Abstract
Background
It is well-known that nanoparticles sediment, diffuse and aggregate when dispersed in a fluid. Once they approach a cell monolayer, depending on the affinity or “stickiness” between cells and nanoparticles, they may adsorb instantaneously, settle slowly – in a time- and concentration-dependent manner - or even encounter steric hindrance and rebound. Therefore, the dose perceived by cells in culture may not necessarily be that initially administered. Methods for quantifying delivered dose are difficult to implement, as they require precise characterization of nanoparticles and exposure scenarios, as well as complex mathematical operations to handle the equations governing the system dynamics. Here we present a pipeline and a graphical user interface, DosiGUI, for application to the accurate nano-dosimetry of engineered nanoparticles on cell monolayers, which also includes methods for determining the parameters characterising nanoparticle-cell stickiness.
Results
We evaluated the stickiness for 3 industrial nanoparticles (TiO2 – NM-105, CeO2 – NM-212 and BaSO4 – NM-220) administered to 3 cell lines (HepG2, A549 and Caco-2) and subsequently estimated corresponding delivered doses. Our results confirm that stickiness is a function of both nanoparticle and cell type, with the stickiest combination being BaSO4 and Caco-2 cells. The results also underline that accurate estimations of the delivered dose cannot prescind from a rigorous evaluation of the affinity between the cell type and nanoparticle under investigation.
Conclusion
Accurate nanoparticle dose estimation in vitro is crucial for in vivo extrapolation, allowing for their safe use in medical and other applications. This study provides a computational platform – DosiGUI – for more reliable dose-response characterization. It also highlights the importance of cell-nanoparticle stickiness for better risk assessment of engineered nanomaterials.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12989-024-00607-4.
Keywords: In vitro nano-dosimetry, Delivered dose, Cell-nanoparticle stickiness, In silico modelling, Graphical user interface
Introduction
The unique physicochemical properties of nanomaterials (NMs) are of interest for a wide range of applications [1, 2]. For this reason, several NMs are now produced industrially and are referred to as engineered nanomaterials (ENMs). In the biomedical field, ENMs are extensively employed in the form of engineered nanoparticles (ENPs) for modulating mechanical characteristics of scaffolds, delivering drugs and genes, and localised sensing, or as nano-transducers for directing cell behaviour in targeted therapy [3–5].
However, the very same features that make ENPs so attractive, also confer them potential toxicity [2, 6]. For this reason, an essential point in nanotoxicology is the characterization of biological effects induced by ENPs in human tissues and organs as a function of their dose, along with the investigation of mechanisms triggering their cytotoxicity. Cell cultures in vitro represent one of the most common methods for dose assessment, although the extrapolation of dose-response behaviour to the in vivo context is still a challenge [7, 8].
A proper definition of the cytotoxic “potential” of a specific ENP would require correlating harmful effects to the cumulative mass of material per unit area effectively interacting with cells during an exposure test, or what we refer to as the “delivered dose”, that is the mass of nanoparticles internalized, adsorbed by or near the cell culture per unit area of the cell-seeded surface [9, 10]. Cells in culture are indeed known to be sensitive to nanoparticles (and their dissolved ions) whether they be internalized, in contact with their surface or in their immediate vicinity. The cytotoxic effects depend on several intrinsic factors, such as nanoparticle (NP) composition, surface chemistry/defects, size distribution, partial solubility and effective density. Cytotoxicity is also conditioned by extrinsic factors (i.e., related to experimental conditions) which ultimately determine the rates of physicochemical phenomena occurring in the system [11]. Although the mechanisms by which NPs induce cellular damage have yet to be fully understood (e.g., uptake kinetics, oxidative stress, complexation phenomena) [12, 13], there is plenty of evidence to suggest that only a fraction of the total amount of material initially administered within the suspension (i.e., the nominal dose, defined as the ratio of the total NP mass in the suspension to the cross-section area of the vial) is perceived by cells (i.e., the delivered dose). While the nominal dose is constant over time and independent of the exposure configuration, the delivered dose is strongly dependent on several parameters, including intrinsic features of NPs such as the agglomeration potential (which in turn determines their size distribution and effective density) [14] as well as extrinsic characteristics: e.g., the NP dispersion preparation method [15], the time of exposure and the geometry of the experimental set-up [9, 16]. It also depends on the affinity or “stickiness” between cells and NPs, a parameter which is still ignored and undervalued. For instance, when cell-NP stickiness is high, NPs adsorbed by the cells are effectively removed from the suspension; consequently, the downward diffusive flux is increased, and this may significantly increase the delivered dose [17]. However, due to experimental difficulties in measuring the delivered dose, most reports on in vitro dosimetry and nanotoxicology refer to the nominal dose. Thus, toxicity may be underestimated (i.e., harmful biological effects are associated with doses higher than those which effectively cause them), making it difficult to properly assess ENP hazard and define reference doses for their safe use [11, 16].
Given this background, in silico models are a crucial tool for estimating the actual amount of ENPs reaching tissues and cells in specific configurations and performing more accurate dose-response characterizations [9, 10]. In particular, the two best-known models in this field are the in vitro sedimentation, diffusion, dissolution and dosimetry (ISD3) model and the distorted grid (DG) model [17, 18].
Both models require the physicochemical characteristics of NPs and the associated stickiness of the cell monolayer as inputs. Since the stickiness is difficult to quantify, ISD3 assumes either instantaneous or zero adsorption (maximal or zero stickiness, respectively) while DG suggests a reflective cell boundary (zero stickiness). The ISD3 and DG models are distributed as MATLAB codes which require a licensed computing environment and basic programming knowledge for entering input parameters and obtaining dose predictions, and only a MATLAB-based cloud application of the DG model has been proposed [19]. As a result, the valuable support that computational models could offer to in vitro dosimetry is unexploited, and their benefits to NM hazard assessment are limited.
To address the lack of methods for assessing and quantifying stickiness and promote the routine use of dosimetry models for accurate prediction of delivered doses in in vitro systems, we have developed an in vitro-in silico pipeline leveraged on DosiGUI, an open-source software platform with an easy-to-use graphical user interface (GUI). DosiGUI integrates the ISD3 and DG models, facilitating their comparison and enabling the identification of the more suitable one for a given NP and experimental configuration. Furthermore, as not all users may know the appropriate value of the model-specific stickiness parameter for a particular setup, DosiGUI includes a knob which allows setting the stickiness at five levels between zero and maximal.
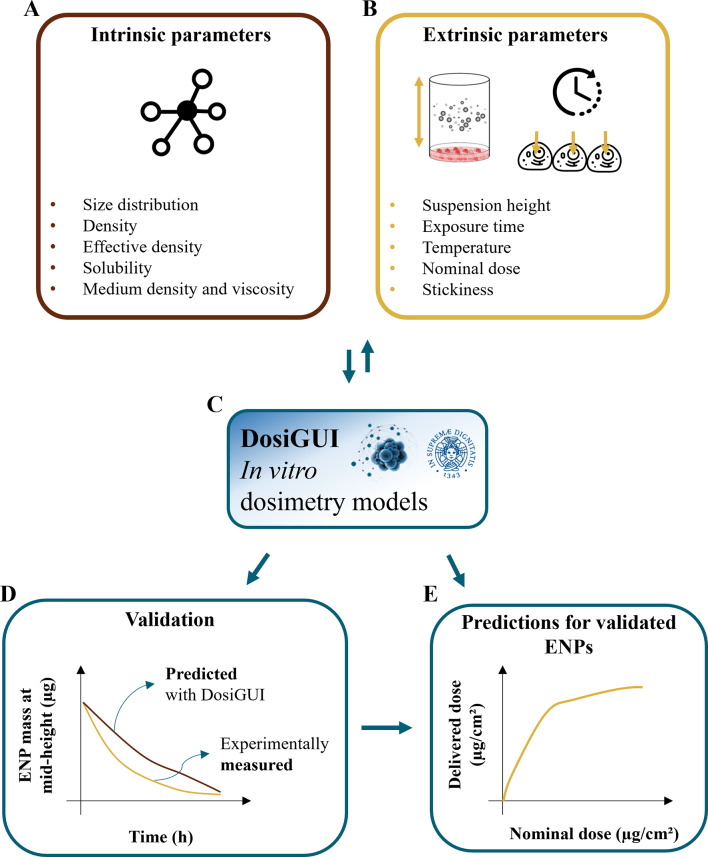
The pipeline (depicted in Fig. 1) includes methods for validating the models for different ENPs, through the use of reference highly sticky and non-sticky bottom surfaces. Following this pipeline, we evaluated DosiGUI’s performance for three ENPs (TiO2 – NM-105, CeO2 – NM-212 and BaSO4 – NM-220) by fitting predictions to experimental data. Finally, as a proof of concept, we applied the pipeline to estimate the dose of the three ENPs delivered to three cell lines – HepG2, A549 and Caco-2 – in a standard exposure scenario. The stickiness of the cell monolayer was identified for each cell-ENP pair using statistical methods for comparing the predicted dose with corresponding experimental data.
Fig. 1.
The in silico-in vitro pipeline leveraging DosiGUI. (A) Physicochemical characterization of ENPs (i.e., intrinsic parameters) using established methods [20, 21, 26, 27] and (B) extrinsic parameters are crucial for optimizing the reliability of delivered dose predictions and improving dose-response characterization. (C) The input dataset is fed to DosiGUI for running simulations. The same dataset can be used for both ISD3 and DG models, and the outcome can be exploited for identifying the ENP-cell stickiness. (D) DosiGUI is validated for specific ENPs by comparing predictions and experimental measurements of a fully characterized test system. (E) Once validated, DosiGUI predictions of delivered dose for a given ENP-cell combination can be extrapolated to various exposure scenarios
Results
DosiGUI validation
The goodness of predictions performed running ISD3 and DG in DosiGUI was assessed as a validation of the software platform. We evaluated to what extent model predictions correlate with corresponding ENP mass fractions measured in cylindrical suspension columns with respectively zero and maximally sticky bottom surfaces (see Materials and Methods, section DosiGUI validation for details).
Table 1 shows how profiles of mid-height mass fraction over time predicted by the two models correlate with experimental data from inductively coupled plasma optical emission spectroscopy (ICP-OES) measurements. Examples of corresponding correlation fittings are reported in Figure A2.
Table 1.
Results of the correlation analysis between measured and simulated mid-height mass fraction profiles over time
| DG | ISD3 | ||||
|---|---|---|---|---|---|
| ENP | Boundary condition |
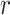
|
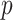
|
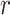
|
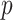
|
| TiO2 (NM-105) | Maximal stickiness | 0.995 | 0.178 | 0.987 | 0.002 |
| TiO2 (NM-105) | Zero stickiness | 0.690 | 0.042 | 0.637 | 0.033 |
| CeO2 (NM-212) | Maximal stickiness | 0.942 | 0.290 | 0.826 | 0.045 |
| CeO2 (NM-212) | Zero stickiness | 0.908 | 0.022 | 0.957 | 0.013 |
| BaSO4 (NM-220) | Maximal stickiness | 0.999 | 0.016 | 0.999 | 0.001 |
| BaSO4 (NM-220) | Zero stickiness | 0.622 | 0.372 | 0.357 | 0.098 |
Correlation analysis for both maximally sticky and non-sticky bottom. The Pearson’s coefficient (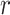 ) and the p-value of the extra sum-of-squares F-test on the slope of the regression line (
) and the p-value of the extra sum-of-squares F-test on the slope of the regression line (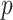 ) are reported as average values of those obtained for the three nominal doses administered in each case (see Figure A2 for examples of the corresponding plots of the correlation fitting)
) are reported as average values of those obtained for the three nominal doses administered in each case (see Figure A2 for examples of the corresponding plots of the correlation fitting)
Previous reports have demonstrated the robustness of ISD3 for silver NPs (Ag – NM-300 K) [10]. However, for the three ENPs studied here, the mass fractions over time at mid-height are more reliably estimated by the DG model. In fact, for both the sticky and reflective boundary conditions, the DG profiles showed a significant correlation (r ≥ 0.6) with measured mid-height mass fraction, along with statistically matching numerical values (p ≥ 0.01).
Cell-ENP stickiness
The DG model implements stickiness as a Langmuir isotherm adsorption (see Eq. (1)) fully characterized by the dissociation constant 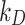 (mol L-1) [17], which is inversely proportional to stickiness. In its original version [18], ISD3 instead accounts for a binary bottom condition (that is instantaneous adsorption or full reflection of NPs), purposely adapted here to allow for a tunable stickiness through the parameter
(mol L-1) [17], which is inversely proportional to stickiness. In its original version [18], ISD3 instead accounts for a binary bottom condition (that is instantaneous adsorption or full reflection of NPs), purposely adapted here to allow for a tunable stickiness through the parameter 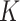 (m) – see Eq. (2). Here too, the higher the parameter value, the lower the NP-cell stickiness.
(m) – see Eq. (2). Here too, the higher the parameter value, the lower the NP-cell stickiness.
The quantity of ENP mass adsorbed over time by cell monolayers in 96-well plates was measured using ICP-OES and compared to that predicted by DG and ISD3 in order to identify meaningful 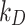 and
and 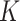 values for all of the ENP-cell type combinations considered. Table 2 shows optimal values of the stickiness parameters (i.e.,
values for all of the ENP-cell type combinations considered. Table 2 shows optimal values of the stickiness parameters (i.e., 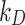 and
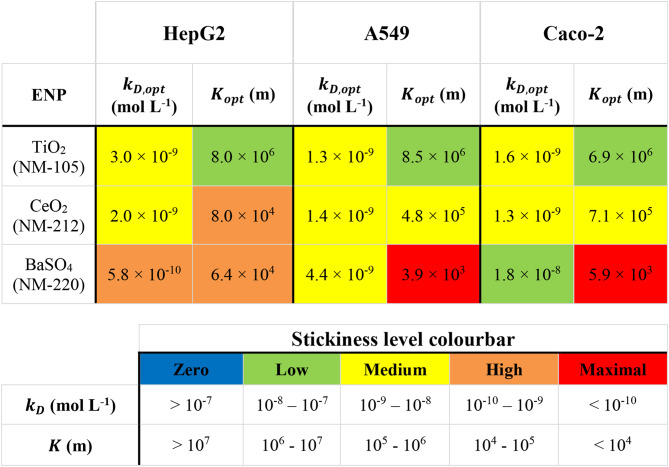
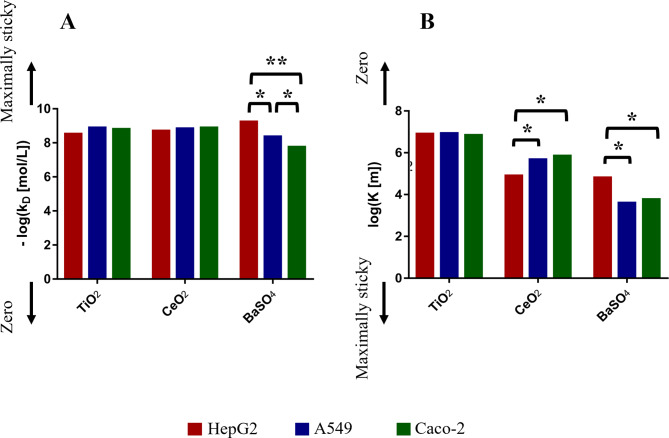
and 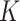 ) and the corresponding stickiness level of tested cell phenotypes with respect to each of the three ENPs analysed. For both models, the stickiness values obtained differ for each ENP, underlining that cell uptake is NP dependent. Vice versa, as clearly depicted in Fig. 2, relevant differences can also emerge among the adsorptive behaviours of the three cell phenotypes for a specific ENP, indicating that stickiness and hence the delivered dose also depends on cell type. Furthermore, it is worth noting that slightly different stickiness “hierarchies” arise based on
) and the corresponding stickiness level of tested cell phenotypes with respect to each of the three ENPs analysed. For both models, the stickiness values obtained differ for each ENP, underlining that cell uptake is NP dependent. Vice versa, as clearly depicted in Fig. 2, relevant differences can also emerge among the adsorptive behaviours of the three cell phenotypes for a specific ENP, indicating that stickiness and hence the delivered dose also depends on cell type. Furthermore, it is worth noting that slightly different stickiness “hierarchies” arise based on 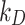 or
or 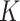 . For instance, using the DG model, Caco-2 cells are the least adsorptive phenotype for BaSO4 (NM-220), while according to the ISD3 model, they have the highest uptake for this ENP (Table 2).
. For instance, using the DG model, Caco-2 cells are the least adsorptive phenotype for BaSO4 (NM-220), while according to the ISD3 model, they have the highest uptake for this ENP (Table 2).
Table 2.
Optimal values for stickiness parameters
Stickiness parameters determining the insoluble ENP adsorption of the three cell phenotypes, identified for the DG (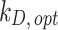 ) and ISD3 (
) and ISD3 (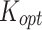 ) models. The colourbar can be used to match the identified parameter values with the five stickiness levels (zero, low medium, high and maximal)
) models. The colourbar can be used to match the identified parameter values with the five stickiness levels (zero, low medium, high and maximal)
Fig. 2.
Optimal values for stickiness parameters describing the ENP adsorption of the three cell phenotypes. Stickiness parameters identified for: (A) the DG model (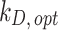 ) and (B) the ISD3 model (
) and (B) the ISD3 model (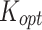 ). For the sake of clarity, bar plots are logarithmically scaled, and differences in terms of stickiness level are highlighted (*one level; **two levels)
). For the sake of clarity, bar plots are logarithmically scaled, and differences in terms of stickiness level are highlighted (*one level; **two levels)
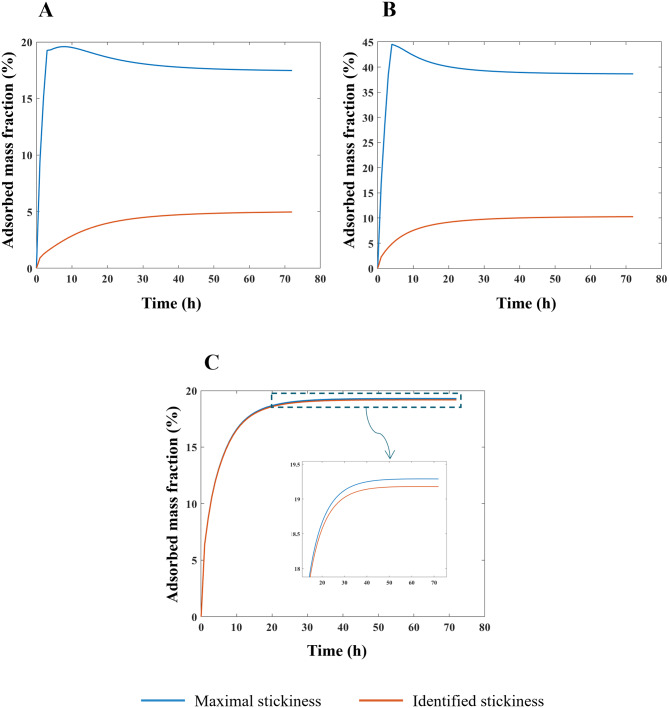
In Fig. 3, the mass fraction adsorbed over time by HepG2 cells for each ENP is shown for two cases: (i) 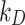 = 0 mol L-1 (maximal stickiness); (ii) optimal
= 0 mol L-1 (maximal stickiness); (ii) optimal 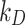 values obtained from the DG model (Table 2). A similar figure is reported in the Additional File 1 (AF) for the ISD3 model (Figure A4). The figures highlight the importance of characterizing the boundary stickiness to obtain meaningful estimates of delivered dose. Using the optimal stickiness, significantly lower adsorbed mass fractions for TiO2 (NM-105) and CeO2 (NM-212) are observed (Fig. 3A and B). On the other hand, the two curves in Fig. 3C for BaSO4 (NM-220) are similar. In fact, as reported in Table 2, this ENP is highly adsorbed on the HepG2 monolayer.
values obtained from the DG model (Table 2). A similar figure is reported in the Additional File 1 (AF) for the ISD3 model (Figure A4). The figures highlight the importance of characterizing the boundary stickiness to obtain meaningful estimates of delivered dose. Using the optimal stickiness, significantly lower adsorbed mass fractions for TiO2 (NM-105) and CeO2 (NM-212) are observed (Fig. 3A and B). On the other hand, the two curves in Fig. 3C for BaSO4 (NM-220) are similar. In fact, as reported in Table 2, this ENP is highly adsorbed on the HepG2 monolayer.
Fig. 3.
Predictions of adsorbed mass fractions over time. Mass fractions adsorbed by HepG2 cells expressed as a percentage of the total amount of ENP mass in the suspension, obtained running the DG model within DosiGUI and assuming a nominally administered dose of 78.1 µg/cm2. Comparison between the case of a maximally sticky bottom (blue line) and the optimal stickiness identified using the pipeline (orange line) for: (A) TiO2 (NM-105), (B) CeO2 (NM-212) and (C) BaSO4 (NM-220)
Prediction of the delivered dose
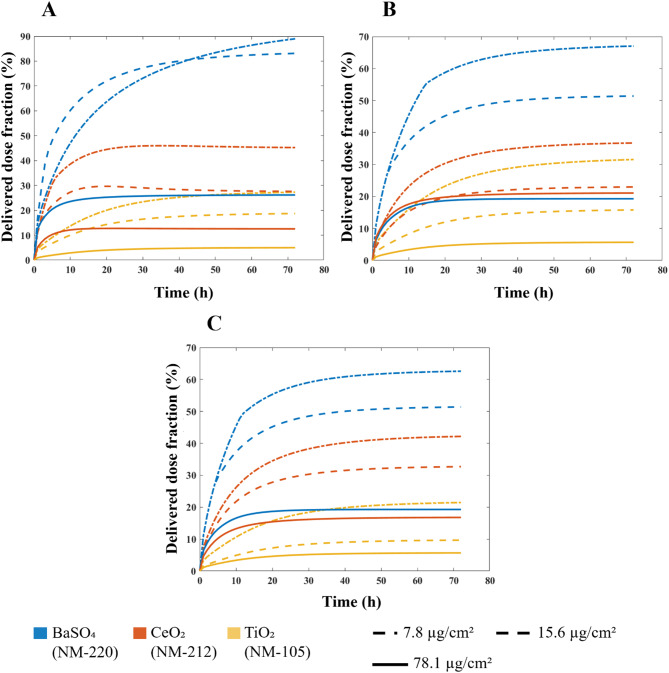
The optimal values of stickiness parameters for each ENP-cell type pair were used to predict the doses delivered to HepG2, A549 and Caco-2 cells, starting from different nominal doses. For the sake of comparison, the delivered dose was expressed as percentage of the nominal one (referred to as delivered dose fraction). Figure 4 reports such predictions; the graphs refer to simulations carried out with the DG model, since it proved to be the more accurate in replicating the dynamics of the three ENPs. Note that, in the nanotoxicology literature, the nominal dose is typically reported, but our data show that, even for long exposure times, cells do not perceive all of the ENPs administered, and the delivered dose reaches a threshold.
Fig. 4.
Predictions of delivered dose. Dose fraction (i.e., delivered dose/nominal dose, percentage) delivered to (A) HepG2 cells, (B) A549 cells and (C) Caco-2 cells over time, obtained running the DG model within DosiGUI
Corresponding predictions by the ISD3 model are shown in the AF (Figure A5). The delivered dose tends to a threshold at a slower rate than using the DG model except for BaSO4 (NM-220), which is the stickiest ENP with respect to any cell type according to the ISD3 model.
In addition, the delivered dose profiles with respect to the nominally administered dose estimated at different time points using the DG and ISD3 models are reported in Figures A6 and A7, respectively. As expected, for each ENP-cell type pair, the delivered dose monotonically increases as a function of the nominal one.
Discussion
Accurately determining the amount of ENPs that cells, tissues or organs are exposed to in specific scenarios (i.e., the actual delivered dose) is a key step towards safely tapping the full potential of ENMs in biomedicine. However, due to technical limitations and the complexity of nano-dosimetry, toxicologists still struggle to provide accurate dose estimations even in typical cell culture experiments. This may result in a misleading characterization of ENP toxicity and consequent hazard assessment.
To tackle these issues, we present an in silico-in vitro pipeline for application to the accurate assessment of delivered doses in monolayer cell cultures. The approach relies on DosiGUI, a purposely developed open-source, multi-model graphical user interface which embeds the two best-known (but unfortunately not widely used) in silico models for computational nano-dosimetry, with the integration of new methods for the identification of parameters quantifying the cell-NP adsorption or stickiness. In fact, the adsorption kinetics of NPs on cells strongly impact on the delivered dose (as highlighted in Fig. 3) but has often been neglected.
As a proof of concept, we demonstrated the robustness of the approach for three ENPs (TiO2 – NM-105, CeO2 – NM-212), BaSO4 – NM-220) with negligible solubility. Starting from physicochemical characterization reported by Wohlleben’s group [20, 21], the reliability of simulations performed using DosiGUI was validated experimentally reproducing two reference boundaries with a maximally adsorptive and a purely reflective surface, respectively. This allows identifying the more suitable model to simulate the adsorption dynamics of each ENP. Following this first validation step, the stickiness parameters (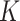 or
or 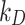 ) describing the affinity between ENPs and three different cell types were identified. The results confirmed that the boundary stickiness is a specific feature of the ENP-cell type combination. In fact, the values of kinetic constants differ with respect to both cell type and ENP, indicating that each cell type interacts differently with each ENP (Table 2). Thus, setting the right stickiness is a crucial step for accurate delivered dose estimation.
) describing the affinity between ENPs and three different cell types were identified. The results confirmed that the boundary stickiness is a specific feature of the ENP-cell type combination. In fact, the values of kinetic constants differ with respect to both cell type and ENP, indicating that each cell type interacts differently with each ENP (Table 2). Thus, setting the right stickiness is a crucial step for accurate delivered dose estimation.
Having characterized the adsorptive behaviour of the three phenotypes, we applied DosiGUI to predict the delivered dose in each ENP-cell type combination in a standard in vitro test configuration. As expected, the simulations show that only a fraction of the administered ENPs interacts with cells, with a threshold effect emerging either for long exposure times or at high nominal doses (Fig. 4 and A4). This is because the adsorption of sedimenting ENPs is a surface occupancy-driven mechanism, which is explicitly modelled in the DG model (see Eq. (1) and section AF3) and may be the reason why its predictions better correlate with experimental data for insoluble ENPs. In fact, for the three ENPs studied in this work, the DG model resulted in a better fit between experimental and computed data (Table 1). However, the stickiness “hierarchies” with respect to both ENP and cell type depend on the model used to identify the empirical constants (e.g., according to the ISD3 model, BaSO4 and Caco-2 cells are maximally sticky, while simulations using the DG model show that the same ENP-cell pair has low stickiness). We underline that such discrepancies are expected since the two models use different analytical formulations for the bottom boundary conditions (see the Materials and Methods, subsection Basic principles of integrated dosimetry models). Thus, 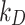 and
and 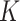 values cannot strictly be compared. In addition, despite its better performance overall, the DG’s Langmuir adsorption function (Eq. (1)) is better suited to short exposure times (i.e., ≤ 24 h), when cell adsorption is effectively limited by the steric occupancy of settling ENPs, which saturate the monolayer surface. For longer time windows, additional mechanisms – leading to mass uptake/internalization – may reduce occupancy, allowing more particles to adhere onto cells. In such a situation, the adsorption kinetics are likely to be better approximated by the ISD3 model. Combining these two approaches for modelling NP adsorption by cells could be used to further improve the accuracy of dose estimation.
values cannot strictly be compared. In addition, despite its better performance overall, the DG’s Langmuir adsorption function (Eq. (1)) is better suited to short exposure times (i.e., ≤ 24 h), when cell adsorption is effectively limited by the steric occupancy of settling ENPs, which saturate the monolayer surface. For longer time windows, additional mechanisms – leading to mass uptake/internalization – may reduce occupancy, allowing more particles to adhere onto cells. In such a situation, the adsorption kinetics are likely to be better approximated by the ISD3 model. Combining these two approaches for modelling NP adsorption by cells could be used to further improve the accuracy of dose estimation.
We should also point out that the contribution of stickiness to delivered dose estimation is likely to be NM dependent. That is, stickiness is less important in ENPs with high effective density and high size values, since gravitational settling will probably predominate over affinity-related cell-material interactions. On the other hand, low-density, small ENPs whose downward motility depends more on diffusion-related phenomena will likely be more influenced by concentration gradients and cell uptake kinetics.
Whenever available, the use of exact numerical values of 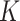 or
or 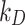 is recommended. However, identifying the stickiness parameters is not a straightforward procedure. Given that different ENP-cell type combinations may exhibit similar adsorption kinetics, in DosiGUI we propose a simplified description of the adsorption behaviour of cells through a knob, which allows choosing among five discrete stickiness levels (zero, low, medium, high, maximal) encompassing the entire range of possible
is recommended. However, identifying the stickiness parameters is not a straightforward procedure. Given that different ENP-cell type combinations may exhibit similar adsorption kinetics, in DosiGUI we propose a simplified description of the adsorption behaviour of cells through a knob, which allows choosing among five discrete stickiness levels (zero, low, medium, high, maximal) encompassing the entire range of possible 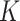 or
or 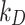 values. We assume therefore that, although the numerical values of these empirical parameters corresponding to different configurations do not exactly match, their stickiness can be reasonably approximated by the same discrete level. Note also that the empirical parameters
values. We assume therefore that, although the numerical values of these empirical parameters corresponding to different configurations do not exactly match, their stickiness can be reasonably approximated by the same discrete level. Note also that the empirical parameters 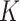 and
and 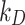 are used to describe the adsorption behaviour because mechanistic knowledge about specific interaction phenomena funneling cellular adhesion and internalization of ENPs is lacking. The integration of tools such as DosiGUI with the development of more comprehensive models of the interaction kinetics between ENPs and cells should be a focus of future, multi-disciplinary studies, allowing mechanistic rather than empirical predictions of ENP-cell interaction.
are used to describe the adsorption behaviour because mechanistic knowledge about specific interaction phenomena funneling cellular adhesion and internalization of ENPs is lacking. The integration of tools such as DosiGUI with the development of more comprehensive models of the interaction kinetics between ENPs and cells should be a focus of future, multi-disciplinary studies, allowing mechanistic rather than empirical predictions of ENP-cell interaction.
This study provides an important contribution to more accurate dose-response characterization of NPs, which can be applied to the improvement of ENP safety and risk assessment. DosiGUI should facilitate the application of in silico tools in nanotoxicology, encouraging more data sharing, cross-laboratory comparisons and an exhaustive characterization of cell-ENP stickiness, which should also be integrated as an essential part of existing ENP databases. As regards further developments, the in silico models embedded within DosiGUI could be further generalized to simulate the dynamics and hence reliably predict cytotoxicity of buoyant (i.e., having effective density smaller than the culture medium density) and hydrophobic (which cannot be dispersed using standard methodologies) NPs [22, 23], as well as extended to three-dimensional configurations for delivered dose estimations in scaffolds, spheroids and organoids. Such models could be used to simulate the dynamics of NPs in more physiologically relevant scenarios and facilitate in vitro-to-in vivo extrapolation.
Materials and methods
Basic principles of integrated dosimetry models
The ISD3 and DG models simulate the temporal and spatial dynamics of NPs suspended in cell culture medium contained in a vial with constant cross-sectional area. They both consider a one-dimensional (1D) model of sedimentation, diffusion and, if applicable, dissolution [17, 18], assuming a stable NP dispersion with unchanged intrinsic parameters over the duration of the simulated exposure experiment. The different processes are described through a series of rate equations as discussed in the AF (section AF2), which are solved numerically to give NP diameter (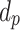 (m)) and number surface density (
(m)) and number surface density (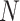 (m-2)) – i.e., the number of NPs having diameter
(m-2)) – i.e., the number of NPs having diameter 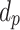 per unit of cross-section area – as functions of time (
per unit of cross-section area – as functions of time (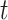 (s)) and the 1D space dimension (
(s)) and the 1D space dimension (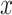 (m), corresponding to height, with the cell monolayer at x = 0). The two models differ in: (i) the method of solving equations, (ii) the way they treat dissolution and (iii) the bottom boundary layer condition, which basically describes the stickiness of the cell monolayer for NPs.
(m), corresponding to height, with the cell monolayer at x = 0). The two models differ in: (i) the method of solving equations, (ii) the way they treat dissolution and (iii) the bottom boundary layer condition, which basically describes the stickiness of the cell monolayer for NPs.
In particular, the DG model accounts for NP dissolution considering a first order rate equation with a rate constant that depends on 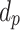 . On the other hand, the ISD3 model implements dissolution as a surface area-driven phenomenon based on a NP-specific kinetic model. This description can be more accurate but less general than that provided by the DG model. It does however require an in-depth characterization of dissolution kinetics for determining an analytical formulation of the associated rate for the NP of interest.
. On the other hand, the ISD3 model implements dissolution as a surface area-driven phenomenon based on a NP-specific kinetic model. This description can be more accurate but less general than that provided by the DG model. It does however require an in-depth characterization of dissolution kinetics for determining an analytical formulation of the associated rate for the NP of interest.
Regarding the bottom boundary, both models allow for tuning the stickiness through a parameter which describes adhesive behaviour ranging from zero to instantaneous adsorption. The DG model accounts for this by means of a Langmuir isotherm adsorption, expressed as in Eq. (1):
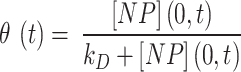 |
1 |
where 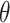 is the surface fraction of the boundary occupied by adsorbed NPs and
is the surface fraction of the boundary occupied by adsorbed NPs and 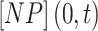 (mol L-1) is the molar concentration of NPs in the vicinity of the boundary (i.e., at x = 0).
(mol L-1) is the molar concentration of NPs in the vicinity of the boundary (i.e., at x = 0). 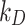 (mol L-1) – the parameter to be set for modulating the boundary stickiness – denotes the equilibrium dissociation constant; it is inversely proportional to the NP-cell stickiness: for a maximally sticky bottom (
(mol L-1) – the parameter to be set for modulating the boundary stickiness – denotes the equilibrium dissociation constant; it is inversely proportional to the NP-cell stickiness: for a maximally sticky bottom (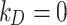 ), the cell monolayer surface is completely occupied by adsorbed NPs (
), the cell monolayer surface is completely occupied by adsorbed NPs (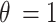 ), while the reflective or zero-stickiness case (
), while the reflective or zero-stickiness case (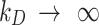 ) implies that the boundary is not at all occupied (
) implies that the boundary is not at all occupied (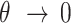 ).
).
The ISD3, on the other hand, assumes instantaneous NP uptake through a Dirichlet boundary condition, i.e., 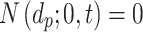 . We modified this condition by parametrizing it with respect to a constant
. We modified this condition by parametrizing it with respect to a constant 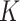 (m), which determines the stickiness of the cell monolayer (Eq. (2)).
(m), which determines the stickiness of the cell monolayer (Eq. (2)).
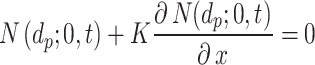 |
2 |
Thus, if 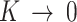 , the equation reduces to maximal stickiness as in ISD3. On the other hand, if
, the equation reduces to maximal stickiness as in ISD3. On the other hand, if 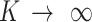 , we have a no-flux or Neumann boundary condition (
, we have a no-flux or Neumann boundary condition (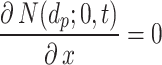 ), implying a purely reflective surface (i.e., zero stickiness). Thus,
), implying a purely reflective surface (i.e., zero stickiness). Thus, 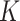 is inversely proportional to the stickiness.
is inversely proportional to the stickiness.
Methods for the identification of the stickiness parameters are reported in the subsection Identification of ENP-cell stickiness parameters.
DosiGUI
DosiGUI was developed as an open-source, standalone desktop application using MATLAB (R2022b, The MathWorks Inc., Boston, Massachusetts). A unique feature of DosiGUI is that only a single input dataset for both ISD3 and DG models is required (Table 3). Thus, the same experimental configuration can be simulated using both models, and the consistency of their predictions can be compared to determine which is the more suitable for estimating the delivered dose of NPs for a given setup. In addition, a stickiness knob allows users to reasonably estimate and set a stickiness level. Alternatively, in the case of already characterized numerical values of 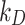 and
and 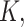 they may also be directly set in the Advanced panels. It is also worth highlighting that, for both DG and ISD3, using default settings and parameters (which correspond to configurations simulated in [17, 18]) in DosiGUI provides exactly the same dose predictions as directly using the original code scripts. A practical guide on how to download, install and use the application is provided in the AF (section AF1).
they may also be directly set in the Advanced panels. It is also worth highlighting that, for both DG and ISD3, using default settings and parameters (which correspond to configurations simulated in [17, 18]) in DosiGUI provides exactly the same dose predictions as directly using the original code scripts. A practical guide on how to download, install and use the application is provided in the AF (section AF1).
Table 3.
DosiGUI input parameters
| Intrinsic parameters | Extrinsic parameters |
|---|---|
ENP size distribution (i.e., diameters (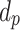 ) and corresponding normalized counts) ) and corresponding normalized counts) |
Suspension height (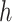 ) ) |
ENP density (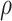 ) ) |
Temperature (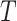 ) ) |
ENP effective density (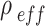 ) ) |
ENM nominal dose (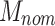 ) ) |
| Dissolution constants 1 | Stickiness parameters (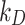 or or 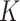 ) ) |
Medium density (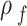 ) ) |
Exposure time (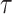 ) ) |
Medium dynamic viscosity (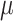 ) ) |
Spatial resolution (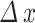 ) ) |
Time resolution (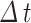 ) ) |
Basic intrinsic and extrinsic input parameters required by DosiGUI for running the ISD3 and DG models
DosiGUI validation
We verified that DG and ISD3 models could correctly account for stickiness by setting up a test system to measure the kinetics of ENP sedimentation in the two extreme boundary conditions (i.e., maximal and zero stickiness, respectively). Then, the test configurations were implemented within DosiGUI for evaluating the goodness of fit with the experimental data.
Experimental setup
The validation was performed using three referenced insoluble ENPs: titanium oxide (TiO2 – NM-105), cerium oxide (CeO2 – NM-212) and barium sulphate (BaSO4 – NM-220), from the European Union’s Joint Research Council (JRC) repository [24]. Protocols used to disperse and characterize them were identical to those previously established in [25], accounting also for specific recommendations by DeLoid et al. [15]. As input colloidal parameters (see the AF, Table A1), we considered effective density values reported in [20, 21], while the ENP size distribution was measured by dynamic light scattering (DLS) of freshly prepared suspensions. In particular, stock dispersions in BSA-water of each ENP were diluted in culture medium and then incubated for 1 h at 37 °C before DLS measurements. The culture medium was Dulbecco’s Modified Eagle Medium (DMEM – Thermo Fisher Scientific, Waltham, USA) supplied with 10% v/v fetal bovine serum (FBS - Thermo Fisher Scientific) and 1% v/v Minimal Essential Medium (Thermo Fisher Scientific).
Briefly, cylindrical plastic vials (base radius of 7.5 mm) containing 8 mL 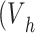 ) of ENP suspension were prepared with a thick layer of gelatin on the base to mimic the maximal stickiness boundary condition, while untreated vials were used to represent zero-stickiness. ENP stock dilutions corresponding to nominal doses of 25, 125 and 250 µg cm-2, respectively, were incubated for 1, 4 and 24 h in both gelatin-coated and uncoated vials. After incubation, 3 mL (
) of ENP suspension were prepared with a thick layer of gelatin on the base to mimic the maximal stickiness boundary condition, while untreated vials were used to represent zero-stickiness. ENP stock dilutions corresponding to nominal doses of 25, 125 and 250 µg cm-2, respectively, were incubated for 1, 4 and 24 h in both gelatin-coated and uncoated vials. After incubation, 3 mL (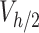 ) was collected from the middle of the suspension column and analysed using inductively coupled plasma optical emission spectrometry (ICP-OES). All experiments were carried out in triplicate. Further details on the experimental protocols are reported in AF6.
) was collected from the middle of the suspension column and analysed using inductively coupled plasma optical emission spectrometry (ICP-OES). All experiments were carried out in triplicate. Further details on the experimental protocols are reported in AF6.
Given that the mid-height and the nominal concentrations refer to different volumes, they must be rescaled as absolute masses to estimate the ENP mass fraction contained in 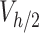 , or
, or 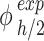 . Eq. (3) was used to convert the outcome of the measurements – a mid-height mass concentration (
. Eq. (3) was used to convert the outcome of the measurements – a mid-height mass concentration (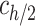 ) – to
) – to 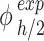 :
:
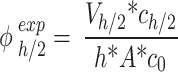 |
3 |
where 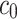 (µg mL-1) is the nominal mass concentration,
(µg mL-1) is the nominal mass concentration, 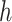 (mm) is the height of the suspension column and
(mm) is the height of the suspension column and 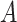 (mm2) its cross-sectional area. Note that since
(mm2) its cross-sectional area. Note that since 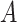 is constant along the liquid column, the ENP mass fraction is equal to the ratio of the corresponding doses (i.e., masses per unit area, see AF2).
is constant along the liquid column, the ENP mass fraction is equal to the ratio of the corresponding doses (i.e., masses per unit area, see AF2).
In silico predictions of mid-height mass fraction
To model the experiments, the dynamics of ENPs along the liquid column was simulated using DosiGUI. The physicochemical properties of the three ENPs were taken as detailed in the previous subsection. The maximally sticky boundary condition in the presence of gelatin was taken into account considering 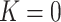 m for ISD3 and
m for ISD3 and 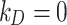 mol L-1 for DG (corresponding stickiness level on the knob: maximal), while a purely reflective bottom was replicated by setting saturating values for both parameters (
mol L-1 for DG (corresponding stickiness level on the knob: maximal), while a purely reflective bottom was replicated by setting saturating values for both parameters (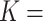 1012 m and
1012 m and 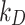 = 10− 5 mol L-1; corresponding stickiness level on the knob: zero). For both models, dissolution was disabled, given the negligible solubility in aqueous media of the three ENPs. All settings used for these simulations are summarized in the AF, Table A2.
= 10− 5 mol L-1; corresponding stickiness level on the knob: zero). For both models, dissolution was disabled, given the negligible solubility in aqueous media of the three ENPs. All settings used for these simulations are summarized in the AF, Table A2.
The results of simulations in terms of ENP mass along the liquid column at the time points of interest (i.e., 1, 4 and 24 h) were rearranged according to Eq. (4) to estimate the fraction of the initially administered mass (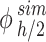 ) contained in
) contained in 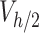 .
.
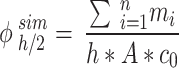 |
4 |
In the equation, 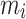 (µg) is the ENP mass in the i-th simulation element at the considered time point, and
(µg) is the ENP mass in the i-th simulation element at the considered time point, and 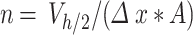 denotes the number of simulation elements in
denotes the number of simulation elements in 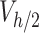 , with height
, with height 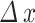 .
.
Then, we evaluated the reliability of simulated data from both ISD3 and DG through correlation analysis between measured (Eq. (3)) and predicted (Eq. (4)) mid-height mass fraction profiles over time (see AF4 for details on the statistical analyses).
Identification of ENP-cell stickiness parameters
Having verified the goodness of model predictions for the three ENMs, we used DosiGUI to characterize the stickiness of three different cell lines – HepG2, A549 and Caco-2 cells (ATCC, Manassas, Virginia, USA) – exposed to each ENP considered through estimation of parameters 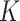 and
and 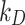 (Eqs. (1) and (2), respectively). The experiments consisted in ICP-OES measurements of the quantity of ENP mass adsorbed by cell monolayers over time. The data were compared through correlation analysis with their corresponding computational predictions in DosiGUI, obtained through parametrization of
(Eqs. (1) and (2), respectively). The experiments consisted in ICP-OES measurements of the quantity of ENP mass adsorbed by cell monolayers over time. The data were compared through correlation analysis with their corresponding computational predictions in DosiGUI, obtained through parametrization of 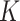 or
or 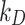 . Using this method, we were able to identify the best value of K and
. Using this method, we were able to identify the best value of K and 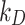 for each cell type-ENP pair.
for each cell type-ENP pair.
Experimental measurements of cell-adsorbed mass fraction
Cells of each phenotype were seeded at confluence in 96-well plates by dispensing 200 µL per well of cell suspension, then incubated overnight before NP exposure (see AF7 for further details on cell maintenance). ENPs were suspended in DMEM (Sigma-Aldrich) supplied with 10% w/v FBS (Sigma-Aldrich). Suspensions containing nominal doses of 7.8, 15.6 and 78.1 µg cm-2 of each ENP were obtained through serial dilutions of a stock batch prepared as described for DosiGUI validation [25]. Then, 100 µL of each suspension was added to the wells. After exposure, the ENP suspension was removed, and wells rinsed twice with phosphate buffered saline (PBS, Lonza, Basel, Switzerland) 1X for eliminating ENPs either deposited (but not adsorbed) onto cells or attached to the walls of the well. Both the overlaying ENP suspension and the washing volume of PBS were analysed to account for all of the initially administered mass. Trypsin-EDTA (Lonza) was then used for detaching cells, and the resulting samples were collected for measuring the cell-adsorbed ENP mass. Two types of controls were used: blank controls (BCs) consisted in cells without ENPs, while positive controls (PCs) were simply 100 µL of the suspension, collected immediately after stock dilution (before the exposure). All experiments were carried out in triplicate for four different exposure times: 4, 8, 24 and 72 h.
The same procedure described in DosiGUI validation was applied, determining the ENP mass concentration in each sample. Note that, for each ENP and nominal dose, the controls BC and PC represent the offset of the measurement and the maximum mass concentration detectable in the samples, respectively. Thus, to get meaningful adsorbed fractions (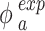 ), the raw data were post-processed as follows:
), the raw data were post-processed as follows:
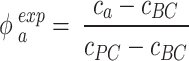 |
5 |
where 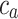 (µg mL-1) is the concentration of ENPs adsorbed by cells as measured by ICP-OES, while
(µg mL-1) is the concentration of ENPs adsorbed by cells as measured by ICP-OES, while 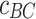 and
and 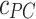 (µg mL-1) are the ENP concentrations detected for the corresponding BC and PC samples, respectively. All the variables in Eq. (5) are expressed as mean ± standard deviation of the triplicate, and standard methods were applied to account for error propagation.
(µg mL-1) are the ENP concentrations detected for the corresponding BC and PC samples, respectively. All the variables in Eq. (5) are expressed as mean ± standard deviation of the triplicate, and standard methods were applied to account for error propagation.
In silico predictions of cell-adsorbed mass fraction and correlation analysis
After setting the input parameters in DosiGUI (geometry and ENP characteristics according to [20]), we ran a parametric sweep for the stickiness varying 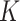 and
and 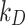 within reasonable ranges (reported in the AF, Table A3). Simulations were carried out for the same nominal doses experimentally administered. The settings for these simulations are summarized in the AF, Table A3.
within reasonable ranges (reported in the AF, Table A3). Simulations were carried out for the same nominal doses experimentally administered. The settings for these simulations are summarized in the AF, Table A3.
The simulated adsorbed mass fraction (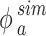 ) for each time point of interest was expressed as:
) for each time point of interest was expressed as:
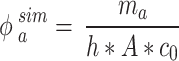 |
6 |
where 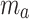 (µg) is the cumulative ENP mass adsorbed at the bottom of the column, and the other parameters are the same as in Eq. (4).
(µg) is the cumulative ENP mass adsorbed at the bottom of the column, and the other parameters are the same as in Eq. (4).
We then performed a correlation analysis between the experimental (Eq. (5)) and simulated (Eq. (6)) mass fractions versus time for identifying the most reliable value of both 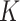 and
and 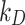 and therefore the associated stickiness level for each of the three ENPs. Specifically, the best fitting regression line for
and therefore the associated stickiness level for each of the three ENPs. Specifically, the best fitting regression line for 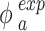 as a function of
as a function of 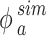 was estimated with a confidence interval of 99% for each model, ENP and nominal dose with respect to each stickiness value considered. The optimal value of
was estimated with a confidence interval of 99% for each model, ENP and nominal dose with respect to each stickiness value considered. The optimal value of 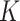 or
or 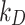 was determined using the same criteria as reported in the section DosiGUI validation. Details on the statistical analyses are provided in AF4.
was determined using the same criteria as reported in the section DosiGUI validation. Details on the statistical analyses are provided in AF4.
Delivered dose prediction
Having established the optimum value of 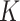 or
or 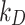 for each ENP-cell type combination, we were able to reliably extend DosiGUI predictions to any exposure scenario involving those combinations. Using the optimum stickiness, we estimated the delivered dose of each of the three ENPs perceived over time by HepG2, A549 and Caco-2 monolayers cultured in 96-well plates. The delivered dose was computed as the mass of ENPs per unit area adsorbed by cells and within the lowest simulation domain (i.e., in the immediate vicinity of the cell culture). An analytical expression for delivered dose and further details of the statistical methods used are given in the AF (Eq. (A6) and AF4).
for each ENP-cell type combination, we were able to reliably extend DosiGUI predictions to any exposure scenario involving those combinations. Using the optimum stickiness, we estimated the delivered dose of each of the three ENPs perceived over time by HepG2, A549 and Caco-2 monolayers cultured in 96-well plates. The delivered dose was computed as the mass of ENPs per unit area adsorbed by cells and within the lowest simulation domain (i.e., in the immediate vicinity of the cell culture). An analytical expression for delivered dose and further details of the statistical methods used are given in the AF (Eq. (A6) and AF4).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Wendel Wohlleben for his insightful advice.
Abbreviations
- NM
Nanomaterial
- ENM
Engineered nanomaterial
- NP
Nanoparticle
- ENP
Engineered nanoparticle
- DG
Distorted grid
- ISD3
In vitro sedimentation, diffusion, dissolution and dosimetry
- GUI
Graphical user interface
- ICP-OES
Inductively coupled plasma optical emission spectrometry
- BC
Blank control
- PC
Positive control
Author contributions
Conceptualization: EB, ALC, AA Methodology: EB, IZ, NG, ALC, AA Software development: EB, PV Investigation: EB, PV, IZ, NG, LF Data analysis and visualization: EB, PV, IZ, DG Supervision: ALC, AA Writing – original draft: EB, AA Writing – review and editing: all authors.
Funding
This work has received funding from the European Union’s Horizon 2020 Research and Innovation Program under grant agreement No. 7601813 PATROLS project. EB’s post-doctoral fellowship was funded by the Next Generation EU project CN00000013 ‘Centro Nazionale 1’ HPC, Big Data and Quantum Computing (CN1, PNRR, Spoke 6: Multiscale modelling and engineering applications).
Data availability
DosiGUI is freely available for download from this open-access GitHub repository: https://github.com/CentroEPiaggio/DosiGUI.git. All experimental and computational data generated and processed in this study are available upon reasonable request to the corresponding author.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hussain CM. Handbook of nanomaterials for industrial applications. Handb. Nanomater. Ind. Appl. Elsevier; 2018.
- 2.Su H, Wang Y, Gu Y, Bowman L, Zhao J, Ding M. Potential applications and human biosafety of nanomaterials used in nanomedicine. J. Appl. Toxicol. John Wiley and Sons Ltd; 2018. pp. 3–24. [DOI] [PMC free article] [PubMed]
- 3.Vieira S, Vial S, Reis RL, Oliveira JM. Nanoparticles for bone tissue engineering. Biotechnol. Prog. John Wiley and Sons Inc.; 2017. pp. 590–611. [DOI] [PubMed]
- 4.Genchi GG, Marino A, Grillone A, Pezzini I, Ciofani G. Remote control of Cellular functions: the role of Smart nanomaterials in the Medicine of the future. Adv Healthc Mater. 2017;6:1700002. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Fan T, Xie Z, Zeng Q, Xue P, Zheng T et al. Advances in nanomaterials for photodynamic therapy applications: Status and challenges. Biomaterials. Elsevier Ltd; 2020. p. 119827. [DOI] [PubMed]
- 6.Soares S, Sousa J, Pais A, Vitorino C. Nanomedicine: principles, properties, and regulatory issues. Front. Chem. Frontiers Media S.A.; 2018. p. 360. [DOI] [PMC free article] [PubMed]
- 7.Sohal IS, O’Fallon KS, Gaines P, Demokritou P, Bello D. Ingested engineered nanomaterials: state of science in nanotoxicity testing and future research needs. Part. Fibre Toxicol. BioMed Central Ltd.; 2018. pp. 1–31. [DOI] [PMC free article] [PubMed]
- 8.Cohen J, Deloid G, Pyrgiotakis G, Demokritou P. Interactions of engineered nanomaterials in physiological media and implications for in vitro dosimetry. Nanotoxicology. 2013;7:417–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Böhmert L, König L, Sieg H, Lichtenstein D, Paul N, Braeuning A, et al. In vitro nanoparticle dosimetry for adherent growing cell monolayers covering bottom and lateral walls. Part Fibre Toxicol. 2018;15:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poli D, Mattei G, Ucciferri N, Ahluwalia A. An Integrated in Vitro–In Silico Approach for Silver Nanoparticle Dosimetry in cell cultures. Ann Biomed Eng. 2020;48:1–10. [DOI] [PMC free article] [PubMed]
- 11.Teeguarden JG, Hinderliter PM, Orr G, Thrall BD, Pounds JG. Particokinetics in vitro: Dosimetry considerations for in vitro nanoparticle toxicity assessments. Toxicol. Sci. Oxford Academic; 2007. pp. 300–12. [DOI] [PubMed]
- 12.Singh RP, Ramarao P. Cellular uptake, intracellular trafficking and cytotoxicity of silver nanoparticles. Toxicol Lett. 2012;213:249–59. [DOI] [PubMed] [Google Scholar]
- 13.Beer C, Foldbjerg R, Hayashi Y, Sutherland DS, Autrup H. Toxicity of silver nanoparticles-nanoparticle or silver ion? Toxicol Lett. 2012;208:286–92. [DOI] [PubMed] [Google Scholar]
- 14.Deloid G, Cohen JM, Darrah T, Derk R, Rojanasakul L, Pyrgiotakis G et al. Estimating the effective density of engineered nanomaterials for in vitro dosimetry. Nat Commun. 2014;5. [DOI] [PMC free article] [PubMed]
- 15.Deloid GM, Cohen JM, Pyrgiotakis G, Demokritou P. Preparation, characterization, and in vitro dosimetry of dispersed, engineered nanomaterials. Nat Protoc. 2017;12:355–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen JM, Deloid GM, Demokritou P. A critical review of in vitro dosimetry for engineered nanomaterials. Nanomedicine. Future Medicine Ltd.; 2015. pp. 3015–32. [DOI] [PubMed]
- 17.DeLoid GM, Cohen JM, Pyrgiotakis G, Pirela SV, Pal A, Liu J, et al. Advanced computational modeling for in vitro nanomaterial dosimetry. Part Fibre Toxicol. 2015;12:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas DG, Smith JN, Thrall BD, Baer DR, Jolley H, Munusamy P, et al. ISD3: a particokinetic model for predicting the combined effects of particle sedimentation, diffusion and dissolution on cellular dosimetry for in vitro systems. Part Fibre Toxicol. 2018;15:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheimarios N, Pem B, Tsoumanis A, Ilić K, Vrček IV, Melagraki G et al. An in Vitro Dosimetry Tool for the Numerical Transport Modeling of Engineered nanomaterials powered by the Enalos RiskGONE Cloud platform. Nanomaterials. 2022;12. [DOI] [PMC free article] [PubMed]
- 20.Keller JG, Quevedo DF, Faccani L, Costa AL, Landsiedel R, Werle K, et al. Dosimetry in vitro – exploring the sensitivity of deposited dose predictions vs. affinity, polydispersity, freeze-thawing, and analytical methods. Nanotoxicology. 2020;15:21–34. [DOI] [PubMed] [Google Scholar]
- 21.Llewellyn SV, Conway GE, Zanoni I, Jørgensen AK, Shah U-K, Seleci DA et al. Understanding the impact of more realistic low-dose, prolonged engineered nanomaterial exposure on genotoxicity using 3D models of the human liver. J Nanobiotechnology 2021 191. 2021;19:1–24. [DOI] [PMC free article] [PubMed]
- 22.Watson CY, DeLoid GM, Pal A, Demokritou P. Buoyant nanoparticles: implications for Nano-Biointeractions in Cellular studies. Small. 2016;12:3172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lizonova D, Trivanovic U, Demokritou P, Kelesidis GA. Dispersion and Dosimetric challenges of Hydrophobic Carbon-based nanoparticles in in Vitro Cellular studies. Nanomaterials. 2024;14:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.JRC Nanomaterials Repository [Internet]. [cited 2023 Sep 18]. https://joint-research-centre.ec.europa.eu/scientific-tools-and-databases/jrc-nanomaterials-repository_en
- 25.Jensen KA. The NANOGENOTOX dispersion protocol for NANoREG. Nrcwe. 2014.
- 26.DeLoid G, Cohen JM, Darrah T, Derk R, Rojanasakul L, Pyrgiotakis G, et al. Estimating the effective density of engineered nanomaterials for in vitro dosimetry. Nat Commun 2014 51. 2014;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treuel L, Eslahian KA, Docter D, Lang T, Zellner R, Nienhaus K et al. Physicochemical characterization of nanoparticles and their behavior in the biological environment. Phys. Chem. Chem. Phys. Royal Society of Chemistry; 2014. pp. 15053–67. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DosiGUI is freely available for download from this open-access GitHub repository: https://github.com/CentroEPiaggio/DosiGUI.git. All experimental and computational data generated and processed in this study are available upon reasonable request to the corresponding author.