Abstract
Background and objective
Genetic polymorphism studies in families and twins indicated the heritability of depression. However, the association between genes with genetic polymorphism and depression provides various findings and remains unclear. Therefore, we conducted a systematic review and meta-analysis to determine the genes with their polymorphism associated with the symptomatic depression known as major depressive disorder (MDD).
Materials and methods
PubMed and Scopus were searched for relevant studies published before May 22, 2023 (1968–2023), and 62 were selected for this review. The study’s bias risk was investigated using the Newcastle–Ottawa scale. Gene functional enrichment analysis was investigated for molecular function (MF) and biological process (BP) and pathways. A meta-analysis of the studied genes that were replicative in the same single nucleotide polymorphism was conducted using a random-effect model.
Results
The 49 genes involved in MDD were studied and engaged in several pathways, such as tryptophan metabolism or dopaminergic and serotonergic synapses. Based on gene overlapping in MF and BP, 13 genes with polymorphisms were identified as related to MDD. Most of them were only studied once. Solute carrier family 6 member 4 (SLC6A4) overlapping between MF and BP and brain-derived neurotrophic factor (BDNF) as unique to BP were replicative studied and used in the meta-analysis. The polymorphism of SLC6A4 SS and LS genotypes increased the occurrence of MDD development but not significantly [odd ratio (OR) = 1.39; 95% confidence interval (CI) = 0.87–2.22; P = 0.16 and OR = 1.13; 95% CI = 0.84–1.53; P = 0.42, respectively]. A similar result was observed for BDNF rs6265 GG (OR = 1.26; 95% CI = 0.78–2.06; P = 0.35) and BDNF rs6265 AA genotypes (OR = 1.12; 95% CI = 0.77–1.64; P = 0.56). These studies indicated low bias and significant heterogeneity.
Conclusion
At least 13 studied genes with polymorphisms were involved in MDD development according to MF and BP, but not significantly. These results suggest that MDD development risk factors might require genetic and other factors for interaction and induction.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12888-024-06195-z.
Keywords: Single nucleotide polymorphism, SLC6A4, BDNF, Major depressive disorder
Introduction
The DSM-V defines major depressive disorder (MDD) as the presence of emotional, neurovegetative, and neurocognitive symptoms for at least two weeks [1, 2]. MDD is a major common mental disorder, a type of depression causing psychological, physical, and social impairments [3, 4]. The lifetime prevalence of MDD is 14–18% worldwide among people suffering from depression [2, 5–7]. Various factors influence the development of depression, including genetics [8–11], viral infection [12–15], substance use [16, 17], chronic illnesses [18, 19], and poor nutrition [20–23].
Currently, clinical examination and subjective assessment of depressive symptoms are the main tools to diagnose MDD; no approved biomarker is available to diagnose psychiatric disorders [24]. Some recently discovered biomarkers, such as BDNF and 5-hydroxy tryptamine (5-HT), can be effectively used for diagnosis and treatment due to their low sensitivity and specificity [24, 25]. Genetic polymorphism in specific gene alleles is a factor influencing depression.
A meta-analysis of family and twin genetic research indicated that depression has a 37% heritability rate [95% confidence interval (CI) = 31−42%] [11]. Numerous studies identified candidate genes involved in the development of depression based on its pathophysiology in different theories, such as the monoamine, neuroendocrine mechanism, neurotrophic, and neuroinflammation theories [2, 26, 27]. Consequently, 100 candidate genes were analyzed to determine potential associations between their alleles and the risk of developing depression [8].
To date, 10 significant common genes with specific variants for MDD based on molecular pathways have been reported. These common genes are present in serotonergic genes; solute carrier family 6 member 4 (SLC6A4), serotonin-transporter-linked promoter region (5-HTTLPR), 5-hydroxytryptamine receptor 1 A (HTR1A) C1019G, SLC6A4 variable-number tandem repeat (VNTR), and tryptophan hydroxylase 1 (TPH1) A218C; dopaminergic genes, such as dopamine transporter 1 (DAT1) 40 bp, dopamine receptor D4 (DRD4) 48 bp, catechol-O-methyltransferase (COMT) Val158Met; vascular genes, such as apolipoprotein E (APOE) e4, angiotensin I converting enzyme (ACE) Ins/Del, methylenetetrahydrofolate reductase (MTHR) C677T, and MTHR A1298C; glutamatergic genes, such as D-amino acid oxidase activator (DAOA) G72/G30 rs3918342; and neurotrophic genes, such as brain-derived neurotrophic factor (BDNF) Val66Met [28]. Nevertheless, most of the study findings on candidate genes for MDD provided conflicting results [9, 28, 29]. Due to different populations, insufficient statistical power in small sample sizes, and different methodologies [30, 31]. Therefore, meta-analysis can relieve the conflict candidate gene finding.
SLC6A4 is one of the most widely identified genes that encode the serotonin transporter (5-HTT) with a 5-HTTLPR polymorphism [32]. A meta-analysis of SLC6A4 SS genotypes revealed significantly associated with an increased MDD risk in the Caucasian population (OR = 1.14; CI = 1.15–1.72) but not in the Asian population (OR = 1.04; 95% CI = 0.51–2.16) [33]. While another reported a significant association with the pool odd ratio (OR) of S allele 1.11 (CI = 1.04–1.19) in mixed-ethnicity [30]. However, the meta-analysis has been inconsistent as well as the single study. MDD is a multifactorial disorder due to genetic factors related to environmental factors that increase the risk of MDD development. There are several environmental have been identified with genetics; childhood adversities, maternal stress, stressful life events, age, and female sex [34–36].
The BDNF Val66Met polymorphism (rs6265) was significantly associated with life stress and depression (P = 0.03), and a significant interaction between stressful life events and the Met variant of BDNF Val66Met was observed (P = 0.01) [37]. On the other hand, some studies indicated that BDNF Val66Met polymorphism was not associated with MDD (OR = 0.96; 95% CI = 0.89–1.05; P = 0.402) [38]. Therefore, identified genetics should be combined with environmental or other factors to increase the association of genetics in MDD. In addition, the candidate genes are also shared with other psychiatric disorders [39]. Consequently, this study aims to determine the common genes with genetic polymorphism of MDD. Identification of common genes based on significant biological process (BP) and molecular function (MF) to enhance the related common genes with MDD in specific terms of BP and MF involved in developing MDD. Moreover, conducted a meta-analysis to investigate the association between gene polymorphism with MDD.
Materials and methods
Literature search
A systematic review and meta-analysis were conducted to determine the genes with genetic polymorphism associated with MDD following the 2020 PRISMA. PubMed and Scopus were searched for relevant studies published before May 22, 2023 (1968–2023) using the following keywords: ‘polymorphism’ OR ‘polymorphisms’ OR ‘mutation’ OR ‘mutations’ AND ‘depression’ OR ‘depressive’ OR ‘major depressive’ NOT ‘review article’ NOT ‘meta-analysis’ NOT ‘book’ OR ‘books’ NOT ‘document’ OR ‘documents’ on PubMed and ‘polymorphism’ OR ‘polymorphisms’ OR ‘mutation’ OR ‘mutations’ AND ‘depression’ OR depressive ‘OR ‘major depressive’ on Scopus.
Selection criteria
The studies were screened based on the following inclusion criteria: (a) case-control study; (b) control subjects satisfying the Hardy–Weinberg equilibrium; (c) providing complete data of cases and controls to calculate the OR with 95% CI; (d) studies on humans. The exclusion criteria were: (a) studies on patients with MDD combined with other diseases; (b) studies on depression types other than MDD; (c) studies on the association between genetics and depression with another risk factor; (d) lack of usable genotype frequency data; (e) non-English language.
Data extraction
Two reviewers (A. S. and T. E.) independently extracted the following information: author, year of publication, country of participants, gene name, method of single nucleotide polymorphism (SNP) test, the number of depression patients and controls, sample type, and outcome measure raw P-values and ORs for genotype frequencies. Inconsistencies were resolved via discussion.
Quality assessment
The quality and bias risk of the studies were assessed by two independent reviewers (A. S. and C. P.) using the Newcastle–Ottawa scale (NOS) for case-control studies. The NOS was assigned for three parts: selection criteria, comparability, and exposure, with an overall score out of 9. Each study was defined as one of three categories: low-quality score of 1–3, medium-quality score of 4–5, and high-quality score of 6–9.
Gene functional enrichment analysis
After being screened with eligibility criteria, genes were analyzed using a web-based gene functional classification tool (DAVID Bioinformatics). Gene ontology (GO) was conducted in three categories: biological process (BP), molecular function (MF), and cellular component (CC). The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was also conducted. The enrichment of GO and KEGG was analyzed statistically with a Benjamini–Hochberg test at P < 0.05.
Statistical analysis
The genes with the most replicated studies were screened with RevMan 5.4 software to analyze the difference in overall genotypes between cases and controls. The heterogeneity between studies was tested using the χ2 test (95% CI) and I². P < 0.05 or I2 > 40% represented heterogeneity between studies, and P > 0.05 or I2 < 40% meant homogeneity.
Results
Literature search and eligible studies
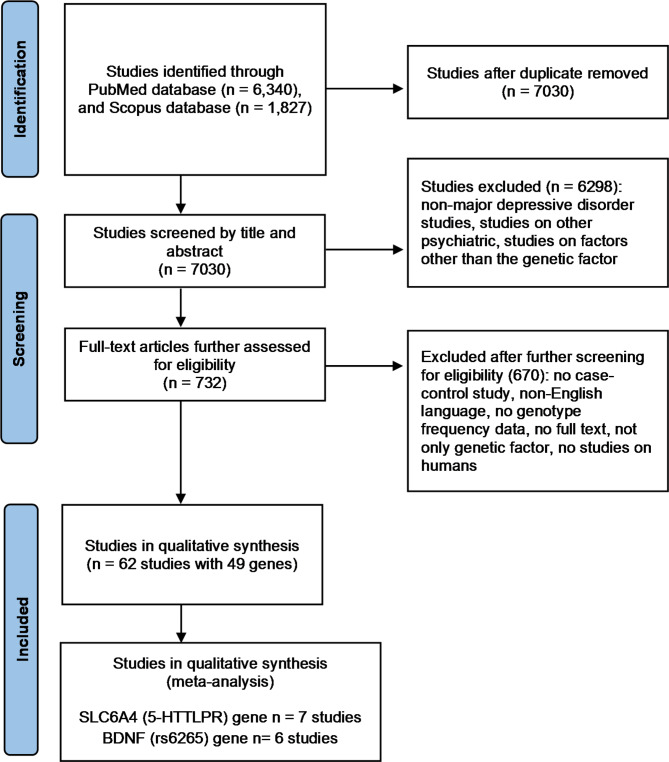
The literature search and eligible studies identified 6,430 studies on PubMed and 1,827 studies on Scopus. As a result, 1,137 studies were duplicated and removed. After screening by title and abstract, 6,298 studies were excluded because they were not depression studies or were studies on other psychiatric factors or factors other than genetic. The remaining 732 studies were assessed as full-text articles for eligibility, and 62 were finally included in the qualitative synthesis of genes with genetic polymorphisms associated with MDD. The flowchart of the literature search is presented in Fig. 1.
Fig. 1.
Flowchart of literature review process [40]
Study quality
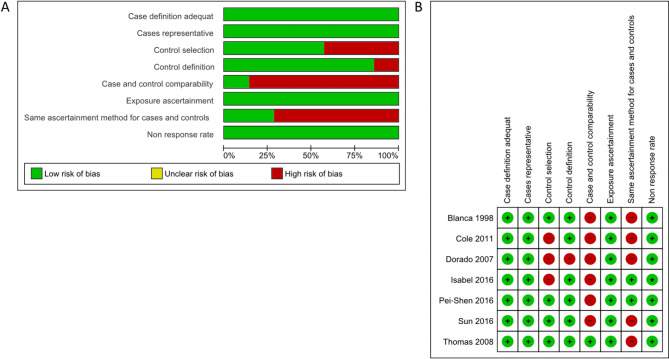
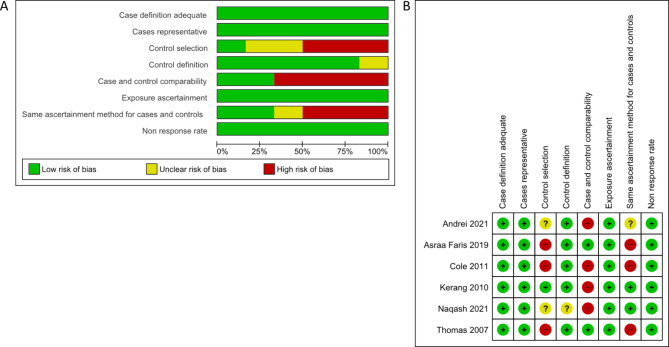
The NOS tool assessed the quality of the 62 case-control studies. Forty-five studies (73%) with a score ˃ 6 were deemed to have a low bias risk, while the remaining 17 studies (27%) obtained a score of 4–5 (Supplementary Table 1). Sixty-two research with 49 gene lists were analyzed for functional enrichment. SLC6A4 and BDNF had more replicative studies in the same SNP and were used for the meta-analysis. Seven SLC6A4 studies displayed a low bias risk in case definition adequate, case representative, control selection, control definition, exposure ascertainment, and non-response rate, but a high bias risk in the case and comparability and same ascertainment method for cases and controls (Fig. 2A, B). In six BDNF studies, a low bias risk of bias was observed regarding case definition sufficient, case representative, control definition, exposure ascertainment, and non-response rate, whereas control selection, case and comparability, and the same ascertainment method for cases and controls displayed a high bias risk (Fig. 3A, B)
Fig. 2.
Bias risk by the NOS assessment tool in seven studies of SLC6A4. (A) Risk of bias graph. (B) Risk of bias summary
Fig. 3.
Risk of bias by the NOS assessment tool in six studies of BDNF. (A) Risk of bias graph. (B) Risk of bias summary
Gene functional enrichment analysis
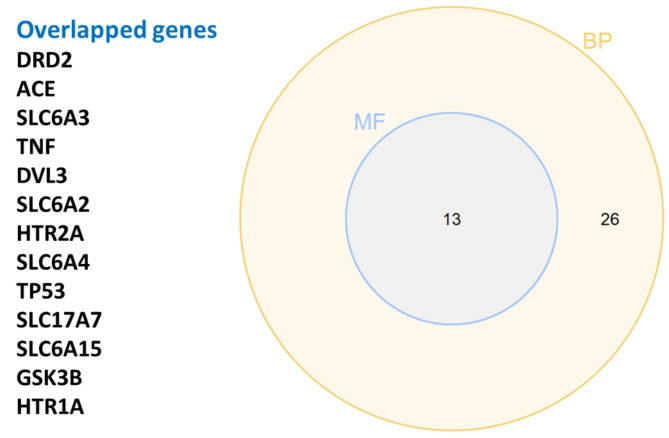
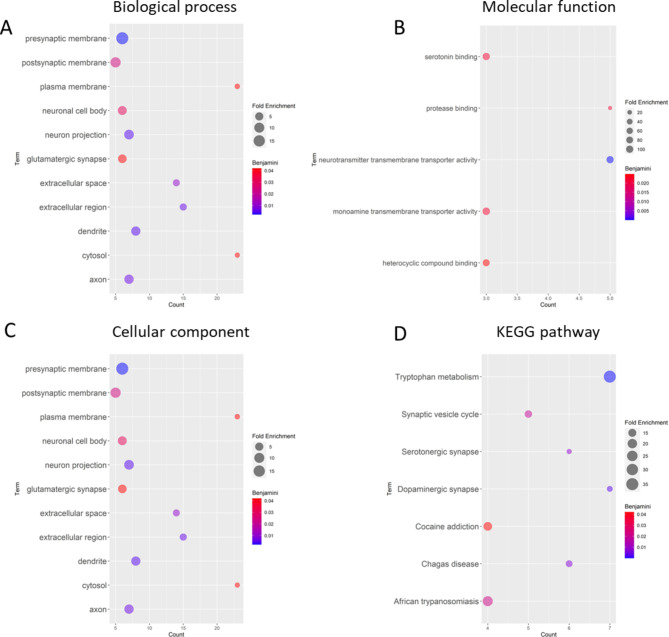
Among the 62 studies, 49 genes were selected and used to perform gene functional enrichment analysis. Several theories were involved with the 49 genes; 22 genes related to the monoamine hypothesis, 13 genes neurotrophic hypothesis, 8 genes neuroinflammation hypothesis, 4 neuroendocrine mechanisms hypothesis, and 6 genes involved in another pathway. There are various sample types were found in 49 genes, and whole blood samples (74%) are most commonly used to identify the gene polymorphism. Furthermore, 55% of the identifications were performed using PCR methods.12 genes with 25 SNPs had significance with MDD (Supplementary Table 3). The GO terms were as follows: 168 BP, 34 MF, and 22 CC. Additionally, the KEGG pathway enrichment was identified in 69 terms. The statistical significance (P < 0.05) in BP and MF was performed to assess the overlapped genes. We identified 13 genes with genetic polymorphisms that could be related to MDD based on MF and BP (Fig. 4). The GO enrichment of BP, which has a high significance, was commonly found in neurotransmitter transport, response to xenobiotic stimulus, and dopamine catabolic process (Fig. 5A). Binding and neurotransmitter transporter activity were commonly found in molecular function (Fig. 5B). The dopaminergic, serotonergic, and tryptophan metabolism synapses were commonly found in the KEGG pathway (Fig. 5D). The functional enrichment indicated that monoamine theories were the target of 49 genes studied.
Fig. 4.
Overlapped genes from MF and BP enrichment analysis
Fig. 5.
GO and KEGG enrichment analysis based on 50 genes. (A) GO enrichment of BP. (B) GO enrichment of MF. (C) GO enrichment of CC. (D) KEGG enrichment analysis
Meta-analysis of SLC6A4 and BDNF
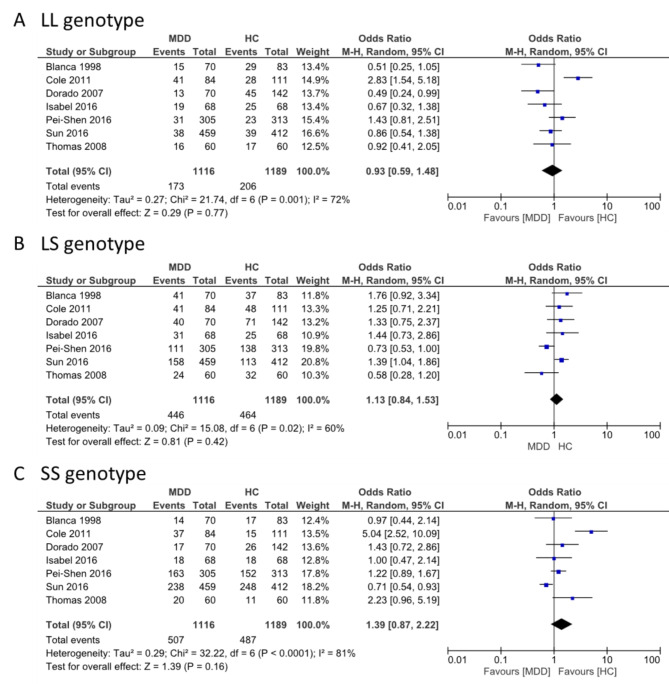
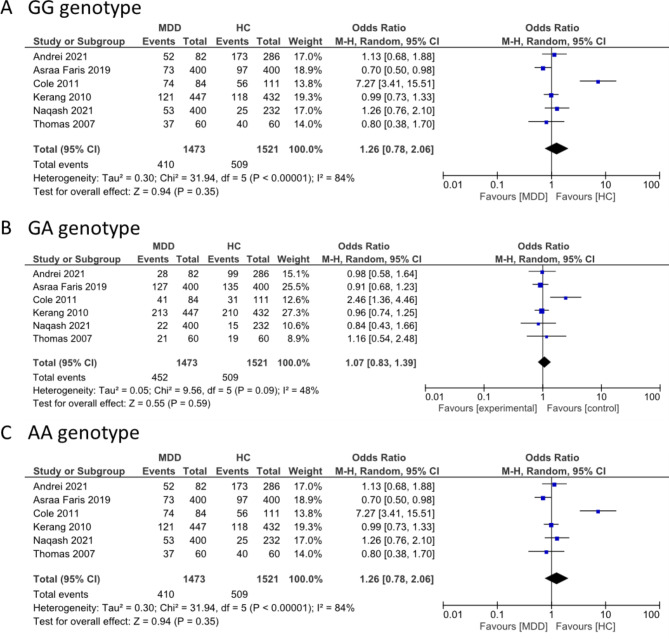
SLC6A4 and BDNF were selected for the meta-analysis because they had more replicative studies in the same SNP. SLC6A4 was found among the overlapped genes between BP and MF. Three genotype frequencies (L/L, L/S, and S/S) of SLC6A4 rs25531 (5-HTTLPR) were compared between MDD cases and controls. The L/L genotype displayed significant heterogeneity (I2 = 72%; P = 0.001). The overall effects test indicated no significant differences in the L/L genotype between MDD and controls (OR = 0.93, 95% CI = 0.59–1.48; P = 0.77). The L/S and S/S genotypes displayed significant heterogeneity (I2 = 60%, P = 0.02 and I2 = 81%, P < 0.0001), and the overall effects test indicated no significant differences between MDD and controls. However, the polymorphism of SLC6A4 SS and LS genotypes increased the occurrence of MDD development, although not significantly (OR = 1.39; 95% CI = 0.87–2.22; P = 0.16 and (OR = 1.13; 95% CI = 0.84–1.53; P = 0.42, respectively) (Fig. 6). BDNF was unique in BP. Three genotype frequencies (G/G, G/A, and A/A) in BDNF rs6265 were compared between MDD and controls. The G/G genotype displayed significant heterogeneity (I2 = 84%, P < 0.00001) and no significant differences in overall effects (OR = 1.26, 95% CI = 0.78–2.06, P = 0.35). The G/A and A/A genotypes displayed no significant heterogeneity (I2 = 48%, P = 0.09), (I2 = 43%, P = 0.12) respectively, and the overall effects test indicated an increase in the occurrence of MDD development but no statistical significance (OR = 1.07; 95% CI = 0.83–1.39, P = 0.59 and OR = 1.12, 95% CI = 0.77–1.64, P = 0.56) (Fig. 7).
Fig. 6.
Forest plot for the association between the polymorphism of SLC6A4 and MDD for the three genotypes (A) LL, (B) LS, and (C) SS
Fig. 7.
Forest plot for the association between BDNF Val66Met polymorphism (rs6265) and MDD for the three genotypes (A) GG, (B) GA, and (C) AA
Discussion
This systematic review and meta-analysis retrieved 62 studies, and 49 genes with genetic polymorphisms associated with MDD were studied. Forty-nine gene polymorphisms were determined for the GO enrichment analysis. Thirteen gene polymorphisms overlapped between (BP) and MF, which could be related to MDD (Fig. 4 and Supplementary Table 2). The replicative studied gene in a similar SNP, SLC6A4, and BDNF were used for meta-analysis and did not display any significant difference between control and MDD cases (Figs. 6 and 7).
The GO enrichment analysis indicated that these 49 studied genes are mainly related to the monoamine pathway, suggesting that most target gene polymorphism was focused only on the monoamine hypothesis. However, several theories of depression development are available, including the monoamine hypothesis, neuroendocrine mechanisms, neurotrophic hypothesis, and neuroinflammation [2, 26, 27]. The monoamine hypothesis was the first formulation to explain the pathogenesis of depression [41–43]; it was based on a deficiency of the neurotransmitters norepinephrine and serotonin in the brain [44]. Additionally, the monoamine theory was used to develop the first antidepressant drugs, which increased monoamine levels in the synaptic cleft [45]. Therefore, many studies have investigated candidate genes for MDD based on pathogenesis, and several candidate genes have been identified. In recent decades, neuroinflammation has been extensively studied regarding the association between inflammation and depression. However, the mechanism of neuroinflammation involved in the cause of depression remains unclear [46–48].
The candidate genes in monoamine theory were identified, SLC6A4 5-HTTLPR polymorphism is the most common. SLC6A4 encodes the serotonin transporter (5-HTT) for the serotonin reuptake from the synaptic cleft to the presynaptic region [49]. The 5-HTTLPR polymorphism study includes short (S) and long (L) alleles. The S allele has been identified as having a higher risk of developing depression [50, 51]. It is also associated with a lower serotonin transporter expression on membranes, indicating a lower ability for serotonin reuptake [50, 52]. Other candidate genes in this theory were identified, including DRD3, DRD4, HTR1A, HTR2A, HTR1B, HTR2C, SLC6A2, SLC6A3, MAOA, TPH1, COMT, and PCLO [49]. A meta-analysis found a significant association between SLC6A3 and DRD4 and MDD [30]. A significant association between MDD and DRD4, HTR1A, SLC6A3, and PCLO was identified by another meta-analysis [28]. In addition, candidate genes in neuroendocrine mechanism theory were identified in MDD: CRHR1 and CRHR2 encode corticotropin-releasing hormone (CRH) receptors, NR3C1 encodes the glucocorticoid receptor [53], and NR3C2 encodes the mineralocorticoid receptor in hypothalamic-pituitary-adrenal systems [54–56]. Previous studies revealed a significant association between depression and CRHR1 rs242939 [53], and a meta-analysis indicated a significant association between CRHR1 variants rs242941, re1876828, and rs242939 with depression [57]. Furthermore, CRHR2 rs37792250 was associated with MDD in the Japanese population [58].
BDNF is commonly involved in the neurogenesis and neuroplasticity theory and is essential in neuronal survival and proliferation [59]. Specifically, the Val66Met polymorphism displays a significant association with decreased levels of BDNF in patients with depressive symptoms [60, 61]. In neuroinflammation, the theory describes the alteration of neuronal and immune interactions by changing cytokine levels [62]; IL-6, IL-10, and TNF-alpha are elevated in depressed patients [62–64]. A meta-analysis of 61 studies revealed a highly significant association between IL-6 and depression [65]. However, a meta-analysis of 10 studies indicated no significant association between the TNF-alpha G-308 A polymorphism and depression. Previous studies investigated the association between the gene polymorphisms of cytokines and depression; IL-10 and IL-6 displayed no statistical significance [66].
This meta-analysis determined the association of the BDNF Val66Met and SLC6A4 (5-HTTLPR) gene polymorphisms with MDD. SLC6A4, located on, 17q11.2, encodes a serotonin transporter [67, 68] expressed in the central nervous system [69]. Our meta-analysis of SLC6A4 indicated that three genotypes (LL, LS, and SS) were not significantly associated with MDD but displayed an increase in the occurrence of MDD development according to the OR values in the SS genotype (OR = 1.39, 95% CI = 0.87–2.22, P = 0.16) and the LS genotype (OR = 1.13, 95% CI = 0.84–1.53, P = 0.42). This finding was inconsistent with the previous study was found significance of the S allele in cases of ICD-10 depressive episodes (OR = 1.24, CI = 1–1.54, x2 = 3.83, P = 0.050) and ICD-10 severe depressive episodes (OR = 1.31, CI = 1.02–1.68, x2 = 4.65, P = 0.031) [70]. Moreover, the studies on the interaction of genes with environmental factors revealed that the SL genotype was significant with the effect of threatening life events on adjusted ZSDS (B = 0.618, SE = 0.211, t = 2.921, P = 0.0038, adjusted R2 = 0.027) and strong with the association in the SS genotype (B = 1.299, SE = 0.333, t = 3.903, P = 0.0001, adjusted R2 = 0.124). In contrast, it was not significant in the LL genotype (B = 0.136, SE = 0.263, t = 0.517, P = 0.6163) [71]. The possibility of negative results in our study is caused by differences in studies such as ethnicity, gender, and age. Our results showed significant heterogeneity since data from the recruited studies are insufficient to conduct a subgroup analysis on environmental factors.
This meta-analysis found no association of the BDNF Val66Met polymorphism in three genotypes (GG, GA, and AA) with MDD. The OR values for the BDNF GG and AA genotypes suggested an increased probability of developing MDD (OR = 1.26, 95% CI = 0.78–2.06, P = 0.35) and (OR = 1.12, 95% CI = 0.77–1.64, P = 0.56), respectively. BDNF rs6265 is located in the protein-coding region of BDNF. This SNP was changed from G to A, resulting in a substitution of valine (Val) to methionine (Met) at codon 66, known as Val66Met, which changes the pro-region of the pro-BDNF protein [72, 73]. Serum BDNF levels for the Met carrier (A allele) were lower than the Val carrier (A allele) (23.08 vs. 26.87; P < 0.002). In other studies, no significant differences were observed in the genotype or allele of the BDNF gene polymorphism in MDD compared to the control, similar to our meta-analysis [74, 75]. Furthermore, the relationship between ethnicity and Val66Met polymorphism was found no significant in MDD [75]. On the other hand, the met al.lele of BDNF was significant between life stress and depression (P = 0.03) [37], and a meta-analysis revealed a significant association between stressful life events and childhood adversity with the Met al.lele in depression (Z = 2.552, P = 0.005 and Z = 1.775, P = 0.03) [76]. The interaction between environmental factors and genetics increases MDD development more than genetic risk factors alone. These reasons lead to this study finding non-significance but showed a high odd ratio in GG and AA genotypes.
The meta-analysis presents some limitations. Firstly, all comparisons of the two gene polymorphisms displayed a significant heterogeneity. Several differences were observed among the studies, including ethnicity, gender, and age. Second, we did not conduct a subgroup analysis for ethnicity, gender, and age due to a lack of data. Third, we cannot construct a funnel plot and Egger’s test for each meta-analysis because the studies included in this meta-analysis are less than 10 according to the PRISMA guideline 2020.
There are studies of systemic review gene polymorphism with MDD but few meta-analysis studies of genetic mutation with overlapping molecular function and biological processes related with MDD. Our study performed a systemic review and conducted a meta-analysis. The creativity of this study used one (SLC6A4) of 13 studied genes that involved between BP and MF of MDD development and a unique gene (BDNF) in BP to conduct a meta-analysis for investigation of the significant gene polymorphism with MDD.
This systemic review and meta-analysis identified 49 common genes with genetic polymorphisms engaged in several pathways, such as tryptophan metabolism, dopaminergic synapse, and serotonergic synapse, and involved in MDD. We identified 13 gene polymorphisms that could be related to MDD based on MF and BP. The SLC6A4 and BDNF polymorphisms increased the occurrence of MDD development but were not statistically significant. Research has demonstrated a wide variation in the association between gene polymorphism and MDD. Nevertheless, the association with MDD is increased when environmental factors and gene polymorphisms are combined. Therefore, the candidate gene polymorphism for diagnostic or therapeutic purposes should be determined with other factors (multiple markers). Genetics didn’t directly affect MDD, but the meta-analysis displayed heterogeneity in other factors such as ethnicity, gender, and age; these factors might be involved with the interaction between genetics and MDD. Therefore, subgroup analysis of genetics and other factors requires further study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to acknowledge Assoc. Prof. Dr. Porjai Pattanittum (Faculty of Public Health, Khon Kaen University, Thailand) and Dr. Kaewjai Thepsuthammarat (Clinical Epidemiology, Faculty of Medicine, Khon Kaen University, Thailand) for their suggestion in the data analysis.
Abbreviations
- 5-HT
5-hydroxy tryptamine
- 5-HTT
Serotonin transporter
- 5-HTTLPR
Serotonin-transporter-linked promoter region
- ACE
Angiotensin I converting enzyme
- APOE
Apolipoprotein E
- BDNF
Brain-derived neurotrophic factor
- BP
Biological process
- CC
Cellular component
- CI
Confidence interval
- COMT
Catechol-O-methyltransferase
- CRH
Corticotropin-releasing hormone
- DAOA
D-amino acid oxidase activator
- DAT1
Dopamine transporter 1
- DRD4
Dopamine receptor D4
- GO
Gene ontology
- HTR1A
5-hydroxytryptamine receptor 1 A
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MDD
Major depressive disorder
- MF
Molecular function
- MTHR
Methylenetetrahydrofolate reductase
- NOS
Newcastle–Ottawa scale
- OR
Odd ratio
- SLC6A4
Solute carrier family 6 member 4
- SNP
Single nucleotide polymorphism
- SSRI
Selective serotonin reuptake inhibitors
- TPH1
Tryptophan hydroxylase 1
- VNTR
SLC6A4 variable-number tandem repeat
Author contributions
Conceptualization, A.S., C.P., N.S., and T.E.; methodology, A.S., C.P., N.S., and T.E; data collection and analysis, A.S., and C.P.; writing—original draft, A.S., T.E, and C.P.; writing—review and editing, A.S., C.P., T.E., N.S., S.A., S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Faculty of Medicine, Khon Kaen University, Thailand (Grant Number IN66093).
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Otte C, et al. Major depressive disorder. Nat Rev Dis Primers. 2016;2(1):1–20. [DOI] [PubMed]
- 2.Malhi GS, Mann JJ. Depress Lancet. 2018;392(10161):2299–312. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe SY, et al. Biological tests for major depressive disorder that involve leukocyte gene expression assays. J Psychiatr Res. 2015;66–67:1–6. [DOI] [PubMed] [Google Scholar]
- 4.Bierut LJ, et al. Major depressive disorder in a community-based twin sample: are there different genetic and environmental contributions for men and women? Arch Gen Psychiatry. 1999;56(6):557–63. [DOI] [PubMed] [Google Scholar]
- 5.Chodavadia P, et al. Prevalence and economic burden of depression and anxiety symptoms among Singaporean adults: results from a 2022 web panel. BMC Psychiatry. 2023;23(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bromet E, et al. Cross-national epidemiology of DSM-IV major depressive episode. BMC Med. 2011;9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fava M, Kendler KS. Major depressive disorder. Neuron. 2000;28(2):335–41. [DOI] [PubMed] [Google Scholar]
- 8.Shadrina M, Bondarenko EA, Slominsky PA. Genetics factors in major depression disease. Front Psychiatry. 2018;9:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levinson DF. The genetics of depression: a review. Biol Psychiatry. 2006;60(2):84–92. [DOI] [PubMed] [Google Scholar]
- 10.Lohoff FW. Overview of the genetics of major depressive disorder. Curr Psychiatry Rep. 2010;12(6):539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157(10):1552–62. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. Meta-analysis of infectious agents and depression. Sci Rep. 2014;4:4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simanek AM, et al. Herpesviruses, inflammatory markers and incident depression in a longitudinal study of Detroit residents. Psychoneuroendocrinology. 2014;50:139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissenborn K, et al. Hepatitis C virus infection and the brain. Metab Brain Dis. 2009;24(1):197–210. [DOI] [PubMed] [Google Scholar]
- 15.Phillips AC, et al. Cytomegalovirus is associated with depression and anxiety in older adults. Brain Behav Immun. 2008;22(1):52–5. [DOI] [PubMed] [Google Scholar]
- 16.McCabe SE, West BT. The 3-year course of multiple substance use disorders in the United States: a national longitudinal study. J Clin Psychiatry. 2017;78(5):e537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng E, et al. Depression and substance use comorbidity: what we have learned from animal studies. Am J Drug Alcohol Abuse. 2017;43(4):456–74. [DOI] [PubMed] [Google Scholar]
- 18.Meng X, D’Arcy C. The projected effect of risk factor reduction on major depression incidence: a 16-year longitudinal Canadian cohort of the National Population Health Survey. J Affect Disord. 2014;158:56–61. [DOI] [PubMed] [Google Scholar]
- 19.Eaton WW, et al. Depression and risk for onset of type II diabetes: a prospective population-based study. Diabetes Care. 1996;19(10):1097–102. [DOI] [PubMed] [Google Scholar]
- 20.Melanson KJ. Nutrition review: relationships of nutrition with depression and anxiety. Am J Lifestyle Med. 2016;1(3):171–4. [Google Scholar]
- 21.Kris-Etherton PM, et al. Nutrition and behavioral health disorders: depression and anxiety. Nutr Rev. 2021;79(3):247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarris J, Schoendorfer N, Kavanagh DJ. Major depressive disorder and nutritional medicine: a review of monotherapies and adjuvant treatments. Nutr Rev. 2009;67(3):125–31. [DOI] [PubMed] [Google Scholar]
- 23.Alpert JE, Fava M. Nutrition and depression: the role of folate. Nutr Rev. 1997;55(5):145–9. [DOI] [PubMed] [Google Scholar]
- 24.Hacimusalar Y, Esel E. Suggested biomarkers for major depressive disorder. Noro Psikiyatr Ars. 2018;55(3):280–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strawbridge R, Young AH, Cleare AJ. Biomarkers for depression: recent insights, current challenges and future prospects. Neuropsychiatr Dis Treat. 2017;13:1245–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subermaniam K, et al. Marine algae as emerging therapeutic alternatives for depression: a review. Iran J Basic Med Sci. 2021;24(8):997–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belmaker RH, Agam G. Major depressive disorder. N Engl J Med. 2008;358(1):55–68. [DOI] [PubMed] [Google Scholar]
- 28.Gatt JM, et al. Specific and common genes implicated across major mental disorders: a review of meta-analysis studies. J Psychiatr Res. 2015;60:1–13. [DOI] [PubMed] [Google Scholar]
- 29.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81(3):484–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-León S, et al. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2008;13(8):772–85. [DOI] [PubMed] [Google Scholar]
- 31.Lohoff FW. Overview of the genetics of major depressive disorder. Curr Psychiatry Rep. 2010;12:539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho PS, et al. Association study of serotonin transporter availability and SLC6A4 gene polymorphisms in patients with major depression. Psychiatry Res. 2013;212(3):216–22. [DOI] [PubMed] [Google Scholar]
- 33.Kiyohara C, Yoshimasu K. Association between major depressive disorder and a functional polymorphism of the 5-hydroxytryptamine (serotonin) transporter gene: a meta-analysis. Psychiatr Genet. 2010;20(2):49–58. [DOI] [PubMed] [Google Scholar]
- 34.Moskvina V, et al. Interrelationship of childhood trauma, neuroticism, and depressive phenotype. Depress Anxiety. 2007;24(3):163–8. [DOI] [PubMed] [Google Scholar]
- 35.Gámez-Guadix M, et al. Longitudinal and reciprocal relations of cyberbullying with depression, substance use, and problematic internet use among adolescents. J Adolesc Health. 2013;53(4):446–52. [DOI] [PubMed] [Google Scholar]
- 36.Gullander M, et al. Exposure to workplace bullying and risk of depression. J Occup Environ Med. 2014;56(12):1258–65. [DOI] [PubMed] [Google Scholar]
- 37.Hosang GM, et al. Interaction between stress and the BDNF Val66Met polymorphism in depression: a systematic review and meta-analysis. BMC Med. 2014;12:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gyekis JP, et al. No association of genetic variants in BDNF with major depression: a meta- and gene‐based analysis. Am J Med Genet Part B: Neuropsychiatric Genet. 2012;162(1):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grotzinger AD, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3(5):513–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page MJ, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunney WE, Davis JM. Norepinephrine in depressive reactions: a review. Arch Gen Psychiatry. 1965;13(6):483–94. [DOI] [PubMed] [Google Scholar]
- 42.Schildkraut JJ. The catecholamine hypothesis of affective disorders: a review of supporting evidence. Am J Psychiatry. 1965;122(5):509–22. [DOI] [PubMed] [Google Scholar]
- 43.Loomer HP, Saunders JC, Kline NS. A clinical and pharmacodynamic evaluation of iproniazid as a psychic energizer. Psychiatric research reports. 1957. [PubMed]
- 44.Hirschfeld RM. History and evolution of the monoamine hypothesis of depression. J Clin Psychiatry. 2000;61(6):4–6. [PubMed] [Google Scholar]
- 45.Perez-Caballero L, et al. Monoaminergic system and depression. Cell Tissue Res. 2019;377:107–13. [DOI] [PubMed] [Google Scholar]
- 46.Troubat R, et al. Neuroinflammation and depression: a review. Eur J Neurosci. 2021;53(1):151–71. [DOI] [PubMed] [Google Scholar]
- 47.Bakunina N, Pariante CM, Zunszain PA. Immune mechanisms linked to depression via oxidative stress and neuroprogression. Immunology. 2015;144(3):365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subermaniam K, et al. Marine algae as emerging therapeutic alternatives for depression: a review. Iran J Basic Med Sci. 2021;24(8):997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shadrina M, Bondarenko EA, Slominsky PA. Genetics factors in major depression disease. Front Psychiatry. 2018;9:382604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lesch K-P, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274(5292):1527–31. [DOI] [PubMed] [Google Scholar]
- 51.Schinka J, Busch R, Robichaux-Keene N. A meta-analysis of the association between the serotonin transporter gene polymorphism (5-HTTLPR) and trait anxiety. Mol Psychiatry. 2004;9(2):197–202. [DOI] [PubMed] [Google Scholar]
- 52.Heils A, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66(6):2621–4. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z, et al. Association of corticotropin-releasing hormone receptor1 gene SNP and haplotype with major depression. Neurosci Lett. 2006;404(3):358–62. [DOI] [PubMed] [Google Scholar]
- 54.DeRijk R, Schaaf M, De Kloet E. Glucocorticoid receptor variants: clinical implications. J Steroid Biochem Mol Biol. 2002;81(2):103–22. [DOI] [PubMed] [Google Scholar]
- 55.Klok MD, et al. Decreased expression of mineralocorticoid receptor mRNA and its splice variants in postmortem brain regions of patients with major depressive disorder. J Psychiatr Res. 2011;45(7):871–8. [DOI] [PubMed] [Google Scholar]
- 56.Schatzberg AF, et al. HPA axis genetic variation, cortisol and psychosis in major depression. Mol Psychiatry. 2014;19(2):220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernandez-Diaz Y, et al. The role of rs242941, rs1876828, rs242939 and rs110402 polymorphisms of CRHR1 gene and the depression: systematic review and meta-analysis. Genes Genomics. 2021;43;1339–49. [DOI] [PubMed]
- 58.Ishitobi Y, et al. Association of CRHR1 and CRHR2 with major depressive disorder and panic disorder in a Japanese population. Am J Med Genet Part B: Neuropsychiatric Genet. 2012;159(4):429–36. [DOI] [PubMed] [Google Scholar]
- 59.Riccio A, et al. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science. 1999;286(5448):2358–61. [DOI] [PubMed] [Google Scholar]
- 60.Lanni C, et al. Depression and antidepressants: molecular and cellular aspects. Cell Mol Life Sci. 2009;66:2985–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Z-Y, et al. Variant brain-derived neurotrophic factor (BDNF)(Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24(18):4401–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Euteneuer F, et al. Depression, cytokines and experimental pain: evidence for sex-related association patterns. J Affect Disord. 2011;131(1–3):143–9. [DOI] [PubMed] [Google Scholar]
- 63.Dowlati Y, et al. A meta-analysis of cytokines in major depression. Biol Psychiatry. 2010;67(5):446–57. [DOI] [PubMed] [Google Scholar]
- 64.Ertenli I, et al. Infliximab, a TNF-alpha antagonist treatment in patients with ankylosing spondylitis: the impact on depression, anxiety and quality of life level. Rheumatol Int. 2012;32:323–30. [DOI] [PubMed] [Google Scholar]
- 65.Howren MB, Lamkin DM, Suls J. Associations of depression with C-reactive protein, IL-1, and IL-6: a meta-analysis. Psychosom Med. 2009;71(2):171–86. [DOI] [PubMed] [Google Scholar]
- 66.Mihailova S, et al. A study of TNF-α, TGF-β, IL-10, IL-6, and IFN-γ gene polymorphisms in patients with depression. J Neuroimmunol. 2016;293:123–8. [DOI] [PubMed] [Google Scholar]
- 67.Ramamoorthy S, et al. Antidepressant-and cocaine-sensitive human serotonin transporter: molecular cloning, expression, and chromosomal localization. Proc Natl Acad Sci. 1993;90(6):2542–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gelernter J, Pakstis A, Kidd K. Linkage mapping of serotonin transporter protein gene SLC6A4 on chromosome 17. Hum Genet. 1995;95:677–80. [DOI] [PubMed] [Google Scholar]
- 69.Hornung J-P. The human raphe nuclei and the serotonergic system. J Chem Neuroanat. 2003;26(4):331–43. [DOI] [PubMed] [Google Scholar]
- 70.Cervilla JA, et al. The 5-HTTLPR s/s genotype at the serotonin transporter gene (SLC6A4) increases the risk for depression in a large cohort of primary care attendees: the PREDICT‐gene study. Am J Med Genet Part B: Neuropsychiatric Genet. 2006;141(8):912–7. [DOI] [PubMed] [Google Scholar]
- 71.Lazary J, et al. New evidence for the association of the serotonin transporter gene (SLC6A4) haplotypes, threatening life events, and depressive phenotype. Biol Psychiatry. 2008;64(6):498–504. [DOI] [PubMed] [Google Scholar]
- 72.Chiaruttini C, et al. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci. 2009;106(38):16481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ventriglia M, et al. Association between the BDNF 196 A/G polymorphism and sporadic Alzheimer’s disease. Mol Psychiatry. 2002;7(2):136–7. [DOI] [PubMed] [Google Scholar]
- 74.Tsai SJ, et al. Association study of a brain-derived neurotrophic-factor genetic polymorphism and major depressive disorders, symptomatology, and antidepressant response. Am J Med Genet B Neuropsychiatr Genet. 2003;123B(1):19–22. [DOI] [PubMed] [Google Scholar]
- 75.Verhagen M, et al. Meta-analysis of the BDNF Val66Met polymorphism in major depressive disorder: effects of gender and ethnicity. Mol Psychiatry. 2010;15(3):260–71. [DOI] [PubMed] [Google Scholar]
- 76.Zhao M, et al. BDNF Val66Met polymorphism, life stress and depression: a meta-analysis of gene-environment interaction. J Affect Disord. 2018;227:226–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.