Abstract
Chloroplasts and photosynthesis are the physiologically fateful arenas of salinity stress. Morphological and anatomical alterations in the leaf tissue, ultrastructural changes in the chloroplast, compromise in the integrity of the three-layered chloroplast membrane system, and defects in the light and dark reactions during the osmotic, ionic, and oxidative phases of salt stress are conversed in detail to bring the salinity-mediated physiological alterations in the chloroplast on to a single platform. Chloroplasts of salt-tolerant plants have evolved highly regulated salt-responsive pathways. Thylakoid membrane remodeling, ion homeostasis, osmoprotection, upregulation of chloroplast membrane and stromal proteins, chloroplast ROS scavenging, efficient retrograde signalling, and differential gene and metabolite abundance are the key attributes of optimal photosynthesis in tolerant species. This review throws light into the comparative mechanism of chloroplast and photosynthetic response to salinity in sensitive and tolerant plant species.
Keywords: halophytes, photosynthetic rate, plastid, salt stress, sensitivity, tolerance
Highlights
Photosynthesis is highly sensitive to salinity stress
An interplay of physiological and molecular determinants exists in the sensitivity of chloroplast to salinity stress
The major tolerance and adaptive mechanisms to salinity are chloroplast membrane remodeling, ion homeostasis, and retrograde signalling
Introduction
Photosynthesis, the process that occurs in the wonderful laboratories of plants namely chloroplasts, dates back its origin to 3,800 million years ago. The Proterozoic era witnessed the origin of photosynthetic cyanobacteria as evident from the microfossils of stromatolites' reefs (Des Marais 2000). With progress in research, it was revealed that despite evolutionary differences between photoautotrophs, the reaction centers and cofactors follow the same three-dimensional structures. Moreover, the dominant carbon fixation process in all oxygenic photosynthetic organisms is the Calvin–Benson (C3) cycle (Blankenship 2010). The process of photosynthesis that reduces gaseous carbon and fixes it to carbohydrates occurs in a biphasic manner. The biphasic mode of photosynthesis occurs in distinct regions of the chloroplast. The light-dependent reactions occur in the grana thylakoids and the carbon-reduction reactions take place in the stroma (Govindjee and Govindjee 1974).
Salinity stress is a major factor determining plant growth and yield, by significantly affecting key physiological processes, such as photosynthesis and energy metabolism (Feng et al. 2014). All the components and processes of photosynthesis mentioned above are directly disturbed by salt stress. Being a primary physiological process, any stress that impairs photosynthesis has a significant bearing on crop yield (Yang et al. 2020). Plants are forced to maintain an optimization between nutrient uptake and restriction of the entry of salinity-associated ions. The genome editing targets for such trade-offs are reported from various plants (Sathee et al. 2022). Salinity stress is a major global environmental challenge that degrades land and impairs production in a vast majority of crops (Soltabayeva et al. 2021). A global analysis of the Earth over 40 years by satellite data and machine learning showed that out of the 120.4 M km2 of non-frigid land, 9.7% is affected by salinity. This area is not small as it encompasses 11.737 M km2 (1,173.7 M ha) of land, of which 16.49 M ha is agricultural land (Hassani et al. 2020). As per the Food and Agriculture Organization (FAO), the total cultivated land is 5,301 M ha (48.01 M km2) (http://www.fao.org/faostat/en/#data/RL) and it needs to feed the burgeoning population (Park et al. 2016). Across the globe, more than 424 M ha of topsoil (0–30 cm) and 833 M ha of sub soil (30–100 cm) are salt-affected (https://www.fao.org/soils-portal/data-hub/soil-maps-and-databases/global-map-of-salt-affected-soils/en/). Approximately, thirty crop species contribute to 90% of plant-based food, most of which show 50–80% yield reduction under conditions of moderate salinity (EC of 4–8 dS m–1) (Zörb et al. 2019). Photosynthetic rate and associated traits, such as stomatal conductance, transpiration rate, internal CO2 concentration, water-use efficiency, and stomatal density have been reported to be the major factors affecting the biomass and grain yield of crops under salinity stress (Lekklar et al. 2019).
In rice, the major cause of grain yield reduction under salinity stress is the photosynthetic limitation and its partitioning to spikelet (Hussain et al. 2017). More than a 65% reduction in photosynthetic rate has been observed in rice genotypes exposed to salinity stress of 8 dS m–1 during the reproductive stage (Lekklar et al. 2019). More than a two-fold reduction in photosynthetic rate has been observed in salt-sensitive and tolerant genotypes of durum wheat under salinity stress of 10 dS m–1 and has been correlated to reduced grain yield and test mass. Interestingly, in durum wheat, photosynthetic rate and stomatal conductance showed a negative correlation at high salinity stress of 15 dS m–1, which suggests a C4-type primary carboxylation using phosphoenolpyruvate carboxylase (PEPcase) (Pastuszak et al. 2022). Salinity-induced reduction in photosynthetic rate, assimilate translocation to developing grains, and sink limitation are the major contributors to low grain yield in maize (Hiyane et al. 2010). The salinity threshold of the majority of vegetable crops ranges from 1 to 2.5 dS m−1. Nearly 80% of the growth reduction in certain vegetable crops such as radish and spinach under saline conditions has been attributed to a decrease in leaf area and photosynthetic rate, and 20% due to a decrease in stomatal conductance (Machado and Serralheiro 2017). Salinity-induced reduction in photosynthesis of legume crops such as soybean and chickpea is not only a result of reduced stomatal conductance, but also due to nonstomatal limitations, such as reduced chlorophyll, chlorophyll fluorescence, and ultrastructural damage (Khan et al. 2015, Nadeem et al. 2019).
With the current annual rate of global soil deterioration, the possibility of famine in near future is looming in the face of mankind. Deciphering how the photosynthetic apparatus gets affected and in turn responds to excess salts could provide new knowledge to enhance agricultural productivity, sustainability, and food security.
Characteristics underlying sensitivity of photosynthesis to salinity in plants
Morphological and anatomical alterations of photosynthetic tissues
Leaf anatomy is a crucial aspect in the maintenance of optimal photosynthesis levels during abiotic stresses (Longstreth and Nobel 1979). The most important anatomical traits of leaf tissue, which are strongly affected by salt stress, are mesophyll cell thickness, area of mesophylls per unit of leaf surface area, a ratio of mesophyll cell surface area to that of leaf surface area, epidermal and leaf thickness (Acosta-Motos et al. 2017). Increased leaf thickness and succulence are linked to a salt-induced decrease in the photosynthesis of salt-sensitive citrus species. This is because it reduces the surface area to volume ratio of mesophyll cells and lowers the intercellular air space, which indirectly affects CO2 assimilation (Romero-Aranda et al. 1998).
The relative contribution of anatomical changes in the leaf to a decrease in photosynthesis is also dependent on the tolerance level of the plant genotype. Navarro and his group studied the anatomical alterations in the leaves of the strawberry tree (Arbutus unedo) under control and varying salt-stress conditions. The cell size of the second layer of palisade cells increased significantly with a concomitant increase in salinity levels, with no change in the cell size of the first palisade layer (Navarro et al. 2007). They also observed a reduction in intercellular spaces in the spongy mesophyll cells under salt stress which was strongly linked to reducing stomatal and mesophyll conductance to CO2. Similarly, henna (Lawsonia inermis L.) plants irrigated with 75 and 150 mM NaCl showed an increase in leaf thickness to leaf area ratio (leaf specific mass) as compared to plants irrigated with normal water. This was confirmed by Fernández-García et al. (2014) as a specific investment by these plants in leaf tissue to increase leaf thickness, leaf specific mass, and thereby maximise photosynthesis under adverse conditions. Wheat plants subjected to varying degrees of salinity displayed an array of anatomical changes, such as decreased wall thickness, a diameter of hollow pith cavity, lower number of vascular bundles with reduced length and width, to survive the adverse condition (Nassar et al. 2020). In general, these anatomical changes under salt stress may not necessarily lead to efficient maintenance of photosynthetic rates, mainly due to a drastic reduction in stomatal conductance. The tolerant genotypes try to survive by anatomical alterations conserving the energy for reproduction.
Ultrastructural changes in the chloroplast
It has been demonstrated that under salt stress, chloroplasts get deformed into irregular shapes with significant compression in the grana thylakoid lamellae (Shu et al. 2015). The separation between cell membranes and chloroplasts reduced grana stacking, while a larger number of osmiophilic granules, giant starch granules, and accumulation of plastoglobules are certain key markers under salt stress (Gao et al. 2015, Guo et al. 2019a). Additionally, stroma thylakoid lamella also gets disrupted due to altered cellular ionic ratio (Shu et al. 2015). Studies from Arabidopsis revealed that chloroplasts of salt-treated seedlings showed deformation, reduced genome copy number, and gene expression. The organelle was enlarged, with reduced grana stacking and larger starch granules (Peharec Štefanić et al. 2013). Oi et al. (2020) described the three-dimensional structural changes in chloroplasts under salt stress in rice. They found that the cells of unstressed plants had elongated meniscus lens-shaped chloroplasts, while the stressed ones were expanded and oval-shaped. Serial sectioning also showed that the plastids in stressed cells were aggregated, but not in physical contact with each other (Oi et al. 2020). Additional observations were found in potato (Gao et al. 2015) and brinjal (Alkhatib et al. 2021), where gradient salinity stress treatment was imposed on seedlings. At the onset of stress, disintegration of outer membrane, and partial dissolution or cavitation of both grana and stroma thylakoids were observed. In the long run (six weeks of stress), there was digestion of starch granules, reduction in chlorophyll content, and the thylakoid membranes became stickier and gradually disappeared.
Compromised integrity of chloroplast membrane system
Cellular membranes are integral to all three major components of salt stress: ion imbalance and associated nutrient stress, osmotic stress, and reactive oxygen species (ROS)-induced oxidative damage. Membrane remodelling has been evident in plants exposed to salt stress via altered permeability and fluidity by associated changes in lipid and protein composition as well as quantity (Guo et al. 2019b). The remodelling is very prompt under salinity stress, whereas in soybean it was shown to occur within 30 min of exposure to salt (Liu et al. 2021). Studies on membrane lipid alterations are largely focused on changes in total membrane lipids with special attention to the plasma membrane. Only very few studies highlighted endomembranous lipid remodelling under salt stress.
The three-membrane chloroplast system including the outer and inner envelope membranes and the thylakoid membrane is significantly affected by altered cellular ion content (Suo et al. 2017). The chloroplast membrane network mainly comprises glycolipids, such as monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and sulfoquinovosyldiacylglycerol (SQDG). The two major salt-mediated responses at the membrane level are changes in the total lipid content and alterations in the ratio of saturated and unsaturated fatty acids components. Salt causes changes in membrane permeability with a reduction in total lipid content and results in excess ion leakage, which is measured as a loss of membrane integrity. The reduction in lipid content and associated loss of integrity along with increased membrane peroxidation levels has been reported to be more prevalent in salt-sensitive plant species/genotypes (Chalbi et al. 2013).
Salt-sensitive plant species commonly show a reduction in MGDG in the chloroplast membranes, resulting in a significant decrease in chlorophyll content (Shu et al. 2012, Yamane et al. 2015). This effect owes to the salt-induced activation of galactolipase and lipoxygenase enzymes (Hasanuzzaman et al. 2014). MGDG plays an important role in forming stable bilayer structures in conjunction with LHCII in the thylakoid membranes. The reduction in MGDG, thus causes serious disruption to the chloroplast membranes by altering the MGDG/LHCII ratio (Simidjiev et al. 2000). Decline in phospholipid (PL) component has been observed in several salt-sensitive plant species (Magdy et al. 1994, Lin and Wu 1996). These PLs can act as precursors for the biosynthesis of glycolipids in chloroplast membranes. A decreased phosphatidylcholine (PC) to phosphatidylethanolamine (PE) ratio has been confirmed as an indicator of salt sensitivity (Liu et al. 2021).
Phosphatidylglycerol (PG), an inevitable lipid of thylakoid membranes has been reported to decrease under salt stress in leaves of Sulla sp. which was speculated to be the major cause of ultrastructural damage to the chloroplast membranes (Bejaoui et al. 2016). A reduced DGDG content was also observed in Catharanthus sp., Arabidopsis, Thellungiella halophila, and Sulla sp. grown under salt stress (Sui and Han 2014, Bejaoui et al. 2016), which is inferred to be a possible reason for loss of membrane integrity and reduced stability of major proteins such as PS1 and LHCII (Boychova Krumova et al. 2010). Arabidopsis displayed a significantly reduced DGDG/MGDG ratio in its chloroplast membranes as compared to salt-tolerant T. halophila. This finding indicates a direct link between this ratio and the integrity of the cellular membrane system (Sui and Han 2014). Omoto et al. (2013) observed that a lesser abundance of MGDG in mesophyll as compared to bundle sheath chloroplast membrane under salt stress was attributed as the major factor of difference between the salt sensitivity of the two cell types in C4 maize.
SQDG is another glycolipid exclusively associated with chloroplast membranes. It contributes towards a more stable protein–lipid configuration in the membrane bilayer and binds with another protein called annexin involved in membrane organisation and fusion (Seigneurin-Berny et al. 2000). In glycophytes, SQDG contents decrease under imposed salinity stress, as evident in maize (Omoto et al. 2016) and Sulla sp. (Bejaoui et al. 2016). Additional findings from salt-sensitive species show that a reduction in SQDG content has been speculated to be due to decreased contents or activity of the rate-limiting enzyme UDP-glucose pyrophosphorylase under stress (Guo et al. 2019b).
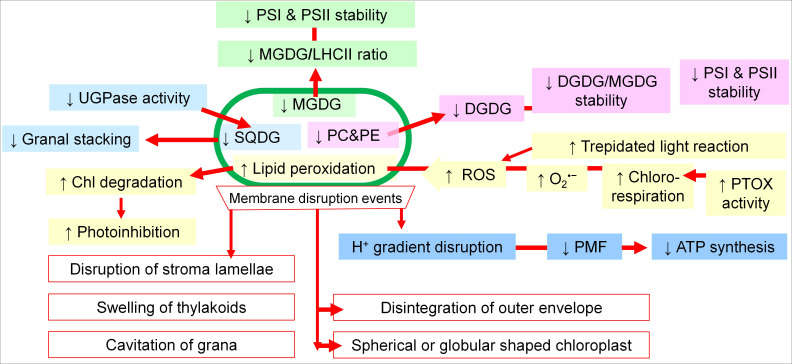
The summarized model, which highlights the major changes in the chloroplasts of salt-sensitive plant species, is depicted in Fig. 1.
Fig. 1. Major routes of chloroplast membrane disruption and remodelling under salt stress in salinity-sensitive plant species. MGDG – monogalactosyldiacylglycerol; PC – phosphatidyl choline; PE – phosphatidyl ethanolamine; UGPase – UDP-glucose pyrophosphorylase; SQDG – sulfoquinovosyldiacylglycerol; LHCII – light-harvesting complex II; PSI – photosystem I; PSII – photosystem II; DGDG – digalactosyldiacylglycerol; PMF – proton motive force; PTOX – plastid terminal oxidase. Each colour represents different functional modules of chloroplast membrane remodelling under salinity stress.
Compromised polypeptide composition of photosystems and light reaction
Chloroplast contains 30–50% of total cellular proteins, among which 80% are proteins embedded in the thylakoid membranes (Lande et al. 2020). Salinity stress mainly impairs photosynthesis by altering the polypeptide composition of the photosystems. This degradative effect was found both in algae as well as higher plants. Salt stress causes a decrease in the number of active PSII sites, damages the oxygen-evolving complex (OEC), and impedes the electron flow to PSI (Kan et al. 2017). The JIP test on salinity-imposed Dunaliella sp. revealed that the water-splitting complex is the first site of damage by excess salts (Ghasemi and Shariati 2012).
Western blot analysis of thylakoid membrane proteins showed that the expression of LHC proteins, such as CP7, CP43, Lhcb1, and Lhcb4 was downregulated in Chlamydomonas reinhardtii under salt stress (Neelam and Subramanyam 2013). Studies have shown that salt stress inhibits the repair of PSII by restraining the activities of the psbA gene that encode the D1 protein in the cyanobacterium Synechocystis sp., both at transcriptional and translational levels (Allakhverdiev et al. 2001). Such damages specific to PSII have also been found in plant systems (Kan et al. 2017). In the susceptible rice cultivar Peta, PSII had lesser content of 33 and 43 kDa polypeptides and a deficiency in 23 kDa polypeptides under salt stress (Wang et al. 2009). Moreover, under salt stress, other proteins related to PSII and PSI, such as PsbP, PsaK, Ycf4, and OHP1 showed decreased abundance in wheat, indicating Na+-mediated damage leading to loss of thylakoids and decreased Fv/Fm (Zhu et al. 2021). Salt stress also damages the transfer of electrons from QA to QB and further to the cytochrome (Cyt) b6f complex (Akhter et al. 2021). In addition, the PetD protein of Cyt b6f complex was found to be degraded under salinity stress in Desmostachya bipinnata (Asrar et al. 2017). The plastoquinone pool remains over-reduced in the process which can lead to the generation of ROS (Akhter et al. 2021). Studies in tomato, rice, etc., have shown degradation of membrane protein components of Cyt b6f and ATP-synthase complex (Li et al. 2018). The physiological outcome of such damages is poor H+ gradient and proton motive force (PMF) in thylakoids and reduced ATP-synthase activity. Chlororespiration is a process occurring in chloroplasts that diverts electrons from PSII via the plastoquinone pool to molecular oxygen, producing water. It is regulated by the enzyme plastid terminal oxidase (PTOX) (Bolte et al. 2020). The transformation of tobacco chloroplast using the PTOX gene from C. reinhardtii showed a light-sensitive response (Ahmad et al. 2012). These effects are the outcome of the pro-oxidant function of the PTOX gene that leads to the formation of superoxide molecules (Krieger-Liszkay and Feilke 2016) (Table 1). These types of negative regulatory genes are being targeted for genome editing by CRISPR/Cas for imparting salinity tolerance in crop plants (Sathee et al. 2022). Thus, the various alterations in membrane proteins contribute to defective photosynthesis in chloroplasts.
Table 1. The major genes involved in imparting susceptibility or tolerance to plants under salinity stress.
| Gene(s) | Protein | Plant species | Salinity stress response | Reference |
| Photosynthetic membrane organisation and PSII activity | ||||
| Fad6 | ω-6 desaturase | Arabidopsis | Seedling stage salinity tolerance | Zhang et al. (2009) |
| MGD | Monogalactosyldiacylglycerol synthase |
Rice (Oryza sativa) |
Salinity tolerance; well-developed thylakoid membrane with improved grana stacking |
Wang et al. (2014) |
|
OEP, OECP, Chla/bBP |
33 kDA oxygen-evolving protein, 23 kDa protein of oxygen-evolving complex of PSII, chlorophyll a/b binding protein |
Rapeseed (Brassica napus) |
Salinity tolerance | Jia et al. (2015) |
| Rrd | Rubredoxin family protein | Alkali grass (Puccinellia tenuiflora) |
Salinity tolerance | Li et al. (2016) |
| RCI | Rare cold-inducible protein | Wheat (Triticum aestivum) |
Salinity tolerance; maintenance of PSII stability and activity |
Khurana et al. (2015) |
| GPAT | Glycerol-3-phosphate acyltransferase | Tomato (Lycopersicon esculentum) |
Enhanced salt tolerance; unsaturation of fatty acids of PG in the thylakoid membrane |
Sun et al. (2010) |
| HCF136 | PSII stability/assembly factor | Wheat (Triticum aestivum) |
Salinity tolerance; maintenance of PSII stability and assembly |
Xu et al. (2016) |
| LHCB6 | Light-harvesting complex protein | Indian mustard (Brassica juncea) |
Salinity tolerance | Singh et al. (2019) |
|
PsbP, Cytb6f, b559-∞ |
23 kDa protein of oxygen-evolving complex of PSII, cytochrome b6f complex associated proteins |
Wheat (Triticum aestivum) |
Salt tolerance | Zhu et al. (2021) |
| PSI P700 | PSI P700 chlorophyll a apoprotein A2 | Cotton (Gossypium
hirsutum L.) |
Salinity tolerance; high electron transfer efficiency |
Gong et al. (2017) |
| Fd, NADHC | Ferredoxin, NAD(P)H dehydrogenase complex |
Wheat (Triticum aestivum) |
Salt tolerance | Zhu et al. (2021) |
| PTOX | Plastid terminal oxidase | Tobacco | Salt sensitivity; increased formation of superoxides |
Ahmad et al. (2012) |
| Photosynthetic dark reactions, other stromal genes, and photorespiration | ||||
| CHL, GSAAT | Magnesium chelatase, glutamate-1-semialdehyde aminotransferase |
Chickpea (Cicer arietinum), Brassica napus |
Salinity tolerance |
Jia et al. (2015), Arefian et al. (2019) |
| FBA | Fructose-1,6-bisphosphate aldolase | Cotton (Gossypium
hirsutum L.) |
Salinity tolerance | Gong et al. (2017) |
| NADP-ME | NADP-malate dehydrogenase | Arabidopsis | Salinity tolerance | Chen et al. (2019a) |
| ALT, AST | alanine aminotransferase, aspartate aminotransferase |
Sesame (Sesamum indicum) |
Salinity tolerance | Zhang et al. (2019) |
| NPR1 | Non-expression of pathogenesis-related genes 1 |
Tobacco (Nicotiana tabacum) |
Salinity tolerance; redox homeostasis |
Seo et al. (2020) |
| GAPB | Glyceraldehyde-3-phosphate dehydrogenase β subunit |
Thellungiella
halophila |
Salinity tolerance | Chang et al. (2015) |
| GO | Glycolate oxidase | Sugar beet (Beta vulgaris) |
Salinity tolerance | Lv et al. (2019) |
| PEPC | Phosphoenolpyruvate carboxylase | Sweet sorghum (Sorghum bicolor) |
Salinity tolerance | Yang et al. (2020) |
| Chloroplast ionic and osmotic homeostasis | ||||
| KEA | K+/H+ antiporter | Arabidopsis | Salinity tolerance; chloroplast osmoregulation, integrity, and pH regulation |
Kunz et al. (2014) |
|
BASS2, PHT4;1, PHT4;4, PHT4;5 |
Bile acid sodium symporter 2, phosphate transporters |
Halophyte species | Salinity tolerance; preferential Na accumulation in the chloroplast |
Bose et al. (2017) |
| CHX23 | Na+ (K+)/H+ exchanger | Arabidopsis | Mutants displayed salt hypersensitivity and impaired photosynthetic performance |
Song et al. (2004) |
| NHD1 | Sodium hydrogen antiporter | Arabidopsis | Mutants displayed salt hypersensitivity and impaired photosynthetic performance |
Müller et al. (2014) |
| OsNHAD | Putative sodium hydrogen antiporter | Rice (Oryza sativa) |
Knockdown led to hypersensitivity to Na+ ions, reduced PSII activity, and altered chloroplast morphology |
Liu et al. (2020) |
| OtsA, OtsB | Trehalose-6-phosphate synthase, Trehalose-6-phosphate phosphatase |
Rice (Oryza sativa) |
Overexpression led to better photosynthetic rate and lower K/Na ratio in root and shoot |
Garg et al. (2002) |
| TPPD | Trehalose 6-phosphate phosphatase | Arabidopsis | Overexpressors were tolerant to high salinity stress |
Krasensky et al. (2014) |
| CodA | Choline oxidase | Tomato (Lycopersicon esculentum) |
Improved ion homeostasis and photosynthesis |
Wei et al. (2017) |
| Oxidative stress and defense | ||||
| Cu/Zn SOD | Copper/zinc superoxide dismutase | Cotton (Gossypium
hirsutum L.) |
Salinity tolerance; ROS scavenging | Luo et al. (2013) |
| GPX | Glutathione peroxidase | Arabidopsis | Salinity tolerance; ROS scavenging | Zhai et al. (2013) |
| γ-TMT | γ-tocopherol methyltransferase | Tobacco (Nicotiana tabacum) |
Salinity tolerance; reduced ROS abundance and membrane injury |
Jin and Daniell (2014) |
| GR3 | Glutathione reductase | Rice (Oryza sativa) |
Knockout led to increased salt sensitivity |
Wu et al. (2015) |
| CAT | Catalase | Cotton (Gossypium
hirsutum L.) |
Salinity tolerance; ROS scavenging | Luo et al. (2013) |
| DHAR | Dehydroascorbate reductase | Rice (Oryza sativa) |
Improved salinity tolerance; altered antioxidant metabolism |
Le Martret et al. (2011) |
| WSL12 | Nucleoside diphosphate kinase | Rice (Oryza sativa) |
Salinity tolerance; increased antioxidant enzyme activities |
Ye et al. (2016) |
Defective dark reaction processes
Several proteomic analyses as well as Western blotting studies have shown the degradation of Calvin cycle enzymes, Rubisco in particular, under salt stress (Asrar et al. 2017, Thagela et al. 2018). The activity of Rubisco was reported to be inhibited both in vivo and in vitro under salinity stress (Gong et al. 2018). Proteomic studies on salt-stressed alfalfa revealed upregulated expression and activity of the Rubisco activase enzyme (RCA) (Xiong et al. 2017). This can be inferred as the survival mechanism adopted by plants to activate the available Rubisco enzymes in the stroma. But under higher doses (600 mM NaCl), RCA content was reduced by 50% as compared to the levels at 400 mM in Leymus chinensis (Li et al. 2017a). Other Calvin cycle enzymes, which are severely affected by excess salts, include glyceraldehyde-3-phosphate dehydrogenase (GAPDH), fructose-1,6-bisphosphatase (FBPase), and ribulose-5-phosphate kinase (RPK) (Acosta-Motos et al. 2017). In C4 plants, phosphoenolpyruvate carboxylase (PEPCase) is more sensitive to salt than Rubisco (Chiconato et al. 2021). The partial or complete closure of stomata also aggravates the effects of salinity stress that limits CO2 availability (Franzisky et al. 2021). The storage of carbohydrates is also hampered by salt stress as evident from the microscopic observations in Ulva prolifera. Under salt stress, starch became smaller and lighter, albeit regained its proper structure during recovery (Huan et al. 2014).
Reduction in copy number of chloroplast DNA
Chloroplast has organellar DNA due to its unique endosymbiotic origin (Dyall et al. 2004). The plant chloroplast genomes or plastomes usually range between 120–200 kbp in size and their copy numbers can vary anywhere from 20 to several hundred depending on the species, developmental stage, and environmental conditions. Salinity stress resulted in a reduction of the average copy number of the chloroplastic genome in Arabidopsis by about 40% (Peharec Štefanić et al. 2013). Intriguingly, chloroplast DNA replication is found to be blocked by inhibition of the photosynthetic electron transport chain (ETC). This suggests the possibility that plastid DNA replication is coupled with photosynthetically supplied reducing power and is redox regulated (Kabeya and Miyagishima 2013). Thus, inhibition of photosynthetic ETC by Na+ ion toxicity in chloroplasts might be a reason for reduced plastome replication. Nevertheless, a more direct effect of salinity stress on molecular factors involved in the rate of replication of the plastid DNA may also play an important role in the reduction (Peharec Štefanić et al. 2013). Indeed, previous studies have shown that the bacterial-like DNA polymerase Pol1A which plays a major role along with DNA Pol1B in the replication of the plastid genome in plants is strongly repressed by osmotic stresses (Morley and Nielsen 2016). However, the exact molecular mechanisms involved in plastome copy number reduction due to stress remains elusive.
Alterations in the transcription of chloroplast-encoded genes
Chloroplast is a semiautonomous organelle, whose genome encodes about 100–250 genes. Proteome studies showed that chloroplasts contain up to 3,000 proteins. Salinity has been shown to drastically alter organellar gene expression (Robles and Quesada 2019). Reduction in the plastome copy number leads to the concomitant decrease in transcript levels of chloroplast-encoded genes involved in photosynthesis. In Arabidopsis, the transcript levels in salt-stressed seedlings were reduced by 50% as compared to the control (Peharec Štefanić et al. 2013). DEAD-box RNA helicases are RNA-binding proteins involved in the formation of functionally processed RNAs in the nucleus, chloroplasts, and mitochondria. In Arabidopsis, ten members of this family are confirmed to be localised in the chloroplasts. In silico analysis of the chloroplast-targeted proteins showed that eight members from rice, seven from maize, and two from wheat are downregulated under salinity stress (Nawaz and Kang 2017).
In plants, mTERFs (mitochondrial transcription termination factors) are a family of nucleus-encoded proteins that are involved in the transcriptional regulation of chloroplastic and mitochondrial genes (Robles and Quesada 2019). Functional genomic approaches in Arabidopsis showed that mTERF6, mTERF10, and mTERF11 function as positive regulators of salinity tolerance. Yet, a few other members, namely, mTERF5 and mTERF9, negatively regulate salinity tolerance as evidenced by enhanced salt tolerance of the mter5 and mter9 mutants (Robles et al. 2018). The expression of chloroplast-encoded photosynthetic genes in Arabidopsis, namely, ATPF (ATP-synthase subunit b, F type), NDHA (NADH dehydrogenase subunit 1), PETB (photosynthetic electron transfer B), and TRNK (tRNA for lysine) were found to be downregulated under salinity stress. In addition, the transcript levels of nuclear-encoded chloroplast-targeted genes, namely, LHCA4 (light-harvesting chlorophyll–protein complex I subunit A4), RBCS1A (ribulose bisphosphate carboxylase small chain 1A), and RCA (Rubisco activase) were also downregulated in salt-stressed sensitive plants (Peharec Štefanić et al. 2013). Recent advances in techniques such as cell fractionation together with the application of advanced next-generation sequencing techniques such as single-cell RNA-Seq, Chloro-Seq, global run-on (GRO)-Seq, global ribosomal profiling (Ribo-Seq) will aid in further understanding of the intricacies involved in retrograde signalling and regulation of transcription in response to salt stress (Leister et al. 2017).
Defects in chloroplast protein biosynthesis
Several studies indicate that salt stress, specifically Na+ ion toxicity, can adversely affect protein synthesis and translational machinery including the functioning of ribosomes (Omidbakhshfard et al. 2012). The levels of chloroplast-encoded tRNA and rRNA were significantly reduced by salinity, indicating defects in the protein synthetic machinery (Peharec Štefanić et al. 2013). Salt stress was found to inhibit the transcription and translation of psbA genes that encode the D1 protein in Synechocystis sp. Northern and Western blotting analysis showed time-dependent reduction in the RNA and corresponding D1 protein. Experiments using [35S] Met labelling also confirmed the reduced translation of various chloroplastic proteins under salt stress (Allakhverdiev et al. 2002).
The Ef-Tu family proteins (eukaryotic translation elongation factor thermo unstable), namely, tufA from pea was found to be involved in protein synthesis in chloroplasts. Salinity stress reduced its transcript abundance which in turn caused alterations in the overall chloroplast protein synthesis (Singh et al. 2004). PSRPs (plastid-specific ribosomal proteins) are a class of proteins involved in protein synthesis, among which PSRP2 (a component of the 30S subunit) of Arabidopsis is chloroplast targeted. PSRP2 overexpression lines displayed sensitivity and mutant lines showed tolerance to salinity. The protein was shown to have RNA chaperone activity and is a negative regulator of stress tolerance and germination (Xu et al. 2013). Salinity-induced attenuation of PSII is associated with the degradation of several important PSII reaction center proteins, mainly the D1 protein (psbA) along with the D2 protein (psbD), and the internal antenna protein CP43 (psbC) which are also essential for its repair (Nagashima et al. 2004).
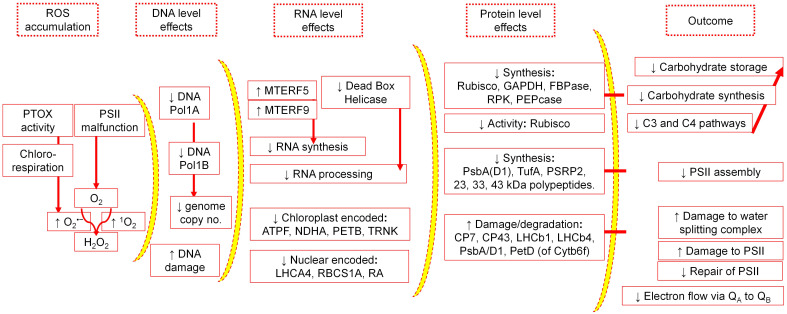
The major components of chloroplast sensitivity to salinity in salt-susceptible plant species are depicted in Fig. 2.
Fig. 2. Major levels of sensitivity to salinity in chloroplasts of salt-sensitive species. ROS – reactive oxygen species; PTOX – plastid terminal oxidase; O2·– – superoxide radical; 1O2 – singlet oxygen; DNA Pol1A – DNA polymerase 1A; DNA Pol1B – DNA polymerase 1B; MTERF – mitochondrial transcription termination factor; ATPF – ATP-synthase subunit b, F type; NDHA – NAD(P)H dehydrogenase subunit 1; PETB – photosynthetic electron transfer B; TRNK – tRNA for lysine; LHCA4 – light-harvesting chlorophyll–protein complex I subunit A4; RBCS1A – ribulose bisphosphate carboxylase small chain 1A; RA – Rubisco activase; GAPDH – glyceraldehyde-3-phosphate dehydrogenase; FBPase – fructose-1,6-bisphosphatase; RPK – ribulose-5-phosphate kinase; PEPCase – phosphoenolpyruvate carboxylase; psbA – photosystem II reaction center protein A; TufA – eukaryotic translation elongation factor thermo unstable A; PSRPs – plastid-specific ribosomal proteins; CP – chlorophyll-binding protein; LhcB – light-harvesting complex protein subunit B; PETD – photosynthetic electron transport subunit D; PSII – photosystem II. Upward and downward arrows indicate upregulation and downregulation, respectively.
How do salt-tolerant plant species sustain photosynthesis under salinity?
Salt-tolerant plant genotypes maintain high photosynthetic rates under salt stress primarily by stabilizing internal CO2 concentration and maintaining the stability of photosynthetic systems (Sui and Han 2014). The adaptive mechanisms at the chloroplast level to cope with salt stress need to be efficiently exploited in the development of salt-tolerant crops. At the cellular level, studies on photosynthetic response under salt stress have focused on retrograde signalling and associated stress responses, chloroplast-specific ion channels and transporters, chloroplasts membrane remodelling, and maintenance of photosynthetic membrane proteins along with CO2-fixing enzymes (Table 1). The succeeding sections deal with all these insights and the differential ability of halophytes to overcome the stomatal and nonstomatal limitations of salt stress.
Maintenance of chloroplast membrane fluidity and stability
Salt-tolerant species like barley showed no lipid reduction under salt stress, along with reduced lipolysis and peroxidation of membrane lipids which helped them maintain normal cellular expansion and growth (Chalbi et al. 2013). Salt stress increased the content of phosphatidyl glycerol (PG) in the photosynthetic membranes of salt-tolerant species such as buffalo grass (Lin and Wu 1996) and many halophytes (Sui and Han 2014, Barkla et al. 2018). These reports are consistent with its critical role in oxygenic photosynthesis and the functioning of PSII and LHCII (Guo et al. 2019b).
Salt-induced increase in glycolipids, especially galactolipids (Wang et al. 2014), along with alteration and incorporation of unsaturated fatty acids into these lipids (Sui and Han 2014) is seen in the photosynthetic membranes of salt-tolerant species. Moreover, a decrease in DGDG to MGDG ratio is common under salt stress, but this decrease is lesser in salt-tolerant species leading to more stable chloroplast membranes (Sui and Han 2014). High contents of SQDG in the chloroplast membranes are often regarded as another unique attribute of salt tolerance as evident from its elevated levels in many tolerant plant species (Ramani et al. 2004, Hamed et al. 2005). Increased levels of unsaturation in the fatty acids of membrane lipids are speculated to be the reason for enhanced salt tolerance in the cyanobacterium Synchococcus sp., which protected the photosynthetic machinery and also activated Salt Overly Sensitive 1 (SOS1) for sodium extrusion from the cell. An increase in plastoglobule size and number has been observed in salt stress (Barkla et al. 2018), and this has been hypothesized to be the outcome of the mobilization of galactolipids in the thylakoid membrane and accumulation of triacylglycerols.
Chloroplast membrane transporters as saviours
Salt stress leads to an increase in Na+ and Cl– concentration and a decrease in K+ concentration inside the chloroplast (Robinson and Downton 1984). In Arabidopsis, K+/H+ exchanger (KEA3) and tandem pore K+ channel (TPK3) are involved in the influx and efflux of K+ into and out of the thylakoid lumen, respectively, along with KEA1 and KEA2 located at the inner chloroplast envelope (Carraretto et al. 2013, Kunz et al. 2014, Finazzi et al. 2015). In salt-tolerant halophytic species, Na+ is preferentially accumulated in the chloroplast (Müller et al. 2014), where it serves other essential functions, too, and this is mediated by Na+-dependent transporters such as bile acid:sodium symporter 2 (BASS2) and phosphate transporters (PHT4;1, PHT4;4, and PHT4;5) (Bose et al. 2017). Nevertheless, excess Na+ beyond a threshold can be detrimental to halophytes as well, and hence these salt-loving species have been speculated to possess transporters that prevent excessive Na+ accumulation in the chloroplasts. In Arabidopsis, CHX23 and NHD1 encode putative Na+/H+ antiporters in the chloroplast envelope, which regulate the entry of Na+ into the chloroplast, and the mutants of which showed significant salt hypersensitivity and impaired photosynthetic performance (Müller et al. 2014). Numerous research groups have also observed preferential K+ and Cl– accumulation in the chloroplasts of halophytes, but the corresponding transporters still need to be unveiled. Recently, a chloroplast membrane-located transporter OsNHAD encoding a putative Na+/H+ antiporter has been identified and correlated to enhanced salt tolerance in rice (Liu et al. 2020).
Salt tolerance and changes in photosynthetic membrane proteins
Several salt-responsive proteins involved in chloroplast membrane organization and associated with photosystem activities have been reported in Arabidopsis, rice, wheat, and tomato (Suo et al. 2017). The Arabidopsis fatty acid desaturase 6 (fad6) mutant displayed salt hypersensitivity, indicating its role in salt tolerance, especially during the early seedling stage (Zhang et al. 2009). An increase in the unsaturation levels of fatty acids in the lipids of the thylakoid membrane helps in accelerated repair and de novo synthesis of D1 protein in PSII (Loll et al. 2007). Another important protein that determines the organisation of the thylakoid membrane is the chloroplast outer envelope membrane-localized MGD. OsMGD gene, overexpressed in tobacco, resulted in increased contents of MGDG and DGDG and the transgenic lines displayed well-developed thylakoid membranes with significant grana stacking (Wang et al. 2014).
Some chloroplast-specific membrane proteins are essential for the normal functioning of PSII. Rubredoxin (Rrd), a non-heme thylakoid membrane protein, when knocked out, showed reduced activity and stability of PSII in green algae, cyanobacteria, and Arabidopsis (Calderon et al. 2013). This is indicative of its role as an electron carrier-mediated ROS scavenger (Li et al. 2016). Similarly, rare cold-inducible (RCI) protein is another candidate chloroplast membrane protein involved in the maintenance of PSII stability and activity under salt stress (Khurana et al. 2015). Two proteins closely associated with PSI and PSII, LHCb6 and 10-kDa PSII polypeptide, were upregulated in the salt-tolerant genotypes of Indian mustard (CS54, variety developed by ICAR-CSSRI), indicating its higher efficiency in photoprotection under salinity (Singh et al. 2019). The abundance of 33-kDa oxygen-evolving protein, 23-kDa protein of OEC of photosystem II and Chl a/b-binding protein also increased under salt stress in salt-tolerant species of Brassica napus (Jia et al. 2015).
Photosystem II stability/assembly factor (HCF136), a hydrophilic protein in the lumen of stroma thylakoids, showed higher abundance in the salt-tolerant wheat genotype, contributing to photosynthetic stability (Xu et al. 2016). Expression of proteins, such as PsaA, PsaB, atpl, and LHC proteins, was found to be elevated initially and then downregulated (Jayakannan et al. 2015) showing that the crop tolerance to salinity stress can enhance the rate of photosynthesis in the initial phases of stress by maximizing light capture. Moreover, PsbP, PsbQ family protein, Cyt b6f complex iron-sulfur subunit, and cytochrome b559-∞ subunit also showed increased abundance in the salt-tolerant wheat isogenic line, indicating better PSII regulation, repair, and PSI assembly under salt stress (Zhu et al. 2020, 2021).
Oxygen-evolving enhancer proteins 1&3 along with plastocyanin-docking protein and PSI subunit VII were also differentially upregulated in tolerant genotypes of Brassica, cotton, and chickpea, indicating better stability of OEC and PSI under salt stress (Yousuf et al. 2016, Gong et al. 2017, Arefian et al. 2019). PSI reaction center subunits such as PsaK and PsaIV have also been reported to increase under salt stress, which helps mediate the binding of antenna complexes to the PSI reaction center (Zhu et al. 2021). A preferential abundance of PSI-P700 chlorophyll a apoprotein A2 (PSI-P700), the primary electron donor of PSI was observed in the salt-tolerant cotton genotype after being exposed to high salt stress for 4 h, indicating higher electron transfer efficiency (Gong et al. 2017). Similarly, an increased abundance of ferredoxin and NAD(P)H-dehydrogenase complex was reported in salt-tolerant wheat (Zhu et al. 2021). ATP synthase alpha and beta subunits got differentially upregulated under saline conditions in the tolerant genotype of common bean and chickpea. This highlights the possible role of these proteins in energy synthesis, as molecular chaperons, and a possible indirect role in the translocation of excess Na+ and Cl– into the vacuole (Arefian et al. 2019).
CEST (chloroplast protein enhancing stress tolerance) is a novel thylakoid membrane-localized protein involved in chloroplast development, suppression of photooxidative damage, growth, and salt tolerance (Yokotani et al. 2011). Transglutaminase, a previously known protein for post-translational modification of other proteins, was found to enhance polyamine signalling and thylakoid stability in tobacco under salt stress (Zhong et al. 2019). Salt stress also increased the abundance of chloroplast-localized small heat shock proteins (sHSPs), sHSP70 in wheat (Zhu et al. 2021), which assists in the import of pre-proteins from the nucleus into the chloroplast and their maturation (Latijnhouwers et al. 2010). Wheat chloroplasts have a very active protein quality control machinery under salt stress via the activation of proteins namely carboxyl-terminal-processing peptidase 3 (CTPA3) and chaperone protein 2 (ClpC2). They are involved respectively in the assembly of PSII proteins and the degradation of damaged proteins (Zhu et al. 2021).
Improved stromal protein levels and activity
Reports on proteomic analysis of chloroplast in response to salinity are limited. Studies on leaf proteome in canola suggested that chloroplast proteins are key indicators of salinity (Iqbal et al. 2019). The abundance of magnesium chelatase and glutamate-1-semialdehyde aminotransferase, key enzymes in chlorophyll biosynthesis increased after exposure to salt stress in chickpea and Brassica napus seedlings, respectively (Jia et al. 2015, Arefian et al. 2019). The large subunit of Rubisco, coded by a single gene in the chloroplast genome is more sensitive to salt stress, but tolerant plant species have been able to maintain its abundance under stress conditions (Arefian et al. 2019, Zhu et al. 2021). Rubisco activase has been reported to be upregulated under salt stress, indicating its vital role in maintaining the active state of Rubisco (Jia et al. 2015). Overexpression of the GAPDH beta subunit protein in the chloroplast stroma prevented the attenuation of ROS-mediated PSII repair and maintained the efficiency of the photosynthetic apparatus in Arabidopsis under salinity (Chang et al. 2015). Concomitantly, fructose-1,6-bisphosphate aldolase activity was differentially upregulated in the tolerant genotypes of upland cotton after 24 h of imposed salt stress (Gong et al. 2017). In addition, a differential abundance of other CO2-fixation enzymes, such as carbonic anhydrase, phosphoribulokinase, phosphoglycerate kinase, sedoheptulose-1,7-bisphosphatase, fructose-bisphosphate aldolase, and ketolase, has been reported in numerous salt-tolerant species or cultivars (Xu et al. 2016, Arefian et al. 2019, Zhu et al. 2021). The expression of NADP-malate dehydrogenase (NADP-ME) is upregulated under salt stress in salt-tolerant cultivars (Chen et al. 2019a). The abundance of amino acids biosynthetic enzymes such as alanine aminotransferase and aspartate aminotransferase also increased under salt stress in the tolerant genotypes of sesame (Zhang et al. 2019).
NPR1 (non-expressor of pathogenesis-related genes 1) protein has been shown to accumulate in the chloroplast stroma with possible roles in redox homeostasis and amelioration of salt-mediated downregulation of photosynthetic capability (Seo et al. 2020). Chloroplast-localized DAD1, phospholipase A, has been speculated to be responsible for the release of linolenic acid from membrane lipids, the precursory step in jasmonic acid (JA) biosynthesis (Ishiguro et al. 2001). In superior salt-tolerant lines of chickpea, tolerance was accredited to the salt-mediated increase of JA biosynthesis (Xu et al. 2016). Cold-regulated (COR) protein was upregulated in Arabidopsis under salt, a potential member in the ABA-dependent salt stress signalling network (Liu et al. 2014). Maturase K, common to all land plant chloroplasts gets activated under salt stress via dephosphorylation (Zörb et al. 2010) and plays a crucial role in primary mRNA processing/splicing (Xu et al. 2016). A multiorganelle-located, conserved protein OsNBL1 has been proposed to interact with plastidic caseinolytic protease OsClpP6 and is essential for enhanced salt tolerance. Nbl1 mutants showed upregulation of several salt-inducible genes, such as HAK1 and HAK5, and were highly salt tolerant. Several chloroplast ribosomal proteins, such as RPL5, RPL10, RPL14, RPL21, and RPL29, got upregulated under salt stress in wheat, which hastened the synthesis of damaged photosynthetic proteins (Zhu et al. 2021).
Post-translational modifications of chloroplast proteins
Post-translational modifications are key aspects of altered protein function, activity, targeting, turnover, and interactions in response to various environmental stresses (Grabsztunowicz et al. 2017). Phosphorylation, glycosylation, carbonylation, nitrosylation, redox modifications, ubiquitination, and SUMOylation are the commonly reported post-translational modifications of chloroplast proteins in response to salt stress (Chang et al. 2012, Liu et al. 2012). Salt stress has been reported to initiate phosphorylation cascades in rice proteins, and a total of 13 phosphorylation sites across eight proteins have been identified, which includes the PSII reaction center protein H and RBCSs (Chang et al. 2012). Phospoproteome studies in maize under salt stress identified several phosphoregulated proteins involved in photosynthesis (Zörb et al. 2010). Phosphorylation of PSII proteins controls the functional folding of photosynthetic membranes in Arabidopsis and helps sustain photosynthetic activity under salinity stress (Fristedt et al. 2009). Moreover, Rubisco activase, RBCLs, chloroplast and mitochondrial ATP-synthase F1 were carbonylated and S-nitrosylated upon exposure to salt stress in Citrus aurantium seedlings (Tanou et al. 2009). In rice, pyruvate phosphate dikinase 1 is ubiquitinated upon salt stress treatment (Liu et al. 2012). Further, the enhanced photosynthetic efficiency of the salt-tolerant wheat introgression line over its parent was attributed to the putative difference in post-translational modification of Rubisco and phosphoribulokinase (Xu et al. 2016). Significant post-translational modifications have also been reported in the chloroplast ROS-scavenging protein, 2-Cys peroxiredoxin, which is strongly correlated with improved stability of the photosynthetic system in salt-tolerant lines (Dietz 2016).
Photorespiration as a protective mechanism under salt stress
Stomatal limitation leads to a deficit in intracellular CO2, which causes the over-reduction of ETC. Rubisco operates as an oxygenase and photorespiration is initiated under saline conditions to channel out the excess light energy (Suo et al. 2017). The osmotic phase of salt stress has been reported to upregulate the expression and activities of photorespiratory genes and enzymes (Bai et al. 2017). Transformed rice with chloroplastic glutamine synthetase showed enhanced salt tolerance by activation of the photorespiratory pathway (Hoshida et al. 2000). In addition, overexpression of the Arabidopsis and bacterial photorespiratory pathway genes, such as serine:glyoxylate aminotransferase (SGAT) and serine hydroxymethyltransferase (HMT), respectively, enhanced the salt tolerance (Waditee-Sirisattha et al. 2017). Sugar beet, a highly salt-tolerant crop species showed increased contents of glycolate and serine along with enhanced expression of glycolate oxidase under salt stress (Lv et al. 2019).
Compatible solutes and osmotolerance
Cellular accumulation of compatible solutes, organic compounds that do not interfere with cellular enzymes and functions, is an important strategy for the protection and survival of plants under salinity. Commonly accumulated compatible solutes include glycine betaine (GB), amino acids such as proline, polyols/sugar alcohols, and quaternary amines (Munns et al. 2020). These solutes play an important role in osmotic adjustment and maintenance of ionic balance by minimizing the entry of salt into plants or by regulating the concentration of salts in the cytoplasm (Shabala et al. 2020). They also act as signalling molecules to increase ABA accumulation, affect gene expression networks, and regulate plant growth under salt stress (Marusig and Tombesi 2020).
Proline serves as one of the major osmolytes that stabilize membranes and protein and also helps in cellular redox homeostasis, ROS scavenging, and maintenance of cytosolic NADP+/NADPH ratio (Reddy et al. 2015). In response to salt stress, chloroplast starch reserve is rapidly catabolized to sucrose and there is a promotion of sugar synthesis and concomitant inhibition of starch synthesis (Kumutha et al. 2008). This build-up of sugars (glucose, sucrose, fructose, and fructans) plays a crucial role in the adaptation to salinity and osmotic stresses (Abideen et al. 2021). Trehalose, a nonreducing disaccharide, is another osmolyte; overexpression of Escherichia coli trehalose biosynthetic genes (otsA and otsB) in rice increased trehalose biosynthesis and enhanced salt tolerance. These transgenic plants were able to maintain a higher K+ ion content over Na+ ion and thus have a lower K+/Na+ ratio both in shoots and roots which helped in ion homeostasis under salt stress. These plants also showed better photosynthetic rates than the wild type (Garg et al. 2002).
Increased contents of methylated inositol and myo-inositol by chloroplast-targeted overexpression of PcINO and McIMT1 improved the growth and photosynthesis of transgenic tobacco (Patra et al. 2010). The quaternary ammonium compounds, such as glycine betaine (GB), choline-O-sulphate, proline betaine, β-alanine betaine, hydroxyproline betaine, dimethyl sulphoniopropionate, and pipecolate betaine, are another group of efficient osmolytes (Ahmad et al. 2013). Amongst these betaine compounds, GB is extensively distributed over different plant species and is accumulated in response to a wide range of abiotic stresses. Apart from mediating osmotic adjustment, GB provides support in Na+/K+ discrimination, antioxidant defense, and protection of membranes, ultimately leading to photosynthetic viability. The exogenous application of GB is also known to protect photosynthetic machinery and improve salt tolerance (Ahmad et al. 2018). Biosynthesis of GB involves the oxidation of choline by choline monooxygenase and betaine-aldehyde dehydrogenase successively in the chloroplastic stroma (Sakamoto and Murata 2002). Overexpression of E. coli betA (choline dehydrogenase) in wheat improved the membrane stability and reduced ion leakage and sodium accumulation (He et al. 2010). Tomato plants overexpressing codA (choline oxidase) gene showed improvement in the rate of photosynthesis, antioxidant enzyme activities, decreased K+ efflux, and increased Na+ efflux in comparison to wild-type plants (Wei et al. 2017).
ROS-scavenging machinery in chloroplasts
Under unfavourable conditions, plants employ metabolic and morphological adjustments to prevent oxidative damage to photosystems. The stress-induced increase in the activity of the enzymes, such as superoxide dismutase (SOD), ascorbate peroxidase (APX), catalase (CAT), peroxidases, and their isoforms, maintain the functional balance between the quenching of ROS and maintenance of H2O2 concentration required for cell signalling (Noctor and Foyer 2016). To resist the oxidative stress caused by excess salts, numerous proteins associated with antioxidant defence were upregulated in wheat chloroplasts that help scavenge ROS and regulate protein turnover under salt stress (Zhu et al. 2021). The H2O2 produced by plastidic Cu/Zn-SOD is scavenged by membrane-bound thylakoid ascorbate peroxidase (tAPX) in a reaction called as water–water cycle (Edreva 2005). Overexpression of plastidic Cu/Zn-SOD improved ROS homeostasis and salinity tolerance in several plant species including Arabidopsis, tobacco, Chinese cabbage, and cotton (Suo et al. 2017). Overexpression of chloroplastic APX imparted salinity tolerance in tobacco (Badawi et al. 2004). Chloroplast targeting of CAT was found to improve salinity tolerance in cotton and Chinese cabbage (Tseng et al. 2007, Luo et al. 2013).
The stromal ascorbate–glutathione (AsA–GSH) cycle reduces H2O2 to H2O in a stromal APX (sAPX) mediated reaction. sAPX utilizes AsA as the electron donor, and AsA is recycled by monodehydroascorbate reductase (MDHAR) or is spontaneously transformed into dehydroascorbate (DHA). DHA is then reduced to AsA by DHAR using reduced glutathione (GSH) as an electron donor. The oxidized glutathione (GSSG) is reduced by glutathione reductase (GR) where NADPH is the electron donor. Expression of sAPX, MDHAR, DHAR, and GR is responsive to salinity. Overexpression of MDHAR and DHAR increased the survival of tobacco plants under salt stress (Suo et al. 2017). The thioredoxin/peroxiredoxin (Trx/Prx) pathway and glutathione peroxidase (GPX) regulates H2O2 contents in chloroplasts. Trx-dependent peroxidase (Prx) scavenges H2O2, and the thioredoxin reductase (TrxR) reduces Trx/Prx system using NADPH as the electron donor. GPX uses GSH as an electron donor and reduces H2O2 using GSH as the electron donor. Transgenic Arabidopsis overexpressing wheat chloroplast GPXs showed increased tolerance to salt and H2O2 (Zhai et al. 2013).
The ROS, ·OH, and 1O2 are scavenged by nonenzymatic antioxidants, such as AsA, GSH, and tocopherol in chloroplasts. Tocopherol is a lipid antioxidant localized in the thylakoid membrane (Suo et al. 2017). Tocopherol biosynthesis is a finely balanced process in chloroplasts. ABC1 (activity of bc1 complex)-like kinases (ABC1K3) and AtSIA1 are associated with salinity tolerance in Arabidopsis (Martinis et al. 2013). Overexpression of γ-tocopherol methyltransferase (γ-TMT) reduced ROS abundance and membrane injury leading to salt stress alleviation in tobacco (Jin and Daniell 2014). Apart from the conventional antioxidant molecules, other regulatory mechanisms have also been documented concerning chloroplast ROS-scavenging under salt stress. Overexpression of chloroplast encoded nucleoside diphosphate kinase 2 (NDPK2) gene from Arabidopsis into sweet potato led to the induction of an array of compounds having antioxidant activity (Kim et al. 2009). The Arabidopsis AtTSPO involved in the transport of photoreactive tetrapyrrole intermediates protected chloroplasts from ROS accumulation in response to 150 mM NaCl (Balsemão-Pires et al. 2011). Methionine sulfoxide reductase reduces the methionine sulfoxide back to methionine during episodes of salinity stress. Overexpression of chloroplast-localized OsMSRA4.1 improved plant survival under salinity (Guo et al. 2009).
Transcriptional regulation of chloroplast genome
Several transcriptomic studies have been undertaken to identify key genes which respond to salinity stress at various stages. The comparative analysis of the transcriptome of a salt-tolerant wheat cultivar (Arg) in comparison to a susceptible variety (Moghan3) revealed several genes involved in photosynthesis such as plastidial chlorophyll-binding proteins (Amirbakhtiar et al. 2021). In salt-tolerant rice, most of the upregulated genes were associated with photosynthetic electron transport (Razzaque et al. 2019). Observations by Fan and coworkers in the response of Salicornia europaea to salinity stress showed the gene expression of several plastidial photosystem-associated proteins, such as PSI and PSII-binding proteins, Cyt b6f complex proteins, and the ATP synthase CF1 subunit (Fan et al. 2013).
In salinity-tolerant sweet sorghum cultivars, PEPC and NADP-ME were upregulated to impart tolerance to the photosynthetic system (Yang et al. 2020). The most significant pathway upregulated in salinity-stress response was the photosynthetic light reaction pathway in a salinity-tolerant chickpea cultivar JG-62 relative to susceptible ICCV-2 (Garg et al. 2016). The tolerant barley genotype (Boulifa) showed differential upregulation of PSI assembly proteins, PSII reaction center proteins, D2, 10-kDa polypeptide, PSI P700 chlorophyll apoproteins, and iron-sulphur center proteins (Ouertani et al. 2021). In a comparative study between the salinity-tolerant peach cultivar GF677 and the susceptible Maotao, the integrity of chloroplast structure and upregulation of photosynthetic genes were identified as critical to salinity tolerance (Sun et al. 2020). In grapes, transcriptome analysis of the moderately salinity-tolerant ‘Thompson seedless’ cultivar showed the overexpression of several chloroplastic genes involved in maintaining photosynthesis, such as APX, Chl b reductase, and Mg-binding proteins among others (Das and Majumder 2019). Ulva compressa is a green alga adapted to a moderately saline environment such as brackish waters. A transcriptomic study of Ulva showed that several plastidial genes associated with photosystem maintenance and photosynthetic acclimation, such as high chlorophyll fluorescence 244 (HCF244), maintenance of photosystem II under high light 2 (MPH2), hypothetical chloroplast reading frame 4 (YCF4), hypersensitive to high light 1 (HHL1), and vesicle-inducing protein in plastids (VIPP) were constantly upregulated in both short- (1, 6 h) and long-term (24, 72 h) salinity treatment. This affirms the critical role of genes associated with maintenance of the photosynthetic assembly in salinity tolerance mechanisms (Xing et al. 2021).
Metabolic reshuffle to optimize photosynthesis under salt stress
Global metabolic changes are a reflection of cellular protein activities and physiological alterations under abiotic stresses in plants. Increased accumulation of organic acids was observed in salt-tolerant barley genotypes (Wang et al. 2019). Similarly, salt-tolerant wild Tibetan barley cultivars (XZ16 and XZ169) accumulated more metabolites related to photosynthesis than their cultivated counterparts (CM72 and Gairdner) (Wu et al. 2013). Decreased glutamic acid was observed in salt-stressed S. persica, which indicates that more of it was channelled for the synthesis of Chl a and Chl b (Kumari and Parida 2018).
On exposure to salinity, plants showed reduced contents of glutamine, D-alanyl-D-alanine, cystine tyrosine, L-alanyl-L-glutamate, D-alanyl-D-serine, and elevated contents of some amino acids, such as phenylalanine, glutamate, methionine, dihydroxyisoleucine, ornithine, L-methionine, 3-methoxytyramine, tetra-homomethionine, 1-(3-aminopropyl)-4-aminobutanal, and L-homomethionine. Moreover, contents of branched-chain amino acids, which act as stress modulators, got increased under salt-stress conditions (Benjamin et al. 2019). Several sugar and sugar alcohols, namely, arabinose, inositol, sorbitol, and mannitol accumulated in the chloroplasts and extra chloroplastic space of salt-stressed sugar beet plants. It reflects that safeguarding the internal osmotic environment of chloroplast is key to survival under salt stress (Lv et al. 2019). Polyamines, such as spermine, putrescine, spermidine, tricaffeoyl spermidine, and coumaroyl spermidine also increased under salt stress in salt-tolerant crops/genotypes (Kumari and Parida 2018). Putrescine interacts with negatively charged thylakoid membranes, increases the lipid accumulation in chloroplasts, and thereby minimises thylakoid membrane degradation (Shu et al. 2012). Secondary metabolites such as terpenes and flavonoids were found to accumulate under salinity stress, which possibly protects the photosynthetic machinery from oxidative stress (Chen et al. 2019b, Qin et al. 2022).
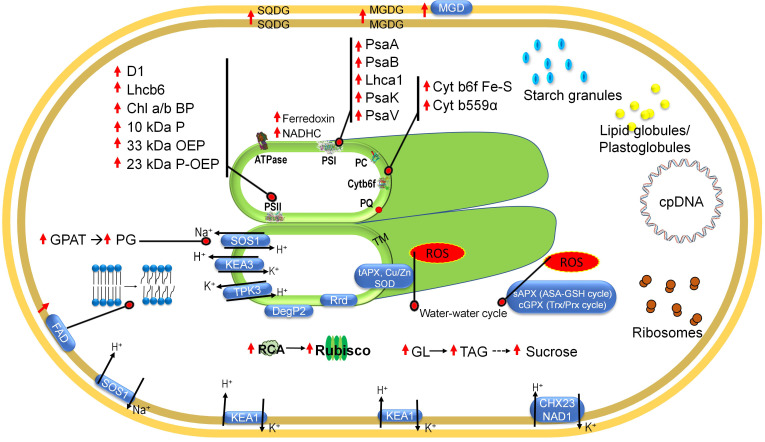
A cumulative model highlighting the major attributes of salt tolerance at the chloroplast level is shown in Fig. 3.
Fig. 3. The major components of tolerance to salinity stress at chloroplast level. MGD – monogalactosyldiacylglycerol synthase; MGDG – monogalactosyldiacylgycerol; SQDG – sulfoquionovosyldiacylglycerol; cpDNA – chloroplast DNA; TM – thylakoid membrane; GPAT – glycerol 3-phosphate acyl transferase; PG – phosphatidylglycerol; ROS – reactive oxygen species; GL – galactolipids; TAG – triacylglycerol; RCA – Rubisco activase; FAD – fatty acid desaturase; NHAD – putative Na+/H+ antiporter; KEA1 & 2 – K+/H+ exchanger; CHX23 & NAD1 – Na+/H+ antiporters; SOS1 – salt overly sensitive 1; TPK – tandem pore K+ channel; DegP2 – trypsin type serine protease; Rrd – rubredoxin; SOD – superoxide dismutase; tAPX – thylakoid membrane-localized ascorbate peroxidase; sAPX – stromal ascorbate peroxidase; AsA–GSH; ascorbate–glutathion; Trx/Prx – thioredoxin/peroxiredoxin; NADHC – NADH dehydrogenase complex; Lhc – light-harvesting complex; Chl a/b BP – chlorophyll a/b-binding protein; OEP – oxygen-evolving protein; Cyt – cytochrome; PSI – photosystem I; PSII – photosystem II; PC – plastocyanin; PQ – plastoquinone. Upward arrows and dotted lines represent upregulation and hypothetical proteins/pathways, respectively.
Salt-mediated retrograde signals from the chloroplast
Chloroplast is probably the most sensitive organelle to salt stress. At the same time, it is also one of the key cellular sensors of climatic fluctuations. Reduction in photosynthetic efficiency and energy supply during salt stress forces the plant to divert energy from growth to stress response. Efficient coordination between organelles and nuclear transcriptional machinery becomes inevitable. This communication, namely retrograde signalling, is highly imperative under stress conditions (Crawford et al. 2018).
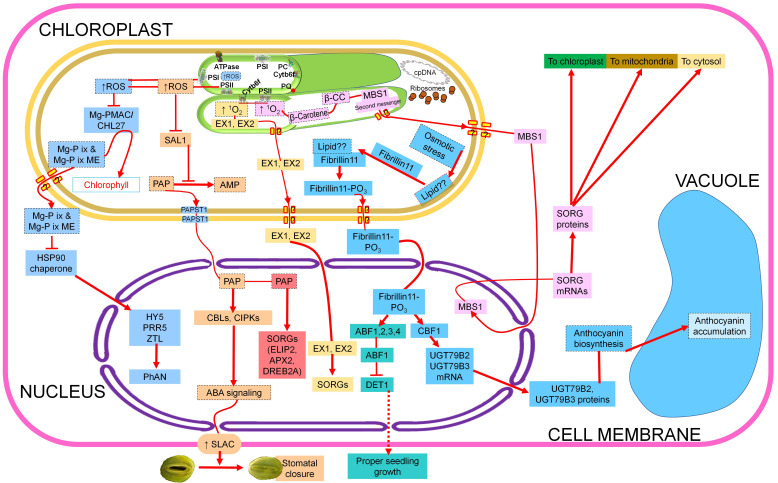
PSII is the main site of 1O2 generation in the chloroplast, which further triggers two different downstream retro-signalling cascades (Cruz de Carvalho 2014). β-carotene quenches singlet oxygen produced in PSII and nonenzymatically gets cleaved in to β-cyclocitral. This secondary messenger interacts with methylene blue sensitivity 1 protein (Shao et al. 2013) and increases the expression of a set of nuclear encoded singlet oxygen-responsive genes (SORGs) (Ramel et al. 2012). This retro-signalling pathway is initiated from the PSII reaction centers of the grana core (Wang et al. 2016). The second pathway operates from the grana margins, with the help of two nuclear-coded, chloroplast-targeted proteins, Executer 1 and 2. They transfer the 1O2-mediated signals to the nucleus and affect the activation of SORGs (Wang et al. 2016). In the nucleus DREB2A, CYT P450, GST 6, WRKY, bHLH, HSPs, and MAP kinase are known to be the common target genes for these two signalling pathways (Fig. 4) (Dogra et al. 2017).
Fig. 4. Major components of salinity-mediated retrograde signalling from the chloroplast. PSI – photosystem I; PSII – photosystem II; PC – plastocyanin; 1O2 – singlet oxygen; β-CC – β-cyclocitral; MBS1 – methylene blue sensitivity 1; SORGs – singlet oxygen-responsive genes; EX1/EX2 – EXECUTER 1/EXECUTER 2; ROS – reactive oxygen species; MgPMAC/CHL27 – Mg-protoporphyrin monomethylester aerobic cyclase; MgPIX/ME – Mg protoporphyrin IX and its monomethyl ester derivative; HY5 – LONG HYPOCOTYL 5; PRR5 – PSEUDO RESPONSE REGULATOR 5; ZTL – ZIETLUPE; PhANGs – photosynthesis-associated nuclear genes; SAL1 – inositol polyphosphate-1-phosphatase; PAP – 3'-phosphoadenosine-5'-phosphate; AMP – adenosine monophosphate; PAPST1 – bidirectional PAP transporter; ELIP2 – EARLY LIGHT-INDUCED PROTEIN 2; APX2 – ascorbate peroxidase 2; DREB2A – DEHYDRATION-RESPONSIVE ELEMENT BINDING 2A; CBLs – calcineurin B-like protein; CIPKs – CBL-interacting protein kinase; ABA – abscisic acid; SLAC – slow anion channel associated 1; Lipid?? – unknown lipid molecule; Fibrillin11 PO3 – phosphorylated fibrillin11; ABF1-4 – ABA-responsive element-binding factor 1-4; CBF1 – dehydration-responsive element binding factor 1; SA – salicylic acid; JA – jasmonic acid; DET1 – DEETIOLATED 1; UGT79B2 & UGT79B3 – Arabidopsis UDP glycosyl transferases. The five different retrograde signalling pathways are shown in different coloured boxes. Further, the branching in individual pathways is also coloured differently. ? depicts unidentified transporters. The unidentified signalling routes are depicted in red dotted arrows and non-protein, non-RNA components are depicted in black dashed boxes. Upward arrows indicate increased accumulation/overexpression.
Another retro-signal from the plastid under any kind of oxidative damage is linked to the tetrapyrroles (e.g., chlorophyll, heme, siroheme, and phytochromobilin) biosynthesis pathway (Crawford et al. 2018). This pathway is highly regulated, and any alteration in the metabolic flux of the pathway has been shown to affect the expression of nuclear-coded photosynthetic genes through retrograde signalling (Ibata et al. 2016). Recent evidence suggests that 3'-phosphoadenosine-5'-phosphate (PAP), a by-product of secondary sulphur metabolism also acts as a second messenger in chloroplast retrograde signalling in response to oxidative stress caused by drought or high light (Estavillo et al. 2011). Oxidative stress inhibits the activity of inositol polyphosphate-1-phosphatase enzyme (SAL1) (Chan et al. 2016), which increases the accumulation of PAP. PAP moves from the plastid to the nucleus via the directional transporter PAPST1 and induces the expression of nuclear-coded stress-response proteins such as early light-induced protein 2 (ELIP2) and ascorbate peroxidase 2 (APX2) (Gigolashvili et al. 2012). The PAP-SAL1 retro-signalling system was shown to interact with the ABA-signalling network to regulate stomatal closure in response to osmotic stress through alternate ABA-signalling components (Fig. 4) (Pornsiriwong et al. 2017).
Another chloroplast protein, fibrillin11 has been shown to play a major role in osmotic stress tolerance during germination as part of retrograde signalling in Arabidopsis. This chloroplast-generated protein signal regulates the expression of ABF (ABA-responsive element-binding protein/factor) and CBF1 (C-repeat binding factor 1) transcription factors in the nucleus (Choi et al. 2021). CBF1 controls the expression of glycosyltransferase genes (UGT79B2 and UGT79B3), involved in anthocyanin synthesis and thereby confers salt tolerance (Fig. 4) (Li et al. 2017b). To elicit an appropriate stress response, these stress-induced retrograde signals must be integrated into stress-responsive pathways in the cytosol. It is hypothesized that the mediator transcriptional co-activator complex in the nucleus might play a key role in integrating stress-signalling pathways operating in different organelles (Crawford et al. 2018).
Photosynthetic apparatus of halophytes and salt tolerance
Though halophytes share salinity tolerance mechanisms with glycophytes, these species have evolved certain additional characteristics to adapt to highly saline soils. These adaptive mechanisms can be exploited to achieve improved salt tolerance in major glycophytes (Volkov 2015). Halophytes maintain their operational and maximum PSII efficiency under progressive salt stress. This is the most important attribute of halophytes which protects them from photoinhibition and associated oxidative stress (Duarte et al. 2014). Nevertheless, a quantitative analysis of halophytic PSII efficiency shows a significant difference between species. For instance, Suaeda fruticosa has a system where PSII can absorb light even under high salt stress. This mechanism is inactive in Halimione portulacoides, leading to lower levels of light absorption and photosynthetic efficiency (Rabhi et al. 2012, Duarte et al. 2014). This put forth an interesting pattern of salt tolerance even across halophytic species of the same family. Moreover, the coordination of both photosystems in terms of the rapid response of the PSII acceptor site and PSI stability contributed towards lowered levels of oxidative stress in the photosynthetic apparatus of halophytic soybean (Yan et al. 2020).
In certain halophytes like Desmostachya bipinnata, moderate salt stress did not significantly affect chlorophyll content, net photosynthetic rate, ETR, and qP. In contrast, these parameters improved under varying degrees of salinity in obligate halophytic species, namely, Sesuvium portulacastrum and Tecticornia indica (Asrar et al. 2017). Halophytes also show spatially different gene expression pattern as evident from the transcriptomic analysis of quinoa. Lowered expression of photosynthesis-related genes in the epidermal bladder cells as compared to mesophyll cells was recorded (Böhm et al. 2018). Another mechanism by which halophytes, such as Suaeda fruticosa and Halimione portulacoides, overcome salt-induced photoinhibition, is the enhanced activation of the xanthophyll cycle as evident from the increased de-epoxidation state index coupled with increased Chl/Car ratio (Wang et al. 2006). Enhanced expression of photosynthetic genes, such as Mn-stabilizing proteins of the oxygen-evolving complex of PSII, CP47 (Chl a/b protein), RBCL, and Rubisco activase has been reported as protective mechanisms in halophytic rice (Porteresia coarctata) and wheat (Thinopyrum ponticum) genotypes (Sengupta and Majumder 2009). The shift in the mode of carbon assimilation is another common mechanism among halophytes under salt stress. For instance, C3 (Mesembryanthemum crystallinum) and C4 (Portulaca oleracea) halophytes have been reported to shift to the CAM pathway under salinity stress (Bose et al. 2014). Moreover, T. halophila showed increased activities of proteins, such as starch synthase, amylase, and starch branching enzymes, complemented by increased starch accumulation in the chloroplasts under salt stress (Wang et al. 2013).
Conclusions and future trends
Photosynthesis is highly sensitive to salt stress. Na+ influences photosynthesis by disrupting the chloroplast membrane system and chloroplast function and by interfering with the stromal CO2-fixing enzymes. Extensive membrane damage disrupts the PMF and causes irregularities at different stages of photophosphorylation. Chloroplasts have evolved highly regulated salt-responsive pathways. This review tried to collate different characteristics of chloroplast sensitivity to salt stress, followed by the tolerance mechanisms, such as thylakoid membrane remodelling, ion homeostasis regulation, chloroplast membrane and stromal proteins and their turnover, osmoprotectants, chloroplast ROS-scavenging system, as well as chloroplast gene expression and metabolic turnover. Halophytes possess unique response salt-responsive traits which help them maintain their photosynthetic levels under high salinity. These provide vital physiological and molecular know-how on chloroplastic salt response and adaptation of photosynthetic machinery to salinity stress. However, the chloroplast salt-response network and its retrograde communication with the nucleus is a too complex process to be fully interpreted by this information. Further investigations on the biological functions of already identified putative chloroplast transporters, genes, proteins, and metabolites along with retrograde signalling components are needed for a better comprehension of the photosynthetic salt response. The genes validated and characterized serve as valuable inputs for sustaining the photosynthesis of sensitive crop cultivars under salt-stress conditions.
Acknowledgments
The director, ICAR – Central Soil Salinity Research Institute, Karnal, Haryana, India is gratefully acknowledged for providing the necessary facilities.
Abbreviations
- APX
ascorbate peroxidase
- AsA–GSH
ascorbate–glutathion
- CAT
catalase
- Chl
chlorophyll
- DGDG
digalactosyldiacylglycerol
- DHA
dehydroascorbate
- DHAR
dehydroascorbate reductase
- ETC
electron transport chain
- FBPase
fructose-1,6-bisphosphatase
- Fv/Fm
maximum photochemical efficiency of PSII
- GADPH
glyceraldehyde-3-phosphate dehydrogenase
- GB
glycine betaine
- GPX
glutathione peroxidase
- GR
glutathione reductase
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- JA
jasmonic acid
- MDHAR
monodehydroascorbate reductase
- MGD
monogalactosyldiacylglycerol synthase
- MGDG
monogalactosyldiacylglycerol
- mTERFs
mitochondrial eukaryotic transcription factors
- NADP-ME
NADP-malate dehydrogenase
- OEC
oxygen-evolving complex
- PAP
3'-phosphoadenosine-5'-phosphate
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PEPcase
phosphoenolpyruvate carboxylase
- PG
phosphatidylglycerol
- PL
phospholipid
- PMF
proton motive force
- PTOX
plastid terminal oxidase
- RBCLs
Rubisco large subunit
- RBCSs
Rubisco small subunit
- RCA
Rubisco activase
- ROS
reactive oxygen species
- RPK
ribulose-5-phosphate kinase
- Rrd
rubredoxin
- sHSPs
small heat shock proteins
- SOD
superoxide dismutase
- SORGs
singlet oxygen-responsive genes
- SQDG
sulfoquinovosyldiacylglycerol
- Trx/Prx
thioredoxin/peroxiredoxin
- UGPase
UDP glucose pyrophosphorylase
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abideen Z., Koyro H.-W., Huchzermeyer B. et al.: Phragmites karka plants adopt different strategies to regulate photosynthesis and ion flux in saline and water deficit conditions. – Plant Biosyst. 155: 524-534, 2021. 10.1080/11263504.2020.1762783 [DOI] [Google Scholar]
- Acosta-Motos J.R., Ortuño M.F., Bernal-Vicente A. et al.: Plant responses to salt stress: adaptive mechanisms. – Agronomy 7: 18, 2017. 10.3390/agronomy7010018 [DOI] [Google Scholar]
- Ahmad N., Michoux F., Nixon P.J.: Investigating the production of foreign membrane proteins in tobacco chloroplasts: expression of an algal plastid terminal oxidase. – PLoS ONE 7: e41722, 2012. 10.1371/journal.pone.0041722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad P., Ahanger M.A., Alyemeni M.N. et al.: Mitigation of sodium chloride toxicity in Solanum lycopersicum L. by supplementation of jasmonic acid and nitric oxide. – J. Plant Interact. 13: 64-72, 2018. 10.1080/17429145.2017.1420830 [DOI] [Google Scholar]
- Ahmad R., Lim C.J., Kwon S.-Y.: Glycine betaine: A versatile compound with great potential for gene pyramiding to improve crop plant performance against environmental stresses. – Plant Biotechnol. Rep. 7: 49-57, 2013. 10.1007/s11816-012-0266-8 [DOI] [Google Scholar]
- Akhter M.S., Noreen S., Mahmood S. et al.: Influence of salinity stress on PSII in barley (Hordeum vulgare L.) genotypes, probed by chlorophyll-a fluorescence. – J. King Saud Univ. Sci. 33: 101239, 2021. 10.1016/j.jksus.2020.101239 [DOI] [Google Scholar]
- Alkhatib R., Abdo N., Mheidat M.: Photosynthetic and ultrastructural properties of eggplant (Solanum melongena) under salinity stress. – Horticulturae 7: 181, 2021. 10.3390/horticulturae7070181 [DOI] [Google Scholar]
- Allakhverdiev S.I., Kinoshita M., Inaba M. et al.: Unsaturated fatty acids in membrane lipids protect the photosynthetic machinery against salt-induced damage in Synechococcus. – Plant Physiol. 125: 1842-1853, 2001. 10.1104/pp.125.4.1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev S.I., Nishiyama Y., Miyairi S. et al.: Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis. – Plant Physiol. 130: 1443-1453, 2002. 10.1104/pp.011114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirbakhtiar N., Ismaili A., Ghaffari M.-R. et al.: Transcriptome analysis of bread wheat leaves in response to salt stress. – PLoS ONE 16: e0254189, 2021. 10.1371/journal.pone.0254189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arefian M., Vessal S., Malekzadeh-Shafaroudi S. et al.: Comparative proteomics and gene expression analyses revealed responsive proteins and mechanisms for salt tolerance in chickpea genotypes. – BMC Plant Biol. 19: 300, 2019. 10.1186/s12870-019-1793-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrar H., Hussain T., Hadi S.M.S. et al.: Salinity induced changes in light harvesting and carbon assimilating complexes of Desmostachya bipinnata (L.). Staph. – Environ. Exp. Bot. 135: 86-95, 2017. 10.1016/j.envexpbot.2016.12.008 [DOI] [Google Scholar]
- Badawi G.H., Kawano N., Yamauchi Y. et al.: Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. – Physiol. Plantarum 121: 231-238, 2004. 10.1111/j.0031-9317.2004.00308.x [DOI] [PubMed] [Google Scholar]
- Bai J., Kang T., Wu H. et al.: Relative contribution of photorespiration and antioxidative mechanisms in Caragana korshinskii under drought conditions across the Loess Plateau. – Funct. Plant Biol. 44: 1111-1123, 2017. 10.1071/FP17060 [DOI] [PubMed] [Google Scholar]
- Balsemão-Pires E., Jaillais Y., Olson B.J.S.C. et al.: The Arabidopsis translocator protein (AtTSPO) is regulated at multiple levels in response to salt stress and perturbations in tetrapyrrole metabolism. – BMC Plant Biol. 11: 108, 2011. 10.1186/1471-2229-11-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla B.J., Garibay-Hernández A., Melzer M. et al.: Single cell-type analysis of cellular lipid remodelling in response to salinity in the epidermal bladder cells of the model halophyte Mesembryanthemum crystallinum. – Plant Cell Environ. 41: 2390-2403, 2018. 10.1111/pce.13352 [DOI] [PubMed] [Google Scholar]
- Bejaoui F., Salas J.J., Nouairi I. et al.: Changes in chloroplast lipid contents and chloroplast ultrastructure in Sulla carnosa and Sulla coronaria leaves under salt stress. – J. Plant Physiol. 198: 32-38, 2016. 10.1016/j.jplph.2016.03.018 [DOI] [PubMed] [Google Scholar]
- Benjamin J.J., Lucini L., Jothiramshekar S., Parida A.: Metabolomic insights into the mechanisms underlying tolerance to salinity in different halophytes. – Plant Physiol. Bioch. 135: 528-545, 2019. 10.1016/j.plaphy.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Blankenship R.E.: Early evolution of photosynthesis. – Plant Physiol. 154: 434-438, 2010. 10.1104/pp.110.161687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm J., Messerer M., Müller H.M. et al.: Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa. – Curr. Biol. 28: 3075-3085, 2018. 10.1016/j.cub.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Bolte S., Marcon E., Jaunario M. et al.: Dynamics of the localization of the plastid terminal oxidase inside the chloroplast. – J. Exp. Bot. 71: 2661-2669, 2020. 10.1093/jxb/eraa074 [DOI] [PubMed] [Google Scholar]
- Bose J., Munns R., Shabala S. et al.: Chloroplast function and ion regulation in plants growing on saline soils: lessons from halophytes. – J. Exp. Bot. 68: 3129-3143, 2017. 10.1093/jxb/erx142 [DOI] [PubMed] [Google Scholar]
- Bose J., Shabala L., Pottosin I. et al.: Kinetics of xylem loading, membrane potential maintenance, and sensitivity of K+-permeable channels to reactive oxygen species: physiological traits that differentiate salinity tolerance between pea and barley. – Plant Cell Environ. 37: 589-600, 2014. 10.1111/pce.12180 [DOI] [PubMed] [Google Scholar]
- Boychova Krumova S., Laptenok S.P., Kovács L. et al.: Digalactosyl-diacylglycerol-deficiency lowers the thermal stability of thylakoid membranes. – Photosynth. Res. 105: 229-242, 2010. 10.1007/s11120-010-9581-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon R.H., García-Cerdán J.G., Malnoë A. et al.: A conserved rubredoxin is necessary for photosystem II accumulation in diverse oxygenic photoautotrophs. – J. Biol. Chem. 288: 26688-26696, 2013. 10.1074/jbc.M113.487629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraretto L., Formentin E., Teardo E. et al.: A thylakoid located two-pore K+ channel controls photosynthetic light utilization in plants. – Science 342: 114-118, 2013. 10.1126/science.1242113 [DOI] [PubMed] [Google Scholar]
- Chalbi N., Hessini K., Gandour M. et al.: Are changes in membrane lipids and fatty acid composition related to salt-stress resistance in wild and cultivated barley? – J. Plant Nutr. Soil Sc. 176: 138-147, 2013. 10.1002/jpln.201100413 [DOI] [Google Scholar]
- Chan K.X., Mabbitt P.D., Phua S.Y. et al.: Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. – P. Natl. Acad. Sci. USA 113: E4567-E4576, 2016. 10.1073/pnas.1604936113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang I.-F., Hsu J.-L., Hsu P.-H. et al.: Comparative phosphoproteomic analysis of microsomal fractions of Arabidopsis thaliana and Oryza sativa subjected to high salinity. – Plant Sci. 185-186: 131-142, 2012. 10.1016/j.plantsci.2011.09.009 [DOI] [PubMed] [Google Scholar]
- Chang L., Guo A., Jin X. et al.: The subunit of glyceraldehyde 3-phosphate dehydrogenase is an important factor for maintaining photosynthesis and plant development under salt stress – based on an integrative analysis of the structural, physiological and proteomic changes in chloroplasts in Thellungiella halophila. – Plant Sci. 236: 223-238, 2015. 10.1016/j.plantsci.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Chen C., Liu H., Wang C. et al.: Metabolomics characterizes metabolic changes of Apocyni Veneti Folium in response to salt stress. – Plant Physiol. Bioch. 144: 187-196, 2019b. 10.1016/j.plaphy.2019.09.043 [DOI] [PubMed] [Google Scholar]
- Chen Q., Wang B., Ding H. et al.: The role of NADP-malic enzyme in plants under stress. – Plant Sci. 281: 206-212, 2019a. 10.1016/j.plantsci.2019.01.010 [DOI] [PubMed] [Google Scholar]
- Chiconato D.A., Costa M.G.S., Balbuena T.S. et al.: Proteomic analysis of young sugarcane plants with contrasting salt tolerance. – Funct. Plant Biol. 48: 588-596, 2021. 10.1071/FP20314 [DOI] [PubMed] [Google Scholar]
- Choi Y.R., Kim I., Kumar M. et al.: Chloroplast localized FIBRILLIN11 is involved in the osmotic stress response during Arabidopsis seed germination. – Biology 10: 368, 2021. 10.3390/biology10050368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford T., Lehotai N., Strand Å.: The role of retrograde signals during plant stress responses. – J. Exp. Bot. 69: 2783-2795, 2018. 10.1093/jxb/erx481 [DOI] [PubMed] [Google Scholar]
- Cruz de Carvalho M.H.: Drought stress and reactive oxygen species: production, scavenging and signaling. – Plant Signal. Behav. 3: 156-165, 2014. 10.4161/psb.3.3.5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P., Majumder A.L.: Transcriptome analysis of grapevine under salinity and identification of key genes responsible for salt tolerance. – Funct. Integr. Genomic. 19: 61-73, 2019. 10.1007/s10142-018-0628-6 [DOI] [PubMed] [Google Scholar]
- Des Marais D.J.: When did photosynthesis emerge on Earth? – Science 289: 1703-1705, 2000. 10.1126/science.289.5485.1703 [DOI] [PubMed] [Google Scholar]
- Dietz K.J.: Thiol-based peroxidases and ascorbate peroxidases: why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? – Mol. Cells 39: 20-25, 2016. 10.14348/molcells.2016.2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra V., Duan J., Lee K.P. et al.: FtsH2-dependent proteolysis of EXECUTER1 is essential in mediating singlet oxygen triggered retrograde signaling in Arabidopsis thaliana. – Front. Plant Sci. 8: 1145, 2017. 10.3389/fpls.2017.01145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte B., Sleimi N., Caçador I.: Biophysical and biochemical constraints imposed by salt stress: learning from halophytes. – Front. Plant Sci. 5: 746, 2014. 10.3389/fpls.2014.00746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall S.D., Brown M.T., Johnson P.J.: Ancient invasions: from endosymbionts to organelles. – Science 304: 253-257, 2004. 10.1126/science.1094884 [DOI] [PubMed] [Google Scholar]
- Edreva A.: Generation and scavenging of reactive oxygen species in chloroplasts: A submolecular approach. – Agr. Ecosyst. Environ. 106: 119-133, 2005. 10.1016/j.agee.2004.10.022 [DOI] [Google Scholar]
- Estavillo G.M., Crisp P.A., Pornsiriwong W. et al.: Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. – Plant Cell 23: 3992-4012, 2011. 10.1105/tpc.111.091033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P., Nie L., Jiang P. et al.: Transcriptome analysis of Salicornia europaea under saline conditions revealed the adaptive primary metabolic pathways as early events to facilitate salt adaptation. – PLoS ONE 8: e80595, 2013. 10.1371/journal.pone.0080595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z.T., Deng Y.Q., Fan H. et al.: Effects of NaCl stress on growth and photosynthetic characteristics of Ulmus pumila L. seedlings in sand culture. – Photosynthetica 52: 313-320, 2014. 10.1007/s11099-014-0032-y [DOI] [Google Scholar]
- Fernández-García N., Olmos E., Bardisi E. et al.: Intrinsic water use efficiency controls the adaptation to high salinity in a semi-arid adapted plant, henna (Lawsonia inermis L.). – J. Plant Physiol. 171: 64-75, 2014. 10.1016/j.jplph.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Finazzi G., Petroutsos D., Tomizioli M. et al.: Ions channels/transporters and chloroplast regulation. – Cell Calcium 58: 86-97, 2015. 10.1016/j.ceca.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Franzisky B.L., Geilfus C.M., Romo-Pérez M.L. et al.: Acclimatisation of guard cell metabolism to long-term salinity. – Plant Cell Environ. 44: 870-884, 2021. 10.1111/pce.13964 [DOI] [PubMed] [Google Scholar]
- Fristedt R., Willig A., Granath P. et al.: Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. – Plant Cell 21: 3950-3964, 2009. 10.1105/tpc.109.069435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H.J., Yang H.Y., Bai J.P. et al.: Ultrastructural and physiological responses of potato (Solanum tuberosum L.) plantlets to gradient saline stress. – Front. Plant Sci. 5: 787, 2015. 10.3389/fpls.2014.00787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.K., Kim J.K., Owens T.G. et al.: Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. – P. Natl. Acad. Sci. USA 99: 15898-15903, 2002. 10.1073/pnas.252637799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R., Shankar R., Thakkar B. et al.: Transcriptome analyses reveal genotype- and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. – Sci. Rep.-UK 6: 19228, 2016. 10.1038/srep19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemi K.M., Shariati M.: [Effect of salt stress on PSII efficiency of Dunaliella bardawil under light and dark conditions.] – J. Cell Tissue 3: 141-151, 2012. [In Persian] http://jct.araku.ac.ir/article_1710.html?lang=en [Google Scholar]
- Gigolashvili T., Geier M., Ashykhmina N. et al.: The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5'-phosphosulfate to the cytosol. – Plant Cell 24: 4187-4204, 2012. 10.1105/tpc.112.101964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D.H., Wang G.Z., Si W.T. et al.: Effects of salt stress on photosynthetic pigments and activity of ribulose-1,5-bisphosphate carboxylase/oxygenase in Kalidium foliatum. – Russ. J. Plant Physiol. 65: 98-103, 2018. 10.1134/S1021443718010144 [DOI] [Google Scholar]
- Gong W., Xu F., Sun J. et al.: iTRAQ-based comparative proteomic analysis of seedling leaves of two upland cotton genotypes differing in salt tolerance. – Front. Plant Sci. 8: 2113, 2017. 10.3389/fpls.2017.02113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindjee G., Govindjee R.: The absorption of light in photosynthesis. – Sci. Am. 231: 68-87, 1974. 10.1038/scientificamerican1274-68 [DOI] [PubMed] [Google Scholar]
- Grabsztunowicz M., Koskela M.M., Mulo P.: Post-translational modifications in regulation of chloroplast function: recent advances. – Front. Plant Sci. 8: 240, 2017. 10.3389/fpls.2017.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Shi Q., Xiong Y., Yin Y. et al.: [Effects of salt-alkaline mixed stress on growth and photosynthetic characteristics of Taxodium hybrid ‘Zhongshanshan 406’.] – J. Nanjing Forest. Univ. 43: 61-68, 2019a. [In Chinese] http://nldxb.njfu.edu.cn/EN/10.3969/j.issn.1000-2006.201805078 [Google Scholar]
- Guo Q., Liu L., Barkla B.J.: Membrane lipid remodeling in response to salinity. – Int. J. Mol. Sci. 20: 4264, 2019b. 10.3390/ijms20174264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Wu Y., Wang Y. et al.: OsMSRA4.1 and OsMSRB1.1, two rice plastidial methionine sulfoxide reductases, are involved in abiotic stress responses. – Planta 230: 227-238, 2009. 10.1007/s00425-009-0934-2 [DOI] [PubMed] [Google Scholar]
- Hamed K.B., Youssef N.B., Ranieri A. et al.: Changes in content and fatty acid profiles of total lipids and sulfolipids in the halophyte Crithmum maritimum under salt stress. – J. Plant Physiol. 162: 599-602, 2005. 10.1016/j.jplph.2004.11.010 [DOI] [PubMed] [Google Scholar]
- Hasanuzzaman M., Alam M.M., Rahman A. et al.: Exogenous proline and glycine betaine mediated upregulation of antioxidant defense and glyoxalase systems provides better protection against salt-induced oxidative stress in two rice (Oryza sativa L.) varieties. – BioMed Res. Int. 2014: 757219, 2014. 10.1155/2014/757219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani A., Azapagic A., Shokri N.: Predicting long-term dynamics of soil salinity and sodicity on a global scale. – P. Natl. Acad. Sci. USA 117: 33017-33027, 2020. 10.1073/pnas.2013771117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Yang A., Zhang W. et al.: Improved salt tolerance of transgenic wheat by introducing betA gene for glycine betaine synthesis. – Plant Cell Tiss. Org. Cult. 101: 65-78, 2010. 10.1007/s11240-009-9665-0 [DOI] [Google Scholar]
- Hiyane R., Hiyane S., Tang A.C., Boyer J.S.: Sucrose feeding reverses shade-induced kernel losses in maize. – Ann. Bot.-London 106: 395-403, 2010. 10.1093/aob/mcq132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida H., Tanaka Y., Hibino T. et al.: Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. – Plant Mol. Biol. 43: 103-111, 2000. 10.1023/A:1006408712416 [DOI] [PubMed] [Google Scholar]
- Huan L., Xie X., Zheng Z. et al.: Positive correlation between PSI response and oxidative pentose phosphate pathway activity during salt stress in an intertidal macroalga. – Plant Cell Physiol. 55: 1395-1403, 2014. 10.1093/pcp/pcu063 [DOI] [PubMed] [Google Scholar]
- Hussain S., Zhang J.H., Zhong C. et al.: Effects of salt stress on rice growth, development characteristics, and the regulating ways: A review. – J. Integr. Agr. 16: 2357-2374, 2017. 10.1016/S2095-3119(16)61608-8 [DOI] [Google Scholar]
- Ibata H., Nagatani A., Mochizuki N.: CHLH/GUN5 function in tetrapyrrole metabolism is correlated with plastid signaling but not ABA responses in guard cells. – Front. Plant Sci. 7: 1650, 2016. 10.3389/fpls.2016.01650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M., Athar H.U.R., Ibrahim M. et al.: Leaf proteome analysis signified that photosynthesis and antioxidants are key indicators of salinity tolerance in canola (Brassica napus L.). – Pak. J. Bot. 51: 1955-1968, 2019. 10.30848/PJB2019-6(38) [DOI] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda K. et al.: The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. – Plant Cell 13: 2191-2209, 2001. 10.1105/tpc.010192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakannan M., Bose J., Babourina O. et al.: Salicylic acid in plant salinity stress signalling and tolerance. – Plant Growth Regul. 76: 25-40, 2015. 10.1007/s10725-015-0028-z [DOI] [Google Scholar]
- Jia H., Shao M., He Y. et al.: Proteome dynamics and physiological responses to short-term salt stress in Brassica napus leaves. – PLoS ONE 10: e0144808, 2015. 10.1371/journal.pone.0144808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Daniell H.: Expression of γ-tocopherol methyltransferase in chloroplasts results in massive proliferation of the inner envelope membrane and decreases susceptibility to salt and metal-induced oxidative stresses by reducing reactive oxygen species. – Plant Biotechnol. J. 12: 1274-1285, 2014. 10.1111/pbi.12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Miyagishima S.Y.: Chloroplast DNA replication is regulated by the redox state independently of chloroplast division in Chlamydomonas reinhardtii. – Plant Physiol. 161: 2102-2112, 2013. 10.1104/pp.113.216291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan X., Ren J., Chen T. et al.: Effects of salinity on photosynthesis in maize probed by prompt fluorescence, delayed fluorescence and P700 signals. – Environ. Exp. Bot. 140: 56-64, 2017. 10.1016/j.envexpbot.2017.05.019 [DOI] [Google Scholar]
- Khan H.A., Siddique K.H.M., Munir R. et al.: Salt sensitivity in chickpea: Growth, photosynthesis, seed yield components and tissue ion regulation in contrasting genotypes. – J. Plant Physiol. 182: 1-12, 2015. 10.1016/j.jplph.2015.05.002 [DOI] [PubMed] [Google Scholar]
- Khurana N., Chauhan H., Khurana P.: Characterization of a chloroplast localized wheat membrane protein (TaRCI) and its role in heat, drought and salinity stress tolerance in Arabidopsis thaliana. – Plant Gene 4: 45-54, 2015. 10.1016/j.plgene.2015.09.005 [DOI] [Google Scholar]
- Kim Y.H., Lim S., Yang K.S. et al.: Expression of Arabidopsis NDPK2 increases antioxidant enzyme activities and enhances tolerance to multiple environmental stresses in transgenic sweetpotato plants. – Mol. Breeding 24: 233-244, 2009. 10.1007/s11032-009-9286-7 [DOI] [Google Scholar]
- Krasensky J., Broyart C., Rabanal F.A., Jonak C.: The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. – Antioxid. Redox Sign. 21: 1289-1304, 2014. 10.1089/ars.2013.5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A., Feilke K.: The dual role of the plastid terminal oxidase PTOX: between a protective and a pro-oxidant function. – Front. Plant Sci. 6: 1147, 2016. 10.3389/fpls.2015.01147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A., Parida A.K.: Metabolomics and network analysis reveal the potential metabolites and biological pathways involved in salinity tolerance of the halophyte Salvadora persica. – Environ. Exp. Bot. 148: 85-99, 2018. 10.1016/j.envexpbot.2017.12.021 [DOI] [Google Scholar]
- Kumutha D., Sairam R.K., Ezhilmathi K. et al.: Effect of waterlogging on carbohydrate metabolism in pigeon pea (Cajanus cajan L.): Upregulation of sucrose synthase and alcohol dehydrogenase. – Plant Sci. 175: 706-716, 2008. 10.1016/j.plantsci.2008.07.013 [DOI] [Google Scholar]
- Kunz H.-H., Gierth M., Herdean A. et al.: Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. – P. Natl. Acad. Sci. USA 111: 7480-7485, 2014. 10.1073/pnas.1323899111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande N.V., Barua P., Gayen D. et al.: Proteomic dissection of the chloroplast: moving beyond photosynthesis. – J. Proteomics 212: 103542, 2020. 10.1016/j.jprot.2019.103542 [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M., Xu X.M., Møller S.G.: Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. – Planta 232: 567-578, 2010. 10.1007/s00425-010-1192-z [DOI] [PubMed] [Google Scholar]
- Le Martret B., Poage M., Shiel K. et al.: Tobacco chloroplast transformants expressing genes encoding dehydroascorbate reductase, glutathione reductase, and glutathione-S-transferase, exhibit altered anti-oxidant metabolism and improved abiotic stress tolerance. – Plant Biotechnol. J. 9: 661-673, 2011. 10.1111/j.1467-7652.2011.00611.x [DOI] [PubMed] [Google Scholar]
- Leister D., Wang L., Kleine T.: Organellar gene expression and acclimation of plants to environmental stress. – Front. Plant Sci. 8: 387, 2017. 10.3389/fpls.2017.00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekklar C., Suriya-Arunroj D., Pongpanich M. et al.: Comparative genomic analysis of rice with contrasting photosynthesis and grain production under salt stress. – Genes-Basel 10: 562-582, 2019. 10.3390/genes10080562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Cui G., Hu G. et al.: Proteome dynamics and physiological responses to short-term salt stress in Leymus chinensis leaves. – PLoS ONE 12: e0183615, 2017a. 10.1371/journal.pone.0183615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Li Y.J., Zhang F.J. et al.: The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. – Plant J. 89: 85-103, 2017b. 10.1111/tpj.13324 [DOI] [PubMed] [Google Scholar]
- Li Y., Liu P., Takano T., Liu S.: A chloroplast-localized rubredoxin family protein gene from Puccinellia tenuiflora (PutRUB) increases NaCl and NaHCO3 tolerance by decreasing H2O2 accumulation. – Int. J. Mol. Sci. 17: 804, 2016. 10.3390/ijms17060804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.F., Zheng Y., Vemireddy L.R. et al.: Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. – BMC Genomics 19: 935, 2018. 10.1186/s12864-018-5279-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Wu L.: Effects of salt stress on root plasma membrane characteristics of salt-tolerant and salt-sensitive buffalograss clones. – Environ. Exp. Bot. 36: 239-254, 1996. 10.1016/0098-8472(96)01025-8 [DOI] [Google Scholar]
- Liu A., Xiao Z., Wang Z. et al.: Galactolipid and phospholipid profile and proteome alterations in soybean leaves at the onset of salt stress. – Front. Plant Sci. 12: 644408, 2021. 10.3389/fpls.2021.644408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.W., Hsu Y.K., Cheng Y.H. et al.: Proteomic analysis of salt-responsive ubiquitin-related proteins in rice roots. – Rapid Commun. Mass Spectrom. 26: 1649-1660, 2012. 10.1002/rcm.6271 [DOI] [PubMed] [Google Scholar]
- Liu D., Hou L., Li W.C. et al.: COR15B expression is affected by chloroplast functionality and its role in response to salt stress in Arabidopsis thaliana. – Biol. Plantarum 58: 667-675, 2014. 10.1007/s10535-014-0451-4 [DOI] [Google Scholar]
- Liu X.S., Feng S.J., Wang M.Q. et al.: OsNHAD is a chloroplast membrane-located transporter required for resistance to salt stress in rice (Oryza sativa). – Plant Sci. 291: 110359, 2020. 10.1016/j.plantsci.2019.110359 [DOI] [PubMed] [Google Scholar]
- Loll B., Kern J., Saenger W. et al.: Lipids in photosystem II: Interactions with protein and cofactors. – BBA-Bioenergetics 1767: 509-519, 2007. 10.1016/j.bbabio.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Longstreth D.J., Nobel P.S.: Salinity effects of leaf anatomy: consequences for photosynthesis. – Plant Physiol. 63: 700-703, 1979. 10.1104/pp.63.4.700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Wu J., Li Y. et al.: Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. – PLoS ONE 8: e54002, 2013. 10.1371/journal.pone.0054002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X., Chen S., Wang Y.: Advances in understanding the physiological and molecular responses of sugar beet to salt stress. – Front. Plant Sci. 10: 1431, 2019. 10.3389/fpls.2019.01431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado R.M.A., Serralheiro R.P.: Soil salinity: effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. – Horticulturae 3: 30, 2017. 10.3390/horticulturae3020030 [DOI] [Google Scholar]
- Magdy M., Mansour F., van Hasselt P.R., Kuiper P.J.C.: Plasma membrane lipid alterations induced by NaCl in winter wheat roots. – Physiol. Plantarum 92: 473-478, 1994. 10.1111/j.1399-3054.1994.tb08838.x [DOI] [Google Scholar]
- Martinis J., Glauser G., Valimareanu S., Kessler F.: A chloroplast ABC1-like kinase regulates vitamin E metabolism in Arabidopsis. – Plant Physiol. 162: 652-662, 2013. 10.1104/pp.113.218644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusig D., Tombesi S.: Abscisic acid mediates drought and salt stress responses in Vitis vinifera – A review. – Int. J. Mol. Sci. 21: 8648, 2020. 10.3390/ijms21228648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley S.A., Nielsen B.L.: Chloroplast DNA copy number changes during plant development in organelle DNA polymerase mutants. – Front. Plant Sci. 7: 57, 2016. 10.3389/fpls.2016.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Kunz H.-H., Schroeder J.I. et al.: Decreased capacity for sodium export out of Arabidopsis chloroplasts impairs salt tolerance, photosynthesis and plant performance. – Plant J. 78: 646-658, 2014. 10.1111/tpj.12501 [DOI] [PubMed] [Google Scholar]
- Munns R., Passioura J.B., Colmer T.D., Byrt C.S.: Osmotic adjustment and energy limitations to plant growth in saline soil. – New Phytol. 225: 1091-1096, 2020. 10.1111/nph.15862 [DOI] [PubMed] [Google Scholar]
- Nadeem M., Li J., Yahya M. et al.: Grain legumes and fear of salt stress: Focus on mechanisms and management strategies. – Int. J. Mol. Sci. 20: 799, 2019. 10.3390/ijms20040799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A., Hanaoka M., Shikanai T. et al.: The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. – Plant Cell Physiol. 45: 357-368, 2004. 10.1093/pcp/pch050 [DOI] [PubMed] [Google Scholar]
- Nassar R.M.A., Kamel H.A., Ghoniem A.E. et al.: Physiological and anatomical mechanisms in wheat to cope with salt stress induced by seawater. – Plants-Basel 9: 237, 2020. 10.3390/plants9020237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Bañón S., Olmos E., Sánchez-Blanco M.J.: Effects of sodium chloride on water potential components, hydraulic conductivity, gas exchange and leaf ultrastructure of Arbutus unedo plants. – Plant Sci. 172: 473-480, 2007. 10.1016/j.plantsci.2006.10.006 [DOI] [Google Scholar]
- Nawaz G., Kang H.: Chloroplast- or mitochondria-targeted DEAD-box RNA helicases play essential roles in organellar RNA metabolism and abiotic stress responses. – Front. Plant Sci. 8: 871, 2017. 10.3389/fpls.2017.00871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam S., Subramanyam R.: Alteration of photochemistry and protein degradation of photosystem II from Chlamydomonas reinhardtii under high salt grown cells. – J. Photoch. Photobio. B 124: 63-70, 2013. 10.1016/j.jphotobiol.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Noctor G., Foyer C.H.: Intracellular redox compartmentation and ROS-related communication in regulation and signaling. – Plant Physiol. 171: 1581-1592, 2016. 10.1104/pp.16.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oi T., Enomoto S., Nakao T. et al.: Three-dimensional ultrastructural change of chloroplasts in rice mesophyll cells responding to salt stress. – Ann. Bot.-London 125: 833-840, 2020. 10.1093/aob/mcz192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard M.A., Omranian N., Ahmadi F.S. et al.: Effect of salt stress on genes encoding translation-associated proteins in Arabidopsis thaliana. – Plant Signal. Behav. 7: 1095-1102, 2012. 10.4161/psb.21218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto E., Iwasaki Y., Miyake H. et al.: Salinity induces membrane structure and lipid changes in maize mesophyll and bundle sheath chloroplasts. – Physiol. Plantarum 157: 13-23, 2016. 10.1111/ppl.12404 [DOI] [PubMed] [Google Scholar]
- Omoto E., Nagao H., Taniguchi M., Miyake H.: Localization of reactive oxygen species and change of antioxidant capacities in mesophyll and bundle sheath chloroplasts of maize under salinity. – Physiol. Plantarum 149: 1-12, 2013. 10.1111/ppl.12017 [DOI] [PubMed] [Google Scholar]
- Ouertani N.R., Arasappan D., Abid G. et al.: Transcriptomic analysis of salt-stress-responsive genes in barley roots and leaves. – Int. J. Mol. Sci. 22: 8155, 2021. 10.3390/ijms22158155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Kim W.-Y., Yun D.-J.: A new insight of salt stress signaling in plant. – Mol. Cells 39: 447-459, 2016. 10.14348/molcells.2016.0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuszak J., Dziurka M., Hornyák M. et al.: Physiological and biochemical parameters of salinity resistance of three durum wheat genotypes. – Int. J. Mol. Sci. 23: 8397, 2022. 10.3390/ijms23158397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B., Ray S., Richter A., Majumder A.L.: Enhanced salt tolerance of transgenic tobacco plants by co-expression of PcINO1 and McIMT1 is accompanied by increased level of myo-inositol and methylated inositol. – Protoplasma 245: 143-152, 2010. 10.1007/s00709-010-0163-3 [DOI] [PubMed] [Google Scholar]
- Peharec Štefanić P., Koffler T., Adler G., Bar-Zvi D.: Chloroplasts of salt-grown Arabidopsis seedlings are impaired in structure, genome copy number and transcript levels. – PLoS ONE 8: e82548, 2013. 10.1371/journal.pone.0082548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornsiriwong W., Estavillo G.M., Chan K.X. et al.: A chloroplast retrograde signal, 3'-phosphoadenosine 5'-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. – eLife 6: e23361, 2017. 10.7554/eLife.23361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Yin Y., Zhao J. et al.: Metabolomic and transcriptomic analysis of Lycium chinese and L. ruthenicum under salinity stress. – BMC Plant Biol. 22: 8, 2022. 10.1186/s12870-021-03375-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi M., Castagna A., Remorini D. et al.: Photosynthetic responses to salinity in two obligate halophytes: Sesuvium portulacastrum and Tecticornia indica. – S. Afr. J. Bot. 79: 39-47, 2012. 10.1016/j.sajb.2011.11.007 [DOI] [Google Scholar]
- Ramani B., Zorn H., Papenbrock J.: Quantification and fatty acid profiles of sulfolipids in two halophytes and a glycophyte grown under different salt concentrations. – Z. Naturforsch. C 59: 835-842, 2004. 10.1515/znc-2004-11-1212 [DOI] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Cuiné S. et al.: Chemical quenching of singlet oxygen by carotenoids in plants. – Plant Physiol. 158: 1267-1278, 2012. 10.1104/pp.111.182394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaque S., Elias S.M., Haque T. et al.: Gene expression analysis associated with salt stress in a reciprocally crossed rice population. – Sci. Rep.-UK 9: 8249, 2019. 10.1038/s41598-019-44757-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S.P., Jogeswar G., Rasineni G.K. et al.: Proline over-accumulation alleviates salt stress and protects photosynthetic and antioxidant enzyme activities in transgenic sorghum [Sorghum bicolor (L.) Moench]. – Plant Physiol. Bioch. 94: 104-113, 2015. 10.1016/j.plaphy.2015.05.014 [DOI] [PubMed] [Google Scholar]
- Robinson S.P., Downton W.J.: Potassium, sodium, and chloride content of isolated intact chloroplasts in relation to ionic compartmentation in leaves. – Arch. Biochem. Biophys. 228: 197-206, 1984. 10.1016/0003-9861(84)90061-4 [DOI] [PubMed] [Google Scholar]
- Robles P., Navarro-Cartagena S., Ferrández-Ayela A. et al.: The characterization of Arabidopsis mterf6 mutants reveals a new role for mTERF6 in tolerance to abiotic stress. – Int. J. Mol. Sci. 19: 2388, 2018. 10.3390/ijms19082388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles P., Quesada V.: Transcriptional and post-transcriptional regulation of organellar gene expression (OGE) and its roles in plant salt tolerance. – Int. J. Mol. Sci. 20: 1056, 2019. 10.3390/ijms20051056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Aranda R., Moya J.L., Tadeo F.R. et al.: Physiological and anatomical disturbances induced by chloride salts in sensitive and tolerant citrus: Beneficial and detrimental effects of cations. – Plant Cell Environ. 21: 1243-1253, 1998. 10.1046/j.1365-3040.1998.00349.x [DOI] [Google Scholar]
- Sakamoto A., Murata N.: The role of glycine betaine in the protection of plants from stress: Clues from transgenic plants. – Plant Cell Environ. 25: 163-171, 2002. 10.1046/j.0016-8025.2001.00790.x [DOI] [PubMed] [Google Scholar]
- Sathee L., Jagadhesan B., Pandesha P.H. et al.: Genome editing targets for improving nutrient use efficiency and nutrient stress adaptation. – Front. Genet. 13: 900897, 2022. 10.3389/fgene.2022.900897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Rolland N., Dorne A.-J., Joyard J.: Sulfolipid is a potential candidate for annexin binding to the outer surface of chloroplast. – Biochem. Bioph. Res. Co. 272: 519-524, 2000. 10.1006/bbrc.2000.2805 [DOI] [PubMed] [Google Scholar]
- Sengupta S., Majumder A.L.: Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: a physiological and proteomic approach. – Planta 229: 911-929, 2009. 10.1007/s00425-008-0878-y [DOI] [PubMed] [Google Scholar]
- Seo S.Y., Wi S.J., Park K.Y.: Functional switching of NPR1 between chloroplast and nucleus for adaptive response to salt stress. – Sci. Rep.-UK 10: 4339, 2020. 10.1038/s41598-020-61379-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S., Chen G., Chen Z.H., Pottosin I.: The energy cost of the tonoplast futile sodium leak. – New Phytol. 225: 1105-1110, 2020. 10.1111/nph.15758 [DOI] [PubMed] [Google Scholar]
- Shao N., Duan G.Y., Bock R.: A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells. – Plant Cell 25: 4209-4226, 2013. 10.1105/tpc.113.117390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S., Guo S.R., Sun J., Yuan L.Y.: Effects of salt stress on the structure and function of the photosynthetic apparatus in Cucumis sativus and its protection by exogenous putrescine. – Physiol. Plantarum 146: 285-296, 2012. 10.1111/j.1399-3054.2012.01623.x [DOI] [PubMed] [Google Scholar]
- Shu S., Yuan Y., Chen J. et al.: The role of putrescine in the regulation of proteins and fatty acids of thylakoid membranes under salt stress. – Sci. Rep.-UK 5: 14390, 2015. 10.1038/srep14390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simidjiev I., Stoylova S., Amenitsch H. et al.: Self-assembly of large, ordered lamellae from non-bilayer lipids and integral membrane proteins in vitro. – P. Natl. Acad. Sci. USA 97: 1473-1476, 2000. 10.1073/pnas.97.4.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.N., Mishra R.N., Agarwal P.K. et al.: A pea chloroplast translation elongation factor that is regulated by abiotic factors. – Biochem. Bioph. Res. Co. 320: 523-530, 2004. 10.1016/j.bbrc.2004.05.192 [DOI] [PubMed] [Google Scholar]
- Singh J., Singh V., Vineeth T.V. et al.: Differential response of Indian mustard (Brassica juncea L., Czern and Coss) under salinity: photosynthetic traits and gene expression. – Physiol. Mol. Biol. Pla. 25: 71-83, 2019. 10.1007/s12298-018-0631-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltabayeva A., Ongaltay A., Omondi J.O., Srivastava S.: Morphological, physiological and molecular markers for salt-stressed plants. – Plants-Basel 10: 243, 2021. 10.3390/plants10020243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.P., Guo Y., Qiu Q. et al.: A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. – P. Natl. Acad. Sci. USA 101: 10211-10216, 2004. 10.1073/pnas.0403709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui N., Han G.: Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. – Acta Physiol. Plant. 36: 983-992, 2014. 10.1007/s11738-013-1477-5 [DOI] [Google Scholar]
- Sun S., Song H., Li J. et al.: Comparative transcriptome analysis reveals gene expression differences between two peach cultivars under saline-alkaline stress. – Hereditas 157: 9, 2020. 10.1186/s41065-020-00122-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J., Zhao Q., David L. et al.: Salinity response in chloroplasts: insights from gene characterization. – Int. J. Mol. Sci. 18: 1011, 2017. 10.3390/ijms18051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanou G., Job C., Rajjou L. et al.: Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. – Plant J. 60: 795-804, 2009. 10.1111/j.1365-313X.2009.04000.x [DOI] [PubMed] [Google Scholar]
- Thagela P., Yadav R.K., Tripathi K. et al.: Salinity induced changes in the chloroplast proteome of the aquatic pteridophyte Azolla microphylla. – Symbiosis 75: 61-67, 2018. 10.1007/s13199-017-0521-4 [DOI] [Google Scholar]
- Tseng M.J., Liu C.W., Yiu J.C.: Enhanced tolerance to sulfur dioxide and salt stress of transgenic Chinese cabbage plants expressing both superoxide dismutase and catalase in chloroplasts. – Plant Physiol. Bioch. 45: 822-833, 2007. 10.1016/j.plaphy.2007.07.011 [DOI] [PubMed] [Google Scholar]
- Volkov V.: Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. – Front. Plant Sci. 6: 873, 2015. 10.3389/fpls.2015.00873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waditee-Sirisattha R., Kageyama H., Tanaka Y. et al.: Overexpression of halophilic serine hydroxymethyltransferase in fresh water cyanobacterium Synechococcus elongatus PCC7942 results in increased enzyme activities of serine biosynthetic pathways and enhanced salinity tolerance. – Arch. Microbiol. 199: 29-35, 2017. 10.1007/s00203-016-1271-z [DOI] [PubMed] [Google Scholar]
- Wang C.-Q., Zhao J.-Q., Chen M., Wang B.-S.: [Identification of betacyanin and effects of environmental factors on its accumulation in halophyte Suaeda salsa.] – J. Plant Physiol. Mol. Biol. 32: 195-201, 2006. [In Chinese] https://pubmed.ncbi.nlm.nih.gov/16622319/ [PubMed] [Google Scholar]
- Wang L., Kim C., Xu X. et al.: Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. – P. Natl. Acad. Sci. USA 113: E3792-E3800, 2016. 10.1073/pnas.1603562113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.L., Hua C., Zhou F., Zhou Q.C.: Effects of NaCl stress on photochemical activity and thylakoid membrane polypeptide composition of a salt-tolerant and a salt-sensitive rice cultivar. – Photosynthetica 47: 125-127, 2009. 10.1007/s11099-009-0019-2 [DOI] [Google Scholar]
- Wang S., Uddin M.I., Tanaka K. et al.: Maintenance of chloroplast structure and function by overexpression of the rice monogalactosyldiacylglycerol synthase gene leads to enhanced salt tolerance in tobacco. – Plant Physiol. 165: 1144-1155, 2014. 10.1104/pp.114.238899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chang L., Wang B. et al.: Comparative proteomics of Thellungiella halophila leaves from plants subjected to salinity reveals the importance of chloroplastic starch and soluble sugars in halophyte salt tolerance. – Mol. Cell. Proteomics 12: 2174-2195, 2013. 10.1074/mcp.M112.022475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zeng X., Xu Q. et al.: Metabolite profiling in two contrasting Tibetan hulless barley cultivars revealed the core salt-responsive metabolome and key salt-tolerance biomarkers. – AoB Plants 11: plz021, 2019. 10.1093/aobpla/plz021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D., Zhang W., Wang C. et al.: Genetic engineering of the biosynthesis of glycinebetaine leads to alleviate salt-induced potassium efflux and enhances salt tolerance in tomato plants. – Plant Sci. 257: 74-83, 2017. 10.1016/j.plantsci.2017.01.012 [DOI] [PubMed] [Google Scholar]
- Wu D., Cai S., Chen M. et al.: Tissue metabolic responses to salt stress in wild and cultivated barley. – PLoS ONE 8: e55431, 2013. 10.1371/journal.pone.0055431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Q., Bi G., Cao M. et al.: Comparative transcriptome analysis provides insights into response of Ulva compressa to fluctuating salinity conditions. – J. Phycol. 57: 1295-1308, 2021. 10.1111/jpy.13167 [DOI] [PubMed] [Google Scholar]
- Xiong J., Sun Y., Yang Q. et al.: Proteomic analysis of early salt stress responsive proteins in alfalfa roots and shoots. – Proteome Sci. 15: 19, 2017. 10.1186/s12953-017-0127-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Lee K., Gu L. et al.: Functional characterization of a plastid-specific ribosomal protein PSRP2 in Arabidopsis thaliana under abiotic stress conditions. – Plant Physiol. Bioch. 73: 405-411, 2013. 10.1016/j.plaphy.2013.10.027 [DOI] [PubMed] [Google Scholar]
- Xu W., Lv H., Zhao M. et al.: Proteomic comparison reveals the contribution of chloroplast to salt tolerance of a wheat introgression line. – Sci. Rep.-UK 6: 32384, 2016. 10.1038/srep32384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Rahman S., Kawasaki M. et al.: Pretreatment with a low concentration of methyl viologen decreases the effects of salt stress on chloroplast ultrastructure in rice leaves (Oryza sativa L.). – Plant Prod. Sci. 7: 435-441, 2015. 10.1626/pps.7.435 [DOI] [Google Scholar]
- Yan K., He W., Bian L. et al.: Salt adaptability in a halophytic soybean (Glycine soja) involves photosystems coordination. – BMC Plant Biol. 20: 155, 2020. 10.1186/s12870-020-02371-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Li J.L., Liu L.N. et al.: Photosynthetic regulation under salt stress and salt-tolerance mechanism of sweet sorghum. – Front. Plant Sci. 10: 1722, 2020. 10.3389/fpls.2019.01722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Hu S., Wu L. et al.: White stripe leaf 12 (WSL12), encoding a nucleoside diphosphate kinase 2 (OsNDPK2), regulates chloroplast development and abiotic stress response in rice (Oryza sativa L.). – Mol. Breeding 36: 57, 2016. 10.1007/s11032-016-0479-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N., Higuchi M., Kondou Y. et al.: A novel chloroplast protein, CEST induces tolerance to multiple environmental stresses and reduces photooxidative damage in transgenic Arabidopsis. – J. Exp. Bot. 62: 557-569, 2011. 10.1093/jxb/erq290 [DOI] [PubMed] [Google Scholar]
- Yousuf P.Y., Ahmad A., Aref I.M. et al.: Salt-stress-responsive chloroplast proteins in Brassica juncea genotypes with contrasting salt tolerance and their quantitative PCR analysis. – Protoplasma 253: 1565-1575, 2016. 10.1007/s00709-015-0917-z [DOI] [PubMed] [Google Scholar]
- Zhai C.Z., Zhao L., Yin L.J. et al.: Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2 tolerances in Arabidopsis. – PLoS ONE 8: e73989, 2013. 10.1371/journal.pone.0073989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.T., Zhu J.Q., Zhu Q. et al.: Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. – Biochem. Bioph. Res. Co. 390: 469-474, 2009. 10.1016/j.bbrc.2009.09.095 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wei M., Liu A. et al.: Comparative proteomic analysis of two sesame genotypes with contrasting salinity tolerance in response to salt stress. – J. Proteomics 201: 73-83, 2019. 10.1016/j.jprot.2019.04.017 [DOI] [PubMed] [Google Scholar]
- Zhong M., Wang Y., Zhang Y. et al.: Overexpression of transglutaminase from cucumber in tobacco increases salt tolerance through regulation of photosynthesis. – Int. J. Mol. Sci. 20: 894, 2019. 10.3390/ijms20040894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Luo F., Zou R. et al.: Integrated physiological and chloroplast proteome analysis of wheat seedling leaves under salt and osmotic stresses. – J. Proteomics 234: 104097, 2021. 10.1016/j.jprot.2020.104097 [DOI] [PubMed] [Google Scholar]
- Zhu J., Fan Y., Shabala S. et al.: Understanding mechanisms of salinity tolerance in barley by proteomic and biochemical analysis of near-isogenic lines. – Int. J. Mol. Sci. 21: 1516, 2020. 10.3390/ijms21041516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zörb C., Geilfus C.-M., Dietz K.-J.: Salinity and crop yield. – Plant Biol. 21: 31-38, 2019. 10.1111/plb.12884 [DOI] [PubMed] [Google Scholar]
- Zörb C., Schmitt S., Mühling K.H.: Proteomic changes in maize roots after short-term adjustment to saline growth conditions. – Proteomics 10: 4441-4449, 2010. 10.1002/pmic.201000231 [DOI] [PubMed] [Google Scholar]