Abstract
In an attempt to isolate mRNA-binding proteins we fractionated Xenopus oocyte lysate by oligo(dT)–cellulose chromatography. A 20 kDa protein was the major component of the eluate. cDNA cloning revealed that this protein is a Xenopus homolog of the cold-inducible RNA-binding protein (CIRP) which was originally identified in mammalian cells as a protein that is overexpressed upon a temperature downshift. This Xenopus protein, termed here xCIRP2, is highly expressed in ovary, testis and brain in adult Xenopus tissues. In oocytes it is predominantly localized in the cytoplasm. By biochemical fractionation we provide evidence that xCIRP2 is associated with ribosomes, suggesting that it participates in translational regulation in oocytes. Microinjection of labeled mRNA into oocytes followed by UV cross-linking of the oocyte lysate led to identification of two major RNA-binding activities. Immunoprecipitation of the RNA-binding proteins demonstrated that one is xCIRP2 and that the other contains FRGY2. FRGY2, which is one of the principal constituents of mRNA storage particles involved in translational masking of maternal mRNA, has an RNA-binding domain conserved to those of bacterial cold shock proteins. Possible implications of the highly abundant expression in oocytes of cold shock RNA-binding proteins of both eukaryotic and prokaryotic types are discussed.
INTRODUCTION
A large body of evidence has revealed that RNA-binding proteins are involved in many aspects of gene expression. Many eukaryotic mRNA-binding proteins function in pre-mRNA processing, mRNA export, mRNA localization, translation and mRNA turnover (1). Some classes of RNA-binding proteins also have structural roles in packaging mRNA. In the nucleus, pre-mRNAs (also called hnRNAs) are complexed with more than 20 species of abundant hnRNP proteins to form hnRNPs (heterogeneous nuclear ribonucleoprotein particles) (2). Some of these hnRNP proteins remain associated with mRNA until nuclear mRNPs (messenger ribonucleoprotein particles) reach the nuclear pore, or even after they are exported to the cytoplasm (3). One example is hnRNP A1 protein, which was originally identified as one of the core proteins of hnRNP and later, along with its homologs, shown to be involved in splicing, to shuttle between the nucleus and cytoplasm and to remain bound to polysomes (2,4–7). Upon mRNA export, cytoplasmic mRNP is formed by dissociation of at least some of the hnRNP proteins and association of mRNP proteins. Y-box proteins are the cytoplasmic mRNP constituents (8–10). In Xenopus laevis oocytes, two Y-box proteins, FRGY2 and its homolog mRNP3 protein, are the major RNA-binding components of the storage mRNPs that store or mask maternal mRNAs in translationally silent states until those mRNAs are recruited to ribosomes for protein synthesis during early development (11–14).
Many RNA-binding proteins share several structural features in common (15–17). One of the most commonly found and best studied is the RNA recognition motif (RRM). The RRM contains two highly conserved short segments (RNP-1 and RNP-2) within which the conserved aromatic amino acids directly interact with RNA (17). Some RNA-binding proteins contain as many as four RRMs which confer the ability to interact with RNA in a sequence-specific or sequence-independent fashion. RRMs are often flanked by other structural features representative of RNA-binding proteins, such as glycine-rich regions or hnRNP K-homology domains that are also involved in interactions with RNA (16).
Mouse CIRP has been identified by cDNA cloning of a RRM-containing RNA-binding protein (18). CIRP consists of a single RRM and a flanking glycine-rich region containing Arg-Gly-Gly (RGG) repeats. Based on its similarity to plant glycine-rich proteins, which are overexpressed during cold shock, the expression of CIRP was examined for its potential increase during cold shock. The levels of CIRP mRNA and protein in cultured mouse cells increase upon a temperature downshift from 37 to 32°C (18). Mouse CIRP is highly expressed in testis within the scrotum, which is maintained at temperatures lower than other parts of the body cavity, while its expression is repressed by exposing the testis to heat stress (19). Furthermore, it is thought that CIRP is responsible for a prolonged G1 phase that has been observed in cells exposed to cold shock, although the mechanisms by which CIRP regulates cell growth are poorly understood (18). Homologs of mouse CIRP have been reported in human, rat, Mexican axolotl and Xenopus cells (20–23). The Xenopus CIRP homolog was found by identifying genes expressed at a particular stage of pronephros formation (23).
In this study we report identification of a novel Xenopus homolog of CIRP. This protein, hereafter referred to as xCIRP2, is highly expressed in oocytes, being predominantly localized in oocyte cytoplasm. xCIRP2 binds to mRNA in vivo and in vitro. Results from density gradient centrifugation show a possible association of xCIRP2 with ribosomes. We propose that xCIRP2 is involved in translational regulation in oocytes through the modulation of ribosomal function.
MATERIALS AND METHODS
Oligo(dT)–cellulose chromatography
Xenopus laevis frogs were maintained at 20°C. Oocytes were isolated from frogs anesthetized in iced water except where otherwise stated. Defolliculated oocytes were prepared by treating frog ovaries with collagenase. Four hundred microliters of oocytes were homogenized in an 800 µl solution of 10 mM Tris–HCl, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 8% glycerol, 1 mM dithiothreitol (DTT) and 100 U/ml RNase inhibitor. The homogenate was centrifuged in a microtube at 15 000 r.p.m. for 10 min at 4°C. The supernatant was adjusted to 250 mM NaCl, mixed with 100 µl of oligo(dT)–cellulose beads (Amersham Pharmacia Biotech) and rotated for 1 h at 4°C. The cellulose beads were then washed twice with binding buffer (10 mM Tris–HCl, pH 7.5, 250 mM NaCl and 2 mM MgCl2) and the bound fraction was eluted with 400 µl of binding buffer containing 25% formamide (24).
cDNA cloning of xCIRP2
A degenerate oligonucleotide set of 5′-TIAAYTTYGAYACIAAYGARGA-3′, encoding amino acids LNFDTNEE, and 5′-ATYTGICKICCRTCIACIGCYTT-3′ (where R = G/A, Y = T/C, K = G/T and I represents inosine), encoding amino acids KAVDGRQI, was used for reverse transcriptase-mediated PCR using Xenopus oocyte mRNAs. The resultant cDNA of 193 bp in length was used to screen a Xenopus oocyte cDNA library (25). Positive clones were plaque purified twice. The cDNA clones were recovered in plasmids using in vivo excision following the manufacturer’s protocol and subsequently sequenced.
Recombinant proteins
To construct xCIRP2 and FRGY2 expression plasmids, PCR was performed using the xCIRP2 cloned cDNA and the FRGY2 coding sequence as the templates, respectively, with primer sets 5′-GGCAGCCATATGTCTGATGAAGGAAAAC-3′ and 5′-AGCCCGCTCGAGCTCGTGTGTAGCATAAC-3′ for xCIRP2 and 5′-GGGAATTCCATATGAGTGAGGCGGAAGCCCAGG-3′ and 5′-AGCCCGCTCGAGTTCTGGGGCAGGTGTATC-3′ for FRGY2. The PCR products were digested with NdeI and XhoI and inserted into the NdeI and XhoI sites of pET24b (Novagen). To obtain recombinant xCIRP2 protein, Escherichia coli strain BL21(DE3) was transformed with the plasmid. Recombinant xCIRP2 was overexpressed by the addition of isopropyl β-d-thiogalactopyranoside to the bacterial culture (800 ml). Bacterial cells were sonicated in 20 ml of sonication buffer consisting of 20 mM Tris–HCl, pH 8.0, and 100 mM NaCl. The cell lysate was centrifuged at 12 000 g for 10 min and the supernatant mixed with an equal volume of sonication buffer containing 8 M urea, followed by application to a TALON metal affinity column (Clontech). 6× Histidine-tagged xCIRP2 protein was eluted with sonication buffer containing 30 mM imidazole and 4 M urea. After dialysis against the sonication buffer containing 4 M urea, the protein was further purified by application to a Ni–NTA Superflow column (Qiagen). The fractions that eluted with the sonication buffer containing 100 mM imidazole and 4 M urea were then pooled and dialyzed against sonication buffer containing 4 M urea and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Recombinant FRGY2 was also expressed in BL21(DE3) cells as described above, but the bacterial cells were sonicated in 20 ml of sonication buffer containing 6 M guanidine–HCl, 10 µg/ml aprotinin, 10 µg/ml leupeptin and 100 µg/ml 4-(2 aminoethyl)benzenesulfonyl fluoride. The soluble proteins were purified by Ni–NTA column chromatography under denaturing conditions. The histidine-tagged FRGY2 was then eluted with sonication buffer containing 6 M urea and 100 mM imidazole and renatured by dialysis against buffer S (50 mM sodium phosphate, pH 7.2, 100 mM NaCl, 5 mM EDTA, 1 mM DTT and 1 mM PMSF). FRGY2 was further purified by SP Sepharose (Amersham Pharmacia Biotech) column chromatography as described (26,27). The purified protein was dialyzed against buffer S.
Antibodies and immunoblotting
For immunization, recombinant xCIRP2 protein was purified (see above) without urea. Polyclonal antisera against xCIRP2 were produced by injecting a rabbit with the recombinant xCIRP2 protein following standard procedures. Another rabbit was immunized with a synthetic peptide, C1 (CGGSYRDSYDSYATH), corresponding to the C-terminal amino acids of xCIRP2 including a cysteine linker which had been coupled to keyhole limpet hemocyanin.
A pool of 10 oocytes was homogenized in a solution of 90 mM HEPES, pH 7.5, 70 mM KCl, 1 mM DTT and 5% sucrose. To obtain Xenopus tissues, adult frogs were anesthetized with ether and killed. Brain and testis were obtained from a male frog, while other tissues were from a female frog. Protein extracts were prepared by homogenizing the tissues in a solution of 10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, 10 µg/ml aprotinin and 10 µg/ml leupeptin. To examine protein localization, nuclei were isolated from oocytes manually. Nuclear or cytoplasmic fractions from 10 oocytes were pooled and analyzed. Protein extracts were then subjected to SDS–PAGE and transferred to a polyvinylidene difluoride membrane. Immunoblotting analysis was performed as described (25). Rabbit polyclonal antisera containing anti-xCIRP2 (1:3000 dilution) or anti-FRGY2 antibodies or a culture supernatant from mouse hybridoma cells that produce monoclonal antibodies against TAF-Iβ were used (25,26).
Northern blotting
RNA was isolated from oocytes and tissues using Trizol (Life Technologies) or Isogen (Nippongene, Tokyo, Japan) and analyzed in an agarose gel containing formaldehyde. The RNA was transferred to a nylon membrane and hybridized to 32P-labeled probe DNA corresponding to the xCIRP2 or Xenopus histone H1 coding sequences.
UV cross-linking
The capped histone H1 and chloramphenicol acetyltransferase (CAT) mRNA was synthesized from linearized plasmids pSPH1.11 and pSPCAT, respectively, by SP6 RNA polymerase in the presence of [α-32P]UTP (28,29). Microinjection of mRNA into Xenopus stage VI oocytes was conducted as described previously (30,31). One hour after injection, oocyte homogenate was irradiated with UV light and digested with RNase A as described (31). Immunoprecipitation of UV cross-linked proteins was performed with polyclonal antibodies against recombinant xCIRP2 or FRGY2. Briefly, the proteins were mixed with protein G–Sepharose (Amersham Pharmacia Biotech) coupled to the antibodies in RIPA buffer [10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40 (NP-40), 0.1% sodium deoxycholate and 1 mM PMSF] for 30 min at 4°C. The Sepharose beads were then washed extensively with RIPA buffer containing 1% SDS. Bound proteins were analyzed by SDS–PAGE and subsequently visualized by autoradiography.
Gel retardation assay
32P-labeled histone H1 mRNA was prepared as described above. Ten femtomoles of RNA probe were incubated with recombinant xCIRP2 and/or FRGY2 proteins at 30°C for 20 min in a 50 µl reaction mixture consisting of 20 mM Tris–HCl, pH 7.5, 100 mM KCl, 4 mM MgCl2, 1 mM DTT and 20 U RNase inhibitor (27). The IgG fraction (0.6 µg) containing the anti-recombinant xCIRP2 antibodies, which had been purified with protein A–Sepharose, was added and the mixtures were then incubated at 30°C for another 10 min. The mixture was electrophoresed in a 1% agarose gel in 0.5× Tris–borate/EDTA (TBE) buffer at room temperature. The gel was then dried and subjected to autoradiography.
Fractionation of oocytes
Xenopus oocytes (500 µl) were homogenized with 9 vol homogenization buffer (10 mM Tris–HCl, pH 8.0, 1 mM DTT and 0.1% NP-40) containing 100 mM NaCl. The homogenates were centrifuged at 12 000 g for 15 min and the supernatant designated ‘total lysate’. For RNase A treatment, RNase A was added to a portion of the total lysate at a final concentration of 0.1 mg/ml and the sample was incubated at 30°C for 10 min. The reaction was terminated by adding RNase inhibitor at a final concentration of 100 U/ml and immediately placed on ice. For high salt concentration treatment oocytes were homogenized with homogenization buffer containing 500 mM NaCl. The total lysates were further centrifuged in a Beckman TLS 55 rotor at 100 000 g for 1 h at 4°C, yielding supernatant (S100) and pellet (P100) fractions. P100 fractions were resuspended in SDS buffer (100 mM Tris–HCl, pH 7.5, 300 mM NaCl, 10 mM EDTA and 2% SDS) and each fraction and the total lysate were analyzed by immunoblotting using anti-xCIRP2 C-terminal peptide antibodies. RNA from each fraction was prepared as described (32).
Sucrose gradient centrifugation
Fifteen stage VI oocytes were homogenized in 150 µl of buffer A (20 mM Tris–HCl, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10 µg/ml cycloheximide and 25 U/ml RNase inhibitor). The homogenate was centrifuged in a microtube at 12 000 g for 10 min. The supernatant was then irradiated with UV light for 30 min on ice. The sample was loaded on a 15–40% sucrose density gradient prepared in buffer A and centrifuged at 55 000 r.p.m. in a Beckman TLS55 rotor for 1 h at 4°C. The samples were collected from the top of the gradient in 19 fractions of 100 µl. For immunoblotting a 5 µl aliquot of each fraction was digested with 0.1 mg/ml RNase A at 37°C for 1 h and analyzed by SDS–PAGE. RNA was prepared from an 80 µl aliquot of every other fraction.
Nicodenz gradient centrifugation
Oocytes (300 µl) were homogenized in 300 µl of buffer N (20 mM HEPES, pH 7.4, 10 mM MgCl2, 200 mM KCl, 100 U/ml RNase inhibitor, 10 µg/ml aprotinin, 10 µg/ml leupeptin and 0.5 mM PMSF) and centrifuged in a microtube at 12 000 g for 10 min. Half of the resulting supernatant was mixed with 0.25 vol 20% formaldehyde, 20 mM HEPES, pH 7.4, 10 mM MgCl2 and 100 mM KCl solution and stored on ice for 20 min. The other half of the supernatant was irradiated with UV light for 30 min on ice. The samples were then loaded on 20–60% Nicodenz gradients prepared in buffer N containing 0.5% NP-40 and centrifuged at 48 000 r.p.m. in a Beckman TLS55 rotor for 20 h at 4°C (33,34). The samples were collected from the top of the gradient in 21 fractions of 90 µl. A 2 µl aliquot of each fraction was digested with RNase A and analyzed by immunoblotting. Forty microliters of each fraction of formaldehyde-fixed sample were digested with 200 µg/ml of proteinase K at 37°C overnight to prepare RNA.
RESULTS
Identification of xCIRP2 in the oocytes
To identify novel mRNA-binding proteins, we fractionated Xenopus oocyte lysate by oligo(dT)–cellulose chromatography following a previously described protocol (24). After extensive washing of the cellulose resin, bound materials were eluted with a buffer containing 25% formamide. Immunoblotting revealed that the eluate contained FRGY2 (also called mRNP4 and p56), shown in Figure 1 as an upper band of a 54–56 kDa doublet (lane 3 and data not shown). The lower band is possibly its homolog mRNP3 (12,14,33). In the same fraction there was a prominent 20 kDa protein (p20). The elution profile of this protein from oligo(dT)–cellulose suggests that p20 is likely bound to poly(A)+ RNA in the oocytes. However, it cannot be ruled out at this point that p20 has a high affinity for oligo(dT). Proteins of similar molecular weight have been detected in previous studies, although to our knowledge further characterization of the proteins has not been reported (24). Therefore, we sought to identify this protein. To this end, p20 was excised from the gel and digested with lysil-endopeptidase. Amino acid sequences of the three resultant peptides (K1–K3) that were microsequenced showed significant homology with the previously reported RNA-binding protein, CIRP (Fig. 2).
Figure 1.

Oligo(dT)–cellulose chromatography. Xenopus oocyte lysate was incubated with oligo(dT)–cellulose. Then after extensive washing with binding buffer, the bound materials were eluted with binding buffer containing 25% formamide. Aliquots of total lysate (lane 1), unbound materials (lane 2) and the eluate (lane 3) were electrophoresed by 12% SDS–PAGE and stained with Coomassie brilliant blue. FRGY2 and the abundant 20 kDa protein (p20) are indicated by arrows and the sizes of molecular weight markers (lane M) are shown on the left.
Figure 2.
cDNA cloning of xCIRP2. The deduced amino acid sequence of cloned xCIRP2 cDNA (Xenopus 2) is aligned with previously reported amino acid sequences, XCIRP (Xenopus 1, accession no. AB007597), human CIRP (D78134), mouse CIRP (D78135) and Mexican axolotl RBP (U71299). ClustalW with minor visual modifications was used to produce the alignments. Conserved amino acids are indicated by asterisks. RNP1 and RNP2 sequences in the RNA recognition motif and RGG sequences are boxed. The sequences of three peptides (K1, K2 and K3) that were microsequenced from gel-purified p20 are indicated. C1 indicates the synthesized peptide sequence used to prepare the polyclonal antibodies.
Using information from the peptide sequences we cloned a 1.3 kb cDNA from a Xenopus oocyte cDNA library. The cDNA contains an open reading frame (ORF) encoding a protein of 166 amino acid residues (Fig. 2). All three peptides were present in the deduced amino acid sequence, thus supporting the identity of p20 and the protein encoded by the cloned cDNA. Since Uochi et al. (23) previously reported cDNA cloning of a Xenopus CIRP homolog, XCIRP, we designated the cloned 20 kDa protein xCIRP2. Alignment of xCIRP2 with other CIRPs revealed 92% identity with XCIRP and 82–83% identity with mammalian CIRPs (Fig. 2). xCIRP2 consists of an RRM containing highly conserved RNP-1 and RNP-2 sequences and a C-terminal glycine-rich region with four RGG repeats.
Until now the occurrence of CIRP protein in oocytes has not been reported in mammals or amphibians. When peptides derived from gel-isolated p20 were fractionated and microsequenced, a small peak of a peptide K2′ (YGRISEVV) was detected next to the peak of peptide K2 (data not shown). The K2′ sequence matched completely that of the deduced XCIRP amino acid sequence (Fig. 2). Since the abundance of K2′ was <10% that of K2, xCIRP2 protein likely comprises the majority of the CIRP expressed in Xenopus oocytes, with a much smaller amount of co-expressed XCIRP.
Expression of xCIRP2 in the oocytes
Next we wished to examine the expression of xCIRP2 in Xenopus by preparing two kinds of polyclonal antibodies against xCIRP2. Firstly, recombinant xCIRP2 protein with a 6× histidine tag was expressed in E.coli, purified utilizing metal affinity resins (Fig. 3A, lane 1) and then injected into a rabbit to raise antibodies against full-length xCIRP2. Secondly, a synthetic peptide corresponding to the C-terminal region of xCIRP2, which differs from that of XCIRP but is conserved in mammalian CIRP (see C1 in Fig. 2), was used to immunize another rabbit. Both rabbit antisera successfully recognized xCIRP2 as a single band in the oocyte lysate by immunoblotting analysis (Fig. 3A, lanes 2 and 3). The antibody against the C-terminal peptide also recognized an 18 kDa protein in HeLa cell lysates whose expression was induced by a downshift in culture temperature from 37 to 33°C, suggesting that it is human CIRP (data not shown).
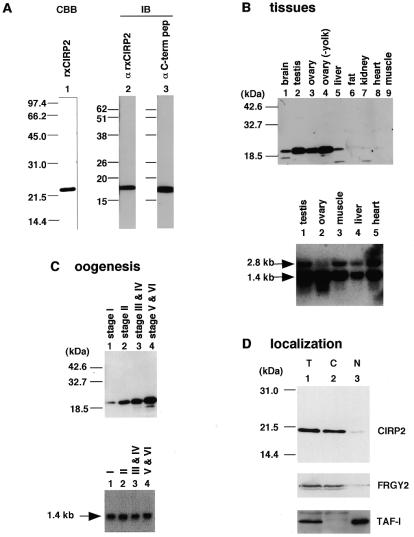
Figure 3.
Expression of xCIRP2. (A) Recombinant xCIRP2 with a 6× histidine tag was prepared. One microgram of the recombinant protein was analyzed by 12% SDS–PAGE and stained with Coomassie brilliant blue along with molecular weight markers (lane 1). Oocyte lysate (2 µg protein) was subjected to immunoblotting with rabbit antisera against recombinant xCIRP2 (lane 2) or against the C-terminal peptide C1 (lane 3; see Fig. 2 for sequence of the peptide). (B and C) The levels of xCIRP2 proteins and mRNAs in Xenopus tissues and oocytes were examined by immunoblotting (top) and northern blotting (bottom), respectively. Five micrograms of protein from tissues (B) or a lysate equivalent to 0.25 oocytes (C) were used for immunoblotting with the antiserum raised against the C-terminal peptide. Lysate from ovaries was extracted with 1,1,2-trichlorotrifluoroethane (Freon) to remove yolk proteins (B, top, lane 4). xCIRP2 mRNA in 10 µg total tissue RNA (B) or in RNA equivalent to one oocyte (C) was examined by northern blotting. Note that the 2.8 kb mRNA was barely detected in the ovaries as well as in the oocytes. (D) Stage VI oocytes were fractionated into nuclei and cytoplasm. The expression of xCIRP2 in total (T), cytoplasmic (C) and nuclear (N) fractions was examined by immunoblotting with anti-xCIRP2 C-terminal peptide antibodies. To confirm the fractionation process, localization of FRGY2 and TAF-I in the same fractions was also examined.
With these antibodies we examined xCIRP2 expression in Xenopus tissues and its accumulation during oogenesis. An adult frog was killed at 20°C and various tissues were isolated. Immunoblotting showed that among the tissues examined xCIRP2 was most abundant in testis, ovary, brain and liver and was faintly detected in kidney and heart (Fig. 3B). xCIRP2 mRNA was detected by northern blotting using the xCIRP2 coding region as a probe. Two mRNAs (2.8 and 1.4 kb) were detected in the tissues tested, the longer being less represented in the ovary. Since the amount of xCIRP2 mRNA was much less variable among tissues relative to that of xCIRP2 protein, our results suggest that xCIRP2 protein levels are post-transcriptionally regulated. Further support for this notion comes from the results of oocyte analyses (Fig. 3C). Xenopus oocytes in the mother’s ovary can be divided into six stages by their size and appearance. xCIRP2 protein apparently accumulated as oocytes grew, while xCIRP2 mRNA levels remained constant. We usually prepare the oocytes from a mother frog anesthetized in ice-water and maintain them at 18°C. No change was observed in xCIRP2 expression in oocytes prepared from frogs anesthetized by ether at 20°C nor from those maintained at lower temperatures (data not shown).
We next determined the localization of the xCIRP2 protein by fractionating an oocyte into cytoplasmic and nuclear fractions (Fig. 3D). xCIRP2 as well as FRGY2 were predominantly found in the cytoplasm (14,28). The possibility of nuclear leakage was excluded by testing the nuclear localization of TAF-I, which is involved in chromatin remodeling (Fig. 3D; 25). However, the nuclear fraction did contain small amounts of xCIRP2 and FRGY2. Their significance is currently unknown (see Discussion).
Major RNA-binding activities in oocyte cytoplasm
Microinjection of labeled RNA into oocytes followed by UV cross-linking allows detection of RNA-binding proteins in vivo (28,31). Previous experiments have demonstrated that when Xenopus histone H1 mRNA is injected, an RNA-binding activity that corresponds to a 60 kDa protein(s) is detected, which was proved by immunoprecipitation to contain FRGY2 (28). We repeated similar experiments, focusing on proteins of low molecular weight (Fig. 4). To this end, 32P-labeled histone H1 mRNA was injected into oocyte cytoplasm. The oocytes were then incubated for 1 h, lysed, irradiated with UV light and digested with RNase. Separation of the resultant products by SDS–PAGE revealed two prominent RNA-binding activities corresponding to 60–65 and 22–25 kDa proteins (Fig. 4A, lane 2, and data not shown). The RNA-binding activity associated with the high molecular weight band was immunoprecipitated with anti-FRGY2 antibodies (lane 4). The 60–65 kDa RNA-binding activity might also involve other proteins such as mRNP3, which has high homology and similar mobility to FRGY2 in SDS–polyacrylamide gels, and p54 RNA helicase (12,14,33,35). Note that protein bands detected by this method appear larger than the actual size of the RNA-binding proteins due to residual RNA molecules that remain cross-linked to proteins even after RNase treatment. We next set out to test whether the 22–25 kDa RNA-binding activity contained xCIRP2, based on the fact that both are localized in the cytoplasm and have similar molecular weights. By immunoprecipitation of the UV cross-linked samples with anti-xCIRP2 antibodies we found that this is indeed the case (lane 3). Furthermore, UV cross-linking of lysates from oocytes microinjected with CAT mRNA yielded similar results, although in these experiments RNA-binding activities corresponding to 90 and 40 kDa were more apparent than those with H1 mRNA (Fig. 4B).
Figure 4.
FRGY2 and xCIRP2 are the major mRNA-binding proteins in Xenopus oocytes. (A) 32P-labeled Xenopus histone H1 mRNA was injected into the cytoplasm of Xenopus oocytes. The oocytes were harvested 1 h later and the oocyte lysate then stored on ice without (lane 1) or with (lanes 2–4) irradiation with UV light and digested with RNase A. Aliquots of UV-irradiated samples were immunoprecipitated with antibodies against the recombinant xCIRP2 protein (lane 3) or anti-FRGY2 antibodies (lane 4). The samples were analyzed by 12% SDS–PAGE and visualized by autoradiography. (B) A similar experiment to that in (A) was performed with CAT mRNA. The UV-irradiated sample (lane 1) was immunoprecipitated with anti-xCIRP2 antibodies (lane 2) or anti-FRGY2 antibodies (lane 3).
By injecting labeled mRNA into the oocytes we found that both FRGY2 and xCIRP2 are major RNA-binding proteins. These experiments, however, do not show whether both proteins bind to the same RNA molecule. To examine this, we performed a gel retardation assay with recombinant proteins, whereby complexes formed by incubating labeled histone H1 mRNA with xCIRP2 or FRGY2 protein were separated by native gel electrophoresis (Fig. 5). The xCIRP2–RNA complexes migrated less far in the presence of higher amounts of xCIRP2, thus demonstrating the stable RNA-binding activity of xCIRP2 (Fig. 5A, lanes 1–4). When a constant small amount of FRGY2 was included in reactions containing different amounts of xCIRP2 the mobility of the xCIRP2–RNA complexes further decreased (compare lanes 2 and 6 or 3 and 7, respectively). These findings suggest that FRGY2 and xCIRP2 are capable of binding to the same RNA molecule. This finding was supported by data from experiments with anti-xCIRP2 antibodies whereby their addition to the RNA-binding reactions resulted in a specific supershift of complexes containing xCIRP2 (Fig. 5B, lane 6). Interestingly, FRGY2–RNA complexes formed in reactions including large amounts of FRGY2 were not affected by the addition of xCIRP2 (Fig. 5A, lanes 9–11) except when more than a 10 molar excess of xCIRP2 over FRGY2 was added (lane 12). The fact that no supershift was observed when anti-xCIRP2 antibodies were added to reactions containing xCIRP2 and FRGY2 in equimolar ratios (Fig. 5B, lane 8) indicates that xCIRP2 fails to bind to stable FRGY2–RNA complexes.
Figure 5.
Gel retardation assay with recombinant xCIRP2 and FRGY2. (A) A gel retardation assay using 32P-labeled histone H1 mRNA (lane 1) was performed with xCIRP2 (18 kDa) and FRGY2 (35 kDa) proteins. RNA was first mixed on ice with 0.07 (lanes 5–8) or 0.7 µg (lanes 9–12) FRGY2 protein and then 0.18 (lanes 2, 6 and 10), 0.9 (lanes 3, 7 and 11) or 3.6 µg (lanes 4, 8 and 12) xCIRP2 protein were added. The mixture was incubated at 30°C for 20 min and electrophoresed in a 1% agarose gel in 0.5× TBE buffer at room temperature. (B) Radiolabeled histone H1 mRNA, 0.36 µg (lanes 5–8) xCIRP2 protein and 0.07 (lanes 3–6) or 0.7 µg (lanes 7 and 8) FRGY2 protein were incubated as in (A). Anti-xCIRP2 antibody (0.6 µg, lanes 2, 4, 6 and 8) was then added and the mixture further incubated at 30°C for 10 min. The reactions were electrophoresed in a 1% agarose gel in 0.5× TBE buffer at room temperature.
Biochemical fractionation of the oocytes
The data presented thus far establish that xCIRP2 and FRGY2 constitute two major RNA-binding proteins in the oocyte cytoplasm. We next wished to determine whether xCIRP2 is associated with large particles in the oocyte cytoplasm. Accordingly, oocytes were subjected to subcellular fractionation by ultracentrifugation at 100 000 g (Fig. 6). When oocytes were homogenized in a buffer containing 100 mM NaCl, xCIRP2 was predominantly found in the pellet (P100) fraction. Most ribosomes also fractionated in P100 under these conditions. By increasing the salt concentration in the oocyte homogenization buffer to 500 mM, xCIRP2 was separated from the sediment fraction in the supernatant (S100) fraction. To elucidate features of the particles with which xCIRP2 is associated, oocyte homogenate prepared in 100 mM NaCl buffer was treated with RNase A and then ultracentrifuged. Following this treatment, rRNA, and presumably mRNA, was digested, thus likely freeing xCIRP2 to fractionate in S100. Taken together, these experiments demonstrate an RNA-dependent association of xCIRP2 with heavily-sedimenting particles in oocytes.
Figure 6.

xCIRP2 protein is associated with heavily sedimenting structures. Oocytes were fractionated as described in Materials and Methods. (Top) Total lysate (T) and the S100 (S) and P100 (P) fractions were analyzed by immunoblotting using anti-xCIRP2 antibodies. Oocytes were homogenized in a buffer containing 100 (lanes 1–3) or 500 mM NaCl (lanes 4–6). For lanes 7–9, total lysate (100 mM NaCl) was digested with RNase A prior to ultracentrifugation. (Bottom) RNA was purified from S100 (lanes 2, 5 and 8) and P100 (lanes 3, 6 and 9), electrophoresed in a 1% agarose gel containing formaldehyde and visualized by staining with ethidium bromide. 28S and 18S indicate rRNAs.
To further characterize the xCIRP2 subcellular localization in oocytes, we conducted density gradient centrifugation. When oocyte lysate was loaded on a sucrose gradient and centrifuged xCIRP2 fractionated exclusively to the top of the gradient (data not shown). This was unexpected, given that it had fractionated in P100 as described above. We reasoned that xCIRP2 association with heavy particles is unstable under our conditions for sucrose gradients. Similarly, Visa et al. (7) found that hrp36 protein, the Chironomus tentans homolog of hnRNPA1 with two RRMs and a C-terminal glycine-rich region, fractionate to the top of the sucrose gradient. They used UV cross-linked cell lysate to observe polysomal association of the protein. We then fixed the oocyte lysate by irradiation with UV light prior to fractionation in the sucrose gradient (Fig. 7A). A small portion of xCIRP2 protein co-fractionated with ribosomes, while a large amount remained at the top of the gradient. The data suggest that at least some of the xCIRP2 protein is associated with ribosomes or particles of similar size in the oocytes. Consistent with previous studies, FRGY2 was distributed to both the mRNP and ribosomal fractions (33,36).
Figure 7.
xCIRP2 co-sediments with ribosomes in density gradients. (A) Oocyte lysate was irradiated with UV light and fractionated through a 15–40% sucrose gradient. The distribution of xCIRP2 and FRGY2 was examined by immunoblotting (western). RNA prepared from every other fraction was electrophoresed in an agarose gel containing formaldehyde and transferred to a nylon membrane. The membrane was stained with methylene blue (Total RNA; 28S and 18S indicate rRNAs) and histone H1 mRNA was detected by northern blotting. The positions of stored mRNP and ribosomes are indicated. (B) Oocyte lysate was fixed either by irradiation with UV light or with formaldehyde. The samples were fractionated through 20–60% Nicodenz gradients. Proteins and RNA were detected as in (A). Positions of free proteins, mRNP and ribosomes are indicated.
In our final experiments oocyte lysate was fractionated through Nicodenz gradients which are used to separate RNA-containing particles by their densities (Fig. 7B; 33,34,37). When the UV cross-linked oocyte lysate was used, xCIRP2 was detected mostly as proteins free of RNA with only small amounts in the fractions co-sedimenting with ribosomes, which is consistent with results from the sucrose gradient. We next fixed the lysate with 4% formaldehyde. Using this fixative xCIRP2 protein co-sedimented with ribosomes in the Nicodenz gradient, while FRGY2 was most abundant in the mRNP fractions. As a control, Xenopus TAF-Iβ remained in the fractions free of RNA after formaldehyde fixation, suggesting that the co-sedimentation of xCIRP2 with ribosomes is specific. Based on these results we concluded that in oocytes xCIRP2 is associated with ribosomes, but that most of the xCIRP2 is released prior to, or through, centrifugation in gradients with no or with inefficient fixation, such as irradiation with UV light.
DISCUSSION
In this paper we have identified xCIRP2 as one of the major RNA-binding proteins in Xenopus oocyte cytoplasm. Data from immunoblotting experiments with recombinant protein show that >80 ng of xCIRP2 protein apparently accumulates in a stage VI oocyte (data not shown). FRGY2 protein is present at up to 5% of the total protein content in a previtellogenic oocyte and its amount remains constant during oogenesis (33,38). These establish that both RNA-binding proteins are abundant in full-grown Xenopus oocytes.
By cellular fractionation we have shown that xCIRP2 is predominantly localized in the oocyte cytoplasm. We confirmed this result by overexpressing tagged xCIRP2 in oocytes (data not shown). In contrast, mammalian CIRP is localized in the nuclei of cultured cells, which was demonstrated by immunocytochemical analysis of cold-induced CIRP and by overexpression of CIRP fused to green fluorescent protein (GFP) (18). There are several possible explanations for this discrepancy. Firstly, it is possible that CIRP localization is dependent on cell type; CIRP might be nuclear in somatic cells while being cytoplasmic in female germ cells. Consistent with this hypothesis, it has been reported that in mouse testis CIRP localizes in the cytoplasm of spermatocytes but in the nuclei of round spermatids (19). Alternatively, mammalian CIRP may possess a nuclear localization signal that is missing in xCIRP2 despite their very high homology. It is important to note, however, that this alternative model does not require that xCIRP2 has a cytoplasmic localization or nuclear export/exclusion signals, since the nucleo-cytoplasmic ratio of Xenopus oocytes is ∼1:25 and since in our fractionation we detected residual amounts of xCIRP2 and FRGY2 in the nuclear fraction (Fig. 3D). Lastly, the difference in localization between mammalian and Xenopus CIRP could be attributed to the detection methods employed, i.e. manual isolation of the oocyte nuclei and microscopic analysis of fluorescent signals. Although other explanations cannot be excluded, we recently observed a nuclear localization of GFP–xCIRP2 overexpressed in HeLa cells (unpublished observation), thus supporting the idea that CIRP localization is cell type-dependent.
By fractionation through density gradients we found an association of xCIRP2 with ribosomes. The fact that only a small part of xCIRP2 co-sedimented with ribosomes in the UV cross-linked lysates raises the possibility that this association takes place during or after preparing the oocyte lysate. However, other data suggest that this possibility is unlikely. First, xCIRP2 was predominantly found in the ribosome-containing P100 fraction when oocyte lysate was fractionated into S100 and P100 fractions. As this method is widely used as an initial step in biochemical fractionation (32,39), the conditions are unlikely to skew the native state. Second, it has been reported that C.tentans hrp36 protein, which shares several structural features with CIRP, behaves similarly (7). Furthermore, additional evidence for ribosome association with hrp36 has been provided by immunoelectron microscopy (7). Finally, following fixation with 4% formaldehyde almost all the xCIRP2 co-sedimented with ribosomes in a Nicodenz gradient. Therefore, it is most probable that xCIRP2 is indeed associated with ribosomes in oocytes.
We found that injection of histone H1 or CAT mRNA into oocyte cytoplasm led to identification of at least two major RNA-binding proteins. Of note, the injected histone H1 and CAT mRNAs are actively translated in oocytes, in contrast to endogenous histone H1 mRNA, which is translationally silenced (28,29). It is thus plausible that the RNA-binding proteins detected in our UV cross-linking experiments are bound to translationally engaged mRNAs. This is consistent with the finding that FRGY2 is associated not only with stored mRNPs but also with ribosome-bound mRNAs (33,36). Rabbit p50, which is the major core protein of mRNPs in somatic cells and structurally homologous to FRGY2, can also be found in free mRNPs as well as ribosome-bound mRNPs (40–42). The molar ratio p50:mRNA is lower in ribosome-bound mRNPs than in free mRNPs. Our gel retardation assay revealed that xCIRP2 is able to bind simultaneously to the same mRNA molecule as FRGY2 unless FRGY2 has formed a stable complex with the mRNA. From these results it seems reasonable to propose that xCIRP2 is associated with or a component of ribosome-bound mRNPs.
Generally, cellular responses to a downshift of environmental temperature are not well understood, although expression of mammalian CIRP is induced by exposing cells to lower temperatures (18). However, studies with bacterial cells have established an intriguing relationship between the protein synthesis machinery and the cold shock response (43,44). Upon a downshift of temperature transient inhibition of overall translation followed by a lag of cell growth results. Polysomes transiently decrease as 70S monosomes and ribosomal subunits accumulate in the cell. During the lag in growth, synthesis of a number of cold shock proteins is induced (44). Some of the cold shock proteins are translation initiation factors or ribosome-associated proteins, which are required for efficient ribosome function for growth resumption after a cell has become adapted to low temperature. In E.coli the major cold shock protein is CspA, which functions as an RNA chaperone (45). Interestingly, inhibitors of protein synthesis, such as chloramphenicol, induce synthesis of CspA concomitant with a decrease in the number of polysomes (46). Thus, it has been proposed that ribosomes are the physiological sensor for the cold shock response (43). In this study we show that xCIRP2 and FRGY2 are major RNA-binding proteins in Xenopus oocytes. FRGY2 consists of a cold shock domain, which is a sequence-specific RNA-binding domain showing 43% identity with E.coli CspA, and a C-terminal tail domain involved in translational repression (8–10). Therefore, xCIRP2, being homologous to a mammalian cold shock RNA-binding protein, might regulate translational efficiency of specific mRNAs by association with ribosomes in Xenopus oocytes, in which translational activity is thought to be kept relatively low by the abundant general translational repressive protein FRGY2. Mammalian CIRP is required for cold-induced growth suppression of cultured cells (18). Therefore, the results described in this paper, together with data describing the behavior of bacterial cold shock proteins, raise the possibility that CIRP affects cell growth by modulating ribosome function in eukaryotic cells.
During the preparation of this manuscript, Saito et al. (47) reported the expression of XCIRP mRNA in an adult frog. They found that the expression of XCIRP in the brain is increased by cold treatment, but that no significant change occurs in the liver. In contrast, we observed no increase in xCIRP2 expression by cold treatment in oocytes (data not shown). Although a possible association of xCIRP2 with ribosomes in oocytes is demonstrated in this paper, cellular functions of xCIRP2 and other CIRPs remain elusive. It is possible that CIRP has distinct functions in the nucleus as well as the cytoplasm of different cell types. It is worth testing whether xCIRP2 binds to (pre-)mRNA in the nucleus and is exported to the cytoplasm associated with mRNA which is then recruited to the ribosomes. Our preliminary experiments to neutralize functions of endogenous xCIRP2 in the oocytes by injecting antibodies were not successful, probably due to the abundance of xCIRP2 protein. By fractionation in a sucrose gradient xCIRP2 was found to be associated with monosomes rather than polysomes. We therefore cannot rule out the possibility that xCIRP2 associates with ribosomes independent of mRNA. The finding that the RGG domain of nucleolin associates with ribosomal proteins argues for this possibility (48). If this were the case, CIRP may modulate ribosome functions by direct association in oocytes. Experiments such as identification of specific RNA sequence preferences of xCIRP2 will better clarify its biological significance.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Alan P. Wolffe for providing anti-FRGY2 antibody and comments on the manuscript. We also thank Dr Kyosuke Nagata for providing anti-TAF-I antibody. We are grateful to The Division of Laboratory Animal Research at RIKEN for preparing the anti-xCIRP2 antibodies. This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan, by a grant for the Bioarchitect Research Program from RIKEN and by a grant from the Nissan Science Foundation.
DDBJ/EMBL/GenBank accession no. AB044535
REFERENCES
- 1.Siomi H. and Dreyfuss,G. (1997) Curr. Opin. Genet. Dev., 7, 345–353. [DOI] [PubMed] [Google Scholar]
- 2.Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 3.Daneholt B. (1997) Cell, 88, 585–588. [DOI] [PubMed] [Google Scholar]
- 4.Mayeda A. and Krainer,A.R. (1992) Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- 5.Izaurralde E., Jarmolowski,A., Beisel,C., Mattaj,I.W., Dreyfuss,G. and Fischer,U. (1997) J. Cell Biol., 137, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinol-Roma S. and Dreyfuss,G. (1992) Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- 7.Visa N., Alzhanova-Ericsson,A.T., Sun,X., Kiseleva,E., Bjorkroth,B., Wurtz,T. and Daneholt,B. (1996) Cell, 84, 253–264. [DOI] [PubMed] [Google Scholar]
- 8.Sommerville J. and Ladomery,M. (1996) FASEB J., 10, 435–443. [DOI] [PubMed] [Google Scholar]
- 9.Graumann P.L. and Marahiel,M.A. (1998) Trends Biochem. Sci., 23, 286–290. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto K. and Wolffe,A.P. (1998) Trends Cell Biol., 8, 318–323. [DOI] [PubMed] [Google Scholar]
- 11.Tafuri S.R. and Wolffe,A.P. (1990) Proc. Natl Acad. Sci. USA, 87, 9028–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deschamps S., Viel,A., Denis,H. and le Maire,M. (1991) FEBS Lett., 282, 110–114. [DOI] [PubMed] [Google Scholar]
- 13.Deschamps S., Viel,A., Garrigos,M., Denis,H. and le Maire,M. (1992) J. Biol. Chem., 267, 13799–13802. [PubMed] [Google Scholar]
- 14.Murray M.T., Schiller,D.L. and Franke,W.W. (1992) Proc. Natl Acad. Sci. USA, 89, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birney E., Kumar,S. and Krainer,A.R. (1993) Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burd C.G. and Dreyfuss,G. (1994) Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 17.Nagai K., Oubridge,C., Ito,N., Avis,J. and Evans,P. (1995) Trends Biochem. Sci., 20, 235–240. [DOI] [PubMed] [Google Scholar]
- 18.Nishiyama H., Itoh,K., Kaneko,Y., Kishishita,M., Yoshida,O. and Fujita,J. (1997) J. Cell Biol., 137, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishiyama H., Danno,S., Kaneko,Y., Itoh,K., Yokoi,H., Fukumoto,M., Okuno,H., Millan,J.L., Matsuda,T., Yoshida,O. and Fujita,J. (1998) Am. J. Pathol., 152, 289–296 [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiyama H., Higashitsuji,H., Yokoi,H., Itoh,K., Danno,S., Matsuda,T. and Fujita,J. (1997) Gene, 204, 115–120. [DOI] [PubMed] [Google Scholar]
- 21.Xue J.H., Nonoguchi,K., Fukumoto,M., Sato,T., Nishiyama,H., Higashitsuji,H., Itoh,K. and Fujita,J. (1999) Free. Radic. Biol. Med., 27, 1238–1244. [DOI] [PubMed] [Google Scholar]
- 22.Bhatia R., Gaur,A., Lemanski,L.F. and Dube,D.K. (1998) Biochim. Biophys. Acta, 1398, 265–274. [DOI] [PubMed] [Google Scholar]
- 23.Uochi T. and Asashima,M. (1998) Gene, 211, 245–250. [DOI] [PubMed] [Google Scholar]
- 24.Darnbrough C.H. and Ford,P.J. (1981) Eur. J. Biochem., 113, 415–424. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto K., Nagata,K., Miyaji-Yamaguchi,M., Kikuchi,A. and Tsujimoto,M. (1999) Mol. Cell. Biol., 19, 6940–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tafuri S.R. and Wolffe,A.P. (1992) New Biol., 4, 349–359. [PubMed] [Google Scholar]
- 27.Bouvet P., Matsumoto,K. and Wolffe,A.P. (1995) J. Biol. Chem., 270, 28297–28303. [DOI] [PubMed] [Google Scholar]
- 28.Bouvet P. and Wolffe,A.P. (1994) Cell, 77, 931–941. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto K., Wassarman,K.M. and Wolffe,A.P. (1998) EMBO J., 17, 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almouzni G. and Wolffe,A.P. (1993) Genes Dev., 7, 2033–2047. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto K., Meric,F. and Wolffe,A.P. (1996) J. Biol. Chem., 271, 22706–22712. [DOI] [PubMed] [Google Scholar]
- 32.Wormington M. (1991) Methods Cell Biol., 36, 167–183. [DOI] [PubMed] [Google Scholar]
- 33.Tafuri S.R. and Wolffe,A.P. (1993) J. Biol. Chem., 268, 24255–24261. [PubMed] [Google Scholar]
- 34.Meric F., Matsumoto,K. and Wolffe,A.P. (1997) J. Biol. Chem., 272, 12840–12846. [DOI] [PubMed] [Google Scholar]
- 35.Ladomery M., Wade,E. and Sommerville,J. (1997) Nucleic Acids Res., 25, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sommerville J. (1990) J. Reprod. Fertil., 42 (suppl.), 225–233. [PubMed] [Google Scholar]
- 37.Wormington M., Searfoss,A.M. and Hurney,C.A. (1996) EMBO J., 15, 900–909. [PMC free article] [PubMed] [Google Scholar]
- 38.Sommerville J. and Ladomery,M. (1996) Chromosoma, 104, 469–478. [DOI] [PubMed] [Google Scholar]
- 39.Siomi M.C., Zhang,Y., Siomi,H. and Dreyfuss,G. (1996) Mol. Cell. Biol., 16, 3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minich W.B., Maidebura,I.P. and Ovchinnikov,L.P. (1993) Eur. J. Biochem., 212, 633–638. [DOI] [PubMed] [Google Scholar]
- 41.Evdokimova V.M., Wei,C.L., Sitikov,A.S., Simonenko,P.N., Lazarev,O.A., Vasilenko,K.S., Ustinov,V.A., Hershey,J.W. and Ovchinnikov,L.P. (1995) J. Biol. Chem., 270, 3186–3192. [DOI] [PubMed] [Google Scholar]
- 42.Davydova E.K., Evdokimova,V.M., Ovchinnikov,L.P. and Hershey,J.W. (1997) Nucleic Acids Res., 25, 2911–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thieringer H.A., Jones,P.G. and Inouye,M. (1998) Bioessays, 20, 49–57. [DOI] [PubMed] [Google Scholar]
- 44.Phadtare S., Alsina,J. and Inouye,M. (1999) Curr. Opin. Microbiol., 2, 175–180. [DOI] [PubMed] [Google Scholar]
- 45.Yamanaka K., Fang,L. and Inouye,M. (1998) Mol. Microbiol., 27, 247–255. [DOI] [PubMed] [Google Scholar]
- 46.Jiang W., Jones,P. and Inouye,M. (1993) J. Bacteriol., 175, 5824–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito T., Sugimoto,K., Adachi,Y., Wu,Q. and Mori,K.J. (2000) Comp. Biochem. Physiol., 125B, 237–245. [DOI] [PubMed] [Google Scholar]
- 48.Bouvet P., Diaz,J.J., Kindbeiter,K., Madjar,J.J. and Amalric,F. (1998) J. Biol. Chem., 273, 19025–19029. [DOI] [PubMed] [Google Scholar]