Abstract
Phosphorylation of nucleic acids is an indispensable process to repair strand interruption of nucleic acids. We have studied the process of phosphorylation using molecular beacon (MB) DNA probes in real-time and with high selectivity. The MB employed in this method is devised to sense the product of a ‘phosphorylation–ligation’ coupled enzyme reaction. Compared with the current assays, this novel method is convenient, fast, selective, highly sensitive and capable of real-time monitoring in a homogenous solution. The preference of T4 polynucleotide kinase (T4 PNK) has been investigated using this approach. The results revealed that a single-stranded oligonucleotide containing guanine at the 5′ termini is most preferred, while those utilizing cytosine in this location are least preferred. The preference of (T)9 was reduced greatly when phosphoryl was modified at the 5′ end, implying that T4 PNK could discern the phosphorylated/unphosphorylated oligonucleotides. The increase of oligonucleotide DNA length leads to an enhancement in preference. A fast and accurate method for assaying the kinase activity of T4 PNK has been developed with a wide linear detection range from 0.002 to 4.0 U/ml in 3 min. The effects of certain factors, such as NTP, ADP, (NH4)2SO4 and Na2HPO4, on phosphorylation have been investigated. This novel approach enables us to investigate the interactions between proteins and nucleic acids in a homogenous solution, such as those found in DNA repair or in drug development.
INTRODUCTION
The phosphorylation of the 5′-hydroxyl termini of nucleic acids plays an important role in nucleic acid metabolism and is indispensable to for the repair of nucleic acids during strand interruption (1). The nucleic acids lesions caused by some endogenous or exogenous agents, such as nucleases, as well as ionizing radiation and chemical agents, usually possess 5′-hydroxyl termini. These situations lead to the failure of 5′-phosphate terminal dependent repair processes for nucleic acids and result in serious consequences (1,2). Phosphorylation of 5′-hydroxyl termini is an inevitable occurrence in many nucleic acid repair mechanisms. Since the first polynucleotide kinase was discovered over 30 years ago, the T4 polynucleotide kinase was found to be the RNA repair enzyme involved in the T4 bacteriophage infection process (2,3). At present, the polynucleotide kinase is known to be widespread in cells and also considered to be a putative DNA repair enzyme that phosphorylates the 5′-hydroxyl of DNA and RNA (1–6). Traditionally, radical isotope 32P-labeling, (denature) PAGE and autoradiography are used to assay the phosphorylation of nucleic acids (1–8). Unfortunately, these approaches are complex, time consuming and discontinuous, and incapable of analyzing a rapid, continuous phosphorylation processes. Therefore, other avenues need to be explored to have a convenient, rapid and accurate method to investigate the phosphorylation of nucleic acids in real time.
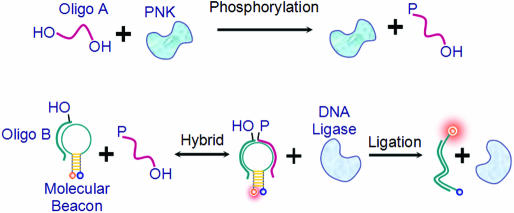
Here, a novel method has been developed for the real-time monitoring of the phosphorylation process using molecular beacon (MB) DNA probes with high sensitivity, excellent selectivity and convenience. MBs are hairpin-shaped nucleic acid probes that possess high selectivity and excellent sensitivity (9–14). MBs have been used in the investigation of genetic disease (15), studies of DNA–protein interactions (16,17), the monitoring of the nucleic acids ligation process (18) and the detection of DNA/RNA hybridization in living cells (19). Our approach for real-time phosphorylation monitoring takes advantage of the inherent single transduction mechanism of MBs as shown in Figure 1.
Figure 1.
Schematic representation for monitoring the phosphorylation of nucleic acids. Top: the Oligo A with 5′-hydroxyl is phosphorylated by polynucleotide kinase. Bottom: the nick formed by Oligo B and phosphorylated Oligo A hybridizing with MB can be sealed by DNA ligase, which results in a fluorescence enhancement of the MB probe.
In Figure 1, the Oligo A, which contains a 5′-hydroxyl group, can be phosphorylated and, together with Oligo B, can be hybridized to the MB to form a ligatable nick. In the presence of DNA ligase, oligonucleotides can be ligated to open the MB, thereby restoring fluorescence. In this assay, the MB plays the role of an information converter, specifically transferring the information that an ‘oligo has been phosphorylated’ into fluorescent signal changes. Thus, phosphorylation of a nucleotide can be monitored in real time using this principle (20). Utilizing the excellent selectivity and high sensitivity of an MB and the rapid ligation feature of DNA ligase, this approach can offer selective, sensitive and simultaneous information about the phosphorylation process.
MATERIALS AND METHODS
Oligonucleotides and reagents
The MBs and oligonucleotides were synthesized by Dalian Takara Bio Inc. (Dalian, China), and T4 DNA ligase, Escherichia coli DNA ligase, T4 polynucleotide kinases were purchased from the same company. Please see the unit definition of these enzymes in Supplementary Material. The index and sequences of the MBs and oligonucleotides are listed in Table 1.
Table 1.
MB and target oligonucleotide sequences
| Index | Sequence (5′–3′) |
|---|---|
| MB1 | (TMR)-CCTCTC CGTGTCTTGTACTTCCCGTCA GAGAGG-(DABCYL) |
| c-DNA | GACGGGAAGTACAAGACAC |
| Oligo A | p-GACGG GAAG |
| Oligo B | TACAA GACAC |
| Oligo C | CACAA GACAC |
| Oligo D | AACAA GACAC |
| Oligo E | GACAA GACAC |
| Oligo F | p-TACAA GACAC |
| Oligo G | p-TTTTTT TTTT |
| Oligo H | TTTTTT TTTT |
| Oligo I | GTTTTT TTTT |
| Oligo J | ATTTTT TTTT |
| Oligo K | CTTTTT TTTT |
| Oligo L | TTTTTT T |
| Oligo M | TTTTTT TTTTT TTTTT |
The stem part of MB1 is spelt out in bold characters. The p in Oligo A, Oligo F and Oligo G represents the phosphate at 5′ end.
Sampling and fluorescence measurement
The two ligation systems employed in our experiments were T4 DNA ligase and E.coli DNA ligase. The buffer for the T4 DNA ligase contained 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2 and 1.0 mM ATP, while the E.coli DNA ligase buffer was composed of 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2 and 0.1 mM NAD. All of the samples were prepared in 80 μl aliquots for all experiments. All fluorescence measurements of the samples were carried out on a fluorometer F2500 (Hitachi, Japan) with excitation at 521 nm and emission at 578 nm for TMR labeling at 5′ end of MB1 as a fluorophore. Each sample was incubated at 37°C for 8 min to obtain equilibrium. After the addition of DNA ligase, the sample was incubated until the fluorescence reached equilibrium again. At this point, T4 PNK was introduced into the solution followed by 4 s of stirring. The fluorescence was recorded simultaneously, and the phosphorylation velocity is represented by the maximum enhancement rate of fluorescence intensity for the assay approach based on an enzyme-coupled reaction.
MB 1 monitoring the phosphorylation process
Three samples were prepared to identify the features of MB1 in a 200 nM concentration: sample A contained MB1 and c-DNA; sample B contained MB1, Oligo A and Oligo B; and sample C contained only MB1. The fluorescence difference between the samples demonstrated the capability of MB1 to discern the different oligonucleotide sequences.
To determine the enzymatic kinetics of the phosphorylation reaction, the aforementioned three samples were incubated at 37°C with the sequential addition of 1.4 U of T4 DNA ligase and 3.0 U of T4 polynucleotide kinase.
The solution for measuring the selectivity of this approach contained 200 nM MB1 and 300 nM Oligo A in E.coli DNA ligase buffer or T4 DNA ligase buffer. The oligonucleotides inspected in this experiment were Oligo B to Oligo E and Oligo H to Oligo M, and each of them were mixed into the sample at a concentration of 300 nM. An aliquot of 25 U of E.coli DNA ligase (or 1.4 U of T4 DNA ligase) and 1.0 U of T4 PNK were used for each sample.
Assay of T4 PNK and investigation of the effects of external factors
In the assay for T4 PNK kinase activity, samples were prepared with 400 nM MB1, 600 nM Oligo A and 600 nM Oligo B. Before the addition of T4 PNK at various concentrations (0.002–10.0 U/ml), T4 DNA ligase (1.4 U) was introduced into each sample. The phosphorylation velocities of each sample were evaluated based on the fluorescence intensity versus time responses.
In the experiments performed to determine the effects of external factors on the phosphorylation enzymatic reaction of T4 PNK, the T4 DNA ligase-based assay system was employed to investigate the effects of (NH4)2SO4 and Na2HPO4. The E.coli ligase-based assay samples were used to explore the effects of NTP and ADP, because NTP is an indispensable cofactor for T4 DNA ligase. All samples contained 200 nM MB1, 300 nM Oligo A, 300 nM Oligo B within corresponding ligase buffer and the desired reagent at the optimal concentrations. The T4 DNA ligase, E.coli DNA ligase and T4 PNK used in these experiments are 1.4, 25 and 0.05 U, respectively, which assured that the activity of the ligase was much greater than T4 PNK.
Investigation of T4 PNK preference for substrates
A series of oligonucleotides (Oligo G to Oligo M) were mixed into the phosphorylation sample at 300 nM to explore the preference of T4 PNK for various substrates. The phosphorylation process was activated by the addition of 0.3 U T4 DNA ligase and 0.015 U T4 PNK. A series of ligation reactions were carried out to determine the effects of various oligonucleotides on the T4 DNA ligase system. The ligation samples were prepared with T4 DNA ligase buffer mixed with 200 nM MB1, 300 nM Oligo A and 300 nM Oligo F. Except for the standard ligation solution, the Oligo G to Oligo M was introduced into the ligation sample at a concentration of 300 nM. T4 DNA ligase (0.3 U) was added into each ligation reaction, and the ligation rates were determined based on the rate of change in the initial fluorescence intensities.
RESULTS AND DISCUSSION
Real-time monitoring of nucleic acids phosphorylation
The fluorescence enhancement of MB1 with c-DNA is ∼28 times higher than MB1 with Oligo A and Oligo B (see Supplementary Figure S-1). The result demonstrated that MB1 has high sensitivity to distinguish oligos before/after ligation upon hybridizing with them. This interesting feature of MB1 establishes the groundwork for monitoring phosphorylation of nucleic acids in homogenous solutions. Three samples designed to test the mechanism were proposed in Figure 1. The fluorescence intensity of the samples was monitored and the time courses were plotted in Figure 2 (Left).
Figure 2.
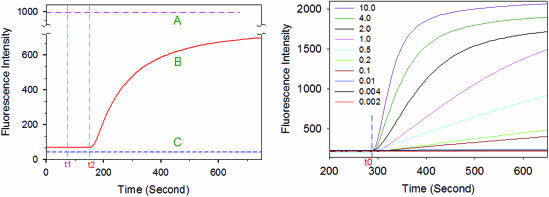
Left: monitoring of the phosphorylation reactions in real-time. The curves A, B and C represent the time courses of samples A, B and C, respectively. At time t1, T4 DNA ligase was added into the samples, while at time t2, T4 polynucleotide kinase was added. Right: time courses of real-time monitoring of the phosphorylation process catalyzed by various concentrations of T4 polynucleotide kinase (in U/ml). T4 polynucleotide kinase was added at time t0.
As illustrated in Figure 2 (Left), after the addition of T4 DNA ligase and T4 polynucleotide kinase, there was no fluorescence change in curves A and C (corresponding to samples A and C, respectively). Even in the presence or absence of the target oligos, MB1's conformation was not affected by the T4 DNA ligase and T4 polynucleotide kinase. The only factor that could affect the conformation of MB1 altering its fluorescence was the presence of the target c-DNA hybridizing with MB1.
In sample B, which is represented by curve B (Figure 2) after the addition of T4 DNA ligase, the fluorescence did not change. After the addition of T4 polynucleotide kinase, the fluorescence was enhanced rapidly, implying MB1's hybridization with the target oligo. According to the mechanism demonstrated in Figure 1, a ‘phosphorylation and ligation’ enzyme-coupled reaction took place, and the newly produced oligo could ‘open’ MB1 by hybridizing with the loop of MB1. Furthermore, the time course of sample B formed an ‘S-shaped’ curve, which is a significant characteristic of an enzyme-coupled reaction. Hence, the phosphorylation rate can be evaluated by the increasing rate of fluorescence change, instead of the initial signal changes. These results indicate that the phosphorylation of nucleic acids can be monitored in real time by this approach.
To determine the optimal T4 polynucleotide kinase (T4 PNK) concentration for the assay, a series of reactions containing varying amounts T4 PNK were investigated. The time courses are shown in Figure 2 (Right). The results indicated that the fluorescence enhancement rate increases with an increasing concentration of T4 polynucleotide kinase. With the enzyme ranging from 0.002 to 4.0 U/ml, the maximum rate of fluorescence increased proportionally to the concentration of T4 polynucleotide kinase, and the detection time was limited to 3 min (see Supplementary Figure S-2). Based on a 3 min reaction time, the detection limits for T4 PNK were determined to be 0.002 U/ml. This assay is convenient, quick and sensitive, and has a wide dynamic range, which makes it a useful approach to analyze the kinase activity of T4 polynucleotide kinase.
High selectivity for substrates
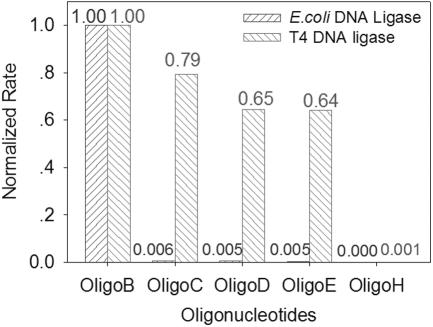
Owing to the high selectivity of the MB and the high fidelity of DNA ligase, the approach should demonstrate a high specificity for its substrates. The selectivity of the assay was evaluated by the maximum rate of fluorescence change for samples with varying substrates. The Oligo B, Oligo C, Oligo D and Oligo E have similar sequences but varying nucleotides at the 5′ termini (Oligo B is complementary to the loop sequence of MB1). The sequences of Oligo H to Oligo M were designed to be non-complementary and un-hybridizable to MB1. They were introduced into the phosphorylation samples based on E.coli DNA ligase or T4 DNA ligase. The results were normalized by data from Oligo B and listed in Figure 3 (Oligo I to Oligo M, data not shown).
Figure 3.
Specificity for substrates in phosphorylation reactions. The phosphorylation rate with various substrates is plotted. All data are normalized according to Oligo B that is complementary to the loop sequence of MB1.
In Figure 3, the phosphorylation rates of Oligo B, Oligo C, Oligo D, Oligo E and Oligo H with E.coli DNA ligase are 1.00, 0.005, 0.006, 0.005 and 0.000, respectively, while the data of samples based on T4 DNA ligase are 1.00, 0.79, 0.65, 0.64 and 0.001. The results of Oligo I to Oligo M were similar to that of Oligo H, so their results were skipped in Figure 3. The assay based on E.coli DNA ligase has more selectivity than the one based on T4 DNA ligase, because the E.coli DNA ligase possesses higher fidelity than T4 DNA ligase. But in random nucleotides that are non-correlative to the assay components, such as Oligo H to Oligo M, the selectivity was very high for both ligation assays. These results suggested that this method could distinguish the phosphorylation of a matching target significantly from three other similar oligos with only one base alteration at the 5′ terminus. High specificity makes this method capable of researching phosphorylation of a target oligo within a homogeneous solution containing diversified oligos in real time, without any separation treatments.
The preference of T4 polynucleotide kinase
When polynucleotide kinase was introduced into a solution containing varied oligonucleotides with a 5′ hydroxyl, the phosphorylation process would occur. The phosphorylation speed and efficiency of these oligonucleotides might be different with varied sequences, length and chemical modifications. Some prior research has revealed the differences in phosphorylation efficiency (21). These results were probably caused by the substrate preference of polynucleotide kinase. We have designed a convenient way to explore the preferences of varied oligonucleotides by employing the high selectivity of this novel monitoring method. When the inspected oligonucleotides were introduced into the phosphorylation solution, the reaction rate was affected. Polynucleotide kinase should work on the added oligonucleotides. The higher the preference for the oligonucleotide, the more time polynucleotide kinase will spend in ligating it; slower phosphorylation rates are expected for less preferential sequences. Therefore, the preference of various oligonucleotides can be investigated by comparing the decrease in the phosphorylation rate. In addition, the effects of the oligonucleotides on ligation can be investigated to calibrate their influence upon a ligation-based assay. The preference can be evaluated by the following formulae:
where the VPi is the phosphorylation rate of a sample mixed with the studied oligonucleotide. The VP0 represents the phosphorylation rate of a standard assay sample (which does not contain the investigated oligonucleotide). The NPi represents the effect ratio on the phosphorylation assay. This novel assay is based on a ‘phosphorylation–ligation’ enzyme-coupled reaction, so the NPi is composed of two fractions: the Pi0 which is the ‘original’ effect of a certain oligonucleotide on the phosphorylation process that has excluded the effect on ligation, and the NLi which characterizes the effect of the substrate on standard ligation. Then:
where the VL0 represents the ligation rate of the standard ligation sample. The VLi is the ligation rate of one sample, and the Pi represents the ratio by which one oligonucleotide decreased the phosphorylation reaction compared with the standard sample. It is apparent that the greater the Pi is, the higher the preference would be for the oligonucleotide. Therefore, Pi was used in this study to characterize the preference of T4 PNK for substrates.
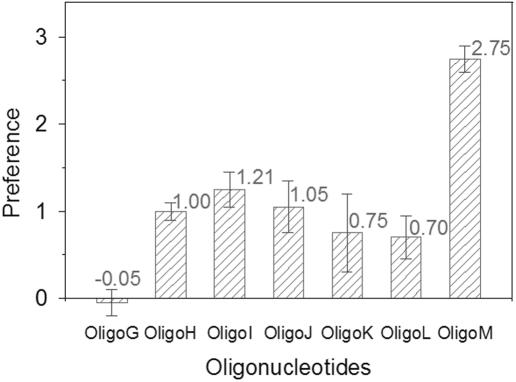
A series of three experiments were designed (i): exploring the preference of oligonucleotides with one base alteration at 5′ terminus (Oligo H to Oligo K), (ii) studying the effects of length on preference (Oligo L, Oligo H and Oligo M) and (iii) investigating the effect of phosphate/hydroxyl at 5′ end (Oligo G and Oligo H). In these experiments, the concentrations of T4 DNA ligase and T4 polynucleotide kinase were 3.7 and 0.2 U/ml, respectively, which meet the requirements for an enzyme-coupled reaction and ensure that the T4 polynucleotide kinase was not in excess for these substrates. The results were normalized by data from Oligo H, which was a common component in all three groups of experiments (Figure 4).
Figure 4.
Preference of T4 PNK for substrates. Preferences of Oligo G to Oligo M are normalized according to the data of Oligo H. The preference value and error bar of each oligonucleotide are indicated at the top of each column.
It can be concluded from Figure 4 that Oligo I, possessing a guanine at the 5′ end, resulted in the highest preference, while Oligo K, having a cytosine at the 5′ end, resulted in the lowest preference. The previous research revealed that the efficiency of phosphorylation by T4 PNK was affected by the base at 5′ end of oligonucleotide. The G end oligo got the highest efficiency and the C end oligo got the lowest (21). The similar patterns observed between preference and phosphorylation efficiency suggested that there might be a correlation between them. These experiments revealed an interesting phenomenon that the preference of Oligo G, which is the same sequence as Oligo H but has a phosphate at the 5′ end, showed great decreased preference than Oligo H. This result implied that the T4 polynucleotide kinase could distinguish the phosphate/hydroxyl at the 5′ end of the nucleotide and skip the phosphorylated oligonucleotide. Oligos H, L and M, which were the same sequence at the 5′ end but varied in length, had different preferences, indicating that the T4 polynucleotide kinase is sensitive to the length of the oligonucleotides. Based on this principle, other factors may affect the preference, such as double-stranded oligonucleotides and hairpin-shaped oligonucleotides, and these can be studied conveniently and precisely. Further research into the preference of T4 PNK may be helpful in understanding the ligation mechanism.
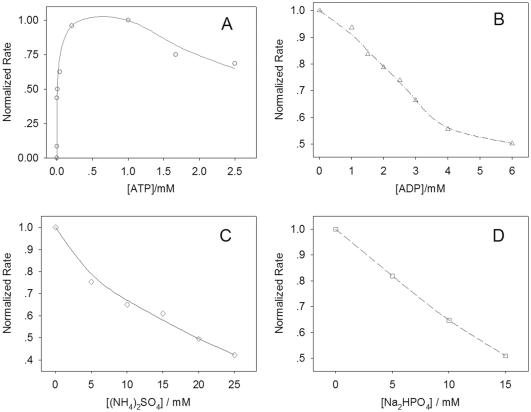
Effects of external factors
The effects of ribonucleoside triphosphates, ADP, sodium hydrogen phosphate and ammonium sulfate on phosphorylation have been studied employing this approach (Figure 5). The E.coli DNA ligase-based assay was employed to explore the effects of ribonucleoside triphosphates, ADP and ammonium sulfate. These chemicals are known to not inhibit E.coli DNA ligase ligation, which may affect the results of the enzyme-coupled reaction. The sodium hydrogen phosphate greatly reduced the ligation catalyzed by E.coli DNA ligase, as well as a moderate inhibition for the T4 DNA ligase-based assay. Hence, the T4 DNA ligase-based assay was chosen for studying the effects of sodium hydrogen phosphate. All of these experiments were performed in excess of DNA ligase to ensure that the results were according to the enzyme-coupled reaction condition (20).
Figure 5.
Effects of external factors on phosphorylation reactions. All data are normalized to the sample that had the highest phosphorylation rate.
The absence of ATP led to the blockage of the phosphorylation process. At a concentration of 125 μM, phosphorylation rates with ATP reached its maximum. While the concentration of ATP was >1.0 mM, a slight decrease in the rate was observed. The CTP, GTP and UTP produced similar effects on phosphorylation (see Supplementary Figure S-3). This slight inhibition caused by higher concentrations of nucleoside triphosphates on the phosphorylation reaction is probably due to a competitive binding reaction, as a result of the nucleoside triphosphates being analogous substances to T4 PNK. As a result, the binding site of T4 PNK is partially blocked. Increasing concentrations of ADP resulted in a decrease of the phosphorylation rate. The presence of 6.0 mM ADP led to a 50% decrease in the phosphorylation rate. This is due to the reversibility of the phosphorylation reaction in the presence of ADP and 5′-phosphoryl nucleic acids. Both ammonium sulfate and sodium hydrogen phosphate greatly inhibited the phosphorylation process. The addition of 20 mM (NH4)2SO4 or 15 mM Na2HPO4 can inhibit phosphorylation by ∼50%. The effect of Na2HPO4 is similar to the phosphate reported in previous research (22). The results showed that this approach can explore the various effects on phosphorylation quickly and accurately.
CONCLUSION
In summary, we have, for the first time, developed an approach for real-time monitoring of nucleic acid phosphorylation based on MB DNA probes. The MB employed in this method is devised to monitor the product of a ‘phosphorylation–ligation’ coupled enzymatic reaction. Compared with current assay methods, this approach is convenient, quick and highly sensitive. It has excellent selectivity and is capable of real-time monitoring in a homogenous solution. The preference of T4 PNK has been studied in detail, and the effects of a few external factors on phosphorylation have been investigated. The monitoring of the oligonucleotide dephosphorylation processes and exchange reactions could also be studied, which would enable the real-time monitoring of the phosphorylation and dephosphorylation processes simultaneously. The approach is a potentially useful tool in researching the interactions between proteins and nucleic acids in homogenous solutions, such as DNA repair, or drug research and development.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dr Timothy Drake for useful discussion and proof reading of the paper. This work was supported by the National Key Basic Research Program (2002CB513100), Pre-Key Project of Basic Research of the Science and Technology Ministry of China (2001-51), Key Project of Natural Science Foundation of People's Republic of China (20135010), Key Project Foundation of China Education Ministry (2000-156), Leading Teacher Foundation of China Education Ministry (2000-65) and Oversea Youth Scholar Co-research Foundation of People's Republic of China (20028506). Funding to pay the Open Access publication charges for this article was provided by Huana University, Changsha 410082, P. R. China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang L.K., Lima C.D., Shuman S. Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme. EMBO J. 2002;21:3873–3880. doi: 10.1093/emboj/cdf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitehouse J., Taylor R.M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D.D., Weinfeld M., Caldecott K.W. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 3.Richardson C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc. Natl Acad. Sci. USA. 1965;54:158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amitsur M., Levitz R., Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meijer M., Busheri F.K., Huang T.Y., Weinfeld M., Young D. Pnk1, a DNA kinase/phosphatase required for normal response to DNA damage by gamma-radiation or camptothecin in Schizosacchramyces pombe. J. Biol. Chem. 2002;277:4050–4055. doi: 10.1074/jbc.M109383200. [DOI] [PubMed] [Google Scholar]
- 6.Wilson D.M., III, Thompson L.H. Life without DNA repair. Proc. Natl Acad. Sci. USA. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L.K., Shuman S. Domain structure and mutational analysis of T4 polynucleotide kinase. J. Biol. Chem. 2001;276:26868–26874. doi: 10.1074/jbc.M103663200. [DOI] [PubMed] [Google Scholar]
- 8.Busheri K., Daly G., Robins P., Canas B., Pappin D.J.C., Sgouros J., Miller G.G., Fakhrai H., Davis E.M., Le Beau M.M., Weinfeld M. Molecular characterization of a human DNA kinase. J. Biol. Chem. 1999;274:24187–24194. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 9.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 10.Perlette J., Tan W.H. Real-time monitoring of intracellular mRNA hybridization onside single living cells. Anal. Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 11.Tan W., Wang K., Drake T. Molecular beacons. Curr. Opin. Chem. Biol. 2004;8:547–553. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Fang X., Li J.J., John P., Tan W., Wang K. Molecular beacons: novel fluorescent probes. Anal. Chem. 2000;72:747A–753A. doi: 10.1021/ac003001i. [DOI] [PubMed] [Google Scholar]
- 13.Nitin N., Santangelo P.J., Kim G., Nie S., Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P., Beck T., Tan W. Design of a molecular beacon DNA probe with two fluorophores. Angew. Chem. Int. Ed. Engl. 2001;40:402–405. doi: 10.1002/1521-3773(20010119)40:2<402::AID-ANIE402>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi S., Bratu D.P., Kramer F.R. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 16.Li J.J., Fang X., Schuster S.M., Tan W. Molecular beacons: a novel approach to detect protein–DNA interactions. Angew. Chem. Int. Ed. Engl. 2000;39:1049–1052. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Li J.J., Geyer R., Tan W. Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-strand DNA. Nucleic Acids Res. 2000;28:e52. doi: 10.1093/nar/28.11.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z., Wang K., Tan W., Li J., Liu L., Guo Q., Meng X., Ma C., Huang S. Real-time monitoring the nucleic acids ligation in homogenous solution using molecular beacon. Nucleic Acids Res. 2003;31:e148. doi: 10.1093/nar/gng146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokol D.L., Zhang X., Lu P., Gewirtz A.M. Proc. Natl Acad. Sci. USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Copeland R.A. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. 2nd edn. New York: Wiley-VCH, Inc.; 2000. [Google Scholar]
- 21.van Houten V., Denkers F., van Dijk M., van den Brekel M., Brakenhoff R. Labeling efficiency of oligonucleotides by T4 polynucleotide kinase depends on 5′-nucleotide. Anal. Biochem. 1998;265:386–389. doi: 10.1006/abio.1998.2900. [DOI] [PubMed] [Google Scholar]
- 22.Lillehaug J.R., Kleppe K. Effect of salts and polyamines on T4 polynucleotide kinase. Biochemistry. 1975;14:1225–1229. doi: 10.1021/bi00677a021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.