Abstract
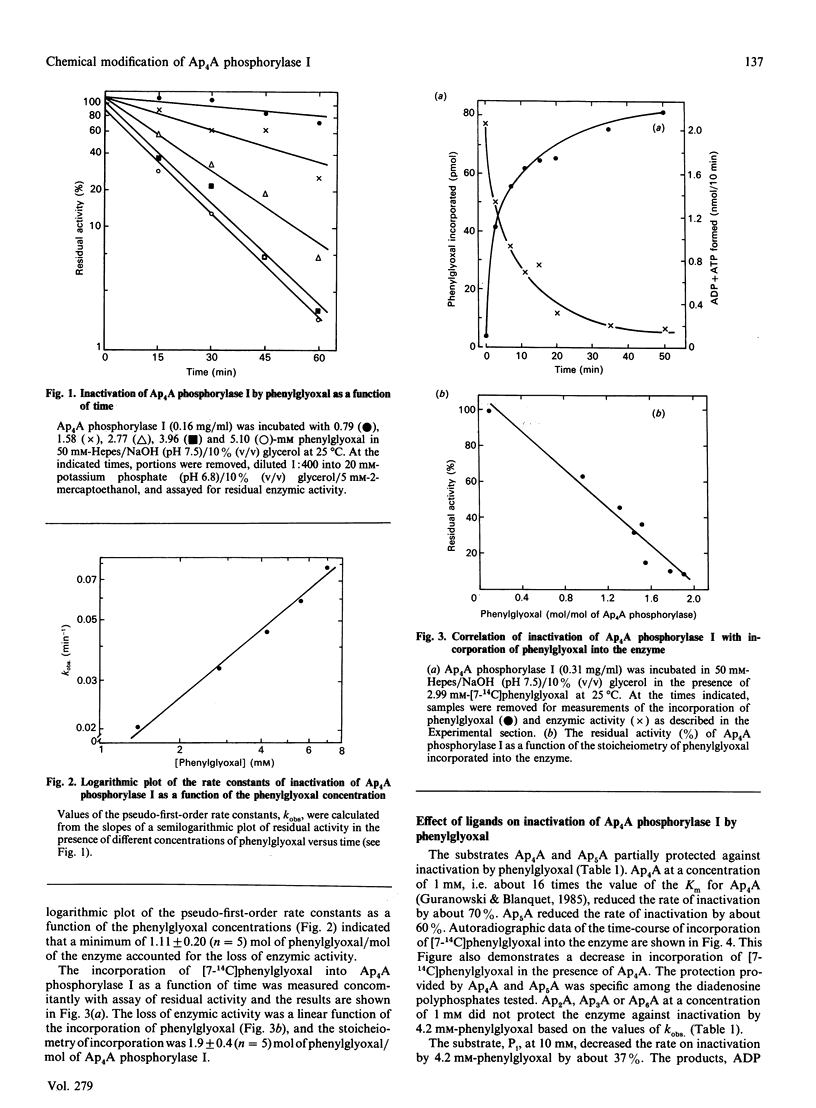
Phenylglyoxal, a reagent with high specificity for arginine residues, inactivated Ap4A phosphorylase I from Saccharomyces cerevisiae in a pseudo-first-order manner. The second-order rate constant was 11.5 +/- 2.5 M-1 min-1. The loss of activity was a linear function of the incorporation of [7-14C]phenylglyoxal. The incorporation of 1.9 +/- 0.4 mol of phenylglyoxal/mol of enzyme accounted for complete loss of activity. The specificity of inactivation by phenylglyoxal was tested in the presence of ApnA (n = 2-6), ADP, ATP and Pi. The substrates, Ap4A, Ap5A and Pi protected the enzyme against inactivation, but Ap2A, Ap3A and Ap6A did not. Ap4A, Ap5A and Pi reduced the rate of inactivation by about 70%, 60% and 37% respectively. The Ap4A phosphorolysis products, ADP and ATP, also partially protected the enzyme against inactivation by phenylglyoxal. Thus Ap4A phosphorylase I probably contains an arginine residue in the binding site for Ap4A.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker J. C., Jacobson M. K. Determination of diadenosine 5',5''',-P1,P4-tetraphosphate levels in cultured mammalian cells. Anal Biochem. 1984 Sep;141(2):451–460. doi: 10.1016/0003-2697(84)90070-8. [DOI] [PubMed] [Google Scholar]
- Barnes L. D., Culver C. A. Isolation and characterization of diadenosine 5',5"'-P1,P4-tetraphosphate pyrophosphohydrolase from Physarum polycephalum. Biochemistry. 1982 Nov 23;21(24):6123–6128. doi: 10.1021/bi00267a015. [DOI] [PubMed] [Google Scholar]
- Barnes L. D., Robinson A. K., Mumford C. H., Garrison P. N. Assay of diadenosine tetraphosphate hydrolytic enzymes by boronate chromatography. Anal Biochem. 1985 Jan;144(1):296–304. doi: 10.1016/0003-2697(85)90120-4. [DOI] [PubMed] [Google Scholar]
- Borders C. L., Jr, Riordan J. F. An essential arginyl residue at the nucleotide binding site of creatine kinase. Biochemistry. 1975 Oct 21;14(21):4699–4704. doi: 10.1021/bi00692a021. [DOI] [PubMed] [Google Scholar]
- Brevet A., Coste H., Fromant M., Plateau P., Blanquet S. Yeast diadenosine 5',5'''-P1,P4-tetraphosphate alpha,beta-phosphorylase behaves as a dinucleoside tetraphosphate synthetase. Biochemistry. 1987 Jul 28;26(15):4763–4768. doi: 10.1021/bi00389a025. [DOI] [PubMed] [Google Scholar]
- Flodgaard H., Klenow H. Abundant amounts of diadenosine 5',5"'-P1,P4-tetraphosphate are present and releasable, but metabolically inactive, in human platelets. Biochem J. 1982 Dec 15;208(3):737–742. doi: 10.1042/bj2080737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison P. N., Barnes L. D. Assay of adenosine 5'-P1-tetraphospho-P4-5"'-adenosine and adenosine 5'-P1-tetraphospho-P4-5"'-guanosine in Physarum polycephalum and other eukaryotes. An isocratic high-pressure liquid-chromatography method. Biochem J. 1984 Feb 1;217(3):805–811. doi: 10.1042/bj2170805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison P. N., Roberson G. M., Culver C. A., Barnes L. D. Diadenosine 5',5"'-P1,P4-tetraphosphate pyrophosphohydrolase from Physarum polycephalum. Substrate specificity. Biochemistry. 1982 Nov 23;21(24):6129–6133. doi: 10.1021/bi00267a016. [DOI] [PubMed] [Google Scholar]
- Guranowski A., Blanquet S. Chromate, molybdate, tungstate and vanadate behave as substrates of yeast diadenosine 5',5'''-p1,p4-tetraphosphate alpha, beta-phosphorylase. Biochimie. 1986 May;68(5):757–760. doi: 10.1016/s0300-9084(86)80170-5. [DOI] [PubMed] [Google Scholar]
- Guranowski A., Blanquet S. Diadenosine 5',5'''-P1, P4-tetraphosphate alpha, beta-phosphorylase from yeast supports nucleoside diphosphate-phosphate exchange. J Biol Chem. 1986 May 5;261(13):5943–5946. [PubMed] [Google Scholar]
- Guranowski A., Blanquet S. Phosphorolytic cleavage of diadenosine 5',5'''-P1,P4-tetraphosphate. Properties of homogeneous diadenosine 5',5'''-P1,P4-tetraphosphate alpha, beta-phosphorylase from Saccharomyces cerevisiae. J Biol Chem. 1985 Mar 25;260(6):3542–3547. [PubMed] [Google Scholar]
- Guranowski A., Jakubowski H., Holler E. Catabolism of diadenosine 5',5"'-P1,P4-tetraphosphate in procaryotes. Purification and properties of diadenosine 5',5"'-P1,P4-tetraphosphate (symmetrical) pyrophosphohydrolase from Escherichia coli K12. J Biol Chem. 1983 Dec 25;258(24):14784–14789. [PubMed] [Google Scholar]
- Guranowski A., Just G., Holler E., Jakubowski H. Synthesis of diadenosine 5',5'''-P1,P4-tetraphosphate (AppppA) from adenosine 5'-phosphosulfate and adenosine 5'-triphosphate catalyzed by yeast AppppA phosphorylase. Biochemistry. 1988 Apr 19;27(8):2959–2964. doi: 10.1021/bi00408a044. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Holler E. Noncovalent complexes of diadenosine 5',5"'-P1,P4-tetraphosphate with divalent metal ions, biogenic amines, proteins and poly(dT). Biochem Biophys Res Commun. 1984 May 16;120(3):1037–1043. doi: 10.1016/s0006-291x(84)80211-9. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Lange L. G., Riordan J. F., Vallee B. L. Identification of a reactive arginyl residue in horse liver alcohol dehydrogenase. Biochem Biophys Res Commun. 1977 Jul 11;77(1):73–78. doi: 10.1016/s0006-291x(77)80166-6. [DOI] [PubMed] [Google Scholar]
- Kaushal V., Avila D. M., Hardies S. C., Barnes L. D. Sequencing and enhanced expression of the gene encoding diadenosine 5',5'''-P1, P4-tetraphosphate (Ap4A) phosphorylase in Saccharomyces cerevisiae. Gene. 1990 Oct 30;95(1):79–84. doi: 10.1016/0378-1119(90)90416-o. [DOI] [PubMed] [Google Scholar]
- Kohlbrenner W. E., Cross R. L. Efrapeptin prevents modification by phenylglyoxal of an essential arginyl residue in mitochondrial adenosine triphosphatase. J Biol Chem. 1978 Nov 10;253(21):7609–7611. [PubMed] [Google Scholar]
- Koland J. G., O'Brien T. A., Gennis R. B. Role of arginine in the binding of thiamin pyrophosphate to Escherichia coli pyruvate oxidase. Biochemistry. 1982 May 25;21(11):2656–2600. doi: 10.1021/bi00540a012. [DOI] [PubMed] [Google Scholar]
- Konishi K., Fujioka M. Chemical modification of a functional arginine residue of rat liver glycine methyltransferase. Biochemistry. 1987 Dec 15;26(25):8496–8502. doi: 10.1021/bi00399a069. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee P. C., Bochner B. R., Ames B. N. AppppA, heat-shock stress, and cell oxidation. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7496–7500. doi: 10.1073/pnas.80.24.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee J. S., Nimmo H. G. Evidence for an arginine residue at the coenzyme-binding site of Escherichia coli isocitrate dehydrogenase. Biochem J. 1989 Jul 1;261(1):301–304. doi: 10.1042/bj2610301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan A. G., Prescott M. Diadenosine 5',5'"-P1,P4-tetraphosphate in developing embryos of Artemia. Nucleic Acids Res. 1984 Feb 10;12(3):1609–1619. doi: 10.1093/nar/12.3.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie A., Antl W. Diadenosine tetraphosphatase from human leukemia cells. Purification to homogeneity and partial characterization. J Biol Chem. 1983 Apr 10;258(7):4105–4109. [PubMed] [Google Scholar]
- Ogilvie A. Determination of diadenosine tetraphosphate (Ap4A) levels in subpicomole quantities by a phosphodiesterase luciferin--luciferase coupled assay: application as a specific assay for diadenosine tetraphosphatase. Anal Biochem. 1981 Aug;115(2):302–307. doi: 10.1016/0003-2697(81)90009-9. [DOI] [PubMed] [Google Scholar]
- Ogilvie A., Lüthje J., Pohl U., Busse R. Identification and partial characterization of an adenosine(5')tetraphospho(5')adenosine hydrolase on intact bovine aortic endothelial cells. Biochem J. 1989 Apr 1;259(1):97–103. doi: 10.1042/bj2590097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plateau P., Fromant M., Brevet A., Gesquière A., Blanquet S. Catabolism of bis(5'-nucleosidyl) oligophosphates in Escherichia coli: metal requirements and substrate specificity of homogeneous diadenosine-5',5'''-P1,P4-tetraphosphate pyrophosphohydrolase. Biochemistry. 1985 Feb 12;24(4):914–922. doi: 10.1021/bi00325a016. [DOI] [PubMed] [Google Scholar]
- Plateau P., Fromant M., Schmitter J. M., Blanquet S. Catabolism of bis(5'-nucleosidyl) tetraphosphates in Saccharomyces cerevisiae. J Bacteriol. 1990 Dec;172(12):6892–6899. doi: 10.1128/jb.172.12.6892-6899.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plateau P., Fromant M., Schmitter J. M., Buhler J. M., Blanquet S. Isolation, characterization, and inactivation of the APA1 gene encoding yeast diadenosine 5',5'''-P1,P4-tetraphosphate phosphorylase. J Bacteriol. 1989 Dec;171(12):6437–6445. doi: 10.1128/jb.171.12.6437-6445.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott M., Milne A. D., McLennan A. G. Characterization of the bis(5'-nucleosidyl) tetraphosphate pyrophosphohydrolase from encysted embryos of the brine shrimp Artemia. Biochem J. 1989 May 1;259(3):831–838. doi: 10.1042/bj2590831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport E., Zamecnik P. C. Presence of diadenosine 5',5''' -P1, P4-tetraphosphate (Ap4A) in mamalian cells in levels varying widely with proliferative activity of the tissue: a possible positive "pleiotypic activator". Proc Natl Acad Sci U S A. 1976 Nov;73(11):3984–3988. doi: 10.1073/pnas.73.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J. F. Arginyl residues and anion binding sites in proteins. Mol Cell Biochem. 1979 Jul 31;26(2):71–92. doi: 10.1007/BF00232886. [DOI] [PubMed] [Google Scholar]
- Rodriguez del Castillo A., Torres M., Delicado E. G., Miras-Portugal M. T. Subcellular distribution studies of diadenosine polyphosphates--Ap4A and Ap5A--in bovine adrenal medulla: presence in chromaffin granules. J Neurochem. 1988 Dec;51(6):1696–1703. doi: 10.1111/j.1471-4159.1988.tb01147.x. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Further studies on the reactions of phenylglyoxal and related reagents with proteins. J Biochem. 1977 Feb;81(2):403–414. doi: 10.1093/oxfordjournals.jbchem.a131472. [DOI] [PubMed] [Google Scholar]
- Takahashi K. The reaction of phenylglyoxal with arginine residues in proteins. J Biol Chem. 1968 Dec 10;243(23):6171–6179. [PubMed] [Google Scholar]
- Vallejo C. G., Lobaton C. D., Quintanilla M., Sillero A., Sillero M. A. Dinucleosidasetetraphosphatase in rat liver and Artemia salina. Biochim Biophys Acta. 1976 Jun 7;438(1):304–309. doi: 10.1016/0005-2744(76)90246-1. [DOI] [PubMed] [Google Scholar]