Abstract
DNA non-homologous end joining, the major mechanism for the repair of DNA double-strands breaks (DSB) in mammalian cells requires the DNA-dependent protein kinase (DNA-PK), a complex composed of a large catalytic subunit of 460 kDa (DNA-PKcs) and the heterodimer Ku70–Ku80 that binds to double-stranded DNA ends. Mutations in any of the three subunits of DNA-PK lead to extreme radiosensitivity and DSB repair deficiency. Here we show that the 283 C-terminal amino acids of Ku80 introduced into the Chinese hamster ovary cell line CHO-K1 have a dominant negative effect. Expression of Ku(449–732) in CHO cells was verified by northern blot analysis and resulted in decreased Ku-dependent DNA end-binding activity, a diminished capacity to repair DSBs as determined by pulsed field gel electrophoresis and decreased radioresistance determined by clonogenic survival. The stable modifications observed at the molecular and cellular level suggest that this fragment of Ku80 confers a dominant negative effect providing an important mechanism to sensitise radioresistant cells.
INTRODUCTION
Ionising radiation induces several types of DNA lesions, including base damage and DNA single- and double-strand breaks (SSBs and DSBs). DSBs occur at a low frequency (40 per Gy/diploid cell) but their induction closely correlates with radiation-induced cell death (1). In yeast homologous recombination is the major mechanism for DSB repair. In contrast, in mammalian cells the predominant DSB repair system is the DNA non-homologous end joining (NHEJ) pathway, which requires no or little sequence homology. One of the main participants in this pathway is the DNA-PK complex, which consists of two components: a 450 kDa catalytic component, DNA-PKcs, and a heterodimeric protein named Ku. Ku is composed of two tightly associated polypeptides of 70 and 80 kDa (Ku70 and Ku80, respectively) and has double strand end-binding activity thereby targeting the complex to DNA ends (2,3). The discovery of several rodent mutants cell lines defective in DSB repair enabled identification of the genes involved in NHEJ. Of these, three ionising radiation complementation groups of rodent cell lines (IR groups 4, 5 and 7) are extremely hypersensitive to γ-rays and defective in DSB repair (4–6).
The Ku protein was found to be the product of the XRCC5 gene by its ability to complement IR group 5 mutants (5,7). Defects in Ku80 and in the other DNA-PK subunits lead not only to hypersensitivity to ionising radiation, but also to altered V(D)J recombination, a process which is involved in the maturation of lymphocytes and involves a site-specific DSB (8–10). Given the crucial role of the DNA-PK complex in determining the response of cells to radiation, targeting the components of this complex represents an appealing possible route to increase the radiosensitivity of mammalians cells.
The purpose of this study was to identify a dominant negative construct of Ku80. Based on data obtained from structure–function analyses of Ku80, we considered that a construct encompassing the C-terminal region of Ku80 might represent a suitable candidate. Here we demonstrate that a 32 kDa C-terminal fragment of Ku80 has dominant negative activity, decreasing Ku-dependent DNA end-binding activity in vivo and increasing the radiosensitivity of CHO-K1 cells.
MATERIALS AND METHODS
Plasmid construction
A 925 bp cDNA fragment encoding the C-terminal hamster Ku80 protein was cloned in the pcDNA3.1 eukaryotic expression vector (Invitrogen, UK). This vector contains the human cytomegalovirus promoter and the neomycin resistance gene for selection of G418-resistant clones.
Cell lines
The parental radioresistant CHO-K1 cell line and Ku80-defective xrs6 cells were routinely cultured in Nunclon plastic flasks containing MacCoy medium (Gibco) supplemented with 10% foetal calf serum and antibiotics.
Transfection
Transfection of CHO-K1 cells by the plasmid constructs was achieved by a standard CaPO4 precipitation method. CHO-K1 cells were transfected with pcDNA3, containing the Ku(449–732) cDNA [CHO-Ku(449–732)], and with pcDNA3 null and pcDNA3 containing the full-length Ku80 cDNA as controls (CHO-T and CHO-Ku80, respectively). Selection of geneticin-resistant colonies was obtained by adding 0.5–1 mg ml–1 G418 (Sigma, St Louis, MO) to the medium.
Northern blotting
Total RNA was prepared using the guanidinium isothiocyanate–CsCl gradient method. Following centrifugation the RNA was washed with 70% ethanol and precipitated with absolute ethanol. An aliquot of 5 µg total RNA was size separated by electrophoresis in a 1% agarose–formaldehyde gel, transferred overnight to a PALL transfer membrane in 0.15 M ammonium acetate, baked at 80°C for 2 h and hybridised under standard conditions with the 32P-labelled 925 bp C-terminal Ku80 cDNA fragment as probe.
Assay for DNA end-binding activity
DNA end-binding assays were carried out essentially as described previously (3,5). In brief, extracts were prepared by a modification of the method of Scholer et al. (11) and the indicated amounts of protein were incubated with γ-32P-labelled double strand oligonucleotide M1/M2 at room temperature for 30 min and DNA–protein complexes resolved on 4% polyacrylamide gels containing 5% glycerol. Anti-Ku80-4 antibodies were used for supershifting experiments.
Irradiation and clonogenic survival assay
Irradiation was performed using a 137Cs γ-ray source at a dose rate of 1.45 Gy min–1 as previously described (12) Exponentially growing cells were trypsinised, plated and irradiated 4 h later. The linear quadratic model was used for fitting survival curves (13).
Kinetics of DSB repair
Cells were prepared for the DSB repair assay as previously described (14) with the notable exception that cells were irradiated in exponential phase. Briefly, cells were irradiated at 4°C with 30 Gy γ-rays using a 137Cs source as described above and incubated at 37°C for varying times. Cells were then trypsinised on ice and agarose plugs of cells containing 2 × 106 cells ml–1 were prepared. The cells embedded in the plugs were lysed in sarkosyl at 50°C for 38 h. The plugs were subjected to pulsed field gel electrophoresis (CHEF DRIII; Bio-Rad, Hercules, CA) at 14°C to allow separation of DNA fragments in the megabase size region. The fraction of activity released (FAR) is calculated from the activity in the lane relative to that in the lane plus the well (14). The results are presented as percentage FAR remaining (FAR at the specified time/FAR at time 0) and were fitted to the variable repair half-time model previously described (15)
RESULTS
Ku(449–732) is expressed in CHO-K1 cells
CHO-K1 cells were transfected with the plasmid pcDNA3.1 alone or containing the terminal 925 bp of Ku80 cDNA, corresponding to the C-terminal 283 amino acids of the Ku80 protein. G418-resistant colonies obtained were designated CHO-T or CHO-Ku(449–732) cells, respectively. Since we could not find any antibody that cross-reacted with a 32 kDa protein, even using an antibody specifically directed toward the C-terminal fragment of Ku80 (Santa Cruz), expression of the protein fragment was examined by northern blot analysis using the Ku(449–732) cDNA fragment as probe. Figure 1 shows that a transcript of the anticipated size was expressed in CHO-Ku(449–732) but not in CHO-T cells. Expression of a full-length Ku80 transcript in xrs6 cells is consistent with previous findings, although the level of expression appeared lower than in CHO-T cells (16). These results show additionally that expression of Ku(449–732) does not impair expression of full-length Ku80.
Figure 1.

Ku(449–732) fragment mRNA expression in transfected cells. Northern blot analysis using Ku(449–732) mRNA as probe. Total RNA was derived from CHO-K1 cells transfected with either empty vector (CHO-T) or the C-terminal 925 bp of Ku80 cDNA [CHO-Ku(449–732)] or from Ku80-defective xrs6 cells. Total RNA was electrophoresed, transferred to PALL membrane and hybridised as described in Materials and Methods. In the bottom panel 18S and 28S bands are shown as homogeneous RNA transfer controls.
Expression of Ku(449–732) sensitises CHO-K1 cells to ionising radiation
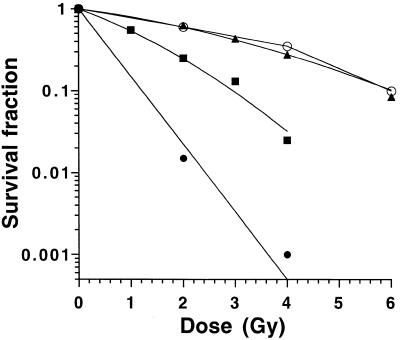
To examine whether expression of the Ku(449–732) fragment impacted upon radiosensitivity, the response of CHO-T, CHO-Ku(449–732) and xrs6 cell lines to radiation was measured using a clonogenic survival assay (Fig. 2). The surviving fractions at 2 Gy (SF2) were 62 and 25% for the CHO-T and CHO-Ku(449–732) cell lines, respectively. The SF2 of the xrs6 cell line, which is known to be extremely radiosensitive, was 1.5%, in agreement with previous published data (5). The radiosensitivity of CHO-Ku(449–732) was measured at various intervals after the initial transfection and cloning and the response to radiation was found to be reproducible after more than 15 cell culture passages (data not shown). We conclude that expression of Ku(449–732) can have a stable dominant negative effect leading to decreased radioresistance of CHO-K1 cells.
Figure 2.
Response of CHO-Ku(449–732) cells to ionising radiation. Cells were exposed to ionising radiation and survival estimated by colony formation after 7 days incubation. CHO-T cells (closed triangles), cell line CHO-Ku(449–732) (closed squares), the Ku80-mutated xrs6 cell line (closed circles) and CHO-K1 cells transfected with full-length Ku80 cDNA (CHO-Ku80, open circles) were used. Experiments were done in quadruplicate and data were fitted to the linear quadratic model. For clarity error bars are not shown.
The survival curve of CHO-K1 cells transfected with full-length Ku80 cDNA is also shown in Figure 2, which shows no variation of radiation sensitivity compared to CHO-T control cells, suggesting that overexpression of Ku80 protein does not change the radioresistance of wild-type cells.
CHO-Ku(449–732) cells have impaired double strand DNA end-binding activity
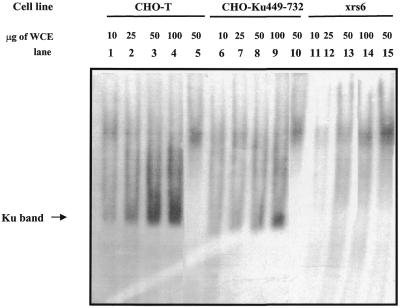
To gain insight into the basis underlying the radiosensitivity of cells, we used an electrophoresis mobility shift assay (EMSA) to examine the double strand DNA end-binding activity. Previous studies have shown that Ku is the major DNA end-binding activity in whole cell extracts. Using CHO-T cells we observed a band representing Ku-dependent DNA end-binding activity since it was supershifted by anti-Ku80 antibodies (Fig. 3, lanes 1–4 and 5 for the supershift) as well as anti-Ku70 antibodies (data not shown). Consistent with this, this band was absent in Ku80-defective xrs6 cells (lanes 11–14). The intensity of this band was decreased in CHO-Ku(449–732) cells compared with equal protein concentrations of extract from CHO-T cells (lanes 6–9). Additionally, there was no evidence for a smaller end-binding product in Ku(449–732)-expressing cells, suggesting that neither Ku(449–732) alone nor a putative Ku(449–732)–Ku70 heterodimer has end-binding activity. We conclude that CHO-Ku(449–732) cells have decreased DNA end-binding activity relative to CHO-T cells, providing an explanation for their increased radiosensitivity.
Figure 3.
DNA end-binding activity in CHO-Ku(449–732) cells. Varying amounts of whole cell extracts (WCE) from CHO-T (lanes 1–5), CHO-Ku(449–732) (lanes 6–10) or xrs6 cells (lanes 11–15) were incubated with [γ-32P]dATP-labelled M1/M2 oligonucleotide probes and separated by polyacrylamide gel electrophoresis. Lanes 1–4, 6–9 and 11–14 represent 10, 25, 50 and 100 µg CHO-T, CHO-Ku(449–732) and xrs6 cell extract, respectively. Lanes 5, 10 and 15 correspond to 50 µg cell extract from CHO-T, CHO-Ku(449–732) and xrs6 cells, respectively, supershifted with anti-Ku80 antibody. To enhance separation the free probe was run to the bottom of the gel and is just visible. The Ku-dependent band, verified by supershifting with anti-Ku80 antibodies, is indicated. There is no evidence of any smaller end-binding proteins in this or other gels run. A weak band of end-binding activity of a size distinct from the Ku band is routinely seen using xrs6 cell extracts. This has the same mobility as the Ku supershifted band. The apparent supershifted band in lane 15 does not represent a supershifted band but this alternative end-binding activity. The magnitude of this supershifted band cannot therefore be attributed solely to supershifted Ku.
DSB rejoining is impaired in CHO-Ku(449–732) cells
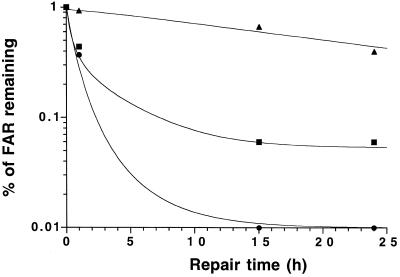
The DSB repair rate of the cells was assessed by measuring the percent FAR remaining as a function of repair time, following exposure to 30 Gy (Fig. 4). After irradiation the percent FAR remaining decreased rapidly up to 1 h, demonstrating fast repair in CHO-T cells, and no unrejoined breaks were detected by 15 h. In contrast, xrs6 cells exhibited a major repair defect leading to 40% residual FAR at 24 h, as previously reported (17). The level of unrejoined breaks in CHO-Ku(449–732) cells appeared to be relatively similar to CHO-T cells after 1 h, whereas it was significantly higher after longer repair times. In particular, when allowing 24 h for repair CHO-Ku(449–732) cells still show a residual FAR of 6%, compared to 0% in the control.
Figure 4.
DSB repair in CHO-Ku(449–732) cells. Cells radiolabelled with [14C]TdR were irradiated with 30 Gy and incubated for varying times and subjected to PFGE. The percentage of FAR remaining in CHO-T (closed triangles), CHO-Ku(449–732) (closed squares) and xrs6 (closed circles) cell lines after up to 24 h for repair is shown. Experiments were done in triplicate and data were fitted to the variable repair half-time model.
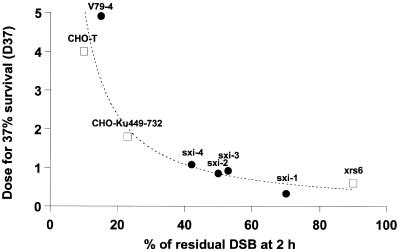
Figure 5 shows that the level of unrejoined DSBs at 2 h post-irradiation correlates with the intrinsic radiosensitivity (dose for 37% survival, D37) when plotted with other radiosensitive rodent mutants such as those of Lee et al. (18) (Fig. 5). This correlation indicates that the D37 is inversely related to the percent of DSBs remaining at 2 h.
Figure 5.
Correlation between intrinsic radiosensitivity and DSB repair defect. Survival data (dose giving 37% survival, D37) were plotted against DSB data (FAR at 2 h) assessed after 2 h for repair. A correlation between D37 and DSB data was obtained with our data (open squares) and those of Lee et al. (18) (closed circles), which involved radiosensitive rodent mutants. Curve fitting was carried out using the inverse function: D37 = 68/percentage remaining DSB (P = 0.04).
DISCUSSION
The purpose of this study was to identify a dominant negative fragment of Ku80 capable of enhancing the radiosensivity of radioresistant cells. Although several studies have described a radiosensitive phenotype due to loss of DNA-PK components in rodent cell lines and knockout mice, the ability to exploit the significant role of DNA-PK in the response to radiation to radiosensitise resistant cells, by expressing a dominant negative construct, has not been previously described. (7,19–21). Several laboratories have undertaken a structure–function analysis of the Ku80 protein. These studies show some discrepancy in characterisation of the region implicated in Ku80–Ku70 heterodimer formation (22–25). Our choice of the Ku80 fragment was influenced by a study carried out using the LexA-based two-hybrid system and an in vitro cDNA expression system, in which a C-terminal 32 kDa fragment of Ku80 was shown to be capable of interacting with Ku70 but the heterodimer formed was incapable of binding to double-stranded DNA ends (22,23). This finding was consistent with another report showing that the minimal region located between amino acids 427 and 531 was sufficient for interaction with Ku70 (24), although it was not in agreement with further work using the GAL4-based two-hybrid system as well as an in vivo system based on immunoprecipitation from transformed cells lacking endogenous Ku80, which identified the central region of Ku80 as responsible for heterodimer formation (23). This latter finding was, however, consistent with the finding that two mutated hamster cell lines (XR-V9B and XR-V15B) have deletions in the central region of Ku80, have altered levels of Ku70 protein and no DNA end-binding activity (20). Finally, a recent study showed that the terminal 178 amino acids were dispensable for interaction with Ku70 and DNA end-binding but were required for the interaction of Ku with the catalytic subunit and for stimulation of kinase activity (25).
Based on these studies, we constructed a cDNA fragment encompassing the terminal 925 bp of Ku80 anticipating that the truncated 32 kDa protein derived might be able to bind the Ku70 subunit, but that the resulting Ku(449–732)–Ku70 complex would be incapable of binding DNA ends. Northern blot analysis verified the expression of Ku(449–732) mRNA in CHO-Ku(449–732) cells (Fig. 1).
Although Ku(449–732) mRNA was reproducibility detectable, we were not able to detect the corresponding Ku(449–732) protein, even when using monoclonal antibodies supposed to bind the C-terminal fragment of Ku80. This lack of detection of Ku(449–732) protein might be due to changes in its conformation, which perhaps would make it undetectable using the available antibodies.
Expression of the Ku(449–732) cDNA in CHO-K1 cells resulted in decreased DNA end-binding activity (Fig. 3), suggesting that at the protein level the Ku(449–732) fragment was able to markedly influence Ku function. This result obtained in vivo is consistent with the in vitro functional analysis of Ku(449–732) done by Wu and Lieber (22). It is notable, however, that some residual DNA end-binding activity remains in CHO-Ku(449–732) cells, suggesting that functional Ku70–Ku80 complexes do form in these cells. Our results suggest that Ku(449–732) competes with Ku80 for Ku70 binding and therefore decreases the level of functional Ku70–Ku80 heterodimers. The impaired DNA end-binding is likely to decrease DNA-PK activity. An attempt was made to address this issue, however, we were not able to demonstrate significant variations in DNA-PK activity. This is in agreement with previous studies showing that rodent cells have low basic DNA-PK activity, making quantitative estimation of DNA-PK activity difficult in these cells (25).
Interestingly, the radiosensitivity of CHO-Ku(449–732) cells was intermediate between CHO-T, expressing the vector alone, and Ku80-defective xrs6 cells (26), which is compatible with the moderate DSB repair deficiency (6% of unrepaired DSB at 24 h). The differences observed in residual DSB joining among a range of cell lines impaired in NHEJ correlates with the cell survival data via an inverse relationship between the amount of unrepaired DSB at 2 h and D37. The CHO-Ku(449–732) cell line is remarkable in that its radiation-induced response is intermediate between controls and hypersensitive cell lines. However, it should also be considered that the DSB assay provides an estimation of unrejoining but not misrepair, which may also account for cell killing.
The intermediate level of radiosensitivity and DSB repair observed in CHO-Ku(449–732) cells is compatible with the hypothesis of the combined presence of both truncated Ku(449–732)–Ku70 heterodimers and residual wild-type Ku80–Ku70 heterodimers. The EMSA analysis did not reveal the presence of a smaller end-binding product in Ku(449–732)-expressing cells, suggesting that the putative Ku(449–732)–Ku70 heterodimer does not have end-binding activity.
One other study has reported a dominant negative construct of Ku80, but only a minor impact on radiation sensitivity of rodent cells was seen and neither in vivo DNA end-binding activity nor DSB repair studies were performed (27).
Our study is the first showing that DSB repair and DNA end-binding activity of Ku80 can be modified by a dominant negative approach.
In conclusion, our results showed that expression of a truncated C-terminal Ku80 fragment in CHO-K1 cells results in decreased DNA end-binding activity in vivo, which impairs DSB rejoining and increases radiosensitivity, thus providing a useful dominant negative construct. We have recently shown that decreased Ku80 protein expression can lead to increased radiosensitivity in human cells (28), thus this dominant negative approach could provide another route to radiosensitise human cancer cells. The ability to enhance radiosensitivity of radioresistant cells has considerable potential application. If tissue-specific expression of such a construct were established, then increased radiosensitivity could be achieved in specific tissues, thus potentially enhancing the efficacy of radiotherapy.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr J. Bénard and Dr E. A. Hendrickson. This work was supported by the Fondazione San Salvatore, Switzerland, Association de la Recherche sur le Cancer (ARC) grant no. 9692 and an individual grant (2000) from the Ligue Nationale Contre le Cancer, GEFLUC, Institut Fédératif de Recherche IFR no. 54, Villejuif, France.
REFERENCES
- 1.MacMillan T.J. and Steel,G.G. (1993) Molecular Aspects of Radiation Biology. Edward Arnold Publishers, Boston, MA, pp. 211–224.
- 2.Dvir A., Peterson,S.R., Knuth,M.W., Lu,H. and Dynan,W.S. (1992) Proc. Natl Acad. Sci. USA, 89, 11920–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottlieb T.M. and Jackson,S.P. (1993) Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- 4.Jeggo P.A., Tesmer,J. and Chen,D.J., (1991) Mutat. Res., 254, 125–133. [DOI] [PubMed] [Google Scholar]
- 5.Taccioli G.E., Gottlieb,T.M., Blunt,T., Priestley,A., Demengeot,J., Mizuta,R., Lehemann,A.R., Alt,F.W., Jackson,S.P. and Jeggo,P.A. (1994) Science, 265, 1442–1445. [DOI] [PubMed] [Google Scholar]
- 6.Thacker J. and Wilkinson,R.E. (1991) Mutat. Res., 254, 135–142. [DOI] [PubMed] [Google Scholar]
- 7.Boubnov N.V., Hall,K.T., Willis,Z., Sang,E.L., Dong,M.H., Benjamin,D.M., Pulaski,C.R., Band,H., Reeves,W., Hendrickson,E.A. and Weaver,D.T. (1995) Proc. Natl Acad. Sci. USA, 92, 890–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taccioli G.E., Rathbun,G., Oltz,E., Stamato,T., Jeggo,P.A. and Alt,F.W. (1993) Science, 260, 207–210. [DOI] [PubMed] [Google Scholar]
- 9.Zhu C.M., Bogue,D.S., Lim,D.S., Hasty,P. and Roth,D.B. (1996) Cell, 86, 379–389. [DOI] [PubMed] [Google Scholar]
- 10.Nussenzweig A., Chen,C., da Costa Soares,V., Sanchez,M., Sokol,K., Nussenzweig,M.C. and Li,G.C. (1996) Nature, 382, 551–555. [DOI] [PubMed] [Google Scholar]
- 11.Scholer H., Haslinger,A., Heguy,A., Holtgreve,H. and Karin,M. (1986) Science, 232, 76–80. [DOI] [PubMed] [Google Scholar]
- 12.Badie C., Iliakis,G. and Foray,N. (1995) Radiat. Res., 144, 26–35. [PubMed] [Google Scholar]
- 13.Fertil B., Dertinger,H., Courdi,A. and Malaise,E.P. (1984) Radiat. Res., 99, 73–84. [PubMed] [Google Scholar]
- 14.Foray N., Priestley,A., Alsbeih,G., Badie,C., Capulas,E.P., Arlett,C.F. and Malaise,E.P. (1997) Int. J. Radiat. Biol., 72, 271–283. [DOI] [PubMed] [Google Scholar]
- 15.Foray N., Badie,C., Alsbeih,G., Fertil,B. and Malaise,E.P. (1996) Radiat. Res., 146, 53–60. [PubMed] [Google Scholar]
- 16.Singleton B.K, Priestley,A., Steingrimsdottir,H., Gell,D., Blunt,T., Jackson,S.P, Lehmann,A.R. and Jeggo,P.A. (1997) Mol. Cell. Biol., 17, 1264–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F., Peterson,S.R., Story,M.D. and Chen,D. (1996) Mutat. Res., 362, 9–19. [DOI] [PubMed] [Google Scholar]
- 18.Lee S.E., Pulasky,C.R., He,D.M., Benjamin,D.M., Voss,M., Um,J. and Hendrickson,E.A. (1995) Mutat. Res., 336, 279–291. [DOI] [PubMed] [Google Scholar]
- 19.Li G.C., Ouyang,X.L., Nagasawa,J.B., Little,J.B., Chen,D.J., Ling,C.C., Fuks,Z. and CordonCardo,C. (1998) Mol. Cell, 2, 1–8. [DOI] [PubMed] [Google Scholar]
- 20.Errami A., Smider,V., Rathmell,W., He,D., Hendrickson,E., Zdzienicka,M. and Chu,G., (1996) Mol. Cell. Biol., 16, 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross G.M., Eady,J.J., Mithal,N.P., Bush,C., Steel,G.G., Jeggo,P.A. and McMillan,T.J. (1995) Cancer Res., 55, 1235–1238. [PubMed] [Google Scholar]
- 22.Wu X. and Lieber,M.R. (1996) Mol. Cell. Biol., 16, 5186–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cary R.B., Chen,F., Shen,Z. and Chen,D.J. (1997) Nucleic Acids Res., 26, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osipovich O., Durum,S.K. and Muegge,K. (1997) J. Biol. Chem., 272, 27259–27265. [DOI] [PubMed] [Google Scholar]
- 25.Singleton B.K., Torres-Arzayus,M.I., Rottinghaus,S.T., Taccioli,G.E. and Jeggo,P.A., (1999) Mol. Cell. Biol., 19, 3267–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeggo P.A. and Kemp,L.M. (1983) Mutat. Res., 112, 313–327. [DOI] [PubMed] [Google Scholar]
- 27.Osipovich O., Duhe,R.J., Hasty,P., Durum,S.K. and Muegge,K. (1999) Biochem. Biophys. Res. Commun., 261, 802–807. [DOI] [PubMed] [Google Scholar]
- 28.Marangoni E., Le Romancer,M., Foray,N., Muller,C., Douc-Rasy,S., Vaganay,S., Abdulkarim,B., Deutsch,E., Barrois,M., Calsou,P., Bernier,J., Salles,B. and Bourhis,J. (2000) Cancer Gene Ther., 7, 339–345. [DOI] [PubMed] [Google Scholar]