identification of the pathogenic bacteria isolated from the BALF have confirmed the presence of an infection.
In recent years, MALDI-TOF MS has emerged as a widely utilized method in microbiology laboratories worldwide, offering a rapid and reliable identification of pathogenic bacteria based on phenotype and specific proteomic characteristics. This technique is valued for its simplicity, high accuracy, and cost-effectiveness. Recent studies have also highlights its significant importance in the classification and identification of specific microbiota [34]. However, despites its numerous advantages, MALDI-TOF MS does have its limitations, particularly in the identification of novel or rare bacterial species [35, 36]. Take our case for example, MALDI-TOF MS failed to accurately identify the isolated bacteria at the species level. Thus, we employed 16S rRNA gene sequencing, a technique widely used for identifying isolates that cannot be determined by conventional methods [37]. Through this approach, the causative organism was finally confirmed as M. antarcticus, highlighting the utility of 16S rRNA gene sequencing as an alternative method in situations where MALDI-TOF MS fails to yield satisfactory results, particularly in the identification of uncommon or rare bacteria. Thus, 16S rRNA gene sequencing emerges as a valuable and effective option for overcoming challenges encountered in microbial identification.
This case report presents the first documented instance of CAP caused by M. antarcticus, with the patient exhibiting satisfactory recovery after receiving appropriate symptomatic treatment. Our findings suggest that the potential emergence of M. antarcticus as a pathogenic agent capable of causing human infections. 16S rRNA gene sequencing facilitated this rare pathogen identification in our case and should be recommended for the identification of unknown bacteria. However, the underlying mechanism of M. antarcticus induced human infections remains unclear. Further research is therefore required to explore the virulence, pathogenicity, and other characteristics of this bacterium in order to gain a better understanding of M. antarcticus.
Acknowledgements
Not applicable.
Abbreviations
- CAP
Community-acquired pneumonia
- BALF
Bronchoalveolar lavage fluid
- MALDI-TOF MS
Matrix-assisted laser desorption ionization/time of flight mass spectrometry
- AST
Antimicrobial susceptibility test
Author contributions
JX, YZ, and ZL conceived and designed the research. LS and HW collected the clinical data. JX and YZ performed the experiments and analyzed the data. MZ performed quality control. JX drafted the primary manuscript. ZL reviewed and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a grant from the Wuhan Municipal Health Commission (Project no. WX21Q42) to YZ.
Data availability
The datasets generated and analyzed during the current study are available in the GenBank under accession number of PP535566.
Declarations
Ethics approval and consent to participate
The study was approved by Medical Ethics Committee of the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology (Project no. WHZXKYL2023-094).
Consent for publication
Written informed consent was obtained from the patient for publication of case report and accompanying images.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jia Xu and Yingmiao Zhang contributed equally to this work.
References
- 1.Liu XY, Wang BJ, Jiang CY, Liu SJ. Micrococcus flavus sp. nov., isolated from activated sludge in a bioreactor. Int J Syst Evol Microbiol. 2007;57(Pt 1):66–9. [DOI] [PubMed] [Google Scholar]
- 2.Lee AY, Chen CH, Liou JS, Lin YC, Hamada M, Wang YT et al. Micrococcus porci sp. nov., isolated from feces of Black Pig (Sus scrofa). Life (Basel). 2022;12(11). [DOI] [PMC free article] [PubMed]
- 3.Chen HH, Zhao GZ, Park DJ, Zhang YQ, Xu LH, Lee JC, et al. Micrococcus endophyticus sp. nov., isolated from surface-sterilized Aquilaria sinensis roots. Int J Syst Evol Microbiol. 2009;59(Pt 5):1070–5. [DOI] [PubMed] [Google Scholar]
- 4.Prakash O, Nimonkar Y, Munot H, Sharma A, Vemuluri VR, Chavadar MS, et al. Description of Micrococcus aloeverae sp. nov., an endophytic actinobacterium isolated from Aloe vera. Int J Syst Evol Microbiol. 2014;64(Pt 10):3427–33. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Xiao Y, Lai W, Jia R, Deng Q, Wang X, et al. Micrococcus lacusdianchii sp. nov., an attached bacterium inhibited by metabolites from its symbiotic algae. J Antibiot (Tokyo). 2024;77(3):163–9. [DOI] [PubMed] [Google Scholar]
- 6.Zhao GZ, Li J, Qin S, Zhang YQ, Zhu WY, Jiang CL, et al. Micrococcus yunnanensis sp. nov., a novel actinobacterium isolated from surface-sterilized Polyspora Axillaris roots. Int J Syst Evolutionary Microbiol. 2009;59(10):2383. [DOI] [PubMed] [Google Scholar]
- 7.Saleh O, Mohamed M, El-Galil MA, El-Kamed A, Sayed H. Isolation and characterization of Micrococcus luteus from Oreochromis niloticus in Egypt. Egypts Presidential Specialized Council for Education and Scientific Research. 2021(2).
- 8.Kloos W, Tornabene T, Schleifer K. Isolation and characterization of Micrococci from Human skin, including two New species: Micrococcus lylae and Micrococcus kristinae1. Int J Syst Evol MicroBiol. 1974;24:79–101. [Google Scholar]
- 9.Zhang JY, Liu XY, Liu SJ. Agrococcus terreus sp. nov. and Micrococcus terreus sp. nov., isolated from forest soil. Int J Syst Evol Microbiol. 2010;60(Pt 8):1897–903. [DOI] [PubMed] [Google Scholar]
- 10.Rieser G, Scherer S, Wenning M. Micrococcus cohnii sp. nov., isolated from the air in a medical practice. Int J Syst Evol Microbiol. 2013;63(Pt 1):80–5. [DOI] [PubMed] [Google Scholar]
- 11.Chittpurna, Singh PK, Verma D, Pinnaka AK, Mayilraj S, Korpole S. Micrococcus lactis sp. nov., isolated from dairy industry waste. Int J Syst Evol Microbiol. 2011;61(Pt 12):2832–6. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Jiang Y, Zhan Y, Wang H, Qin T, Lu Z. First case report of human infection with Micrococcus yunnanensis identified by 16S rRNA gene sequencing: a case report. Med (Baltim). 2022;101(48):e32108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Xu Y, Ma Y, Zhou P. Characterization of Micrococcus antarcticus sp. nov., a psychrophilic bacterium from Antarctica. Int J Syst Evol Microbiol. 2000;50(Pt 2):715–9. [DOI] [PubMed] [Google Scholar]
- 14.Fan HX, Miao LL, Liu Y, Liu HC, Liu ZP. Gene cloning and characterization of a cold-adapted β-glucosidase belonging to glycosyl hydrolase family 1 from a psychrotolerant bacterium Micrococcus antarcticus. Enzyme Microb Technol. 2011;49(1):94–9. [DOI] [PubMed] [Google Scholar]
- 15.Miao LL, Fan HX, Qu J, Liu Y, Liu ZP. Specific amino acids responsible for the cold adaptedness of Micrococcus antarcticus β-glucosidase BglU. Appl Microbiol Biotechnol. 2017;101(5):2033–41. [DOI] [PubMed] [Google Scholar]
- 16.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67(5):1613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein J, CONFIDENCE LIMITS ON. PHYLOGENIES: AN APPROACH USING THE BOOTSTRAP. Evolution. 1985;39(4):783–91. [DOI] [PubMed] [Google Scholar]
- 20.Eucast T, Mic H, Mic E, Ip NA. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 14.0, 2024. http://www.eucast.org
- 21.Stackebrandt E, Koch C, Gvozdiak O, Schumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int J Syst Bacteriol Int J Syst Bacteriol. 1995;45(4):682–92. [DOI] [PubMed] [Google Scholar]
- 22.Wieser M, Denner EB, Kämpfer P, Schumann P, Tindall B, Steiner U, et al. Emended descriptions of the genus Micrococcus, Micrococcus luteus (Cohn 1872) and Micrococcus lylae (Kloos et al. 1974). Int J Syst Evol Microbiol. 2002;52(Pt 2):629–37. [DOI] [PubMed] [Google Scholar]
- 23.Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022;50(D1):D801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erbasan F. Brain abscess caused by Micrococcus luteus in a patient with systemic lupus erythematosus: case-based review. Rheumatol Int. 2018;38(12):2323–8. [DOI] [PubMed] [Google Scholar]
- 25.Zhu M, Zhu Q, Yang Z, Liang Z. Clinical characteristics of patients with Micrococcus luteus Bloodstream infection in a Chinese tertiary-care hospital. Pol J Microbiol. 2021;70(3):321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buonsenso D, Lombardo A, Fregola A, Ferrari V, Piastra M, Calvani M, et al. First Report of Micrococcus luteus native Valve endocarditis Complicated with Pulmonary Infarction in a Pediatric patient: Case Report and Literature Review. Pediatr Infect Dis J. 2021;40(7):e284–6. [DOI] [PubMed] [Google Scholar]
- 27.Guerra JMM, Asenjo MM, Martín CR. Bacteraemia by Micrococcus luteus in an immunocompromised patient. Med Clínica (English Edition). 2019;152(11). [DOI] [PubMed]
- 28.Song SH, Choi HS, Ma SK, Kim SW, Shin JH, Bae EH. Micrococcus aloeverae - a Rare cause of peritoneal Dialysis-related Peritonitis confirmed by 16S rRNA gene sequencing. J Nippon Med Sch. 2019;86(1):55–7. [DOI] [PubMed] [Google Scholar]
- 29.Qin T, Zhou H, Ren H, Meng J, Du Y, Mahemut M, et al. Incidence, etiology, and environmental risk factors of Community-Acquired Pneumonia requiring hospitalization in China: a 3-Year, prospective, Age-Stratified, Multicenter Case-Control Study. Open Forum Infect Dis. 2021;8(11):ofab499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu YN, Zhang YF, Xu Q, Qiu Y, Lu QB, Wang T, et al. Infection and co-infection patterns of community-acquired pneumonia in patients of different ages in China from 2009 to 2020: a national surveillance study. Lancet Microbe. 2023;4(5):e330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Wang J, Cai H, Lin M, Zhang Y, Huang L. Enhanced hemolytic activity of Mesophilic Aeromonas salmonicida SRW-OG1 is brought about by elevated temperatures. Microorganisms. 2022;10(10). [DOI] [PMC free article] [PubMed]
- 32.Mandviya P, Ghanwate N, Thakare P. First case report of isolation of Micrococcus lylae from urinary catheter of a 50-year-old woman suffering from malignancy. J Infect Dev Ctries. 2023;17(7):1041–6. [DOI] [PubMed] [Google Scholar]
- 33.Almirall J, Serra-Prat M, Bolíbar I, Balasso V. Risk factors for community-acquired pneumonia in adults: a systematic review of Observational studies. Respiration. 2017;94(3):299–311. [DOI] [PubMed] [Google Scholar]
- 34.Chen L, Gao W, Tan X, Han Y, Jiao F, Feng B, et al. MALDI-TOF MS is an effective technique to classify specific microbiota. Microbiol Spectr. 2023;11(3):e0030723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha SM, Kim CK, Roh J, Byun JH, Yang SJ, Choi SB, et al. Application of the whole genome-based bacterial identification system, TrueBac ID, using clinical isolates that were not identified with three matrix-assisted laser Desorption/Ionization time-of-flight Mass Spectrometry (MALDI-TOF MS) systems. Ann Lab Med. 2019;39(6):530–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau SK, Tang BS, Teng JL, Chan TM, Curreem SO, Fan RY, et al. Matrix-assisted laser desorption ionisation time-of-flight mass spectrometry for identification of clinically significant bacteria that are difficult to identify in clinical laboratories. J Clin Pathol. 2014;67(4):361–6. [DOI] [PubMed] [Google Scholar]
- 37.Lao HY, Ng TT, Wong RY, Wong CS, Lee LK, Wong DS, et al. The clinical utility of two high-throughput 16S rRNA gene sequencing workflows for taxonomic assignment of unidentifiable bacterial pathogens in Matrix-assisted laser desorption ionization-time of Flight Mass Spectrometry. J Clin Microbiol. 2022;60(1):e0176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the GenBank under accession number of PP535566.