Abstract
Gene activation in eukaryotes is inherently combinatorial depending on cooperation between different transcription factors. An example where this cooperation seems to be directly exploited for regulation is the Bas1p/Bas2p couple in yeast. Bas1p is a Myb-related transcription factor that acts together with the homeodomain-related Bas2p (Pho2p) to regulate purine and histidine biosynthesis genes in response to extracellular purine limitation. We show that fusion of the two factors abolished adenine repression, suggesting that what is regulated by adenine is the Bas1p–Bas2p interaction. Analysis of Bas1p deletions revealed a critical domain (Bas1p interaction and regulatory domain, BIRD) acting in two-hybrid assays as an adenine-dependent Bas1p–Bas2p interaction domain. BIRD had a dual function, as an internal repressor of a centrally located Bas1p transactivation domain on the ADE1 promoter and as a Bas2p-dependent activator on the HIS4 promoter. This promoter-dependent behavior reflected a differential binding to the two promoters in vivo. On ADE1 Bas1p bound the promoter efficiently by itself, but required adenine limitation and Bas2p interaction through BIRD for derepression. On HIS4 efficient promoter binding and derepression required both factors and adenine limitation. We propose a promoter-dependent model for adenine regulation in yeast based on controlled Bas1p–Bas2p interactions through BIRD and exploited differentially by the two promoters.
INTRODUCTION
Transcriptional activation in eukaryotes is inherently combinatorial. A common view is that gene-specific regulation is obtained because each promoter is utilizing a unique combination of transcription factors for its activation. It is therefore not surprising that interactions between transcription factors are directly exploited by cells to regulate gene expression. Members of the Myb family of transcription factors often activate their target genes in close cooperation with DNA-binding proteins of other classes (1). This cooperation is probably crucial for the proper function of c-Myb itself, since it appears to be perturbed during oncogenic activation (2,3). The AMV v-Myb contains point mutations abolishing its cooperation with C/EBP-β (4) and the E26 v-Myb encodes a fusion between truncated Myb and Ets transcription factors (3,5). The Myb-related transcription factor Bas1p in yeast also activates its target genes in close cooperation with a member of another class of transcription factors, the homeodomain protein Bas2p (Pho2p). In Saccharomyces cerevisiae the transcription of all the genes of the de novo purine pathway (ADE genes) requires the cooperative action of these two transcription factors (6). The same is true for three genes involved in the histidine biosynthesis pathway (HIS1, HIS4 and HIS7) as well as for the genes GLN1, SHM2 and MTD1 (7–11). Activation of these target genes is repressed by adenine in the growth medium through an unknown response pathway that leads to down-regulation of the activity of the Bas1p/Bas2p couple (6,8,10,11). It has been proposed that adenine repression operates by directly affecting the interaction between Bas1p and Bas2p, an interaction believed to unmask a latent activation function in Bas1p (12).
In the present work we have addressed this model of gene regulation through modification of transcription factor interactions. We demonstrate that a covalent fusion of the two factors led to loss of adenine repression of ADE genes, strongly supporting the hypothesis that adenine repression operates through a modification of the interaction between Bas1p and Bas2p. A C-terminal deletion approach led us to identify a regulatory domain, termed BIRD, in Bas1p. In the absence of Bas2p, this BIRD domain acted as a repressor of an internal transactivation domain of Bas1p on all promoters tested. However, in the presence of Bas2p BIRD changed into a positive acting domain in a promoter-dependent manner. We provide two-hybrid evidence that BIRD acts as an adenine-dependent Bas1p–Bas2p interaction domain. We finally show that a fine tuned activation of target genes is obtained due to differential binding of Bas1p and Bas2p to the promoters of their target genes.
MATERIALS AND METHODS
Yeast strains and media
Yeast strains used in this study were Y329 (MATα, leu2-3,112, ura3-52, gcn4-2, bas1-2), Y330 (MATα, leu2-3,112, ura3-52, gcn4-2, bas2-2), L4233 (MATα, leu2-2, ura3-52, gcn4-2, bas1-2, bas2-2) and Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3,112, gal4Δ, met–, gal80Δ, URA3::GAL1UAS-GAL1TATA-LacZ; Clontech). Yeast were grown in SD or SC medium as described (13). SD-CASA medium is SD supplemented with 0.2% (w/v) casamino acids (Difco Laboratories).
BAS1 expression plasmids
The effector vector YPL was constructed to express deletion mutants of Bas1p. The vector is CEN4, LEU2 and has a PGK promoter/terminator. The different Bas1p deletion mutants were made by PCR using genomic DNA from S.cerevisiae as a template. The BAS1 fragments were cloned into the vector YPL using SpeI and MluI in the polylinker between the PGK promoter/terminator. The suffix in the different effector plasmid names indicates the last amino acid residue remaining in the deleted protein. A common N-terminal PCR-primer 5′-TCTCTTACTAGTATGTCTCACCACCACCACCACCACGGTTCGAATATAAGTACCAAAGAT-3′ was used for YPL-BAS1[FL] and the different C-terminal deletion mutants of Bas1p. The individual C-terminal primers used were 5′-TTCTCTACGCGTCTATCAACTAGGATTCAGTGGCAGG-3′ for YPL-BAS1[FL], 5′-TTCTCTACGCGTCTATCATTGATGTACCACTTGAGG-3′ for YPL-BAS1[ΔC750], 5′-TTCTCTACGCGTCTATCAGCCGTTACTTCTTACTGA-3′ for YPL-BAS1[ΔC664], 5′-TTCTCTACGCGTCTATCACAAATAGTTGAAATGCGCTA-3′ for YPL-BAS1[ΔC630], 5′-TTCTCTACGCGTCTATCATGACATACTATTAGCACTAACG-3′ for YPL-BAS1[ΔC591], 5′-TTCTCTACGCGTCTATCATTGTGAACCAAGTTCATTTGC-3′ for YPL-BAS1[ΔC550] and 5′-TTCTCTACGCGTCTATCATTTTACTAATTTTGTGCTGTC-3′ for YPL-BAS1[ΔC432]. In all plasmids the BAS1 gene corresponding to the PCR fragment was fully verified by sequencing. YPL-BAS1[ΔBIRD], expressing Bas1p with an internal deletion of the BIRD domain (Δ631–664), was constructed in several steps. First, AflII sites were introduced just after the BAS1 630 (PCR1) and 664 (PCR2) codons by two distinct PCR reactions performed with genomic DNA as template and with oligonucleotides 5′-TCTAACGGAACTATTGACAGCACA-3′ and 5′-GGACCTTGGAGATCTAACCGCGGAACTTAAGTAGTTGGAAATGCGCAATTCTATTAGGACCTAC-3′ for PCR1 or 5′-TGGAGGATTTGGATTCGTAGTAGC-3′ and 5′-GCTTGGCCACTTAAGTAAAACTAAGAAAAAAGAAAAAAGAAAAAGTGAATCATCTCAG-3′ for PCR2. These PCR products were both cloned in pBluescriptSKII+ opened by SmaI, giving plasmids B219 and B221 (for PCR1 and PCR2, respectively). Then, the AflII–BglII fragment of B219 was replaced by the AflII–BglII fragment from B221, resulting in plasmid B229. Finally, YPL-BAS1[ΔBIRD] was obtained by replacement of the AccI–BglII fragment of YPL-BAS1[FL] by the AccI–BglII fragment from B229. The BAS1–VP16 and BAS1–BAS2 fusions were constructed to be under the control of the BAS1 promoter, as the same constructs in the YPL vector were toxic in yeast. These fusions were therefore inserted in the P79 plasmid (CEN, URA3) harboring the entire BAS1 gene including its promoter (14). The control plasmid without BAS1 is named B836 (from the laboratory collection of G. Fink). The BAS1–VP16 fusion was constructed in several steps. First, the SalI–ClaI fragment from P79 was transferred to pBluescriptSKII+ opened by SalI and ClaI (B170). Then, the BamHI–BglII cassette containing the VP16 transactivation domain sequence and a stop codon (from plasmid pDBD11-R2R3; 15) was introduced in B170 opened by BglII to give plasmid B174. The final plasmid containing BAS1 fused to VP16 (B184) was obtained by replacing the SalI–ClaI fragment of P79 by a SalI–ClaI fragment from B174. The BAS1–BAS2 fusion was constructed in two steps. A BamHI–BamHI cassette containing the full BAS2 open reading frame (obtained by PCR with oligonucleotides 5′-CTCTTAGGATCCCCCGGGATGATGGAAGAATTCTCGTACGAT-3′ and 5′-TCCTGGATCCTCATCATATCCATCTATGCTCG-3′ on yeast genomic DNA as template and fully sequenced) was introduced into plasmid B170 opened by BglII to give plasmid B272. The final plasmid containing the BAS1–BAS2 fusion (B273) was obtained by replacing the SalI–ClaI fragment of P79 by the SalI–ClaI fragment from B272. The predicted fusion protein, Bas1p[1–704]–Bas2p[1–559], consisted of a C-terminally truncated Bas1p fused to full-length Bas2p. Truncation of the last 107 residues of Bas1p had no apparent effect on Bas1p function. The GAL4 DNA-binding domain (Gal4p DBD) fusions were inserted in the CEN6 TRP1 bait vector pDBT (16). The different BIRD–Gal4p DBD fusions were made by PCR using p79 as template. The N-terminal primers used were 5′-TTCTCTTAGATCTCTCCACCAACTATACGGCCTCATTTAG-3′ for pDBT-BAS1-631–664, 5′-TTCTCTTAGATCTCTCCGAATTTCAATGGAACAAATGGCA-3′ for pDBT-BAS1-592–664 and 5′-TTCTCTTAGATCTCTAGCAATAGAGAAACAAACAGCCCGT-3′ for pDBT-BAS1-551–664. The C-terminal primer was the same for all the BAS1–BIRD constructs (5′-AGTTCTCTGCAGGCGGCCGCGGATCCTATCAGCCGTTACTTCTTACTGAATTTGG-3′). The BIRD fragments were cloned into the vector pDBT using NotI and BglII in the polylinker. In all plasmids the BAS1 gene corresponding to the PCR fragment was fully verified by sequencing. To construct pDBT BAS1FL, a new MCS in the pDBT vector was made by annealing the oligos 5′-AATTCTAACTAGTCCACGCGTGAGCT-3′ and 5′-CACGCGTGGACTAGTTAG-3′ and ligating the product into the pDBT vector digested with EcoRI and SacI. The new vector was called pDBT-SM. The BAS1FL fragment was isolated from the YPL-BAS1[FL] vector digested with SpeI and MluI and inserted into the pDBT-SM vector digested with the same enzymes.
BAS2 expression plasmids
A HindIII–HindIII fragment carrying the entire BAS2 gene was inserted into the CEN4, LEU2 vector YCplac111 (17). The resulting plasmid was named P1262. The plasmid expressing BAS2 fused to the VP16 transactivation domain has already been described (18).
β-Galactosidase measurements
For β-galactosidase (β-Gal) assays, yeast cells (Y329, Y330 and Y4233) were co-transformed with two plasmids, one reporter carrying the LacZ fusion and one centromeric effector carrying either wild-type, fused to VP16 or deletion mutants of BAS1 or BAS2. The following reporter plasmids were used: a series of ADE–LacZ fusions already described (6,8) and pHIS4–LacZ, which is a URA3 2µ plasmid where LacZ is activated by a promoter derived from HIS4 in S.cerevisiae. It contains both the proximal and distal binding sites for Bas1p and also the binding sites for Bas2p and Rap1p. A similar plasmid, but with a LEU2 marker, was obtained using the pUL9 swapping plasmid as described (19). When reporter activation in this system was compared with activation in an isogenic gcn4BAS1BAS2 strain we observed that effector-encoded Bas1p closely mimicked the behavior of Bas1p encoded by the genomic locus, both with respect to levels of transactivation and adenine repression (data not shown). For the two hybrid experiment the Y187 strain was transformed with a centromeric plasmid expressing the ADE2 gene (to allow reporter assays in the absence of adenine) with either the Bas2–VP16 plasmid or an empty vector as a ‘prey’ plasmid and with a plasmid expressing the Gal4p DBD protein (pDBT) fused or not to the BIRD fragments or to full-length Bas1p as ‘bait’ plasmid. In all experiments six clones of each transformation were grown overnight in SC medium and then diluted to 0.1 OD600 in the same medium supplemented or not with 0.15 mM adenine. After 6 h at 30°C the β-Gal assays were performed as already described (14).
Generation and purification of antibodies
Anti-Bas1p antibodies were raised against an N-terminal Bas1p subdomain corresponding to amino acid residues 1–272. This truncated protein was expressed and purified from Escherichia coli as described (20) and used for immunization using the antibodies service of Eurogentec Bel S.A. Antibody purification was performed as described (21) on immunoaffinity resins obtained by aqueous coupling of N-terminal Bas1p(1–272) on affi-Gel®10 resin as described by the manufacturer (Bio-Rad). Purified antibodies were concentrated on protein A–Sepharose (Pharmacia) and stored as previously described (22).
Total yeast protein extracts and western blot analyses
Y329 and L4233 cells transformed with YPL plasmids containing the wild-type or deleted BAS1 genes were grown in SC medium lacking leucine to an OD600 of 1. Cells (2 ml of each culture) were harvested by centrifugation, rinsed with 1 ml of water and resuspended in 500 µl of 5% TCA before addition of 500 mg of glass beads. Cells were disrupted by vigourous vortexing for 15 min at room temperature and incubated for 15 min on ice. Supernatants were transferred to clean tubes and centrifuged for 5 min at 20 000 g. Supernatants were discarded and pellets were neutralized with 5 µl of 1 M Tris–HCl, pH 8, resuspended in 20 µl of Laemmli sample buffer (23) and incubated at 100°C for 3 min. Proteins were separated by 12.5% Tris–glycine SDS–PAGE (23) and transferred to a polyvinylidene difluoride membrane (Amersham). Western blot analysis was performed with the ECL Plus™ western blot kit (Amersham) with purified anti-Bas1p (1 µg purified IgG/ml) as primary antibody and peroxidase-conjugated anti-rabbit IgG (diluted 1:6000; Pierce) as secondary antibody.
RESULTS
Adenine repression of ADE genes acts through modulation of the interaction between the transcription factors Bas1p and Bas2p
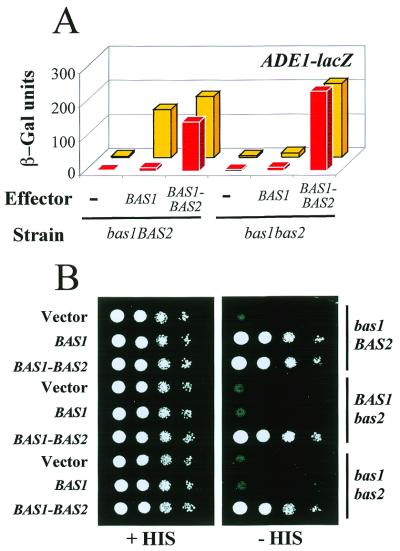
From studies of reporter activation using lexA–Bas1p and lexA–Bas2p fusions, Zhang et al. proposed a model for adenine repression of ADE genes in S.cerevisiae stating that complex formation between the two transcription factors Bas1p and Bas2p is a critical regulated step in this response (12). We reasoned that if this model is correct, a covalent fusion between the two factors should make the Bas1p/Bas2p couple constitutively active and cause the ADE genes to be fully expressed in the presence of adenine. The results presented in Figure 1A show that this is indeed the case. Here adenine repression was monitored in a gcn4bas1BAS2 strain using an ADE1–LacZ reporter. While adenine repression was normal with an introduced BAS1 plasmid, it was totally abolished when the strain was transformed with the BAS1–BAS2 fusion plasmid (Fig. 1A, left). In the latter case the ADE1–LacZ reporter was activated to a similar level whether or not adenine was present in the growth medium. Control experiments confirmed that both partners of the expressed Bas1p–Bas2p covalent fusion were functional, as judged from the ability of the fusion to activate the ADE1–LacZ reporter in the absence of Bas2p (Fig. 1A, right) and from its ability to restore growth in the absence of histidine in three yeast mutant strains. The gcn4 strains used require both Bas1p and Bas2p for growth in the absence of histidine (Fig. 1B; 11). Similar loss of adenine repression was observed when activation of a HIS4–LacZ reporter was measured (data not shown). These observations are consistent with a model of regulated factor interaction explaining adenine repression in S cerevisiae. Consequently, we set out to map in more detail the functional domains in Bas1p responsible for this adenine repression.
Figure 1.
Fusion of Bas1p and Bas2p leads to constitutive expression of their target genes. (A) In vivo effect of a Bas1p–Bas2p fusion on ADE1–LacZ reporter gene activatiom. Y329 (gcn4bas1BAS2) and L4233 (gcn4bas1bas2) cells were co-transformed with a plasmid carrying the ADE1–LacZ fusion and either the B836 plasmid (empty vector, lane –) or the P79 plasmid encoding wild-type Bas1p (BAS1) or the B273 plasmid encoding the Bas1p–Bas2p fusion (BAS1–BAS2). β-Galactosidase (β-Gal) assays were performed on exponentially growing cells in the presence (red boxes) or absence (yellow boxes) of adenine as described in Materials and Methods. (B) In vivo effect of the Bas1p–Bas2p fusion on endogenous HIS gene expression. Y329, Y330 (gcn4BAS1bas2) and L4233 cells were transformed with either the B836 plasmid (vector), the p79 plasmid or the B273 plasmid. Serial dilutions of the cells (104, 103, 102 and 10 cells) were spotted on SC medium lacking uracil and supplemented (left) or not (right) with histidine. Cells were grown for 36 h at 30°C.
Identification of a Bas1p interaction and regulatory domain (BIRD)
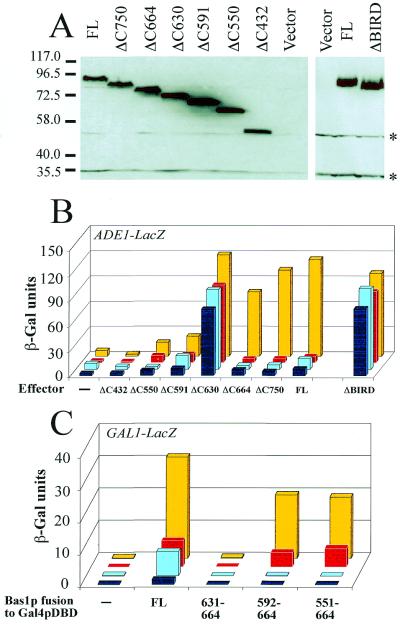
A Bas1p-dependent effector–reporter system was constructed to dissect the functional domains in Bas1p. To identify critical regions in Bas1p six C-terminal deletions were made, all expressed at comparable levels (Fig. 2A) and all retaining the nuclear localization previously demonstrated for Bas1p (14) (GFP data not shown).
Figure 2.
Bas1p contains a small domain (BIRD) with interaction and regulatory functions. (A) Expression level of the deletion mutants of Bas1p in the bas1bas2 yeast strain. The bas1bas2 (L4233) strain was transformed with the empty control plasmid (vector) or YPL-BAS1 plasmids carrying the full-length (FL) or the deletion version of the BAS1 gene. Total yeast protein extracts were prepared as described in Materials and Methods. Proteins were subjected to SDS–PAGE and electroblotted on a polyvinylidene difluoride membrane. The blot was then incubated with anti-Bas1p purified antibodies (1 µg IgG/ml) followed by horseradish peroxidase-labeled IgG (1/6000 dilution) as secondary antibodies and, finally, with luminescent substrate before exposure to film. The cross-reacting bands are marked by asterisks. Comparable levels of the different Bas1p proteins were also observed in a gcn4bas1BAS2 strain (not shown). (B) In vivo effects of the deletions in Bas1p assayed with the ADE1–LacZ reporter. The bas1BAS2 (Y329, yellow and red boxes) and bas1bas2 (L4233, light and dark blue boxes) cells were co-transformed with a plasmid carrying the ADE1–LacZ fusion and the different YPL-BAS1 plasmids carrying the full-length (FL) or deleted BAS1 gene. The lane ‘Effector –’ corresponds to transformation of yeast strains with the centromeric vector not carrying the BAS1 gene (YPL). Transformed cells were grown in the presence (red and dark blue boxes) or absence (yellow or light blue boxes) of adenine and β-Gal assays were performed as described in Materials and Methods. (C) In vivo role of BIRD in the Bas1p–Bas2p interaction monitored by a two-hybrid approach. The Y187 yeast strain containing an ADE2-expressing plasmid (pAZ11) (27) was transformed with a centromeric bait plasmid expressing Gal4p DBD fusions (pDBT) (16) and with a second prey plasmid expressing (yellow and red boxes) or not (light and dark blue boxes) the Bas2p–VP16 chimera. Cells were grown in the presence (red and dark blue boxes) or absence (yellow and light blue boxes) of adenine in SC medium lacking histidine and tryptophan. β-Gal assays were performed as described in Materials and Methods.
The deletions were first examined for their ability to activate the ADE1 promoter, since ADE1 is one of the strongly adenine repressed genes. The effector with the largest deletion (ΔC432) was not able to transactivate the ADE1 reporter since it gave β-Gal levels similar to the empty vector (Fig. 2B). Transactivation was just detectable with the ΔC550 and ΔC591 deletions, but became fairly high with the ΔC630 deletion. This high activation was observed whether or not Bas2p was present and indicates that a bona fide transactivation domain is present in the central part of Bas1p and requires a region N-terminal to residue 630 for activity. This transactivation domain by itself showed no significant adenine dependence, since very similar β-Gal-activities were measured both in the presence and absence of adenine. Notably, when 34 more residues were included (ΔC664), transactivation was totally lost in three out of four conditions tested, suggesting that the region between residues 630 and 664 acted as an internal repressor domain restraining the action of the central activation domain under most conditions. The only condition where ΔC664 remained active was in the absence of adenine and presence of Bas2p, exactly the conditions where adenine repression is normally relieved. To simplify the discussion, we will designate the 630–664 region BIRD (for Bas1p interaction and regulatory domain) due to its properties, analyzed below.
To confirm the importance of this BIRD region for the regulatory properties of Bas1p, we constructed an internal BIRD deletion, which was expressed at the same level as full-length Bas1p (Fig. 2A). As expected, the BIRD-deleted Bas1p had lost its repressor activity on the ADE1 promoter, which caused constitutive activation of the ADE1–LacZ reporter and no response to adenine (Fig. 2B). Relative to ΔC664, no further changes were observed with the more extended Bas1p variants (ΔC750 and FL). Similar behavior was found for all the Bas1p deletion mutants when they were tested for their abilities to activate an ADE17–LacZ fusion (data not shown). Altogether, these results immediately suggested a simple model for the adenine response where BIRD in Bas1p acts as an internal repressor of a neighboring transactivation domain, but in a way that is signal-responsive and relieved in the presence of Bas2p when adenine is limiting.
The Bas2p dependence further suggested that BIRD could be implicated in the interaction between Bas1p and Bas2p. To directly test this hypothesis, we developed a two-hybrid approach to monitor in vivo the possible role of BIRD in the Bas1p–Bas2p interaction. The bait plasmids expressed different Bas1p regions all including BIRD fused to the DNA-binding domain of Gal4p (Gal4p DBD) as well as a Bas1p full-length control fused to Gal4p DBD. The prey plasmid expressed a Bas2p–VP16 fusion and the interaction was monitored in a yeast strain harboring an integrated GAL1–LacZ reporter. No reporter activation was observed with the 34 residue BIRD fused to Gal4p DBD (Fig. 2C). However, in the presence of an N-terminally extended version of BIRD (residues 592–664) we observed an activation of the GAL1–LacZ reporter (Fig. 2C). This activation was observed only in the presence of the prey Bas2p–VP16, strongly suggesting an interaction between the enlarged BIRD and the Bas2p–VP16 protein. Most important, this interaction was clearly adenine dependent, being stronger in the absence than in the presence of adenine. This suggests that a 73 residue region including BIRD in fact acts as an adenine-dependent Bas1p–Bas2p interaction domain and that this domain is able to transfer adenine-regulatory properties to Gal4p DBD in the presence of Bas2p–VP16. Figure 2C also includes a control with Bas1p FL fused to Gal4p DBD that showed normal adenine repression of reporter activation, validating that Bas1p remained regulated as a Gal4p DBD fusion (Fig. 2C). The weak reporter activation observed with Bas1p FL in the absence of the prey Bas2p–VP16 (‘autoactivation’ in two-hybrid terminology) is probably caused by the presence of a transactivation domain in Bas1p. That this weak autoactivation is also adenine responsive is what is expected, since endogenous Bas2p is present in the two-hybrid strain used for the experiment.
Promoter-dependent behavior of Bas1p
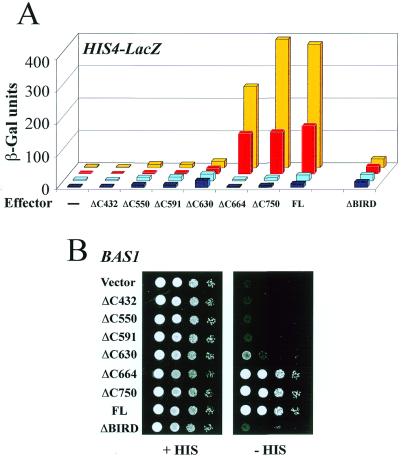
To determine if the model of adenine regulation deduced for the two ADE genes analyzed (ADE1, Fig. 2B; ADE17, data not shown) was generally valid for most Bas1p target genes, we extended the analysis by studying the activation of a HIS4–LacZ reporter by the same series of deletions of Bas1p. Here we got an unexpected result (Fig. 3). Although the profile of transactivation measured in the absence of Bas2p resembled the profile measured with the ADE1–LacZ reporter (except for lower values, Fig. 3A, dark and light blue boxes), the profile of transactivation changed dramatically in the presence of Bas2p (Fig. 3A, red and yellow boxes). Now, inclusion of the small BIRD region caused a marked increase in activity in addition to conferring adenine responsiveness, as if BIRD were a strong transactivation domain, not a repressor domain. This behavior was not a special phenomenon observed with the plasmid reporter construct only, because the deletions caused a corresponding pattern of growth in a gcn4 background, essentially reflecting expression of the endogenous HIS4 gene (24): no or very weak growth was observed for the smallest deletions (up to Δ630), but the addition of BIRD increased growth to a level comparable to those obtained for wild-type Bas1p (Fig. 3B). Thus, while BIRD mediated the adenine response on both promoters, it acted in a promoter-dependent fashion as an internal repressor domain on the ADE1 promoter and as an activator domain on the HIS4 promoter. The BIRD-deleted version of Bas1p also exhibited this promoter-specific behavior. In contrast to its derepression effect on the ADE1 promoter, BIRD-deleted Bas1p had lost its activator properties on the HIS4 promoter, which here caused inability to activate the HIS4–LacZ reporter (Fig. 3A) and the endogenous HIS genes (Fig. 3B). This promoter-dependent change in behavior of Bas1p merited further studies to unravel its mechanism.
Figure 3.
BIRD acts as an activation domain on the HIS4 promoter. (A) In vivo effect of the deletions in Bas1p on HIS4–LacZ reporter gene activation. The bas1BAS2 (Y329, yellow and red boxes) and bas1bas2 (L4233, light and dark blue boxes) cells were co-transformed with a plasmid carrying the HIS4–LacZ fusion and the different plasmids YPL-BAS1 as in Figure 2B. Transformed cells were grown in the presence (red and dark blue boxes) or absence (yellow or light blue boxes) of adenine and β-Gal assays were performed as described in Materials and Methods. (B) In vivo effects of the deletions in Bas1p on endogenous HIS genes expression. Y329 (bas1BAS2) cells were transformed with either the YPL empty control plasmid (vector) or the YPL-BAS1 plasmids and were treated as described in the legend to Figure 1B.
The promoter-dependent behavior reflects a differential binding of the Bas1p/Bas2p couple to the promoters
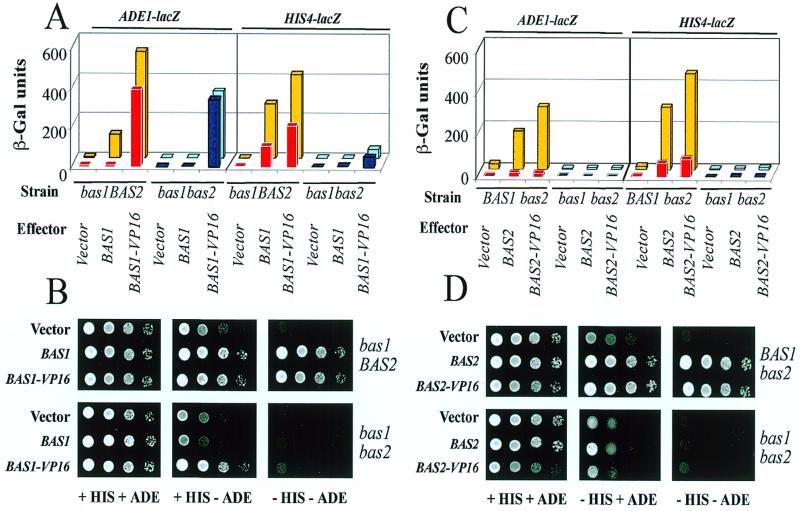
Reporter activation of course reflects both DNA binding of Bas1p and its transactivation properties and both could be affected by the cooperating Bas2p and adenine repression. To obtain a direct read-out of DNA binding to the two promoters in vivo, we fused Bas1p to the strong and constitutive transactivation domain VP16 from Herpes simplex virus. We reasoned that if Bas1p bound efficiently by itself to the promoter analyzed, Bas1p–VP16 should activate the reporter in the absence of Bas2p and independent of adenine status. When this analysis was performed, a clear promoter difference in DNA binding was indeed observed. The ADE1–LacZ reporter was efficiently activated by the Bas1p–VP16 fusion in the absence of Bas2p and independent of adenine status (Fig. 4A). The HIS4–LacZ reporter, on the other hand, was poorly activated by the Bas1p–VP16 fusion in the absence of Bas2p (Fig. 4A). Here, the presence of Bas2p caused an important increase in transactivation (10-fold in the presence of adenine and 7-fold in its absence), which we attribute to Bas2p-assisted recruitment of Bas1p to the promoter. Again, these observations were confirmed by growth phenotypes reflecting endogenous gene activation (Fig. 4B). On all media tested the growth observed for the bas1BAS2 strain containing the BAS1–VP16 fusion gene was similar to those observed for wild-type BAS1. In contrast, in the absence of Bas2p we observed informative differences. Loss of HIS genes activation in the absence of Bas2p, seen as lack of growth on His– media, was not restored by Bas1p–VP16 (Fig. 4B). This is consistent with our conclusion that Bas1p binds poorly to the HIS4 promoter by itself and requires Bas2p to be recruited to this promoter. Furthermore, when adenine was removed (His+ Ade– medium) reduced growth was observed for the bas1bas2 strain due to limiting activation of endogenous ADE genes in the absence of Bas2p (Fig. 4B). However, this reduction in growth was not observed in the presence of Bas1p–VP16, suggesting efficient binding of Bas1p–VP16 to endogenous ADE genes independent of Bas2p. Taken together, we conclude that Bas1p is able to bind productively to the ADE1 promoter on its own, but by itself causes little or no transactivation, while the factor alone binds much less efficiently to the HIS4 promoter in vivo, despite its efficient binding to HIS4-derived DNA fragments in vitro (14,20). On this promoter it appears that Bas2p is needed to recruit Bas1p to the promoter.
Figure 4.
Binding of Bas1p or Bas2p to promoters in vivo monitored by VP16 fusions. (A) Activation of ADE1–LacZ or HIS4–LacZ fusions by Bas1p–VP16 in vivo. Yeast cells were co-transformed with a plasmid carrying the LacZ fusion and a plasmid carrying one of the following genes: BAS1 (P79), BAS1–VP16 (B184) or the corresponding empty vector (B836). Transformed cells were grown in the presence (red and dark blue boxes) or absence (yellow or light blue boxes) of adenine and β-Gal assays were performed as described in Materials and Methods. (B) Activation of ADE or HIS genes by Bas1p–VP16 in vivo. Y329 (bas1BAS2) and L4233 (bas1bas2) cells were transformed with either the empty control plasmid or plasmids carrying the wild-type or VP16 version of the BAS1 gene and were treated as described in the legend to Figure 1B. (C) Activation of ADE1–LacZ or HIS4–LacZ fusions by Bas2p–VP16 in vivo. Yeast cells were co-transformed with a plasmid carrying the LacZ fusion and a plasmid carrying one of the following genes: BAS2 (P1262), BAS2–VP16 (18) or the corresponding empty vector Ycplac111. Transformed cells were grown in the presence (red and dark blue boxes) or absence (yellow or light blue boxes) of adenine and β-Gal assays were performed as described in Materials and Methods. (D) Activation of ADE or HIS genes by Bas2p–VP16 in vivo. Y330 (BAS1bas2) and L4233 (bas1bas2) cells were transformed with either the empty control plasmid or plasmids carrying the wild-type or VP16 version of the BAS2 gene and were treated as described in the legend to Figure 1B.
From an analogous series of experiments with a Bas2p–VP16 fusion we concluded that Bas2p is unable by itself to be recruited to either the HIS4 or ADE promoter and requires the cooperative action of Bas1p to do so. This is seen by very poor activation of the ADE1–LacZ and the HIS4–LacZ reporters in the bas1bas2 strain after introduction of the Bas2p–VP16 fusion (Fig. 4C). Furthermore, growth in the same strain was abolished on His– media and reduced on Ade– media, but in no cases were these growth defects restored by Bas2p–VP16 (Fig. 4D). It is noteworthy that the recruitment of Bas2p–VP16 to the ADE1 promoter is highly adenine dependent, again suggesting that adenine regulation operates through an alteration of the Bas1p–Bas2p interaction.
Having found that two Bas1p target genes behaved differently with respect to recruitment of transcription factors, we examined a more extensive series of established target genes for their activation by Bas1p–VP16 in the presence or absence of Bas2p. Six additional LacZ reporters were used which were activated by one of the following Bas1p-responsive promoters: ADE2, ADE4, ADE5,7, ADE8, ADE13 or ADE17. Table 1 shows the β-Gal values obtained as well as ratios showing the Bas2p dependence of activation on the various promoters (ratios of reporter activity using Bas1p–VP16 in the absence relative to in the presence of Bas2p). As expected, a very low ratio (0.11–0.16) was obtained with HIS4–LacZ, showing a high dependence on Bas2p for HIS4 promoter binding in vivo, while the high ratio (0.71–0.97) observed with ADE1–LacZ shows a weak dependence on Bas2p for ADE1 promoter binding. Most of the other ADE promoters (ADE2, ADE4, ADE8, ADE13 and ADE17) behaved like ADE1, giving ratios >0.7. None had the same high Bas2p dependence for promoter binding as HIS4. One promoter appeared to be in an intermediate position. While the ADE5,7 promoter resembled the other ADE promoters in the presence of adenine (ratio 0.73) it showed a 3-fold difference in reporter activity in the absence of adenine, dependent on whether Bas2p was present or not (ratio 0.35).
Table 1. Bas2p dependence of Bas1p–VP16 recruitment to different ADE genes.
| – ADE | + ADE | |||||
|---|---|---|---|---|---|---|
| Fusion | + Bas2pa | – Bas2pb | Ratioc | + Bas2pa | – Bas2pb | Ratiod |
| HIS4–LacZ | 412 | 44 | 0.11 | 187 | 31 | 0.16 |
| ADE1–LacZ | 535 | 379 | 0.71 | 389 | 379 | 0.97 |
| ADE2–LacZ | 99 | 70 | 0.71 | 80 | 75 | 0.94 |
| ADE4–LacZ | 438 | 406 | 0.93 | 390 | 371 | 0.95 |
| ADE5,7–LacZ | 176 | 62 | 0.35 | 97 | 71 | 0.73 |
| ADE8–LacZ | 112 | 101 | 0.90 | 99 | 98 | 0.99 |
| ADE13–LacZ | 175 | 138 | 0.79 | 113 | 110 | 0.97 |
| ADE17–LacZ | 652 | 513 | 0.78 | 520 | 423 | 0.81 |
The yeast strain Y329 (gcn4bas1BAS2) or L4233 (gcn4bas1bas2) was transformed with a plasmid carrying one of the LacZ fusions listed and either the B836 empty vector or the plasmid carrying the BAS1–VP16 gene. Six to 12 clones of each transformation were grown overnight in SC medium lacking uracil and leucine and then diluted to 0.1 OD600 in the same medium supplemented (+ ADE) or not (– ADE) with adenine. After 6 h incubation at 30°C the β-galactosidase (β-Gal) assays were performed as described in Materials and Methods. Results given in the table correspond to β-Gal units obtained with the BAS1–VP16 plasmid subtracted from those obtained with the control plasmid (B836).
aβ-Galactosidase units measured in Y329 strain.
bβ-Galactosidase units measured in L4233 strain.
cRatio –Bas2p/+Bas2p in the absence of adenine.
dRatio –Bas2p/+Bas2p in the presence of adenine
DISCUSSION
We have in the present work addressed the molecular mechanism underlying the regulatory properties of Bas1p, a Myb family transcription factor in S.cerevisiae. The regulatory properties of this factor were found to be intimately linked to its ability to interact with the cooperating factor Bas2p through a specific region of Bas1p designated BIRD. This domain confers on Bas1p a complex behavior that depends on the presence or absence of the cooperating factor Bas2p, varies with the promoter to which the factors are recruited and responds to the presence or absence of adenine in the growth medium.
A central feature of BIRD was that it made Bas1p signal responsive and able to change activity dependent on the presence or absence of adenine in the growth medium. While Bas1p[1–630] activated the ADE1–LacZ and HIS4–LacZ reporters with the same efficiency in the presence or absence of adenine, inclusion of BIRD in Bas1p[1–664] resulted in a clear adenine-repressible activity similar to that observed for the full-length protein (Figs 2 and 3).
Another striking feature was the dual function of BIRD, acting either as a repressor or activator domain depending on the context. On all promoters tested BIRD behaved as an internal repressor domain in the absence of Bas2p (Figs 2 and 3). In contrast, BIRD behaved phenotypically as a transactivation domain in the presence of Bas2p on the HIS4 promoter (Fig. 3A and B). It is noteworthy that this activator property of BIRD was only observed in the presence of Bas2p and only on the HIS4 promoter, to which Bas1p binds poorly by itself. We therefore think that the activator properties of BIRD could be explained if this region acted as a Bas1p–Bas2p interaction domain. Direct evidence for such a function was provided by a two-hybrid approach, showing that a small region including BIRD fused to Gal4p DBD resulted in reporter activation in the presence of Bas2p–VP16. Even more important, this interaction was strengthened in the absence of adenine. The observation that a Gal4p DBD–BIRD fusion was adenine regulated shows that adenine regulation is a transferable property and strongly suggests that the Bas1p–Bas2p interaction through BIRD is regulated by adenine.
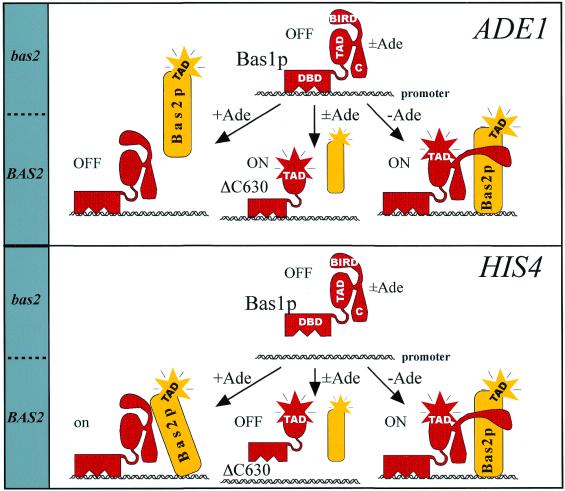
The identification of BIRD function in Bas1p allows us to propose a specific model for adenine regulation of Bas1p target genes, as illustrated in Figure 5. This model hypothesizes that the critical step regulated by adenine is the protein–protein interaction between Bas1p and Bas2p. Our two-hybrid data as well as the finding that a covalent fusion between the two factors totally abolished adenine repression strongly support this aspect of the model (Figs 1 and 2). The second element of the model is that BIRD plays a critical role in the mechanism of adenine repression, most probably by being itself a dual interaction domain engaged both in internal contacts within Bas1p (repression) and in external contacts with Bas2p (activation) (Figs 2 and 3). Based on these two elements we propose the following mechanism. In the absence of Bas2p, BIRD is assumed to interact with and functionally block the central transactivation domain of Bas1p. When Bas2p is present, we hypothesize that BIRD can interact with this cooperating factor in a regulated fashion, moderately in the presence of adenine and strongly in its absence. The BIRD–Bas2p interaction then controls the fine tuned equilibrium between free and bound Bas1p and Bas2p, leading to regulated promoter recruitment. On promoters like ADE1 only Bas1p is recruited in the presence of adenine (Fig. 4) and the repression function of BIRD will silence the bound factor. When adenine is removed, some kind of modification occurs that leads to a strengthened BIRD–Bas2p interaction, sufficient to also recruit Bas2p to the promoter. Since Bas2p does not bind the promoter by itself, it needs to be recruited and this recruitment occurs through an adenine-regulated Bas1p–Bas2p interaction. We hypothesize that the Bas1p–Bas2p interaction will lead to a conformational change in Bas1p and expose its hidden transactivation domain. The final outcome will be exposed transactivation domains of both factors, resulting in a strong synergistic activation of the ADE gene.
Figure 5.
A model for promoter-specific adenine regulation of the Bas1p/Bas2p couple. DBD, TAD, BIRD and C in the Bas1p drawings designate its DNA-binding domain, transactivation domain, Bas1p interaction and regulatory domain and C-terminal end, respectively. The ΔC630 version of Bas1p refers to the C-terminal deletion mutant of Bas1p spanning residues 1–630. The presence or absence of external adenine is indicated by + Ade, – Ade or ± Ade.
The factor–promoter equilibrium must be different on the HIS4 promoter. Our VP16 data suggest that neither Bas1p nor Bas2p were able to bind efficiently by themselves on this promoter (Fig. 4). For Bas1p this may be related to the chromatin packing of the Bas1p recognition sites since these sites bind Bas1p efficiently in vitro (20,25), but require the Rap1p factor to become accessible in chromatin (26). Since now both factors need to be recruited to the promoter and recruitment depends on an adenine-regulated Bas1p–Bas2p interaction, we observe that both Bas1p–VP16 and Bas2p–VP16 are sensitive to adenine. To explain the high basal activity on HIS4 in the presence of adenine we must assume that the moderately strong BIRD–Bas2p interaction is sufficient to tilt the equilibrium towards recruitment of both factors to the HIS4 promoter, leading to substantial levels of transactivation even in the presence of adenine. We do not know exactly what causes this equilibrium difference, but since the overall equilibrium is a function of the combined affinities of the Bas1p–DNA, Bas2p–DNA and Bas1p–Bas2p interactions, it would have been sufficient that the Bas2p–HIS4 interaction was stronger than the Bas2p–ADE1 interaction to explain the differences in the overall equilibrium. Finally, our model predicts that in the absence of adenine a strengthening of the BIRD–Bas2p interaction affects the conformational change in Bas1p, leading to better exposure of its transactivation domain and enhanced overall transactivation.
In conclusion, the present work shows that the Bas1p/Bas2p couple is an interesting and simple model for a regulatory mechanism that operates at the level of combinatorial transcription factor interaction. It is noteworthy that several intriguing parallels exist between yeast Bas1p and vertebrate c-Myb. The possibility of combining a genetic and biochemical approach in the study of the Myb family factors in yeast will hopefully provide conclusions that may also inspire directions of research on vertebrate c-Myb.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Svava Wetzel for skillful assistance with parts of the work. We are grateful to F. R. Cross, G. R. Fink, C. R. Goding and P. Recordon for providing plasmids and yeast strains. B.P. was supported by a long-term fellowship from the Federation of European Biochemical Societies (FEBS). This work was supported by The Norwegian Research Council, The Norwegian Cancer Society and the Anders Jahres Foundation.
REFERENCES
- 1.Lipsick J.S. (1996) Oncogene, 13, 223–235. [PubMed] [Google Scholar]
- 2.Ganter B. and Lipsick,J.S. (1999) Adv. Cancer Res., 76, 21–60. [DOI] [PubMed] [Google Scholar]
- 3.Lipsick J.S. and Wang,D.M. (1999) Oncogene, 18, 3047–3055. [DOI] [PubMed] [Google Scholar]
- 4.Kowenz-Leutz E., Herr,P., Niss,K. and Leutz,A. (1997) Cell, 91, 185–195. [DOI] [PubMed] [Google Scholar]
- 5.Metz T. and Graf,T. (1991) Cell, 66, 95–105. [DOI] [PubMed] [Google Scholar]
- 6.Denis V., Boucherie,H., Monribot,C. and Daignan-Fornier,B. (1998) Mol. Microbiol., 30, 557–566. [DOI] [PubMed] [Google Scholar]
- 7.Denis V. and Daignan-Fornier,B. (1998) Mol. Gen. Genet., 259, 246–255. [DOI] [PubMed] [Google Scholar]
- 8.Daignan-Fornier B. and Fink,G.R. (1992) Proc. Natl Acad. Sci. USA, 89, 6746–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolfes R.J., Zhang,F. and Hinnebusch,A.G. (1997) J. Biol. Chem., 272, 13343–13354. [DOI] [PubMed] [Google Scholar]
- 10.Springer C., Krappmann,S., Kunzler,M., Zmasek,C. and Braus,G.H. (1997) Mol. Gen. Genet., 256, 136–146. [DOI] [PubMed] [Google Scholar]
- 11.Arndt K.T., Styles,C. and Fink,G.R. (1987) Science, 237, 874–880. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F., Kirouac,M., Zhu,N., Hinnebusch,A.G. and Rolfes,R.J. (1997) Mol. Cell. Biol., 17, 3272–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman F., Fink,G.R. and Hicks,J.B. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Pinson B., Sagot,I., Borne,F., Gabrielsen,O.S. and Daignan-Fornier,B. (1998) Nucleic Acids Res., 26, 3977–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ording E., Bergholtz,S., Brendeford,E.M., Jamin,N. and Gabrielsen,O.S. (1996) Oncogene, 13, 1043–1051. [PubMed] [Google Scholar]
- 16.Navarro P., Durrens,P. and Aigle,M. (1997) Biochim. Biophys. Acta, 1343, 187–192. [DOI] [PubMed] [Google Scholar]
- 17.Gietz R.D. and Sugino,A. (1988) Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- 18.Hirst K., Fisher,F., McAndrew,P.C. and Goding,C.R. (1994) EMBO J., 13, 5410–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross F.R. (1997) Yeast, 13, 647–653. [DOI] [PubMed] [Google Scholar]
- 20.Høvring P.I., Bostad,A., Ording,E., Myrset,A.H. and Gabrielsen,O.S. (1994) J. Biol. Chem., 269, 17663–17669. [PubMed] [Google Scholar]
- 21.Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Pinson B., Pillois,X., Brethes,D., Chevallier,J. and Napias,C. (1996) Eur. J. Biochem., 239, 439–444. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U.K. (1970) Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 24.Arndt K.T., Styles,C.A. and Fink,G.R. (1989) Cell, 56, 527–537. [DOI] [PubMed] [Google Scholar]
- 25.Tice-Baldwin K., Fink,G.R. and Arndt,K.T. (1989) Science, 246, 931–935. [DOI] [PubMed] [Google Scholar]
- 26.Devlin C., Tice-Baldwin,K., Shore,D. and Arndt,K.T. (1991) Mol. Cell. Biol., 11, 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stotz A. and Linder,P. (1990) Gene, 95, 91–98. [DOI] [PubMed] [Google Scholar]