Abstract
Background
Fusarium head blight (FHB), a devastating disease of wheat production, is predominantly elicited by Fusarium graminearum (Fg). The tetraploid Thinopyrum elongatum is a tertiary gene resource of common wheat that possesses high affinity and displays high resistance traits against multiple biotic and abiotic stress. We aim to employ and utilize the novel FHB resistance resources from the wild germplasm of common wheat for breeding.
Results
Durum wheat-tetraploid Th. elongatum amphiploid 8801 was hybridized with common wheat cultivars SM482 and SM51, and the F5 generation was generated. We conducted cytogenetically in situ hybridization (ISH) technologies to select and confirm a genetically stable 7E(7D) substitution line K17-1069-5, which showed FHB expansion resistance in both field and greenhouse infection experiments and displayed no significant disadvantage in agronomic traits compared to their common wheat parents in the field. The F2 segregation populations (K17-1069-5 × SM830) showed that the 7E chromosome conferred dominant FHB resistance with dosage effect. We developed 19 SSR molecular markers specific to chromosome 7E, which could be conducted for genetic mapping and large breeding populations marker-assisted selection (MAS) during selection procedures in the future. We isolated a novel Fhb7 allele from the tetraploid Th. elongatum chromosome 7E (Chr7E) using homology-based cloning, which was designated as TTE7E-Fhb7.
Conclusions
In summary, our study developed a novel wheat-tetraploid Thinopyrum elongatum 7E(7D) K17-1069-5 substitution line which contains stable FHB resistance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05703-3.
Keywords: Fusarium head blight, Tetraploid Thinopyrum elongatum, Common wheat substitution line, Fhb7 gene resistance
Background
Fusarium head blight (FHB), caused by multiple pathogenic fungi Fusarium spp., seriously damages wheat yield and quality in temperate regions worldwide. It reduced more than half of production during the epidemic years [1, 2]. In China, FHB caused 10–50% wheat yield losses from moderate to severe epidemic years, which undermined about 10 million hectares of wheat production [3]. The FHB-infected spikes are deficient in nutrients, cause reddish grain color, reduce grain size, and produce mycotoxins in the seeds, which are hazardous to human and animal bodies [4, 5]. Deoxynivalenol (DON) is a prevalent mycotoxin and potentially restrain protein synthesis. It can induce diarrhea, anorexia, vomiting, and gastrointestinal bleeding in animals, and it is difficult to eliminate through chemical and physical treatments or temperature changes with long-term storage [6, 7].
To maintain grain quality and productivity, resistant varieties and new sources of resistance are necessary to identify and develop in germplasm. Fhb1-Fhb9 were currently nine formally nominated FHB resistance genes [8–16]. Fhb1, Fhb2, Fhb4, Fhb5, and Fhb8 are resistance fungal expansion genes (type II resistance) and were identified in wheat cultivar Sumai 3 and/or landrace Wangshuibai [8–12], Fhb9 was derived from Ji5265 [13], Fhb3, Fhb6, and Fhb7 were originated from wild Triticeae species Leymus racemosus (Lam.) Tzvelev, Campeiostachys kamoji (Ohwi) B.R.Baum, J.L.Yang & C.Yen (Synonyms Elymus tsukushiensis Honda), and Thinopyrum poticum (Podp.) Barkworth & D.R.Dewey, respectively [14–16]. Currently, Fhb1 and Fhb7 have been successfully identified based on the positional cloning. They present different FHB resistance mechanisms, of which Fhb1 encodes a histidine-rich calcium-binding protein, and Fhb7 encodes a glutathione S-transferase [16–18]. Because of this exceptional rarity of wheat main FHB resistance QTLs, the discovery of novel FHB-resistant gene resources is very important for wheat disease resistance breeding.
Plentiful FHB resistance from wild Triticeae relatives has been evaluated, such as Thinopyrum, Roegneria, Leymus, Pseudoroegneria, and Agropyron [19, 20]. Thinopyrum elongatum (Host) D. R. Dewey (Triticeae, Poaceae), with E as the designed genome symbol, contains 2×, 4×, and 10× ploidys [21]. According to whole genome comparison, the common wheat D sub-genome and the Th. elongatum E genome has high genetic affinity, indicating the potential that Th. elongatum could be used in wheat breeding through chromosome engineering [16]. Through distant hybridization and chromosome engineering, the alien segments of the Thinopyrum species with desirable genes deployed successfully in wheat over the last few decades. For instance, the wheat × Th. elongatum derivative lines have been reported carrying genes showed FHB resistance (Fhb7), rust resistance (Sr24 and Lr24), and powdery mildew resistance (Pm51), as well as the abiotic stresses resistance to salt salinity, chilling, and drought [22–29]. There are many successful examples of transferring chromosome segments of diploid Th. elongatum and decaploid Th. ponticum to wheat [27, 30, 31], however, the FHB resistance and other excellent agronomy traits in the tetraploid Th. elongatum still needs further exploration and utilization.
Previously, we hybridized durum wheat-tetraploid Th. elongatum amphiploid 8801 with common wheat cultivar Shumai 482 (hereafter SM482), and crossed the F1 with common wheat cultivar Shumai 51 (SM51). The generation was advanced to the F5 generation. The present study aims to (1) select a wheat-tetraploid Th. elongatum substitution line and characterize the genome constitution; (2) investigate the FHB resistance phenotype and the agronomic characters of the substitution line; (3) generate and verify the specificity of molecular markers for the alien chromosome; (4) analyze the genetic effect of the alien chromosome.
Results
Characterizing the genomic constitution and cytogenetic stability of the 7E(7D) substitution line
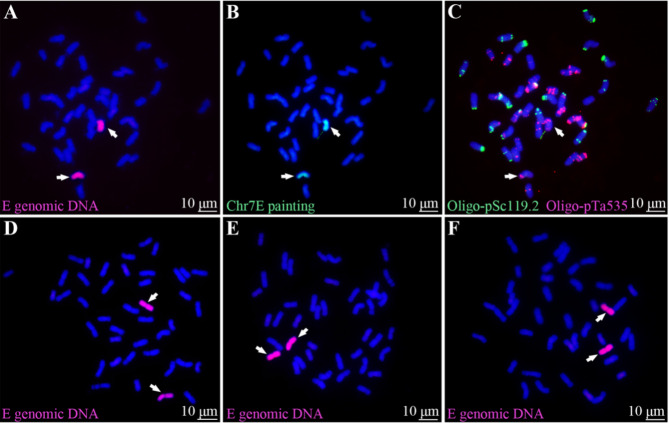
To generate the 7E(7D) substitution line, we performed hybridization between durum wheat-tetraploid Th. elongatum amphiploid 8801 (♀ parent) and SM482 (♂ parent) and the F1 plants were further hybridized with SM51 and selfied to F5 generation. The substitution line was selected from the F5 generation based on GISH analysis using the gDNA of tetraploid Th. elongatum as probes. The result revealed that one plant K17-1069-5 possessed 42 chromosomes, including two E genome signals (in magenta) (Fig. 1A). To distinguish the homoeology of E chromosomes in common wheat genetic background, oligo-FISH painting probes Chr1E–Chr7E was performed. It displayed that the alien chromosomes were labeled by the Chr7E probe (in green) (Fig. 1B). The chromosomal composition of K17-1069-5 was characterized with non-denature FISH (ND-FISH), by comparing with the cytogenetic karyotype of CS [32] and tetraploid Th. elongatum [33]. The result showed that K17-1069-5 was absent in a 7D chromosome pair of common wheat and replaced by a 7E chromosome pair (Fig. 1C). Thus, we confirmed that K17-1069-5 was a 7E(7D) substitution line. The cytological stability of K17-1069-5 was evaluated using 40 selfed seeds of K17-1069-5 randomly selected for GISH analysis. All 40 plants contained the same genomic constitution as K17-1069-5 (Fig. 1D–F). Therefore, K17-1069-5 was identified as a genomic-stable 7E(7D) substitution line.
Fig. 1.
Characterization of the genome composition of the 7E(7D) substitution line and its offspring by using cytogenetic techniques. A, Genomic in situ hybridization (GISH) analysis of K17-1069-5 using tetraploid Th. elongatum gDNA as the E genome probe (magenta). B Fluorescence in situ hybridization (FISH) painting using Chr7E probe (green). C FISH analysis using Oligo-pTa535 (magenta) and Oligo-pSc119.2 (green) probes. D-F Cytogenetic stability identification of selfed seeds of K17-1069-5 using GISH (E genome in magenta). White arrows display the alien chromosomes
The FHB resistance and agronomic characters of 7E(7D) substitution line
The 7E(7D) substitution line and common wheat parents (SM482 and SM51) were infected with one of the major pathogen Fusarium graminearum (Fg) strain PH-1 to assess the severity level of FHB from 2022 to 2023 in the greenhouse and field experimental conditions. After 14 days post-inoculation (dpi) and 21 dpi, it displayed that the susceptible control Roblin and common wheat parents were FHB highly susceptible and the percentage of diseased spikelets (PDS) exceeded 72%, while Sumai3 (resistance control), 8801, and 7E(7D) substitution lines displayed immunity to FHB with expanding resistance to adjacent spikelets and rachis in both greenhouse and field conditions (Fig. 2). In summary, wheat-tetraploid Th. elongatum 7E(7D) substitution line displayed excellent Type II FHB resistance to Fg strain PH-1.
Fig. 2.
The FHB resistant evaluation of 7E(7D) substitution line and their common wheat parental materials SM482 and SM51
Agronomic traits investigation exhibited that the 7E(7D) substitution line had a longer flag leaf than the parents. It possessed similar plant height, tiller numbers, spike length, and spikelet numbers to common wheat parents SM482 and SM51, while significantly better than the alien parent amphiploid 8801 (Fig. 3; Additional file 1: Fig. S1; Additional file 2: Table S1). In summary, the tetraploid Th. elongatum 7E chromosome displayed less negative effect on common wheat in terms of agronomic characteristics.
Fig. 3.
Agronomic performance of the 7E(7D) substitution line, common wheat parental materials SM482 and SM51
Generation and verification of SSR molecular markers specific for 7E chromosome
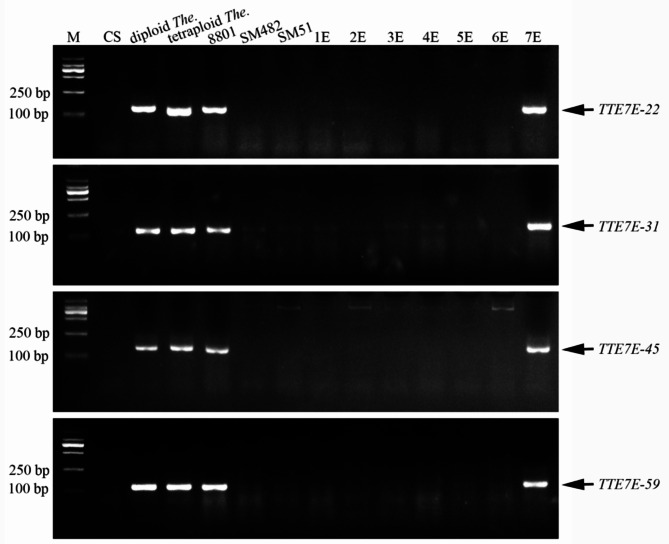
To accurately identify the 7E chromosome from a common wheat genetic background, we developed Simple Sequence Repeat (SSR) markers specific to Chr7E which can be applied in the alien chromosome segment identification for large-scale of marker-assisted selection (MAS). First, the diploid Th. elongatum 7E reference genome was aligned with Chr1E–Chr6E and reference CS (v2.1), and, 1999 SSR markers were identified specific to Chr7E. In total, 60 SSR primer pairs were arbitrarily picked to confirm their specificity by PCR. A total of 19 SSRs displayed the targeted amplify bands in diploid and tetraploid Th. elongatum, 8801, and 7E(7D) substitution line, while absent in common wheat CS, SM482, SM51, and 1E–6E substitution lines. Consequently, these 19 SSRs could be used to detect the Chr7E under a common wheat background (Fig. 4; Table 1).
Fig. 4.
Generation and verification of the 7E-chromosome unique SSR primers. SSR molecular markers TTE7E-22, TTETE-31, TTETE-45, and TTETE-59
Table 1.
The specific SSR primer pairs of the Chr7E identified in this study
| Primers | Sequences of the Special Primers (5’–3’) | |
|---|---|---|
| Forward | Reverse | |
| TTE7E-17 | ACCTACTCTGACCGCCTGAA | GAAATATCATGCCGGGAGAC |
| TTE7E-20 | AAAAGGAGTGGGAGGAGGAG | CGGCGCTTACCTAAACTTTG |
| TTE7E-21 | GTCAACTCGCCAGGTCTCTC | CAAAACCTCCGCCTTAGGTA |
| TTE7E-22 | CTGCGCTACCAGCTACCTCT | TAGCGCTCTTCCGCTACTTC |
| TTE7E-23 | ATGCCTCGATCCATACCTTG | GACGGTGGTAGAGCTTCTGG |
| TTE7E-26 | TATATCGCCTGCTCGAACG | ATTTATGTGCGGCAAAAAGG |
| TTE7E-27 | CCAGGCCAGTTTACTCAAAGA | GAGAGGATGGATGCCACTTT |
| TTE7E-30 | GTGAGGTTCCACCGGTTTT | CGTATTTGGAAAAGTTTCAGTGG |
| TTE7E-31 | AGCCGGAGCCTATTCTTTTT | GACGCAGCTGATCAATAGGA |
| TTE7E-34 | TTGTTGTTGCAAAATGCACTC | CGGAAGGCAACTTCTTCTTG |
| TTE7E-38 | TCCGGTGGCTTCTTCTTCT | TGACCAGCACGGAACAATAG |
| TTE7E-41 | AAGGCGTAAAAAGTGGAGCA | TACCCATTGCAGCCACTAAA |
| TTE7E-43 | CCTCACTCCCTCTCTGATCG | GCAACCTTGACCATGTTCCT |
| TTE7E-45 | TTATGTCATCACGCGCCTAC | TCAATACGCGATGAACATTTTT |
| TTE7E-46 | GAAATTGAACCACGCATCCT | AGTTCCTCGTTGGCATCACT |
| TTE7E-47 | TCAAAAGCGCCCTACTCATT | ATCCTCTAGCCTCGCCTTTT |
| TTE7E-50 | CATGGAAGGGTTCGTAAGGA | TCCACTCCCAAAATCCACTC |
| TTE7E-58 | GAACTAGATGGGCGTCTCCA | AGGGCAAGGAAACTGTCTCA |
| TTE7E-59 | TCTGGCAATGGATGAATGAA | ATATACCCTCCCCGGCACTA |
Transmission and genetic effects of Chr7E and FHB resistance in common wheat genetic background
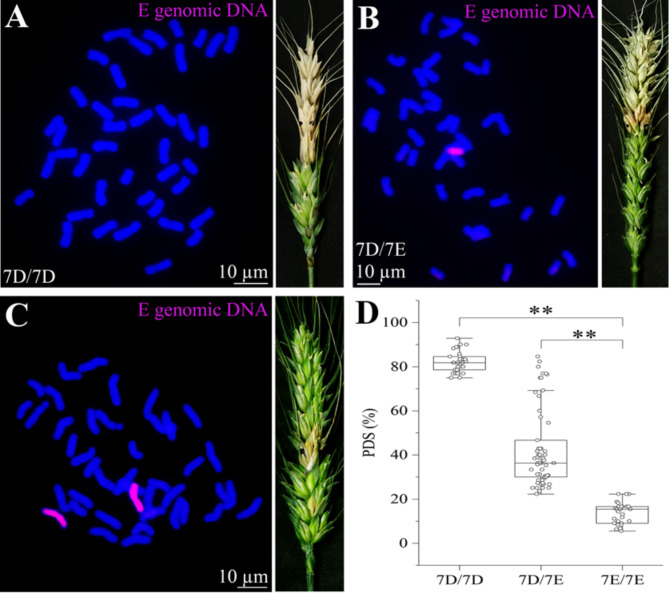
To analyze the pyramid effect on FHB resistance contributed by Chr7E, we constructed an F2 population using FHB resistance 7E(7D) substitution line and FHB susceptible wheat SM830. We genotyped 122 F2 individuals using the Chr7E-specific marker TTE7E-31 (Fig. 5A–C; Additional file 2: Table S2). Afterwards, we performed cytogenetic GISH analysis to quantity the alien chromosome number. We found a reasonable separation ratio (7D/7D: 7D/7E: 7E/7E) of roughly 1:2:1 in the F2 population with 36, 59, and 27 plants respectively (Fig. 5D). The FHB severity of F2 plants was examined with PH-1 inoculation in the greenhouse at the flowering stage (Additional file 2: Table S2). The common wheat parents and all F2 plants with 7D/7D genotypes were highly susceptible to FHB. In the 7D/7E genotype group, 45 of 59 were resistant to FHB, while 14 of them were susceptible. All the 27 plants in the 7E/7E genotype were highly resistant to FHB. This result implied that the high FHB resistance was provided by the alien Chr7E of tetraploid Th. elongatum which considerably reduced the proportion of infected spikelets. Consequently, we deduced that the Chr7E has a dosage-dependent dominant type II FHB resistance locus (s).
Fig. 5.
Genetic effects analysis of Chr7E in common wheat background. A-C GISH identification of three F2 genotypes and FHB resistance (A 7D/7D, without alien E chromosome, B 7D/7E, containing one 7E chromosome, and C 7E/7E, containing two 7E chromosomes) with tetraploid Th. elongatum whole genome DNA probe (magenta). D The PDS percentage for different genotypes. The statistical significance differences were evaluated using a Student’s t-test (* p < 0.05, ** p < 0.01, ns not significant)
The Chr7E of tetraploid Th. elongatum contains a novel Fhb7 allele
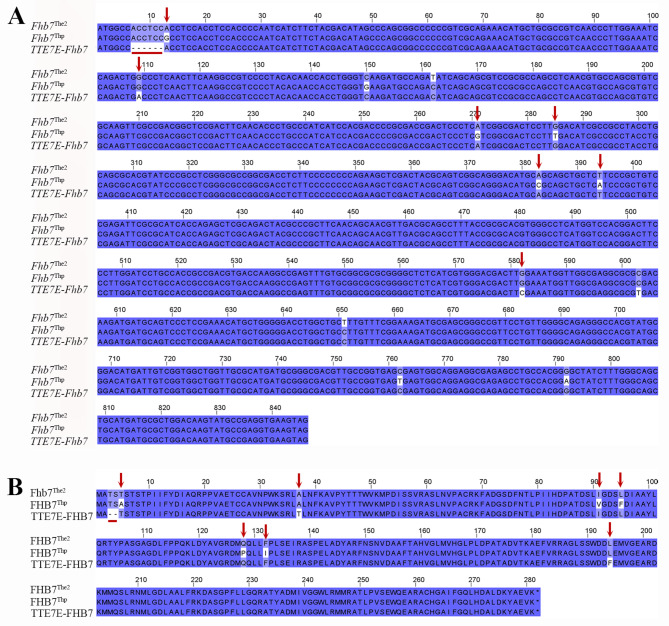
To detect whether the 7E(7D) substitution line contained Fhb7 from the 7E of diploid Th. elongatum or its homologs, we used two functional markers of Fhb7 (Fhb7Thp and Fhb7The2). The result showed that the 7E(7D) substitution line might contain a functional Fhb7 allele. Then we used the homology-based cloning method to clone the Fhb7 allele from tetraploid Th. elongatum, which were named as TTE7E-Fhb7. Nucleotide alignment displayed that the sequence in the 7E(7D) substitution line displayed 6 bp deletion compared to both Fhb7The2 and Fhb7Thp, as well as 5 and 10 SNPs compared to Fhb7The2 and Fhb7Thp, respectively (Fig. 6A). Thus, we confirmed that TTE7E-Fhb7 in the tetraploid Th. elongatum 7E possessed an Fhb7 homologous gene. Comparative analysis of protein primary sequences indicated that TTE7E-Fhb7 lack a serine and threonine at the N-terminal, in addition, two nonsynonymous mutants from SNPs differences made alanine to serine, leucine to phenylalanine (Fig. 6B). However, whether these sequence differences could influence the function of gene or not needs further exploration.
Fig. 6.
The detection of the functional marker (GST) of Fhb7 to 7E(7D). A Nucleotide sequence alignment. B Amino acid sequences alignment. The Fhb7-Thp sequence from the BAC clone NODE_28_length_58203_cov_9148.280322 [16]; The Fhb7The2 and Fhb7Thp sequence from the Tel7E01T1020600.1. The blue arrow pairs indicate the initiation codon and termination codon, respectively. The red lines below indicate deletion. Arrows above the sequences display anonymous SNP mutants
Discussion
Employment of wild germplasm resources in wheat breeding
Interspecific hybridization and chromosome engineering are two crucial methods for wheat breeders to introduce new traits from wild germplasm into elite modern cultivars. The 1RS·1BL and 6VS·6AL are two translocation lines that are effectively employed in wheat breeding program with desirable disease resistance and great agronomic performance [34, 35]. Genes from wheatgrasses (Thinopyrum spp.) have been transferred into cultivated Triticum spp by employing amphidiploids, and even small alien fragment introgressions. In this study, the 7E(7D) substitution line presented less negative effect on common wheat, and hence it is a potential germplasm for FHB resistance breeding.
To introduce the alien material with FHB resistance gene(s) into the wheat genetic background and reduce the redundancy traits on alien chromosomes, several chromosome engineering methods have been developed: the use of Chinese Spring ph1b mutant (CS ph1b) [36, 37], gamma irradiation [38, 39], and breakage-fusion mechanism [40, 41]. By using these methods translocations and/or deletions can be induced in the substitution line, and can narrow down the physical interval of alien segments, to have a better compensation effect and genetic balance for breeding [34, 35, 42]. All these three strategies could further explore the FHB resistance TTE7E-Fhb7 haplotype and mechanism, and deploy FHB resistance from the tetraploid Th. elongatum 7E chromosome to wheat breeding.
The application of Th. elongatum in common wheat disease resistance
It has been proved that the chromosome 7E group included the genes/loci resistance to biotic and abiotic stress [43, 44]. For instance, gene Lr19 was resistance to leaf rust [45], gene Sr25 was stem rust resistant to Ug99 races [46, 47], and a yellow pigment content (YPC) Yp gene which are closely linked and localization at the distal region of 7E#l chromosome [43, 44]. Tounsi et al. [48] reported that the 7E chromosome enhanced the tolerance to salinity stress in the durum wheat. More recently, two quantitative trait loci (QTLs) for FHB resistance were identified from the homologous group 7 in different Thinopyrum species [16, 49]. In this research, we generated an FHB expansion-resistant resource7E(7D) substitution line, without negatively compromising the agronomic characteristics of common wheat. Hence, Thinopyrum spp. has the potential to be utilized for wheat FHB resistance breeding as an appropriate bridge breeding material.
Wheat FHB resistance is considered a quantitative trait, that governed by one or several QTLs (probably major or minor effects), additionally, it is associated with genotypes and environment interactions [50]. It has been reported that the average percentage of infected spikes of the lines carrying resistance genes was lower than lines without the resistance genes, and the effect was most obvious in the case of gene Fhb1 and the combination of Fhb1 + Fhb2 and Fhb1 + Fhb2 + Fhb4 [51]. The FHB resistance was stronger when Fhb1 and Fhb9 were combined than the single Fhb1 or Fhb9 gene, indicating a positive dosage effect between the two QTLs [13]. In this research, F2 segregation population plants with single alien 7E chromosomes varied for FHB resistance from resistance to susceptible, while homozygous 7E chromosome plants were highly resistant to FHB, indicating the significant dosage effect.
High allelic multiplicity of resistance genes differing in pathogen resistance in plants
Plants recognize pathogens through nucleotide-binding leucine-rich repeat receptors (NLRs) intracellularly [52]. Variations have led to multiple functional alleles within the same locus in syntenic regions, likely enabling the recognition of different effector alleles to confer resistance or exhibit different resistance magnitudes. There are multiple examples of allelic diversity that give rise to functional diversity in resisting various pathogens. For example, powdery mildew (Pm) resistance CNLs Pm60a, Pm60, and Pm60b cloned from Triticum urartu are allelic to PmG16/MlIW18/MlIW172 from wild emmer wheat [53]. In Arabidopsis, abundant intraspecies diversity possesses highly variable NLRs (hvNLRs), provides new pathogen-recognition specificities, and is associated with allelic diversity and genome at a population level [54]. Consistent with our previous wheat-Elymus repens 7St FHB-resistance translocation line work [55], in this research, we confirmed that the FHB expansion resistance was conferred by TTE7E-Fhb7 loci in the Chr7E of tetraploid Th. elongatum. Meanwhile, three Fhb7 alleles alignment revealed that the sequence of TTE7E-Fhb7 exhibits 6 bp deletion as well as 5 and 10 SNPs with Fhb7The2 and Fhb7Thp, respectively. This indicates multiple FHB resistance alleles may exist in this homoeologous group. Whether TTE7E-Fhb7 has the same detoxification function as Fhb7Thp, and the relationship between Fhb7The2 and Fhb7Thp need further study.
Strategies unveiling the candidate genes associated with aimed traits
Wild relatives are crucial germplasm resources for wheat improvement and breeding. Traditionally, cytogenetic ISH experiments were conducted to discriminate the alien chromosomes. Recently, a newly developed oligo-painting FISH technique has been designed for chromosome-specific identification, which can be applied to alien homoeologous chromosome groups [56, 57]. Generally, cytogenetic-based methods directly visualize the alien segments introduced into the wheat genetic background [58]. Nevertheless, it costs labor and time, especially with small alien fragments or sequence introgression that may display undetectable fluorescence signals due to highly condensed mitotic chromosomes and the limited resolution of the microscope.
Molecular markers were conducted to precisely track alien sequences with targeted traits in a wheat genetic background, and significantly improve the efficiency of the MAS during the breeding cycle [59, 60]. In the past, a series of molecular markers in Thinopyrum elongatum Chr1E–7E have been continuously generated, such as expressed sequence tags (EST), PCR-based landmark unique gene (PLUG) markers, specific-locus amplified fragment sequencing (SLAF-seq), GBS (genotyping-by-sequencing). Gong et al. [61] confirmed the three 4E chromosome long arm translocation lines at the centromeric region by 55 K SNP arrays since the most of the SNPs in the translocation regions were detected as deletions.
More and more released reference genome and re-sequencing data offer an advantage for detecting alien genetic information. Based on the sequencing system, Deng et al. [62] generated a wheat–Thinopyrum elongatum array for genotyping alien E chromosome of Thinopyrum species and derivative. The chip array assists in the cytological characterization of chromosome engineering breeding. Kompetitive allele-specific PCR (KASP) markers provides efficient approach for large population genotyping during breeding [63]. In this study, we generated and verified Chr7E-specific SSR markers for MAS. We plan to obtain a small 7E chromosome translocation line and develop KASP co-dominant markers for identifying the alien fragments with FHB resistance.
More recently, target resistance genes (R genes) and specific genomic regions have been efficiently characterized using next-generation sequencing (NGS). However, due to abundant genetic diversity and variations, aimed R genes in wild germplasm may not be included in the published reference haplotypes. Additionally, target R genes may be suppressed for chromosome recombination and linkage redundancy. Multiple strategies, such as chromosome sorting, chromosome microdissection, bulked segregation analysis, MutMap, QTL-seq, reference genome assembling, or primary long-read sequencing, offer targeted gene cloning and exploring gene mechanisms [64, 65]. To clone and characterize the FHB resistance gene in the Chr7E of tetraploid Th. elongatum, we will sequence the partial amphiploid Trititrigia 8801 parental by long-read sequencing technology.
Methods
Materials
The tetraploid Triticum durum-tetraploid Th. elongatum partial amphiploid Trititrigia 8801 (2n = 6x = 42, genome constitution AABBEE) was provided by Dr. George Fedak from Ottawa Research and Development Centre, Agriculture and Agri-Food Canada. It displayed broad-spectrum resistance to biotic stress such as rust, FHB, and powdery mildew, and tolerance to abiotic stress such as drought, chilling, and salinity. Triticum aestivum cv. Shumai 482 (SM482) and Shumai 51 (SM51) in Sichuan province were susceptible to FHB, which were used as parental lines for 7E(7D) substitution line generation. The F2 population of the substitution line and Shumai 830 (SM830) was constructed for genetic effect analysis of the alien 7E chromosome. The GISH analysis was conducted using the gDNA of tetraploid Th. elongatum (2n = 4x = 28, EEEE) as the probe, and the fragmented gDNA of T. aestivum cv. Chinese Spring ‘CS’ (2n = 6x = 42, AABBDD) as the block. T. aestivum cv. Sumai 3 and Robblin were applied as the FHB resistance and susceptible control group, respectively.
Preparation of chromosome spreads
The mitotic metaphase chromosome spreads were carried out according to Aliyeva-Schnorr et al. [66] with adjustments. Shortly, the fixed root-tip meristems were digested for 60 min at 37 °C using enzyme solution (cellulase onozuka R-10: 2% pectolyasein Y-23 = 4: 2). This was followed by two times washing in 75% ethanol, followed by the addition of 10–15 µL 90% acetic acid (per meristem) in place of the ethanol, and crushing into meristems suspension. The suspension was spread onto slides with a humidity range of 50–55%. Using phase-contrast microscopy, slides with expected chromosome morphology and number were selected once they had dried, and they were kept at − 20 °C until used for sequential ISH.
Sequential ISH analyses and microscopy
GISH protocol was conducted according to Wu et al. [67]. The whole gDNA of tetraploid Th. elongatum was labeled using dUTP-ATTO-550 (Jena Bioscience, Jena, Germany), meanwhile, the CS gDNA was applied as a block and fragmented by autoclave (121 ℃ for 5 min). The probe: block equals 1:150. In total, 20 µL of hybridization solution was hybridized with each prepared slide (16 µL hybridization mixture, 100 ng/µL labeled probe, 15000 ng/µL block).
To distinguish alien chromosomes of tetraploid Th. elongatum and substitutional common wheat chromosomes, we conducted mc-FISH with probes Oligo-pSc119.2 (6-FAM-5’) and Oligo-pTa535 (Tamra-5’), and compared with cytogenetic karyotype of wheat and Th. elongatum [33, 34].
To identify the homology of the E chromosome, oligo-FISH probes Chr1E–Chr7E were conducted as sequential bulk oligo-FISH painting [57, 68]. In total, a 10 µL hybridization mixture consisted of 3 µL of mc-FISH probes (200–300 ng), 4 µL of 50% dextran sulfate sodium salt, 2 µL of 20× SSC, and 100% formamide. Sequential in situ hybridization protocol was referred to by Wu et al. [67].
The 4,6-diamino-2-phenylindole solution (DAPI; Vector Laboratories, Burlingame, CA, USA) was applied as a counterstain for in situ hybridization. We observed and recorded the fluorescence signals using a fluorescence microscope Olympus BX63 (Olympus, Tokyo, Japan) equipped with a DP80 CCD camera.
Phenotyping of FHB resistance
The Fg strain PH-1 was applied for FHB inoculation. Type II resistance, indicating the ability to resistance fungal spread, is evaluated using single floret inoculation (percentage of damaged florets among infected spikes) in the greenhouse (22–25 °C) and the experimental field of Sichuan Agricultural University (Wenjiang, Sichuan, China), with ten plant replicates per wheat lines. In total, 10 µL mixed Fg (1 × 103/mL) conidial suspension was inoculated into two flowering florets in the middle spikelet with a pipette gun or syringe. We sprayed sterile water on the clingfilm and wrapped the inoculated spike for 48 h to ensure inoculation with sufficient humidity. The PSD was recorded 14 days post-inoculation (dpi) and 21 dpi in the greenhouse and field, respectively.
Evaluation of agronomic performance
Investigations were conducted on agronomic traits of amphiploid 8801, 7D (7E) substitution line, SM482, and SM51 in the field (Chengdu, China), including plant height, tillering, flag leaf length, flag width, spike length, grain number per spike, and spikelet number per spike were investigated. Each material was planted in four rows, 15 grains per row (0.3 m* 1.5 m = space* length). Six individual plants of each sample were selected for agronomic traits investigation. We used OriginPro 2021 (OriginLab Corp., Northampton, MA, USA) software to calculate the mean and standard deviation of each group of agronomic traits. One-way ANOVA (p < 0.01) was conducted to determine if the differences between the 7D (7E) substitution line and parental material were significant, and was visualized in a bar chart.
Locus-specific molecular marker development
MISA (http://pgrc.ipk-gatersleben.de/misa/download/misa.pl) was used to obtain SSR molecular markers from the reference diploid Th. elongatum (ASM1179987v1) genome Chr7E. Using bedtools (V2.28.0), we pulled the 150 bp flanking sequences surrounding the SSR sites. To select SSR markers with target fragments, we then performed a mock e-PCR specifical amplification. Afterward, we aligned SSR sequences of the above amplification with Chr1E–Chr6E and the reference ‘Chinese Spring’ (IWGSC RefSeq v2.1) and eliminated homologous sequences. We randomly selected SSR primers and synthesized them in Sangon Biotech (Chengdu) Co., Ltd. Using 3% agarose gel, we detected the specificity of PCR amplification products. Primers that showed positive amplification in Trititrigia 8801 and 7E(7D) substitution lines, yet no amplification in SM482 and SM51, were selected as 7E-specific primers.
Pedigree analysis
We constructed F2 population (7E(7D) substitution line× SM830 F2, 122 F2 plants) to analyse the genetic effect of 7E(7D) substitution line on FHB resistance. Three genotypes will be segregated in the F2 population, 7E/7E (7EL-homozygous), 7E/7D (7E/7D -heterozygous) and 7D/7D (7E-null). To accurately genotype F2 plants, 7E-specific SSR marker TTE7E-31 detection was followed by GISH identification to quantify the alien 7E chromosome number. The phenotypes of FHB resistance of all F2 plants were identified in the greenhouse according to the 7E(7D) substitution line above. The statistical significance of differences in PDS means between genotypes was analyzed using a two-sample t-test (*, p < 0.05; **, p < 0.01; ns, p > 0.05) by OriginPro 2021 (OriginLab Corp., Northampton, MA, USA) software and was visualized with a boxplot.
Homology-based cloning of Fhb7 allele and sequence alignment
To determine the Fhb7 homologous gene in the 7E(7D) substitution line, we conducted PCR amplification using Fhb7 diagnostic markers, GST-F/R and 26,102-F/R [16]. PCR was performed in a 50 µL reaction mixture: 25 µL 2×Taq PCR PreMix, 2.5 µL per primer, 200 ng/µL DNA, and ddH2O to the final volume. The PCR amplification was checked using a 1.5% (w/v) agarose gel and cloned into the pMD19-T vector (TaKaRa) according to the manufacturer’s instructions. Six randomly chosen positive clones were sequenced in Sangon Biotech (Chengdu) Co., Ltd. The nucleotide and CDS sequences data were blasted with DNAMAN 9.0. The Fhb7 was assigned the designation Fhb7The2 in Th. elongatum and Fhb7Thp in Th. ponticum [69].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Additional file 1: fig. S1 Statistical analysis of 7E(7D) substitution and their parental lines for eight agronomic traits. a Plant height (cm). b Tiller number. c Spike length (cm). d Spikelets per spike. e Grains per spike. f Flag leaf length (cm). g Flag leaf width (cm). h 1000-grain weight (g). Asterisks indicate the level of significance: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Supplementary Material 2: Additional file 2: table S1 Agronomic traits of 7E(7D) substitution line and their parents. Table S2 Genetic effects of 7E chromosome on FHB resistance in F2 population.
Acknowledgements
Not applicable.
Abbreviations
- CS
ChineseSpring
- DON
Deoxynivalenol
- dpi
Days post-inoculation
- EST
Expressed Sequence Tags
- FHB
Fusarium Head Blight
- FISH
Fluorescence In Situ Hybridization
- GBS
Genotyping-By-Sequencing
- GISH
Genome In Situ Hybridization
- KASP
Kompetitive Allele-Specific PCR
- MAS
Marker-Assisted Selection
- NGS
Next-Generation Sequencing
- NLRs
Leucine-Rich Repeat Receptors
- PLUG
PCR-based Landmark Unique Gene
- Pm
Powdery Mildew
- PDS
Percentage of Diseased Spikelets
- QTLs
Quantitative Trait Loci
- SLAF-seq
Specific-Locus Amplified Fragment Sequencing
- SM482
Shumai 482
- SM51
Shumai 51
- SM830
Shumai 830
- SSC
Saline Sodium Citrate
- SSR
Simple Sequence Repeats
- YPC
Yellow Pigment Content
Author contribution
Conceptualization, H.K; Methodology, D.W. and F.W.; Validation, Y.L., W.Z., J.Z., L.C., and Y.W.; Formal Analysis, L.S., X.F., H.Z., and Y.Z.; Investigation, F.W., Y.M., Y.Z., and W.Z.; Resources, J.Z. and Y.C.; Data Curation, H.Z; Writing – Original Draft Preparation, D.W., Y.H., and Y.X.; Writing – Review & Editing, D.W., Y.L., and H.K.; Visualization, F.W. and Y.M.; Funding Acquisition, D.W and H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (32200180), the Special Projects of the Central Government in Guidance of Local Science and Technology Development (2023ZYD0088), and the Science and Technology Department of Sichuan Province (2023NSFSC1995, 2024NSFSC1327).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dandan Wu and Fei Wang contributed equally to this work.
References
- 1.Bottalico A, Perrone G. Toxigenic fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur J Plant Pathol. 2002;108:611–24. [Google Scholar]
- 2.Buerstmayr M, Steiner B, Buerstmayr H. Breeding for Fusarium head blight resistance in wheat—progress and challenges. Plant Breed. 2020;139:429–54. [Google Scholar]
- 3.Ma ZQ, Xie Q, Li GQ, Jia HY, Zhou JY, Kong ZX, et al. Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor Appl Genet. 2020;133:1541–68. [DOI] [PubMed] [Google Scholar]
- 4.Logrieco A, Bottalico A, Mulé G, Moretti A, Perrone G. Epidemiology of toxigenic fungi and their associated mycotoxins for some Mediterranean crops. Epidemiology of mycotoxin producing fungi: under the aegis of COST action 835 ‘agriculturally important toxigenic fungi 1998–2003, EU project (QLK 1-CT-1998–01380). 2003;645–67.
- 5.Desjardins AE. Fusarium mycotoxins: chemistry, genetics, and biology. American Phytopathological Society (APS; 2006.
- 6.Terzi V, Tumino G, Stanca AM, Morcia C. Reducing the incidence of cereal head infection and mycotoxins in small grain cereal species. J Cereal Sci. 2014;59:284–93. [Google Scholar]
- 7.Thapa A, Horgan KA, White B, Walls D. Deoxynivalenol and zearalenone-synergistic or antagonistic agri-food chain co-contaminants? Toxins. 2021;13:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuthbert PA, Somers DJ, Thomas J, Cloutier S, Brule-Babel A. Fine mapping Fhb1, a major gene controlling Fusarium head blight resistance in bread wheat (Triticum aestivum L). Theor Appl Genet. 2006;112(8):1465–72. [DOI] [PubMed] [Google Scholar]
- 9.Cuthbert PA, Somers DJ, Brûlé-Babel A. Mapping of Fhb2 on chromosome 6BS: a gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L). Theor Appl Genet. 2007;114:429–37. [DOI] [PubMed] [Google Scholar]
- 10.Xue SL, Li GQ, Jia HY, Xu F, Lin F, Tang MZ, et al. Fine mapping Fhb4, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L). Theor Appl Genet. 2010;121(1):147–56. [DOI] [PubMed] [Google Scholar]
- 11.Xue SL, Xu F, Tang MZ, Zhou Y, Li GQ, An X, et al. Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L). Theor Appl Genet. 2011;123(6):1055–63. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Li GQ, Jia H, Cheng R, Zhong JK, Shi JX, et al. Breeding evaluation and precise mapping of Fhb8 for Fusarium head blight resistance in wheat (Triticum aestivum). Plant Breed. 2024;143(1):26–33. [Google Scholar]
- 13.Zhang FP, Zhang HJ, Liu JL, Ren XM, Ding YP, Sun FY, et al. Fhb9, a major QTL for Fusarium head blight resistance improvement in wheat. J Integr Agr. 2024a. 10.1016/j.jia.2024.03.045. [Google Scholar]
- 14.Qi LL, Pumphrey MO, Friebe B, Chen PD, Gill BS. Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor Appl Genet. 2008;117(7):1155–66. [DOI] [PubMed] [Google Scholar]
- 15.Cainong JC, Bockus WW, Feng YG, Chen PD, Qi LL, Sehgal SK, et al. Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor Appl Genet. 2015;128(6):1019–27. [DOI] [PubMed] [Google Scholar]
- 16.Wang HW, Sun SL, Ge WY, Zhao LF, Hou BQ, Wang K, et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368:eaba5435. [DOI] [PubMed] [Google Scholar]
- 17.Li GQ, Zhou JY, Jia HY, Gao ZX, Fan M, Luo YJ, et al. Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat Genet. 2019;51(7):1106–12. [DOI] [PubMed] [Google Scholar]
- 18.Su ZQ, Bernardo A, Tian B, Chen H, Wang S, Ma HX, et al. A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat Genet. 2019;51:1099–105. [DOI] [PubMed] [Google Scholar]
- 19.Wan YF, Yen C, Yang JL. The diversity of head-scab resistance in Triticeae and their relation to ecological conditions. Euphytica. 1997;97:277–81. [Google Scholar]
- 20.Oliver RE, Cai X, Xu SS, Chen X, Stack RW. Wheat-alien species derivatives: a novel source of resistance to Fusarium head blight in wheat. Crop Sci. 2005;45:1353–60. [Google Scholar]
- 21.Yen C, Yang JL. Triticeae biosystematics; Chinese Agricultural Press: Beijing, China, 2013; Volume 5.
- 22.Dvorák J, Edge M, Ross K. On the evolution of the adaptation of Lophopyrum elongatum to growth in saline environments. Proc Natl Acad Sci U S A. 1988;85:3805–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fedak G. Molecular aids for integration of alien chromatin through wide crosses. Genome. 1999;42:584–91. [Google Scholar]
- 24.Mago R, Bariana HS, Dundas IS, Spielmeyer W, Lawrence GJ, Pryor AJ, et al. Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor Appl Genet. 2005;111:496–504. [DOI] [PubMed] [Google Scholar]
- 25.Ceoloni C, Forte P, Kuzmanović L, Tundo S, Moscetti I, Vita PD, et al. Cytogenetic mapping of a major locus for resistance to Fusarium head blight and crown rot of wheat on Thinopyrum elongatum 7EL and its pyramiding with valuable genes from a th. Ponticum homoeologous arm onto bread wheat 7DL. Theor Appl Genet. 2017;130:2005–24. [DOI] [PubMed] [Google Scholar]
- 26.Wang YZ, Cao Q, Zhang JJ, Wang SW, Chen CH, Wang CY, et al. Cytogenetic analysis and molecular marker development for a new wheat–Thinopyrum ponticum 1Js (1D) disomic substitution line with resistance to stripe rust and powdery mildew. Front Plant Sci. 2020;11:1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korostyleva TV, Shiyan AN, Odintsova TI. The genetic resource of Thinopyrum elongatum (Host) D.R. Dewey in breeding improvement of wheat. Russ J Genet. 2023;59:983–90. [Google Scholar]
- 28.Zeng J, Zhou CL, He ZM, Wang Y, Xu LL, Chen GD, et al. Disomic substitution of 3D chromosome with its homoeologue 3E in tetraploid Thinopyrum elongatum enhances wheat seedlings tolerance to salt stress. Int J Mol Sci. 2023;24:1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng XF, Xiao Y, Wang LH, Yang XY, Deng PC, Zhao JX, et al. Cytogenetic characterization and and molecular marker development of a novel wheat-Thinopyrum ponticum 5E (5D) disomic substitution line with resistance to powdery mildew and stripe rust. J Integr Agric. 2024. 10.1016/j.jia.2024.04.012. [Google Scholar]
- 30.Fedak G, Han FP. Characterization of derivatives from wheat-Thinopyrum wide crosses. Cytogenet Genome Res. 2005;109(1–3):360–7. [DOI] [PubMed] [Google Scholar]
- 31.Baker L, Grewal S, Yang CY, Hubbart-Edwards S, Scholefield D, Ashling S, et al. Exploiting the genome of Thinopyrum elongatum to expand the gene pool of hexaploid wheat. Theor Appl Genet. 2020;133:2213–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang ZX, Yang ZJ, Fu SL. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet. 2014;55:313–8. [DOI] [PubMed] [Google Scholar]
- 33.Li DY, Li TH, Wu YL, Zhang XH, Zhu W, Wang Y, et al. FISH-based markers enable identification of chromosomes derived from tetraploid Thinopyrum elongatum in hybrid lines. Front Plant Sci. 2018;9:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing L, Yuan L, Lv Z, Wang Q, Yin C, Huang Z, et al. Long-range assembly of sequences helps to unravel the genome structure and small variation of the wheat–Haynaldia villosa translocated chromosome 6VS.6AL. Plant Biotechnol. J. 2021;19:1567–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlegel R. Current list of wheats with rye and alien introgression. Germany: Gatersleben; 2022. http://www.rye-gene-map.de/rye-introgression/. [Google Scholar]
- 36.Sears ER. Genetic control of chromosome pairing in wheat. Annu Rev Genet. 1976;10:31–51. [DOI] [PubMed] [Google Scholar]
- 37.Fan CL, Hao M, Jia ZY, Neri C, Chen X, Chen WS, et al. Some characteristics of crossing over in induced recombination between chromosomes of wheat and rye. Plant J. 2021;105:1665–76. [DOI] [PubMed] [Google Scholar]
- 38.Sears ER. The transfer of leaf rust resistance from Aegilops umbellulate into wheat. In genetics in plant breeding. Brookhaven symposia in biology No. 9; Brookhaven National Laboratory: Upton, NY, USA. 1956;9:1–21.
- 39.Bhat TA, Khalid RH, editors. Biotechnologies and genetics in plant mutant breeding. Volume 1. Waretown, NJ, USA: Apple Academic; 2023. [Google Scholar]
- 40.Friebe B, Zhang P, Linc G, Gill BS. Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis ii. Cytogenet Genome Res. 2005;109:293–7. [DOI] [PubMed] [Google Scholar]
- 41.Prieto P. Chromosome manipulation for plant breeding purposes. Agronomy. 2020;10(11):1695. [Google Scholar]
- 42.Han BH, Wang X, Sun YY, Kang XL, Zhang M, Luo JW, et al. Pre-breeding of spontaneous robertsonian translocations for density planting architecture by transferring Agropyron cristatum chromosome 1P into wheat. Theor Appl Genet. 2024;137:110. [DOI] [PubMed] [Google Scholar]
- 43.Ceoloni C, Forte P, Gennaro A, Micali S, Carozza R, Bitti A. Recent developments in durum wheat chromosome engineering. Cytogenet Genome Res. 2005;109:328–44. [DOI] [PubMed] [Google Scholar]
- 44.Ceoloni C, Kuzmanović L, Forte P, Gennaro A, Bitti A. Targeted exploitation of gene pools of alien Triticeae species for sustainable and multi-faceted improvement of the durum wheat crop. Crop Pasture Sci. 2014;65:96–111. [Google Scholar]
- 45.Gennaro A, Koebner RMD, Ceoloni C. A candidate for Lr19, an exotic gene conditioning leaf rust resistance in wheat. Funct Integr Genom. 2009;9:325–34. [DOI] [PubMed] [Google Scholar]
- 46.Li HJ, Wang XM. Thinopyrum ponticum and Th. intermedium: the promising source of resistance to fungal and viral diseases of wheat. Genet Genom. 2009;36:557–65. [DOI] [PubMed] [Google Scholar]
- 47.Liu S, Yu LX, Singh RP, Jin Y, Sorrells ME, Anderson JA. Diagnostic and co-dominant PCR markers for wheat stem rust resistance genes Sr25 and Sr26. Theor Appl Genet. 2010;120:691–697. [DOI] [PubMed]
- 48.Tounsi S, Giorgi D, Kuzmanović L, Jrad O, Farina A, Capoccioni A, et al. Coping with salinity stress: segmental group 7 chromosome introgressions from halophytic Thinopyrum species greatly enhance tolerance of recipient durum wheat. Front Plant Sci. 2024;15:1378186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo XR, Wang M, Kang HY, Zhou YH, Han FP. Distribution, polymorphism and function characteristics of the GST-encoding Fhb7 in Triticeae. Plants. 2022;11:2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radecka-Janusik M, Piechota U, Piaskowska D, Goral T, Czembor P. Evaluation of Fusarium head blight resistance effects by haplotype-based genome-wide association study in winter wheat lines derived by marker backcrossing approach. Int J Mol Sci. 2022;23:14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang QQ, Liu FF, Wan YX, Cao WX, Li Y, Li Y, et al. Identification of resistance to Fusarium head blight and gene effect analysis of new wheat lines. J Triticeae Crops. 2024b;44(5):567–76. [Google Scholar]
- 52.Chen J, Zhang XX, Rathjen JP, Dodds PN. Direct recognition of pathogen effectors by plant NLR immune receptors and downstream signalling. Essays Biochem. 2022;66(5):471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou SH, Shi WQ, Ji JH, Wang HM, Tang YS, Yu DZ, et al. Diversity and similarity of wheat powdery mildew resistance among three allelic functional genes at the Pm60 locus. Plant J. 2022;110(6):1781–90. [DOI] [PubMed] [Google Scholar]
- 54.Sutherland CA, Prigozhin DM, Monroe JG, Krasileva KV. High allelic diversity in Arabidopsis NLRs is associated with distinct genomic features. EMBO Rep. 2024;25(5):2306–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang F, Zhao X, Yu XH, Zhu W, Xu LL, Cheng YR, et al. Identification and transfer of resistance to Fusarium head blight from Elymus repens chromosome arm 7StL into wheat. J Integr Agr. 2024. 10.1016/j.jia.2024.03.027. [Google Scholar]
- 56.Li GR, Zhang T, Yu ZH, Wang HJ, Yang EN, Yang ZJ. An efficient Oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J. 2020;105:978–93. [DOI] [PubMed] [Google Scholar]
- 57.Chen C, Han YS, Xiao H, Zou BC, Wu DD, Sha LN, et al. Chromosome-specific painting in Thinopyrum species using bulked oligonucleotides. Theor Appl Genet. 2023;136:177. [DOI] [PubMed] [Google Scholar]
- 58.Han GH, Liu SY, Wang J, Jin YL, Zhou YL, Luo QL, et al. Identification of an elite wheat-rye T1RS·1BL translocation line conferring high resistance to powdery mildew and stripe rust. Plant Dis. 2020;104:2940–8. [DOI] [PubMed] [Google Scholar]
- 59.Liu LQ, Luo QL, Li HG, Li B, Li ZS, Zheng Q. Physical mapping of the blue-grained gene from Thinopyrum ponticum chromosome 4Ag and development of blue-grain-related molecular markers and a FISH probe based on SLAF-seq technology. Theor Appl Genet. 2018;131:2359–70. [DOI] [PubMed] [Google Scholar]
- 60.An DG, Ma PT, Zheng Q, Fu SL, Li LH, Han FP, et al. Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor Appl Genet. 2019;132:257–72. [DOI] [PubMed] [Google Scholar]
- 61.Gong BR, Chen LF, Zhang H, Zhu W, Xu LL, Cheng YR, et al. Development, identification, and utilization of wheat–tetraploid Thinopyrum elongatum 4EL translocation lines resistant to stripe rust. Theor Appl Genet. 2024;137:17. [DOI] [PubMed] [Google Scholar]
- 62.Deng PC, Du X, Wang YZ, Yang XY, Cheng XF, Huang CX, et al. GenoBaits®WheatplusEE: a targeted capture sequencing panel for quick and accurate identification of wheat–Thinopyrum derivatives. Theor Appl Genet. 2024;137:36. [DOI] [PubMed] [Google Scholar]
- 63.Wu DD, Zhao X, Xie YQ, Li LY, Li YH, Zhu W, et al. Cytogenetic and genomic characterization of a novel wheat-tetraploid Thinopyrum elongatum 1BS·1EL translocation line with stripe rust resistance. Plant Dis. 2024;108:2065–72. [DOI] [PubMed] [Google Scholar]
- 64.Tribhuvan KU, Sandhya, Kumar K, Sevanthi AM, Gaikwad K. MutMap: a versatile tool for identification of mutant loci and mapping of genes. Ind J Plant Physiol. 2018;23:612–21. [Google Scholar]
- 65.Li YH, Wei ZZ, Sela H, Govta L, Klymiuk V, Roychowdhury R, et al. Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69. Plant Commun. 2024;5(1):100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aliyeva-Schnorr L, Beier S, Karafiátová M, Schmutzer T, Scholz U, Dolezel J, et al. Cytogenetic mapping with centromeric bacterial artificial chromosomes contigs shows that this recombination-poor region comprises more than half of barley chromosome 3H. Plant J. 2015;84:385–94. [DOI] [PubMed] [Google Scholar]
- 67.Wu DD, Ruban A, Fuchs J, Macas J, Novák P, Vaio M, et al. Nondisjunction and unequal spindle organization accompany the drive of Aegilops speltoides B chromosomes. New Phytol. 2019;223(3):1340–52. [DOI] [PubMed] [Google Scholar]
- 68.Shi PY, Sun HJ, Liu GQ, Zhang X, Zhou JW, Song RR, et al. Chromosome painting reveals inter-chromosomal rearrangements and evolution of subgenome D of wheat. Plant J. 2022;112:55–67. [DOI] [PubMed] [Google Scholar]
- 69.Zhang W, Danilova T, Zhang MY, Ren SF, Zhu XW, Zhang QJ, Zhong SB, Dykes LD, Fiedler J, Xu S, Frels K, Wegulo S, Boehm J, Cai XW. Cytogenetic and genomic characterization of a novel tall wheatgrass-derived Fhb7 allele integrated into wheat B genome. Theor Appl Genet. 2022;135:4409–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Additional file 1: fig. S1 Statistical analysis of 7E(7D) substitution and their parental lines for eight agronomic traits. a Plant height (cm). b Tiller number. c Spike length (cm). d Spikelets per spike. e Grains per spike. f Flag leaf length (cm). g Flag leaf width (cm). h 1000-grain weight (g). Asterisks indicate the level of significance: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Supplementary Material 2: Additional file 2: table S1 Agronomic traits of 7E(7D) substitution line and their parents. Table S2 Genetic effects of 7E chromosome on FHB resistance in F2 population.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.