Abstract
Widespread occurrence of a separate small RNA derived from the 5′-end of 23S rRNA and of an intervening sequence (IVS) which separates this domain from the main segment of 23S rRNA in the α-proteobacteria implies that processing reactions which act to excise the IVS are also maintained in this group. We previously characterized the first example of processing of this IVS in Rhodopseudomonas palustris, which is classified with the Bradyrhizobia. In this case, IVS excision occurs by a multistep process and RNase III appears to act at an early step. Here, we characterize in vivo and in vitro IVS processing in two other related, but phenotypically distinct, Bradyrhizobia. We also examine in vivo and in vitro processing of rRNA precursors from a more distantly related α-proteobacterium, Rhodobacter sphaeroides which produces a separate 5′ 23S rRNA domain but has different sequences in the 5′ 23S rRNA IVS. The details of the in vivo processing of all of the Bradyrhizobial rRNAs closely resemble the R.palustris example and in vitro studies suggest that all of the Bradyrhizobia utilize RNase III in the first step of IVS cleavage. Remarkably, in vivo and in vitro studies with R.sphaeroides indicate that initial IVS cleavage uses a different mechanism. While the mechanism of IVS cleavage differs among these α-proteobacteria, in all of these cases the limits of the internal segments processed in vivo are almost identical and occur far beyond the initial cleavage sites within the IVSs. We propose that these bacteria possess common secondary maturation pathways which enable them to generate similarly processed 23S rRNA 5′- and 3′-ends.
INTRODUCTION
Bacterial 23S rRNAs form highly conserved tertiary structures as a result of conservation of the length and position of RNA helices and single stranded loops (1,2). The sequence variation within these RNA helices may be significant but is almost always the result of base changes on one side of a helix which are accompanied by compensating base changes in the other member of a base pair on the opposite side of the helix (2). Based on sequence comparisons of closely related species, discontinuities in the linear sequence have been detected in certain cases resulting from insertions in the DNA called intervening sequences (IVSs) (3). In most cases, the RNA segment corresponding to the DNA insertion is removed by an RNA processing event, resulting in a break in the covalent structure of the RNA backbone which does not disrupt the RNA tertiary structure because the overall base pairing is conserved (3). Existence of these insertions has been attributed to insensitivity of certain positions in 23S rRNA to insertion and cleavage of the rRNA at these sites has no effect on its function.
The presence of IVSs and their processing from bacterial rRNAs has been described as a ‘sporadic’ phenomenon because there appears to be no phylogenetic link between the studied cases (3). Additionally, certain Salmonella strains have been found to contain multiple rrn operons with some 23S rRNA genes containing, and others lacking, an IVS (3) and in some cases with different IVS sequences at the same position within the rRNA (4). Certain positions in 23S rRNA are known to be especially prone to insertion/deletion processes, and insertions at these sites can be found in unrelated species and with distinctly different IVS sequence content (5–7). Lack of any evidence of evolutionary linkage among species bearing the 23S rRNA IVSs suggests there is little or no adaptive advantage of IVS insertion at these positions (3,4). In one study, recombinant DNA technology was used to place a Salmonella IVS within the 23S rRNA of Escherichia coli, a species which does not normally contain one (8). In agreement with the ideas above, presence of the IVS and ability or failure to process it gave no detectable phenotype (9).
‘Sporadic’ occurrence of bacterial IVSs contrasts with the ubiquitous presence and processing of internal transcribed spacer 2 (ITS2) observed in formation of the cytoplasmic 5.8S rRNA and 5′-end of the 25S–28S rRNA (10,11). In general, it is not possible to trace phylogeny based on the sequence of ITS2 DNA insertions despite the fact that these are found at a common position in the genes for all eukaryotic cytoplasmic rRNA precursors. However, a recent study suggests that the ITS2 processing signals in organisms as distantly related as vertebrates and yeast have common RNA structures (12). Studies in several systems suggest a common processing mechanism involving initial endonuclease cleavages followed by 3′→5′ riboexonuclease activity of the ‘exosome’, which is used to remove this RNA segment from the rRNA precursor (13–15). The importance of ITS2 processing is unclear. Mutations within, or complete deletion of ITS2 affected cell viability and blocked accumulation of the structural rRNAs linked in cis (16,17). However, studies with mutants in genes for enzymes involved in this process have shown that 5.8S rRNAs with unprocessed 3′ or 5′ extensions can still be incorporated into functional ribosomes (18).
Based on the existence of a small DNA insertion in Rhodobacter sphaeroides 23S rRNA genes, presence of an IVS at a site close to, but not identical to, that of the eukaryotic ITS2 was first proposed (19). Our own experiments (20) and those of others (7,21) have demonstrated the widespread occurrence of a processed 5′ 23S rRNA domain of almost identical size in α-proteobacteria as diverse as Rhodopseudomonas sp., Agrobacterium sp., Bradyrhizobia sp., Rhizobium sp. (in the α-2 group) and Rhodobacter sp. (in the α-3 group). All of these species have 5′ IVSs in the 23S rRNA genes. In the case of Rhodopseudomonas palustris 23S rRNA, we have found that purified E.coli RNase III or an R.palustris post-ribosomal supernatant (PRS) which contains the analogous activity, can excise a 35–40 nt segment by cleavage of an RNA duplex within the IVS. However, a much larger segment is removed in vivo, suggesting the existence of secondary maturation processes. This is the first example of an RNase III-initiated processing event within a eubacterial IVS which also removes conserved rRNA sequences that flank it.
Ubiquitous presence of the 5′ IVS in 23S rRNA genes of α-proteobacteria provides a counter-example to the sporadic IVS insertions observed in the γ-proteobacteria and suggests the possibility that the phylogeny of the 5′ 23S rRNA IVS might be traced and that at least some of the reactions involved in processing the IVS are conserved among the α-proteobacteria. Here, we begin this analysis by providing details of this process in other α-proteobacteria. Together, the data suggest that 5′ 23S rRNA IVS processing is widely distributed in α-proteobacteria and while it may not necessarily be initiated by an RNase III-dependent mechanism, secondary maturation processes are likely to produce similar processed ends in diverse species.
MATERIALS AND METHODS
PRS preparation, RNA isolation and analysis
Total RNA was isolated by acidic hot phenol extraction from saturated 100 ml cultures of R.palustris, R.sphaeroides, Rhodobacter capsulatus or Rhodospirillum rubrum cells grown aerobically in YT media or from Bradyrhizobium japonicum, Bradyrhizobium USDA 4362 or USDA 4377 cells grown in minimal media plus yeast extract and mannitol. Methylobacterium extorquens RNA was prepared as above from 250 ml cultures grown in PBY media supplemented with 0.5% methanol. Preparation of a PRS from R.sphaeroides was as previously described for R.palustris (20) while for Bradyrhizobium USDA 4362, PRS was prepared in the same way from 500 ml of cells grown aerobically in minimal media plus yeast extract and mannitol with constant shaking. Electrophoresis, transfer of RNA from denaturing acrylamide gels and northern blotting experiments were as previously described (20). Hybridization was carried out as described (20) at 20°C below the calculated melting temperature with 5′-32P-labeled oligonucleotide probes complementary to positions 44–61 or 503–523 in the R.palustris 23S rRNA sequence (22). Detection of hybridization to labeled probe was with a Fujix BAS2000 Imaging System.
Primer extension
Primer extension and subsequent gel analysis was performed as described previously (20), with 21mer primers at 23S rRNA positions 305–325 and 503–523 complementary to the R.palustris sequence, except that total RNA was prepared from Bradyrhizobia USDA 4362 and USDA 4377 as described above.
S1 mapping
Rhodobacter sphaeroides cosmid 8148 (19) was digested with PvuII and a 3.2 kb fragment containing the rrnA 23S rRNA gene was subcloned into the pBSSK(+) vector (Stratagene, San Diego, CA) which had been EcoRV-digested and dephosphorylated. Probes were obtained by AvrII cleavage of this plasmid and 3′-end labeling with [α-32P]dATP using DNA polymerase Klenow fragment. After cleavage with EcoRV, a 129 bp probe fragment which corresponded to sequences from just within the 5′-end of the 23S rRNA and extending into the 5′ IVS was purified from acrylamide gels by electroelution. Other procedures involving S1 nuclease were as described (20).
RNA linker-mediated cDNA cloning
The technique, including RNA linker, reverse primer sequences and DNA primer used for PCR synthesis of the strand complementary to the cDNA, was performed as described (20) with the modification that total RNA from USDA 4362 and USDA 4377 was used as template. Attempts to apply the same technique to total RNA or the purified 23S rRNA 5′ domain from R.sphaeroides were unsuccessful. Cloning of PCR products was in the pGEM-T vector using the protocol described by the manufacturer (Promega Biotech, Madison, WI).
DNA sequencing
Inserts from 35–40 isolates of each RNA linker-mediated cDNA cloning experiment were sequenced using an ABI377 automated DNA sequencer and dye primer sequencing kit (ABI-Perkin Elmer, Foster City, CA). For PCR-generated clones derived from genomic DNA of Bradyrhizobium USDA 4362, USDA 4377 and M.extorquens, 8–10 isolates were sequenced on both strands, as above.
Purification of RNase III from R.sphaeroides
The purification used 1 l of cells grown aerobically to an OD660 of 1.3 in YE-acid media. Cells were broken by sonication in extraction buffer (23) for 10 min using a 50 s on, 10 s off cycle in an ice-water bath. The sonicate was centrifuged at 10 000 r.p.m. for 10 min and the supernatant was saved. The pellet was re-extracted by resuspension in extraction buffer and a further 6 min of sonication and then centrifuged as above. Supernatants were pooled and then centrifuged at 14 000 r.p.m. for 20 min. The supernatant from this step was centrifuged again at 39 000 r.p.m. for 2.5 h in a Hitachi Type 40 rotor and Hitachi CP100α ultracentrifuge and the resulting PRS served as the starting material for RNase III purification. Ammonium sulfate fractionation and DEAE cellulose column chromatography were as described (23). Fractions containing active RNase III were found in both the low-salt DEAE cellulose column wash and 50 mM KCl elution steps as described (23). Fractions obtained from the low-salt column wash contained significantly lower levels of unrelated nuclease contaminants and these were used for RNase III assays.
In vitro RNA synthesis and processing
For processing in vitro with PRSs of USDA 4362 or R.palustris, 23S rRNA precursor from R.palustris was synthesized from a pSP12 template using SP6 RNA polymerase (Takara) as previously described (20). For comparative studies, genomic DNAs were prepared from 5 ml cultures of R.palustris, M.extorquens, R.sphaeroides, B.japonicum type strain, USDA 4362 and USDA 4377 using a GenomicPrep Kit (Amersham Pharmacia). Internal fragments of 23S rDNA were synthesized by PCR using a forward primer corresponding to positions 43–62 and a reverse primer corresponding to positions 503–523, in the R.palustris 23S rRNA sequence. These primer binding sites are highly conserved in all of the known α-proteobacterial 23S rRNA sequences and were effective for amplification in each case. Plasmid clones were analyzed by restriction mapping and selected to ensure that each was in the same orientation for transcription by T7 RNA polymerase. Several isolates of each were sequenced as described above. Sequences of DNA segments from R.palustris, B.japonicum and R.sphaeroides obtained by this method were identical to previously reported sequences (19,22). To produce the templates for rRNA precursor synthesis, each plasmid was cleaved at a conserved ScaI restriction site found at the same relative position in the 23S rRNA genes of each of these species (position 486 in the R.palustris 23S rRNA sequence). Templates were extracted with phenol and then chloroform, ethanol precipitated, dried and then resuspended in sterile DEPC-treated water. In vitro transcription was carried out with T7 RNA polymerase (Takara) in a 30 µl volume for 1 h using the buffer supplied by the manufacturer. Transcription products were resuspended in DEPC-treated water and pooled. rRNA precursors were purified from 6% acrylamide 8 M urea gels after excision of ethidium bromide-stained bands by shaking overnight at room temperature in 0.5 ml of 0.15 M sodium acetate pH 5.0/1mM EDTA. Processing reactions were carried out for varying times in a 10 µl volume of a buffer containing 20 mM Tris–HCl pH 7.6, 10 mM MgCl2, 100 mM NH4Cl, 5% glycerol at 37°C, as described (20), with the addition of 0.5–1.0 µl his-tagged E.coli RNase III, Bradyrhizobium USDA 4362 or R.palustris PRS or R.sphaeroides RNase III. Reactions were terminated by adding an equal volume of formamide sequencing dyes and heating to 90°C for 3 min. Analysis of processed products on 6.25–7.5% acrylamide gels containing 8 M urea and 0.5× TBE was as previously described (20). For Bradyrhizobium USDA 4362 and USDA 4377 RNAs, primer extension was carried out using the primer complementary to R.palustris 23S rRNA positions 305–325 on RNA processing reactions which were scaled up 4-fold and stopped, after 0 or 30 min, by phenol extraction and ethanol precipitation.
Nucleotide sequence accession numbers
Nucleotide sequences of 23S rRNA segments from Bradyrhizobium USDA 4362, USDA 4377 and M.extorquens have been submitted to GenBank and have been assigned the accession numbers AF23946, AF184910 and AF23945, respectively. Accession numbers of other 23S rRNA sequences from the database are as follows, R.palustris, EMBL X71840, B.japonicum, EMBL X71939, R.sphaeroides, EMBL X53853 (rrnA), R.rubrum, X87290 and E.coli GenBank U00096 (25002–27905, rrlH).
RESULTS
Distribution of the small 5′ 23S rRNA domain in α-proteobacteria
The purple non-sulfur bacteria R.palustris is closely related to the root-nodulating Bradyrhizobia based on 16S rRNA analysis (24,25). Remarkably, a group of bacteria isolated from stem nodules of the host plant and having both photosynthetic and symbiotic characteristics have also been described as Bradyrhizobia (26,27). Unlike R.palustris, the phototrophic Bradyrhizobia carry out aerobic photosynthesis (28). We have recently characterized 16S rRNAs from the phototrophic Bradyrhizobium sp. USDA 4362 and USDA 4377, in order to better understand the phylogenetic affinity of R.palustris to phototrophic and symbiotic Bradyrhizobia (25).
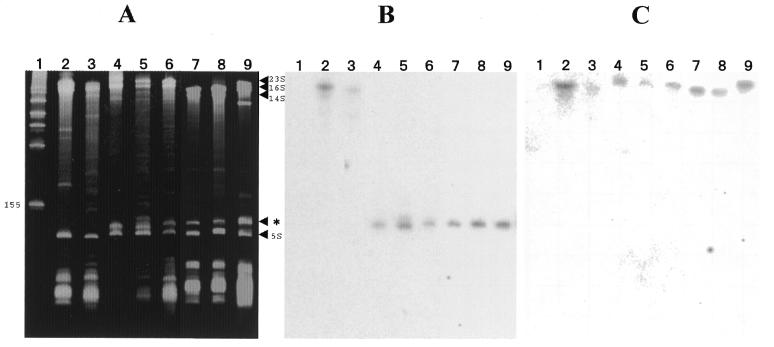
We examined low molecular weight RNA of several strains of α-proteobacteria, including the two species of phototrophic Bradyrhizobia for presence of a separately processed 5′ 23S rRNA domain by denaturing acrylamide gel electrophoresis (Fig. 1A). Total RNA from these species was transferred to a filter and then probed with an oligonucleotide homologous to a conserved region present in the 5′-end of the 23S rRNAs of all eubacteria (Fig. 1B). A low molecular weight RNA is found at the same position (∼125 nt) for each of the Bradyrhizobia that reacts with this probe (Fig. 1A and B). The same result is found with total RNA from R.sphaeroides, R.capsulatus and M.extorquens. Existence of this band and its hybridization with the probe clearly distinguishes the ability of these α-2 and α-3 proteobacteria to process the small 5′ 23S rRNA domain. In contrast, the same probe reacts with the uncleaved 23S rRNAs of the γ-proteobacterium E.coli and R.rubrum, an α-1 proteobacterium. Neither of these species contains an IVS in the 5′-end of its 23S rRNA gene. Failure of high molecular weight rRNAs from the α-2 and α-3 proteobacteria to hybridize to this probe does not indicate that these RNAs were poorly transferred since another downstream probe gives good hybridization with the high molecular weight rRNA from each of these species (Fig. 1C). Rather, as previously suggested for R.palustris (20), in each case the in vivo processing is very rapid and efficient.
Figure 1.
Acrylamide gel electrophoresis and northern hybridization analysis of RNA from E.coli and α-proteobacteria. Lane 1, marker RNA; lane 2, E.coli total RNA; lane 3, R.rubrum total RNA; lane 4, R.palustris purified LSU ribosomes; lane 5, USDA 4362 total RNA; lane 6, USDA 4377 total RNA; lane 7, R.sphaeroides total RNA; lane 8, R.capsulatus total RNA; lane 9, M.extorquens total RNA. (A) 6.5% acrylamide denaturing gel. Positions of 23S rRNA, 16S rRNA, 14S rRNA and 5S rRNA are indicated on the right margin as well as the 23S rRNA 5′ domain (*). Position of the lowest molecular weight marker band is also indicated on the left margin (155 nt). (B) Northern hybridization of a 5′-32P-labeled oligonucleotide complementary to positions 44–61 in R.palustris 23S rRNA to RNA transferred to a nitrocellulose filter from the gel in (A). (C) Northern hybridization of a 5′-32P-labeled oligonucleotide complementary to positions 503–523 in R.palustris 23S rRNA to RNA transferred to a nitrocellulose filter from a gel identical to that in (A). 23S rRNA sequences chosen for synthesis of complementary oligonucleotides in experiments of (B) and (C) are completely conserved in all of the bacterial 23S rRNAs studied here.
Sequence comparison of IVSs from Bradyrhizobia and other species
Hybridization studies in Figure 1 and previous studies (7,20,21) demonstrate that α-proteobacteria that produce separate 5′ 23S rRNA domains possess 5′ 23S rRNA IVSs. In order to study the generality of this phenomenon in all of the species examined in Figure 1 we cloned the IVSs from phototrophic Bradyrhizobia USDA 4362 and USDA 4377, as well as M.extorquens using PCR with oligonucleotides complementary to conserved sequences that flank the IVS and then sequenced the corresponding segments. In the same PCR experiment, as controls, we cloned and confirmed the previously determined sequences of the analogous IVS-containing regions from R.palustris, B.japonicum (22) and R.sphaeroides (19).
Figure 2A shows the alignment of the sequences from the Bradyrhizobia. All of the sequences show strong homologies in the regions flanking the IVS. The IVS is defined as the sequence within the limits of the RNA segment processed by RNase III. The limits of this piece were determined in the previous study with R.palustris (20) and sequences up to this point are nearly identical in the other Bradyrhizobia. Divergence occurs within this segment in R.palustris, B.japonicum and Bradyrhizobium USDA 4377 and USDA 4362.
Figure 2.
(A) Sequence alignment of internal DNA segments from Bradyrhizobial 23S rRNAs cloned by PCR. The origins of the sequences are as indicated on the left side of each line of sequence and the positions within 23S rRNA are shown at the beginning and end of each line. The numbering of bases for Bradyrhizobium USDA 4362 and USDA 4377 23S rRNAs is arbitrary since complete sequences have not been determined in these cases. Identical bases are marked (*). Arrowheads indicate positions of RNase III cleavage sites determined here and from studies with R.palustris (20). In each case, adjacent partially homologous sequences of 11 nt on the left of the RNase III cleavage sites are boxed to indicate potential binding sites for RNase III. On the top and bottom, sets of sequence arrows bracket the sequence representing the range of endpoints (left and right limit IPE) found in the in vivo RNA as determined in this and the previous study. (B) Sequence alignment of internal segments of 23S rRNAs from E.coli and representative α-proteobacteria containing and lacking 5′ IVSs. The numbering for M.extorquens 23S rRNA is arbitrary since a complete sequence has not been determined in this case. Limits of the IPE as determined from studies with R.palustris are shown by brackets and the putative RNase III binding sites are boxed. Also boxed are sequences denoted as L1, R1 and R2 where homology occurs in M.extorquens, R.palustris and R.sphaeroides but not the other species. Arrowheads indicate RNase III cleavage sites determined by primer extension.
Figure 2B shows alignment of sequences from R.palustris which bear the smallest and best characterized IVS in the Bradyrhizobial group and those of more distantly related species. Again, all of the sequences can be aligned based on homology in the regions which flank the R.palustris IVS. The region which corresponds to the R.palustris IVS in M.extorquens is similar in size, but the sequence is unrelated. In the same region, R.sphaeroides bears an even smaller segment of unrelated sequence. In the cases of both R.rubrum and the γ-proteobacterium E.coli, the small size of the corresponding sequences precludes presence of a 5′ 23S IVS. Thus, unlike the Bradyrhizobia (Fig. 2A), it is not possible to discern common sequence features within the regions corresponding to the IVSs in these sequences, even when there is sufficient sequence to propose that an IVS is actually present. Sequences from the 5′-ends of 23S rRNAs from species with the smallest and largest IVSs studied here, R.sphaeroides and B.japonicum type strain, have been modeled (2) and are shown in Figure 3.
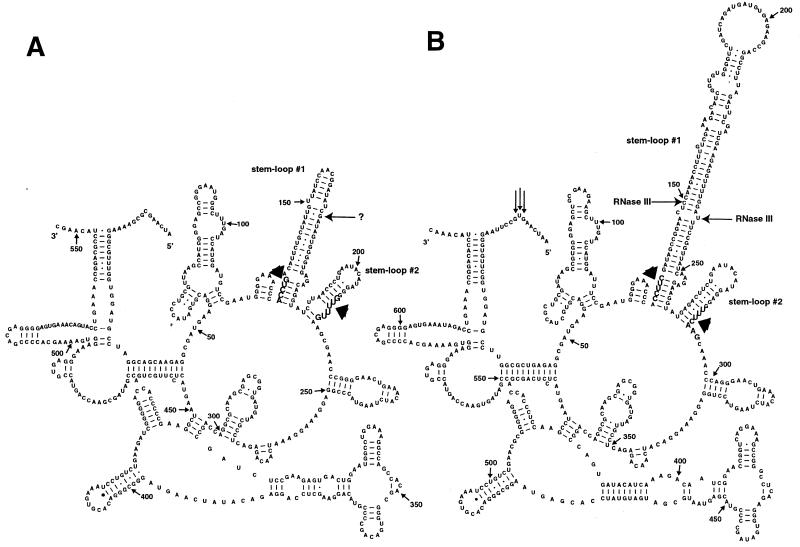
Figure 3.
Structural prediction of RNA segments from the 5′-ends of (A) R.sphaeroides and (B) B.japonicum 23S rRNAs. Shown are 553 and 635 nt from the 5′-ends of R.sphaeroides and B.japonicum 23S rRNAs, respectively, and the 5′- and 3′-ends are marked. Nucleotide positions are indicated numerically with small arrows at 50 base intervals. The 5′-end(s) of the separate 5′ domain determined by primer extension in R.palustris (20) are marked by vertical arrows. Bases indicated with wide arrows in large font and in boldface denote the boundaries of the internally processed segments removed in vivo, as determined by primer extension, S1 mapping and RNA linker ligation-mediated cDNA cloning and sequencing, which includes two stem–loop structures (#1 and #2). In vitro cleavages by E.coli RNase III on the B.japonicum 23S rRNA precursor are shown by bold horizontal arrows on both sides of stem–loop structure #1 in (B). The in vitro RNase III cleavage detected on the right side of stem–loop structure #1 is also the first detectable intermediate in processing of USDA 4362 and USDA 4377 rRNAs. In vivo cleavage by an unspecified nuclease in the R.sphaeroides 5′ IVS is indicated by an arrow (?) on the right side of stem–loop structure #1 in (A). The model used is described previously (2).
Determination of the limits of the internally processed elements in the Bradyrhizobia
Previous results from R.palustris suggest that an RNA segment far larger than the region removed by RNase III is excised by secondary maturation processes (20). We distinguish the smaller IVS from this larger segment which we term an internally processed element (IPE) because the IPE includes conserved sequences which might be predicted to be involved in ribosome function or assembly yet are removed by the processing. We determined the limits of the segments processed in vivo in USDA 4362, USDA 4377 and R.sphaeroides to ascertain if these were similar to each other and also to the case of R.palustris (20).
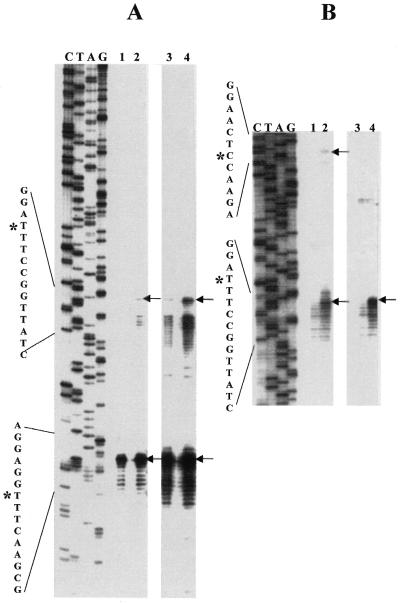
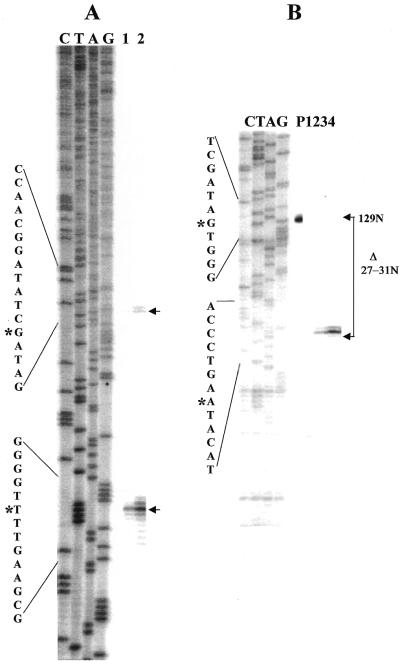
The 5′-ends of the main segments of the 23S rRNAs were characterized by primer extension using a primer complementary to the rRNA segment predicted to be close to the fragmentation point, based on the previous study. The 3′-ends of the small 5′ 23S rRNA domains from the Bradyrhizobia were characterized by RNA linker-mediated cDNA synthesis from total RNA and cloning of PCR fragments followed by DNA sequencing of these clones. Primer extension results for USDA 4377 and USDA 4362 are nearly identical (Fig. 4A, lanes 1 and 2, lower arrow) and demonstrate major end points at identical positions and minor endpoints spanning a region of ∼10 nt downstream of this. This region contains sequences homologous to the previously determined 5′-end of the R.palustris main segment 23S rRNA (20). Interestingly, if the primer extension portion of the gel is overexposed, minor 5′-ends can be detected 35–45 nt upstream of these clusters (Fig. 4A, lanes 3 and 4, upper arrow). Some minor ends are detected within stem–loop #2 but the most 5′ in vivo cleavage coincides with the site cleaved in vitro by E.coli RNase III on the right side of stem–loop #1 (Fig. 4B, lanes 2 and 4, lower arrows). In vivo cleavage at the site predicted to be the left-side RNase III cleavage point in stem–loop #1 is not detected, though weak cleavage can be detected here in vitro (Fig. 4B, lane 2, upper arrow).
Figure 4.
(A) Primer extension identifies the right side of the Bradyrhizobial IVS and also the extent of the IPE. Primer extension analysis of total RNA from phototrophic Bradyrhizobium USDA 4362 and USDA 4377 using a 5′-32P-labeled primer at 23S rRNA positions 305–325 complementary to the R.palustris 23S rRNA sequence. Lefthand lanes (C, T, A, G) are dideoxy sequence marker generated with the same primer and 23S rDNA-containing plasmid DNA from USDA 4377. Since the sequences of USDA 4362 and USDA 4377 are identical in this region, only the USDA 4377 dideoxy sequence marker is presented. Lanes 1 and 2 from the same gel are intentionally overexposed (lanes 3 and 4) to show the presence of low abundance rRNA 5′-ends derived from partially processed 23S rRNA intermediates. DNA sequence around the processing sites is presented on the left margin, with position of the cleavage which generates the first detectable intermediate (*) on the upper sequence and the predominant cleavage which generates completely processed 23S rRNA (*) on the lower sequence. (B) Primer extension was performed on a mixture of rRNA precursors partially processed in vitro with E.coli RNase III. From Bradyrhizobium USDA 4377 (lanes 1 and 2) and Bradyrhizobium USDA 4362 (lanes 3 and 4). In lanes 1 and 3, the RNA processing reactions were terminated immediately by phenol extraction while in lanes 2 and 4, processing reactions were allowed to proceed for 30 min. The first detectable in vivo cleavage [upper arrows in (A)] corresponds precisely to the site cleaved in vitro by RNase III on the right side of stem–loop structure #1 in the Bradyrhizobium USDA 4362 and USDA 4377 rRNA precursors [lower arrows in (B)]. In the case of the Bradyrhizobium USDA 4362 RNase III processing reaction, primer extension products corresponding to the cleavage at the left side of stem–loop structure #1 migrate more slowly and have not entered this portion of the gel.
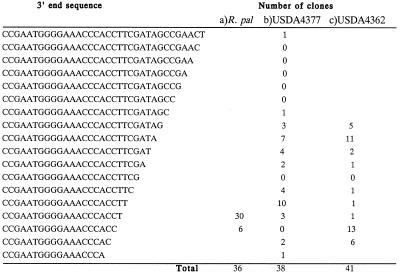
The sequences of RNA linker-mediated RT–PCR derived clones from the 3′-ends of the small 5′ 23S rRNA domain from Bradyrhizobium USDA 4362 and Bradyrhizobium USDA 4377 are shown in Figure 5. In each case we find RNA with major 3′ endpoints corresponding closely to that determined with gel-purified RNA from LSU ribosomes of R.palustris (ACC/ACCT). We believe this represents the fraction derived from ribosomes. We also find endpoints further downstream indicative of incompletely processed RNA from both USDA 4362 and USDA 4377, with 5–8 base 3′ extensions. If the furthest extended 3′-ends are considered, these are very close to the RNase III cleavage site on the left side of stem–loop structure #1. Thus, unlike the previous experiments which employed highly purified RNA from R.palustris ribosomes for characterization, with an unfractionated sample, rRNA precursors are found indicative of processing at both the 5′-ends of the main 23S rRNA piece and at the 3′-end of the small 5′ domain.
Figure 5.
3′-End determination of the small separate 5′ 23S rRNA domain from sequence analysis of RNA linker ligation-mediated cDNA cloning using total RNA from Bradyrhizobium USDA 4362 and USDA 4377 or gel-purified low molecular weight RNA from LSU ribosomes of R.palustris. (a) For R.palustris, a larger sample of clones was examined than in the initial analysis (20) though the same highly purified RNA preparation was used as template. (b) Total RNA from USDA 4377 and (c) USDA 4362 was used as template. Two major clusters of RNA 3′ endpoints are detected in each of these cases corresponding to incompletely and completely processed rRNA. The incompletely processed RNA 3′-ends regions lie close to the RNase III cleavage sites on the left side of stem–loop structure #1 mapped previously by primer extension using in vitro processed R.palustris precursors (20). Putative mature 3′-ends from USDA 4377 and USDA 4362 RNAs correspond closely to those from RNA isolated directly from R.palustris LSU ribosomes (A).
Primer extension employing the same primer as above enables detection of major and minor cDNA products indicative of the mature 5′-ends of the main segment of R.sphaeroides 23S rRNA and incompletely processed products cleaved within the 5′ IVS, respectively (Fig. 6A). Unlike the results with Bradyrhizobia, minor IVS cleavage products were sufficiently abundant to be detected without long exposure of the autoradiograph. For reasons that are not understood, the RNA linker-mediated RT–PCR method could not be applied to R.sphaeroides. S1 mapping was therefore used for 3′-end determination of the small 5′ rRNA domain (Fig. 6B). The results show a cluster of major 3′-ends spanning a 5-nt region.
Figure 6.
Defining the limits of the internally processed segment in R.sphaeroides. (A) Primer extension identifies 5′-end of the main segment of R.sphaeroides 23S rRNA and also the location of a processing site in the IVS. Primer extension analysis of total RNA from R.sphaeroides using a 5′-32P-labeled primer at 23S rRNA positions 305–325 complementary to the R.palustris 23S rRNA sequence. Lefthand marker lanes (C, T, A, G) are as described above except R.sphaeroides rDNA was used as template for synthesis. In lanes 1 and 2, 0.5 and 1.5 µg of total RNA were used as template for cDNA synthesis. DNA sequence around the processing sites is presented on the left margin, with position of the cleavage which generates the first detectable intermediate marked by asterisks on the upper sequence and the predominant cleavage which generates completely processed 23S rRNA by an asterisk on the lower sequence. (B) S1 mapping of the 3′-end of the small 5′ 23S rRNA domain. S1 cleavage products are aligned with the same sequence marker as in (A) which is used here only as a size standard. Position of the full-length probe within the sequence is shown by an asterisk in the upper portion of the sequence on the left margin and the position of the most abundant S1 cleavage product by an asterisk on the lower sequence. Brackets on the right margin show the change in size of probe due to S1 cleavage (Δ27–31 nt). Lanes are P, undigested probe; 1, no RNA added; 2, 0.25 µg purified 5′ domain added; 3, 0.5 µg purified 5′ domain added; 4, 1.0 µg purified 5′ domain added.
In Figures 2 and 3, the cleavage sites mapped by primer extension, S1 mapping, RNA linker ligation/cDNA cloning and sequencing with 23S rRNAs from R.sphaeroides, R.palustris and the other Bradyrhizobia sp. are indicated.
Processing of IVSs in vitro with RNase III
Earlier studies in the γ-proteobacteria (3,4,9) have demonstrated that RNase III alone can completely process IVSs by cleavage in duplex RNA segments both in vitro and in vivo. Our previous study with R.palustris (20) suggests that RNase III cleavage of the IVS is only the first step needed to process the separated 5′ 23S rRNA domain and that the cleavage specificity of the E.coli RNase III enzyme was almost identical to that of a PRS of R.palustris. We assume that the cleavage specificity of PRSs from the other Bradyrhizobia will also mimic that of E.coli RNase III since identical or nearly identical sequences to those found in the R.palustris 5′ IVS are found at analogous positions in the IVSs of these species (Fig. 2A). In support of this hypothesis, we have found that a 5′ IVS-containing 23S rRNA precursor from R.palustris can be cleaved with a PRS derived from Bradyrhizobium USDA 4362 to give a pattern of RNA fragments nearly indistinguishable from the homologous extract and closely resembling that found using purified E.coli RNase III (data not shown).
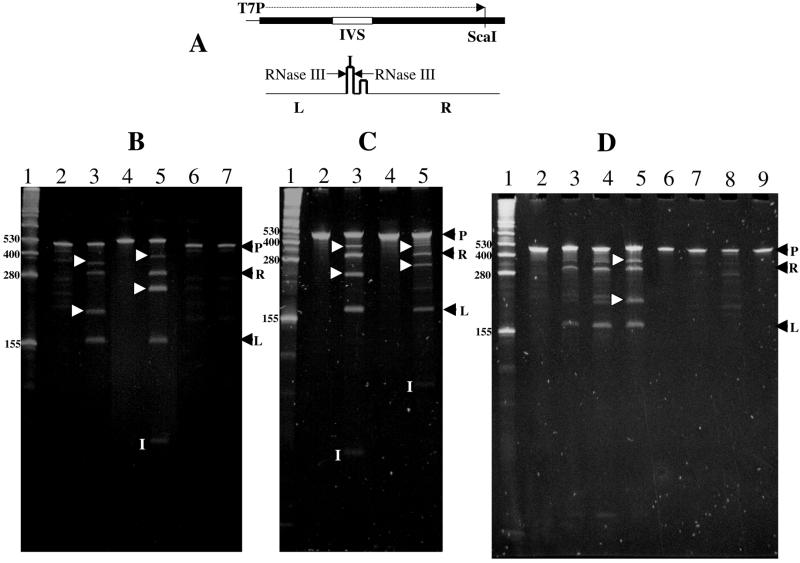
We asked whether precursor RNAs from several of the IVS-bearing species examined in Figure 1 might be specifically cleaved by RNase III. Cloning of the PCR fragments from various α-proteobacteria in the same orientation in the pGEM-T vector downstream of the T7 promoter allows precursor RNAs containing the 5′ IVSs to be transcribed in vitro (Fig. 7A). Templates were cut with the restriction enzyme ScaI at a site conserved within all of the respective 23S rRNA genes and, with the exception of a small 5′ RNA segment derived from T7, run off transcripts containing only internal segments of 23S rRNA were produced. RNA precursors were gel purified and a pure preparation of E.coli RNase III was used as a diagnostic reagent to study specific cleavage of each precursor in Figure 7B and C. Incubations with RNase III were carried out for short periods to generate intermediates as well as products. Identity of each band was established by carrying digestions to completion (data not shown).
Figure 7.
RNA processing of 23S rRNA precursors of Bradyrhizobia and R.sphaeroides in vitro using RNase III purified from E.coli or R.sphaeroides. (A) Structure of DNA region and transcripts derived from plasmids bearing cloned PCR fragments from 23S rRNA genes of various α-proteobacteria. Upper portion shows the structure of a representative template containing an internal segment from 23S rRNA which bears the 5′ IVS. PCR fragments were made with primers complementary to conserved regions of 23S rRNA and are represented by the broad black line. DNA fragments derived from PCR synthesis on R.palustris, Bradyrhizobium USDA 4362, R.sphaeroides, Bradyrhizobium USDA 4377 and B.japonicum genomic DNA templates were cloned in the pGEM-T vector. Plasmid DNAs were cleaved at the ScaI site found in all of these 23S rRNA genes and transcribed in vitro from the T7 promoter on the vector portion (T7P) using T7 RNA polymerase. Lower line shows the proposed in vitro processing pathway of the IVS-containing precursor in those cases where RNase III cleavage occurs. Non-concerted cleavage by RNase III at sites indicated by the arrows in stem–loop #1 produces RNA fragments from the left side (L), right side (R) and the IVS (I). Intermediates (L+I) and (R+I) are produced in unequal amounts in each case due to variable cleavage on either side of stem–loop #1. (B) Ethidium bromide-stained 6.25% acrylamide gel showing electrophoretic separation of the products of processing reactions carried out for 0 h (lanes 2, 4 and 6) and 2 h (lanes 3, 5 and 7) in the presence of 0.5 µl his-tagged E.coli RNase III. Lane 1, RNA marker (Gibco-BRL); lanes 2–3, R.palustris 23S rRNA precursor; lanes 4–5, Bradyrhizobium USDA 4362 rRNA precursor; lanes 6–7, R.sphaeroides 23S rRNA precursor. (C) Ethidium bromide-stained 7.5% acrylamide gel showing electrophoretic separation of the products of processing reactions carried out for 0 h (lanes 2 and 4) and 2 h (lanes 3 and 5) in the presence of 0.5 µl his-tagged E.coli RNase III. Lane 1, as above; lanes 2–3, Bradyrhizobium USDA 4377 rRNA precursor; lanes 4–5, B.japonicum 23S rRNA precursor. (D) Ethidium bromide-stained 7.5% acrylamide gel showing electrophoretic separation of the products of processing reactions carried out for 0 min (lanes 2 and 6), 10 min (lanes 3 and 7) and 30 min (lanes 4 and 8) in the presence of 1.0 µl partially purified R.sphaeroides RNase III or 30 min with 1.0 µl his-tagged E.coli RNase III (lanes 5 and 9). In (B), (C) and (D), RNA marker sizes are indicated on the left margin of the gel and completely processed products are indicated on the right margin (R and L). These are derived by cleavage as shown in (A). P, position of the rRNA precursor in each case; I [in white within (B), (C) and (D)], IVS-containing cleavage products from USDA 4362, USDA 4377 B.japonicum and R.palustris from left to right, respectively. Products proposed to be processing intermediates resulting from non-concerted RNase III cleavage (L+I and R+I) based on their size, run at positions between bands L and R and between R and P, respectively, and are marked by white arrowheads on the side of lanes 3 and 5 in (B) and (C) and lane 5 in (D).
All of the RNA substrates are cleaved by E.coli RNase III except for that of R.sphaeroides (Fig. 7B) and in every case where cleavage occurs, very similar gel patterns are observed. The RNase III cleavage pattern resembles that previously determined on an R.palustris rRNA precursor synthesized by SP6 RNA polymerase from another template (20). Due to variability in the size of the DNA insertion within the boundaries of the IVS, variable sized IVS-containing fragments (I in Fig. 7B and C) of ∼90, 85 and 60 nt are produced from B.japonicum, USDA 4362 and USDA 4377 precursors, respectively, by RNase III cleavage. The sizes of these fragments are in close agreement with the predicted sizes of the IVSs based on sequence alignment (Fig. 2A). The IVS-containing fragment from R.palustris is too small (40 nt) to be visualized on these gels but its presence can be demonstrated indirectly by sizing of intermediate cleavage products (see below) and with primer extension reactions performed on a mixture of these intermediates (20). Two RNA fragments of constant size (∼160 and 295 nt) are also produced by cleavage of each of the rRNA precursors (L and R in Fig. 7A–D). This is easily understood since each 23S rDNA was amplified using primers complementary to a common sequence present in each 23S rRNA and the RNase III cleavage sites lie at the same relative position within each precursor. Other variable sized bands are also produced whose size is the sum of the right or left end fragments plus the IVS fragment (R+I or L+I in each case).
The presence of these additional variable size fragments and their non-stoichiometric yield indicates that non-concerted cleavage occurs at RNase III sites which are located at the same positions with respect to the ends of each of the precursor RNAs. Non-concerted cleavage by RNase III at the cleavage site within the R.palustris IVS has previously been characterized in detail (20). Other characterized cases where non-concerted RNase III cleavage occurs have implicated unpaired bases in the RNA duplex around the site of cleavage (29) and this also true for all of the Bradyrhizobia, where the site of mispairing is conserved in these sequences (Figs 2A and 3B).
Computer modeling of the R.sphaeroides IVS sequence (Fig. 3A) reveals that the size of stem–loop structure #1 is close to the predicted limit of substrates bound by RNase III (29). Failure of E.coli RNase III to cleave within this 5′ IVS might therefore be explained by failure to bind. Alternatively, this enzyme might possess a different specificity versus R.sphaeroides RNase III. To address the question of cleavage specificity, we purified RNase III (23) from a PRS of R.sphaeroides. Purification was necessary because the crude PRS caused non-specific breakdown of the R.sphaeroides (and other) rRNA precursors. The R.sphaeroides enzyme preparation was used to study cleavage of rRNA precursors from R.sphaeroides and R.palustris (Fig. 7D). We find that the heterologous R.palustris precursors are cleaved by the R.sphaeroides enzyme preparation to yield fragments almost identical to that generated by pure E.coli RNase III (Fig. 7D, lanes 2–5), but those from R.sphaeroides are not (Fig. 7D, lanes 6–9). With the R.sphaeroides rRNA precursor, only minor non-specific cleavage products, not derived from the IVS, are obtained. Changing the ionic conditions did not alter the specificity (data not shown).
DISCUSSION
In this paper, we have presented evidence for a common mechanism of 5′ 23S rRNA IVS processing in a group of closely related, but phenotypically distinct Bradyrhizobia. All of these species have conserved RNA duplex sequences in stem–loop #1 and a conserved site of mispairing at a position previously determined to be the RNase III cleavage point in the R.palustris 23S rRNA precursor (20). Presence of mispaired bases at the cleavage point has been demonstrated to strongly affect the mechanism of cleavage without influencing binding of RNase III (30). We believe that all of these species employ an RNase III enzyme of similar or identical specificity to cleave at the homologous sites. In agreement with this hypothesis, cleavage specificity on an R.palustris rRNA precursor of PRSs from USDA 4362, R.palustris and purified E.coli RNase III are similar (data not shown). In addition, minor rRNA cleavage products are detected in vivo at precisely the same points determined for in vitro cleavage with E.coli RNase III (Fig. 4). These observations suggest that the first step in Bradyrhizobial 23S rRNA 5′ IVS processing is duplex cleavage by RNase III similar to that described in several other cases for IVSs excised from other positions in 23S rRNA in other bacterial groups (3,6,8,9).
Hybridization studies shown in Figure 1 demonstrate that the small RNA from M.extorquens, R.sphaeroides and R.capsulatus which co-migrates with the 5′ 23S rRNA domain of R.palustris represents the cognate 5′ 23S rRNA domain from these species. The sequence and structure of the 5′ IVS in the Rhodobacter species (19,31) and M.extorquens is distinctly different from that of the Bradyrhizobia (Figs 2B, 3A and data not shown). In the case of R.sphaeroides the RNA duplex of stem–loop structure #1 is small (Fig. 3A) and is not cleaved by purified E.coli RNase III (Fig. 7B) nor by RNase III from R.sphaeroides (Fig. 7D). RNase III enzymes generally cleave both sides of an RNA duplex of sufficient length at a well defined site with a defined polarity (29). In vivo cleavage of the R.sphaeroides 5′ IVS (Fig. 3A) occurs on the right side, close to the top of stem–loop #1, at a cleavage site that is atypical of RNase III enzymes based on cleavage of other similar stem–loop structures (29). A heterologous RNase III enzyme has been claimed to cleave in vitro, at a position close to this in a related R.sphaeroides 23S rRNA precursor (21). Our observations with the homologous enzyme suggest that R.sphaeroides 23S rRNA 5′ IVS cleavage is accomplished by an activity different from RNase III.
Due to the large internal RNA segment which is removed in vivo in the case of R.palustris, we have previously proposed the existence of secondary RNA processing events which act to mature the RNA ends after RNase III cleavage at an early step (20). We find that regardless of the size of the insertion within the boundaries of the RNase III-like cleavage sites and of potentially highly structured RNA within this segment, all of these rRNAs undergo a similar secondary maturation process, resulting in removal of the intervening RNA. The nature of these secondary maturation events is unclear and the enzymology of the process has so far not been studied. Low levels of in vivo rRNA precursors are detected by primer extension around the position of the cleavage sites which produce the primary 5′-ends of the main 23S rRNA segments (Figs 4A and 6A). This suggests that the secondary processing reaction(s) initiated at this site are likely to be very fast and efficient. An unusual and discontinuous fine structure is seen in the primer extension results with the Bradyrhizobia suggesting differential susceptibility of RNA within the IPE to the secondary processing enzyme(s). We speculate that strict conservation of mispairing at the Bradyrhizobial RNase III cleavage sites provides a substrate for subsequent secondary processing events which might utilize single-stranded RNA ends.
Several consistent phylogenetic trees of the α-proteobacteria have been presented based on 16S rRNA sequences (32,33). The purple non-sulfur bacterium R.rubrum (α-1) has been proposed to be representative of a primitive branch of this subdivision and is thought to be a progenitor of all other members, which include the later arising Azospirillum (α-1) and Rhodobacter (α-3) groups. The major species of the α-2 group including R.palustris and M.extorquens are proposed to have evolved later from a Rhodobacter-like ancestor. Based on this scheme, Rhodobacter is the earliest arising species bearing the 5′ 23S rRNA IVS and as shown in Figure 1, both R.capsulatus and R.sphaeroides 23S rRNAs are processed at the 5′-end. The more primitive R.rubrum bears no 5′ 23S rRNA IVS (Fig. 2B) and, as expected (Fig. 1), the 23S rRNA is not processed at the 5′-end. Sequences, sizes and structures of the 5′ 23S rRNA IVSs show great diversity among α-proteobacterial species. Failure of RNase III from R.sphaeroides to cleave its own 5′ 23S rRNA IVS could indicate that this bacterium employs an RNase III-independent mechanism for cleavage of this IVS. Acquisition of a site susceptible to RNase III cleavage within the 5′ IVS may have evolved later in the α-2 proteobacteria. There may be other, perhaps more ancient, activities in α-3 proteobacteria and perhaps also in α-2 proteobacteria which can also cleave in or around the 5′ IVS. These might be among the same activities which carry out secondary maturation in the Bradyrhizobia.
We searched for common sequence motifs and potential secondary structures within the IPE, outside of the limits of the region cleaved by RNase III, which might serve as signals in the secondary maturation processes. Interestingly, there are two clusters of bases which are conserved in all of these species (Fig. 2B) and in all α-proteobacteria which have been demonstrated to process the 5′ IVS (data not shown). These clusters, L1 and R1 (Fig. 2B) form the duplex at the base of stem–loop structure #1. Rhodospirillum rubrum, an α-proteobacteria which does not have a 5′ IVS and does not produce a processed 5′ 23S rRNA domain has a different sequence here but still is predicted to form an analogous stem–loop. In addition, a second site of mispairing exists in the base of stem–loop structure #1 (Fig. 3A and B) which consistently presents a loop of 3–4 bases on the right side. On the right side of the IPE, a 5-nt sequence, R2 (GGUUU) is found in the right side of stem–loop structure #2. Weak base pairing or unpaired bases are also found at the base of this stem–loop structure in those α-proteobacteria which process the IPE (Fig. 3A and B and data not shown). This contrasts with the situation in E.coli and R.rubrum where perfect base pairing is found at the same position. The determinants of 5′ 23S rRNA IVS processing beyond cleavage of the RNA duplex of stem–loop structure #1 are unclear since the sequence differences between those species that carry out the processing and those that do not are quite small.
When in vitro synthesized RNA precursors are treated with PRSs prepared from various species of Bradyrhizobia, only the RNase III-like activity is manifest and complete processing to the endpoints observed in vivo is not observed (20 and data not shown). This could suggest that some of the maturation occurs during the process of ribosome assembly. Studies of secondary rRNA maturation processes at the 5′-end of 23S rRNA in E.coli (34) have presented the idea that the true substrate of these reactions is a ribonucleoprotein complex. Early studies on feedback regulation of ribosomal protein synthesis hypothesized that RNA degradation activities were associated with these proteins but this idea was later discarded (35,36). We speculate that the minor sequence differences present in the excised 5′ segments from α-2 and α-3 proteobacterial 23S rRNAs are conserved because they are somehow essential to RNA cleavage processes involved in ribosome assembly. These sequences, perhaps combined with novel properties of the ribosomal proteins in the same species of α-proteobacteria, may contribute either directly or indirectly to the RNA cleavage reactions.
While a DNA glycosylase activity has been described for the eukaryotic S3 protein (37), ribonuclease activities associated with other ribosomal proteins have not been described. Database searches reveal that ribosomal protein L24, which is known to bind in this region of the 23S rRNA (38), has a C-terminal extension of seven amino acids in the α-2 proteobacteria R.palustris and Rickettsia prowazekii, as well as other unusually conserved amino acids at the C-terminus. Determining whether these residues play a role in RNA cleavage, linking RNA segments separated by the cleavage or both, will be an experimental challenge for the future.
Acknowledgments
ACKNOWLEDGEMENTS
Dr R. Gutell provided Postscript files of projections of the eubacterial LSU rRNA structures. We thank Dr A. Nicholson for providing clones of his-tagged E.coli RNase III and helpful discussions and S. Shimokawa for help with DNA sequencing. Drs J. H. Roh and S. Kaplan provided R.sphaeroides cosmid 8148. This work was supported by the New Energy and Industrial Technology Development Organization.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +81 774 75 2308; Fax: +81 774 75 2321; Email: yukawa@rite.or.jp AF23946, AF184910, AF23945
REFERENCES
- 1.Egeberg J., Larsen,N. and Garrett,R.A. (1990) In Hill,W.E., Dahlberg,A., Garrett,R.A., Moore,P.B., Schlessinger,D. and Warner,J.R. (eds), The Ribosome: Structure, Function and Evolution. American Society for Microbiology, Washington, DC, pp. 168–179.
- 2.Gutell R. (1996) In Zimmerman,R.A. and Dahlberg,A.E. (eds), Ribosomal RNA, Structure, Evolution, Processing and Function in Protein Biosynthesis. CRC Press, Boca Raton, FL, pp. 111–128.
- 3.Burgin A.B., Parados,K., Lane,D.J. and Pace,N.R. (1990) Cell, 60, 405–414. [DOI] [PubMed] [Google Scholar]
- 4.Mattatall N.R. and Sanderson,K.E. (1996) J. Bacteriol., 178, 2272–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trust T., Logan,S., Gustafson,C.E., Romaniuk,P.J., Kim,N.W., Chan,V.L., Ragan,M.A., Guerry,P. and Gutell,R.R. (1994) J. Bacteriol., 176, 4597–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kordes E., Jock,S., Fritsch,J., Bosch,F. and Klug,G. (1994) J. Bacteriol., 176, 1121–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selenska-Pobell S. and Evguenieva-Hackenberg,E. (1995) J. Bacteriol., 177, 6993–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory S.T., O’Connor,M. and Dahlberg,A.E. (1996) Nucleic Acids Res., 24, 4918–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattatall N.R. and Sanderson,K.E. (1998) FEMS Microbiol. Lett., 159, 179–185. [DOI] [PubMed] [Google Scholar]
- 10.Walker T.A. and Pace,N.R. (1983) Cell, 33, 320–322. [DOI] [PubMed] [Google Scholar]
- 11.Gray M.W. and Schnare,M.N. (1990) In Hill,W.E., Dahlberg,A., Garrett,R.A., Moore,P.B., Schlessinger,D. and Warner,J.R. (eds), The Ribosome: Structure, Function and Evolution, American Society for Microbiology, Washington, DC, pp. 589–597.
- 12.Joseph N., Krauskopf,E., Vera,M.I. and Michot,B. (1999) Nucleic Acids Res., 27, 4533–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell P., Petfalski,E. and Tollervey,D. (1996) Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 15.Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999) Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeney R. and Yao,M.C. (1989) EMBO J., 8, 4169–4175. [Google Scholar]
- 17.Musters R.H., Planta,R.J., van Heerikhuizen,H. and Raue,H.A. (1990) In Hill,W.E., Dahlberg,A., Garrett,R.A., Moore,P.B., Schlessinger,D. and Warner,J.R. (eds), The Ribosome: Structure, Function and Evolution. American Society for Microbiology, Washington, DC, pp. 435–442.
- 18.Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dryden S.C. and Kaplan,S. (1990) Nucleic Acids Res., 18, 7267–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahn K., Inui,M. and Yukawa,H. (1999) Nucleic Acids Res., 27, 2741–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evguenieva-Hackenberg E. and Klug,G. (2000) J. Bacteriol., 182, 4719–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Springer N., Ludwig,W. and Hardarson,G. (1993) Syst. Appl. Microbiol., 16, 468–470. [Google Scholar]
- 23.Robertson H.D. (1990) Methods Enzymol., 181, 189–202. [DOI] [PubMed] [Google Scholar]
- 24.Yanagi M. and Yamasato,K. (1993) FEMS Microbiol. Lett., 107, 115–120. [DOI] [PubMed] [Google Scholar]
- 25.Inui M., Roh,J.H., Zahn,K. and Yukawa,H. (2000) Appl. Environ. Microbiol., 66, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young J.P.W., Downer,H.L. and Eardly,B.D. (1991) J. Bacteriol., 173, 2271–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleischman D.E., Evans,W.R. and Miller,I.M. (1995) In Blankenship,R.E., Madigan,M.T. and Bauer,C.E. (eds), Anoxygenic Photosynthetic Bacteria. Kluwer Academic, Dordrecht, pp. 123–136.
- 28.Ladha J.K. and So,R.B. (1994) Int. J. Syst. Bacteriol., 44, 62–73. [DOI] [PubMed] [Google Scholar]
- 29.Nicholson A.W. (1996) Prog. Nucleic Acid Res. Mol. Biol., 52, 1–65. [DOI] [PubMed] [Google Scholar]
- 30.Li H. and Nicholson,A.W. (1996) EMBO J., 15, 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 31.Hopfl P., Ludwig,W. and Schleifer,K.H. (1988) Nucleic Acids Res., 16, 2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woese C.R. (1987) Microbiol. Rev., 51, 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen G.J., Woese,C.R. and Overbeek,R. (1994) J. Bacteriol. 176, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King T.C. and Schlessinger,D. (1987) In Niedhardt,F.C. (ed.), Escherichia coli and Salmonella typhimurium, Vol. 1. American Society for Microbiology, Washington, DC, pp. 703–718.
- 35.Yates J.L. and Nomura,M. (1980) Cell, 21, 517–522. [DOI] [PubMed] [Google Scholar]
- 36.Nomura M. (1990) In Hill,W.E., Dahlberg,A., Garrett,R.A., Moore,P.B., Schlessinger,D. and Warner,J.R. (eds), The Ribosome: Structure, Function and Evolution. American Society for Microbiology, Washington, DC, pp. 3–55.
- 37.Wilson D.M. III, Deutsch W.A. and Kelley,M.R. (1994) J. Biol. Chem., 269, 25359–25364. [PubMed] [Google Scholar]
- 38.Østergaard P., Phan,H., Johansen,L.B., Egeberg,J., Østergaard,L., Porse,B.T. and Garrett,R.A. (1998) J. Mol. Biol., 284, 227–240. [DOI] [PubMed] [Google Scholar]