Abstract
Background:
Systematic evidence maps are increasingly used to develop chemical risk assessments. These maps can provide an overview of available studies and relevant study information to be used for various research objectives and applications. Environmental epidemiological studies that examine the impact of chemical exposures on various ‘omic profiles in human populations provide relevant mechanistic information and can be used for benchmark dose modeling to derive potential human health reference values.
Objectives:
To create a systematic evidence map of environmental epidemiological studies examining environmental contaminant exposures with ‘omics in order to characterize the extent of available studies for future research needs.
Methods:
Systematic review methods were used to search and screen the literature and included the use of machine learning methods to facilitate screening studies. The Populations, Exposures, Comparators and Outcomes (PECO) criteria were developed to identify and screen relevant studies. Studies that met the PECO criteria after full-text review were summarized with information such as study population, study design, sample size, exposure measurement, and ‘omics analysis.
Results:
Over 10,000 studies were identified from scientific databases. Screening processes were used to identify 84 studies considered PECO-relevant after full-text review. Various contaminants (e.g. phthalate, benzene, arsenic, etc.) were investigated in epidemiological studies that used one or more of the four ‘omics of interest: epigenomics, transcriptomics, proteomics, and metabolomics. The epidemiological study designs that were used to explore single or integrated ‘omic research questions with contaminant exposures were cohort studies, controlled trials, cross-sectional, and case-control studies. An interactive web-based systematic evidence map was created to display more study-related information.
Conclusions:
This systematic evidence map is a novel tool to visually characterize the available environmental epidemiological studies investigating contaminants and biological effects using ‘omics technology and serves as a resource for investigators and allows for a range of applications in chemical research and risk assessment needs.
Keywords: Systematic evidence map, Environmental epidemiology, ‘Omics
1. Introduction
There is a vast array of chemicals and contaminants that humans are potentially exposed to, of which the majority lack the substantial toxicity data needed to perform comprehensive human health assessments. Traditional environmental epidemiology has aimed to characterize how individual or mixtures of exposures are associated with one or several apical health outcomes; however, it seldom sufficient to characterize all environmentally-associated biological or health outcomes (Kyrtopoulos, 2013). Technological advancements in ‘omics (e.g., genomics, transcriptomics, proteomics, and metabolomics) have resulted in improved capabilities of generating high-dimensional molecular data (e.g., hundreds to thousands of genes, methylations, proteins, and metabolites) that are informative about internalization of exposures and perturbations to physiological activities (Kyrtopoulos, 2013). Information from ‘omics analyses can be used as a complement to traditional environmental epidemiology and expand understanding of the impacts of chemicals on health and on disease etiology.
The field of human health risk assessment is utilizing practices of evidence mapping in order to systematically identify relevant studies to a given topic of interest (Bragge et al., 2011; Wolffe et al., 2019). A systematic evidence map may provide an overview of available scientific studies that can be used to identify data gaps in the topic of interest and provide relevant information such as the number of studies on the topic, study design, and study characteristics (Bragge et al., 2011; Miake-Lye et al., 2016; Wolffe et al., 2019). Moreover, evidence mapping can be helpful for gathering relevant toxicity and mechanistic data for legacy and emerging chemicals forms an increasingly vital part of risk assessment, and advances in analytical techniques and scientific understanding continue to broaden the scope of available data beyond from those of the traditional in vivo or in vitro toxicity testing (Wolffe et al., 2019).
A number of environmental epidemiological studies have examined the impact of various chemical or contaminant exposures on certain tissue-based (usually blood) ‘omic profiles in human populations. Information from these studies can help identify biological profile changes related to known or suspected adverse effects associated with the exposures of interest. These human population based studies using ‘omics analyses can also be informative of exposure and early biological effect biomarkers as well as molecular and cellular events that are indicative of modes-of-action or key events in adverse outcome pathways (Espín-Pérez et al., 2014). We developed a systematic evidence map (SEM) of environmental epidemiological studies examining chemical or contaminant exposures with ‘omics analyses in order to characterize the extent of available studies and for future research needs as well as potential future applications in chemical risk assessments. Single or multi-omic integration in epidemiological studies can provide a significant opportunity to increase the understanding of health and disease with respect to biological mechanism, molecular targets, and biomarkers (Karczewski and Snyder, 2018). Thus, the ‘omics data from these epidemiological studies have the potential to inform various aspects of risk assessment such as mechanism of action, exposure assessment, toxicokinetics, and dose-response assessment (Yu et al., 2016).
2. Methods
We developed a PECO statement (Participants, Exposure, Comparator, and Outcomes) to define the scope of the SEM:
Participants: Any population and lifestage (occupational or general population, including children and other sensitive populations).
Exposure: Any chemical and/or environmental contaminant
Comparator: Comparison or reference population exposed to lower or no levels of chemical/contaminant to more highly exposed population; or humans who serve as their own control by comparing before-and-after outcomes following chemical/contaminant exposure
Outcome: Molecular analyses from use of the following ‘omics:
Transcriptomics: gene expression changes (RNA transcripts)
Metabolomics: metabolites produced by cell, tissue, or organism
Proteomics: protein functions and interactions
Epigenomics: epigenetic modifications (e.g., DNA methylation, histone modification, microRNA) that influence gene expression.
To identify relevant literature, we developed a comprehensive search strategy. The primary databases searched were PubMed, Web of Science, Toxline, and Toxic Substances Control Act Test Submissions (TSCATS). Additional resources outside of the four bibliographic databases were also used such as the reference list of studies and reviews screened as meeting PECO criteria after full-text review, and publicly available study and data information from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). We curated specific search terms from relevant vocabularies (Table 1) for each category of ‘omics (transcriptomics, epigenomics, metabolomics, and proteomics), excluding terminology that was too broad to be useful. For example, we did not include “cell” and “array” as terms as they were too general, and fold-change was reported in various ways to express quantification of expression (not just for ‘omics) so was not included in the search terms. However, we included specific terms such as “differential expression,” “differential expressed genes,” or “differential gene expression” that consistently produced on-topic results. Our strategy was further refined (Table S1) to recall ‘omics studies meeting two criteria points: studies had to first focus on human populations, and then they must also contain information relevant to chemical exposure.
Table 1.
Relevant Search Terms.
| Subject | Terms |
|---|---|
| Chemical Exposure | inhalation, ingestion, skin contact, chemical exposure, molecular epidemiology |
| Metabolomics | metabolite, biofluid, tissue, metabolome, metabolism, molecular phenotype, sugar, lipid, amino acid, fatty acid, phenolic compound, alkaloids, small molecules, biomarker |
| Epigenomics | epigenome, DNA, gene expression, DNA methylation, histone modification, microRNA, miRNA, Illumina 450 K, Infinium HumanMethylation, Illumina 850 K |
| Transcriptomics | RNA, mRNA, mRNA expression, microarray, high-throughput sequencing, HTS, transcriptome, next-generation sequencing, NGS, RNA-Seq, differentially expressed genes, differential genes, differential gene expression |
| Proteomics | proteomes, protein expression, protein activity, protein degradation, protein production, steady-state abundance, post-translational modifications, PTM, mass spectrometry |
| ToxNet Narrowing Terms | biological effect, exposure, biomarker |
| Human Health Terms | development*, skin, tissue, derm*, human health, health, epidemiology, child*, teenager, adolescen*, pregnan*, adult*, general population, population, blood, serum, birth |
| Exclusion Terms | social, in vitro, in-vitro, animal, mouse, mice, rat*, beagle*, rodent*, rabbit*, dog*, cat*, guinea pig*, primate*, monkey*, pig*, fish*, bird*, frog*, in vivo, in-vivo, cell line, yeast, tick, ecotoxicity, nematode, mosquito*, drosophila, mite*, reptile, parasit*, poultry, mussel*, parasite*, murine, protozoa*, fungi, fungus, cetacean, canine, feline |
2.1. Initial literature searches
The initial literature search was performed in October 2019 and considered studies from 1995-present. The starting year for this search was chosen with consideration to keystone publications, namely the first microarray expression analysis described in the developing era of genomics (Schena et al., 1995). An update search was performed in December 2020 to integrate references published since the initial search, and title/abstract and full-text screening were performed (details discussed in literature screening and data extraction section) on the resulting group of studies.
2.2. Validation searches
The initial primary searches were designed to be broad and capture a wide array of information, but there were concerns that certain types of studies may have been missed. Thus, we performed additional focused validation searches (October 2021) to identify specific categories of potentially missed relevant information. First, we aimed to identify non-English studies by performing SQL querying against the reference database. The query enables us to check against the language field in the host database. Any values other than English were considered for this validation step. Searches were also performed to identify references specifically regarding arsenic and polychlorinated biphenyls (PCBs) (Table S2), as they were examples of chemicals investigated across all ‘omics platforms (epigenomics, transcriptomics, metabolomics, and proteomics) from studies evaluated through initial title/abstract screening. Additionally, we performed searches targeted to identify studies of “chemical” exposure and phthalate exposure (Table S3). Phthalates were chosen as a broad chemical class that may have missing studies from the initial search, which would indicate that the initial search terms need to be re-evaluated. Validation searching for these studies occurred in two parts. The first keyword set identified individual phthalates using common names and abbreviations. The second keyword set identified phthalates categorically (e.g., phthalate esters (PAEs), plasticizers). The resulting group of studies from the validation searches were then also put through a title/abstract screening effort.
2.3. Literature screening and data extraction
Because the number of references retrieved was large from the search efforts, the results were first imported into SWIFT-Review software (https://www.sciome.com/swift-review/) to remove duplicate and off-topic references. It has pre-set literature search filters (health outcome and evidence stream) that can be applied to identify studies that are more likely to be useful for identifying human health content from those that likely do not. Using the evidence stream filter in SWIFT-Review, we selected for human studies only, and tagged for animal or plant studies, in-vitro studies, ecotoxicity, physical chemistry, and environmental fate to be excluded. Using the health outcomes filter, we selected all relevant health outcomes (e.g., hematological and immune, developmental, cancer, respiratory, endocrine, reproductive, renal, hepatic, cardiovascular, musculoskeletal, neurological, nutritional and metabolic, mortality, skin and connective tissue, and ocular and sensory) and excluded studies with tags for physiological based pharmacokinetic (PBPK) modeling and simulation, or that had no tags.
Title/abstract screening of relevant studies was then performed in SWIFT-Active Screener (https://www.sciome.com/swift-activescreener). To be considered for inclusion, studies needed to meet the PECO criteria. If the studies were included, information was noted on the type(s) of ‘omics (transcriptomics, epigenomics, metabolomics, or proteomics) examined in the study. in general, reviewers can save time by reviewing titles and abstracts in SWIFT-Active Screener until an estimated 95% recall since it uses “active” machine learning in which real time screening decisions help to prioritize unscreened studies for relevance (Howard et al., 2020). For this systematic evidence map process, reviewers screened all 100% of included studies in SWIFT-Active Screener in order to ensure no pertinent environmental epidemiological studies were missed.
Full text screening and data extraction of the included studies were performed concurrently in DistillerSR (https://www.evidencepartners.com/products/distillersr-systematic-review-software). Both title/abstract and full-text screening processes using SWIFT-Review, SWIFT-Active Screener, and DistillerSR were conducted by two independent reviewers. The data extraction consisted of information on study population (pregnant women, occupational, general population-adults, children/adolescents), study design (case-control, cohort, controlled-trial, cross-sectional, other), years of data collection, study sample size, country of study/population, exposure measurement (chemicals analyzed, exposure levels, and matrix), availability of data (e.g., whether data was publicly available or accessible), potential confounders, type of ‘omics and platform, and biological matrix used for ‘omics. We then created an evidence map summarizing the available and extracted data. The code book is available in the Supplemental Materials. Visualizations were generated using Microsoft Excel and Tableau.
3. Results
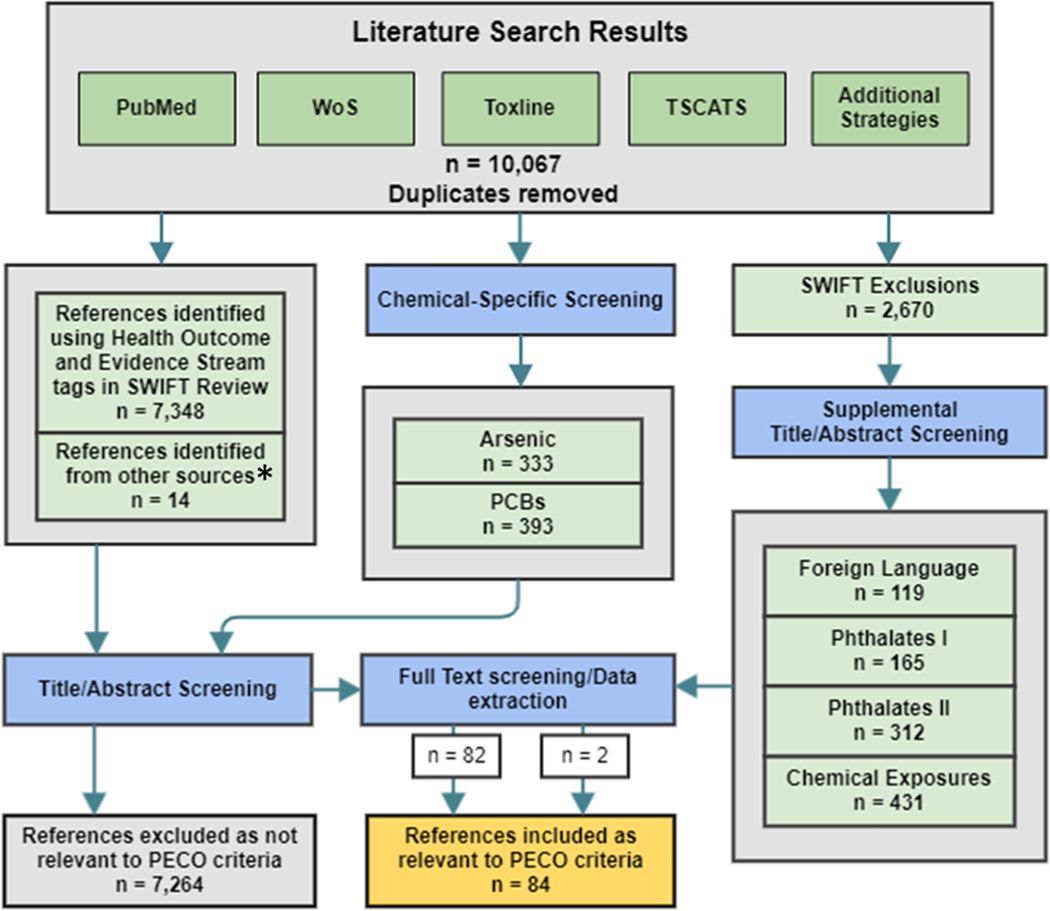
Our searches spanning from January 1, 1995 until October 13, 2021 across the databases retrieved 10,067 total records (Fig. 1). 7,397 studies were identified after undergoing pre-set literature filters, and 7,348 unique studies were selected for title/abstract screening after removal of duplicates and off-topic references (Fig. 1). As part of validation searches, we performed chemical-specific searches for arsenic and PCBs as many of the identified studies from the initial search examined these two chemicals with the four ‘omics of interest. We identified 333 studies for arsenic and 393 studies for PCB (Fig. 1). We also performed validation searches on 2,670 studies that were initially excluded through the filters in SWIFT-Review to check that relevant references were not missed and used additional search terms to identify any relevant studies for foreign language (119 studies), chemical exposure (431 studies), and two keyword sets for phthalates (165 studies for set I and 312 studies for set II) (Figure S1). From filtering and screening in SWIFT-Active Review and Screener of studies identified from initial and validation searches as well as through reference lists and GEO database, 84 unique relevant environmental epidemiological studies were identified (Fig. 1).
Fig. 1. Study Selection Diagram (Literature Search Results).
*References identified from other sources (n = 14) were collected from reviews, abstracts, and suggested work in screened studies.
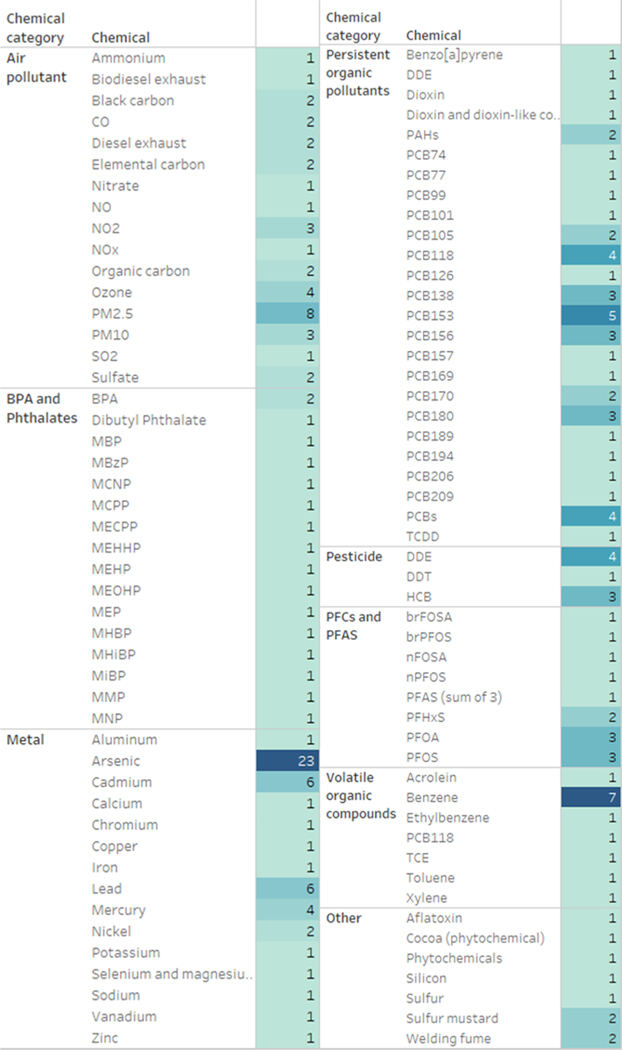
The chemicals investigated in the identified epidemiology ‘omics studies are shown in Fig. 2. We group the chemicals into categories of air pollutants, BPA and phthalates, metals, persistent organic pollutants, pesticides, PFCs and PFAS, volatile organic compound, and other. The top five chemicals that were often examined in epidemiology ‘omics studies were arsenic, PCBs (grouped), particulate matter (PM) 2.5, benzene, and metals (cadmium, lead).
Fig. 2. Summary of chemical exposures of included studies*.
*Some studies may appear in multiple categories for examining one or more chemicals.
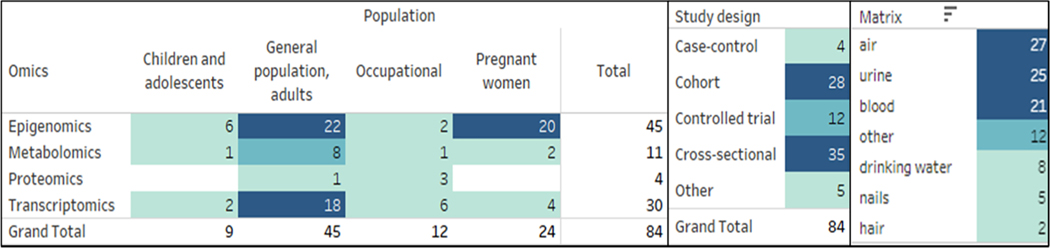
Cross-sectional (n = 35) and cohort studies (n = 28) were the most common study designs across all of the ‘omics epidemiology studies, followed by controlled exposure trials (n = 12) and case-control studies (n = 4) (Fig. 3). Omics investigations are often cross-sectional in nature with samples collected at a single time point due to limited availability of data or post-hoc additions to existing studies (Chu et al., 2019). Various exposure matrix were examined in the epidemiological ‘omics studies (Fig. 3), and air (n = 27), urine (n =25), and blood (n = 21) were mostly used to measure exposure levels (Fig. 3). For most ‘omics analyses, studies used serum, placenta, cord blood, or urine to extract the biological markers (e.g., RNA, DNA, metabolites, or proteins) (Table S4). Different populations such as children and adolescents < 18 years old, adults, occupational workers, and pregnant women were all investigated across studies using epigenomics, metabolomics, or transcriptomics for chemical exposure effects (Fig. 3). Studies that used proteomics investigated chemical exposures among mostly occupational (n = 3) and adult general population(n = 1) (Fig. 3). Studies of pregnant women most often used epigenomics (n = 20) to investigate effects from prenatal chemical exposures, which is of interest because prenatal exposure can dysregulate the fetal epigenome with potential consequences for subsequent adverse health effects manifesting in childhood, over lifetime, or transgenerationally (Perera and Herbstman, 2011).
Fig. 3. Summary of study design and population of included studies*.
*Some studies may appear in multiple categories for examining one or more ‘omics.
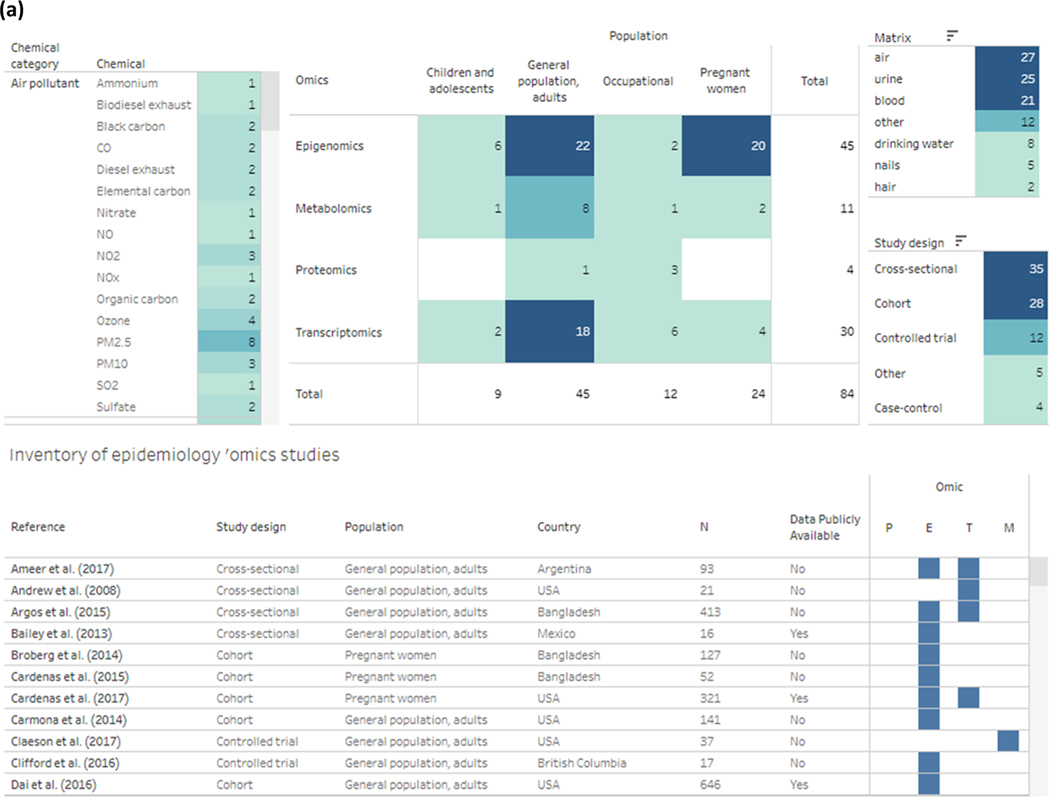
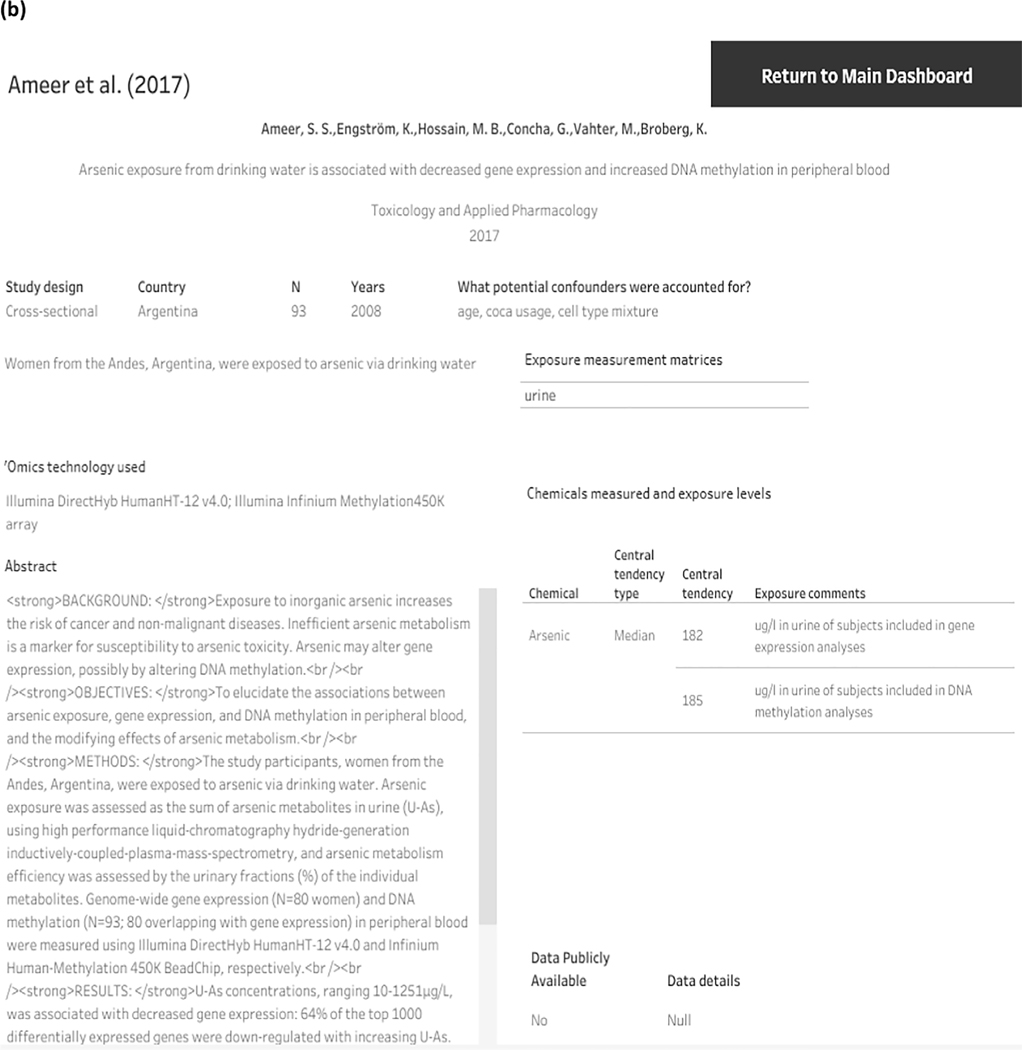
An interactive dashboard of the data in the systematic evidence map is available at: https://public.tableau.com/app/profile/literature.inventory/viz/Omics-epi-SEM/Omics_Inventory, and information of the 84 included environmental epidemiology studies are summarized below. The dashboard can be filtered by chemical, omics type, population category, study design, and matrix (snapshot of dashboard shown in Fig. 4a). The sample size of the 84 studies in this systematic evidence map ranged from 5 to 2,411 participants (see interactive dashboard, Fig. 4a). Overall, studies were often conducted in United States (n = 24) as well as in countries such as China (n = 11), Mexico (n = 8), and Bangladesh (n = 6) (see interactive dashboard, Fig. 4a). Studies most often used epigenomics (n = 45), transcriptomics (n = 30), and metabolomics (n = 11) but used less of proteomics (n = 4) when examining associations with chemical exposures (see interactive dashboard, Figs. 3 and 4a). There were several epidemiological studies (n = 6) that examined more than one ‘omics, such as transcriptomics and epigenomics (see interactive dashboard, Fig. 3 Fig. 4a).
Fig. 4.
Snapshots from interactive dashboard that display extracted information of the identified epidemiological ‘omics studies. (See above-mentioned references for further information.).
When an individual study is selected from the interactive systematic evidence map, the reader can access additional study details such as years of data collection, data availability, ‘omics technology platform, potential confounders the study accounted for, and exposure levels (includes information on the central tendency type, value, and additional information such as units of measurement) (see interactive dashboard). An example of an individual study and information is shown in Fig. 4b. Thirty-five studies (42%) had publicly available data and provided database (such as GEO or dbGAP) accession numbers (see interactive dashboard). Additionally, most of the studies (n = 73, 87%) indicated potential confounders and performed restriction, matched the study subjects with respect to potential confounders, or adjusted for them in the statistical analyses (Fig. 4) (see interactive dashboard).
4. Discussion
The systematic evidence map presented here summarizes the available body of environmental epidemiological studies investigating molecular effects from chemical exposures by using ‘omics technology. Key information on study design, study population characteristics, type of ‘omics and technology, and chemical exposure are available, allowing the user to inspect and analyze the available studies. This evidence map serves as an inventory and starting place for further investigations. It can be used to inform future research or the design of environmental epidemiological studies examining chemical exposures and ‘omics by identifying the relevant information in which the presented studies have and also identifying potential data gaps.
Environmental epidemiological studies that examine ‘omics need to consider unique potential sources of bias related to sampling of tissues and analyzing of high-throughput data. This SEM extracted some information relevant to bias (e.g., matrix, exposure levels, adjustment for confounding, population description), but risk of bias evaluation was not performed. Researchers using this data should consider potential sources of bias as relevant to their research question. The challenges associated with ‘omics data require researchers to consider sources of bias in traditional epidemiological studies, such as confounding, selection bias, measurement error, and reverse causation, but also consider unique biases such as cellular to tissue heterogeneity or technical variability (Everson and Marsit, 2018; Rockett et al., 2004). Statistical power is important to consider when trying to detect true associations from ‘omics data; thus, power calculations must be performed to estimate appropriate sample sizes (Everson and Marsit, 2018). Additionally, environmental epidemiological studies using ‘omics data should have information on quality control, filtering processes, normalization and appropriate statistical methods. As various ‘omics platforms exist, there are potential challenges to applying and analyzing ‘omics in epidemiological studies (see examples listed in Table S5); and thus, the generated data need to be carefully analyzed and interpreted (Franks and Pomares-Millan, 2020; Krassowski et al., 2020). Moreover, environmental epidemiologic studies often examine multiple -omics within the population of interest and so study designs that involve repeated sampling on study subjects to generate multiple-omics are particularly useful for biological inference (Chu et al., 2019). Thus, thoughtful incorporation of different study design principles, data filtering and analysis, validation of possible biomarkers in independent populations, and further analytical development of integrative ‘omics methods are necessary to continue the understanding of relevant biological processes inferred from ‘omics in these environmental epidemiological studies.
There are several strengths and limitations to our systematic evidence map. As mentioned above, we did not examine perform risk of bias evaluation, and thus it is not known whether the included studies are of high or low quality. As part of our future research applications of using this systematic evidence map, we are working to create data quality metrics and study evaluation criteria on domains such as population selection, exposure assessment, and risk of bias for these environmental epidemiological studies examining chemical exposures with ‘omics. A major strength of our systematic evidence map is that we searched diverse sources of evidence by using multiple databases and reviewing reference lists of included studies and reviews to identify relevant references, which we believe has resulted in a comprehensive inventory but with potential for missing relevant studies. Another strength is that we worked with an expert librarian to develop and optimize our search terms and targeted strategies. Moreover, using the extracted information from our systematic evidence map, others can adapt for quantifiable risk assessment or research needs such as developing analytic strategies based on the study design, types of ‘omic data available, and exposure information
In general, data derived from epidemiological research has certain advantages over animal or in-vitro experimental studies when assessing exposure-outcome associations for risk assessments. For example, the target species (human) is directly relevant and does not require for interspecies or high to low dose extrapolations (Burns et al., 2019). Technological advancements in ‘omics have resulted in improved capacity to measure molecular changes in biological samples that are informative about internalization of exposures and physiological perturbations (Everson and Marsit, 2018). These advances have expanded researchers’ capabilities to examine the underlying etiology of environmentally-associated diseases. Using analysis from ‘omics in environmental epidemiological studies as a complement to traditional epidemiology and experimental studies can further our understanding of the direct impacts of chemicals on human health and inform relevant human data for risk assessment. For instance, ‘omics data in epidemiological studies are being used to better characterize molecular initiating events and provide evidence of key events at different levels of molecular processes in adverse outcome pathways (Brockmeier et al., 2017). The ‘omics data from epidemiological studies can provide mechanistic evidence to support chemical read-across, weight of evidence for certain mechanisms, understanding of biological networks, and potential quantitative development of point of departures; and we are focusing on the latter application for future work using this systematic evidence map. Hence, qualitative and quantitative approaches and considerations to using various ‘omics data sets in risk assessments or other regulatory landscapes continue to be explored (Boverhof and Zacharewski, 2006; Buesen et al., 2017; D. Ghosh et al., 2018; Pennie et al., 2004).
5. Conclusions
‘Omics data have generally been used in chemical risk assessments to provide information about the mode or mechanism of action and are now transitioning to using these data to potentially derive human health relevant toxicity values. For example, a recent study using an epidemiological cohort estimated inorganic arsenic doses that corresponded to changes in transcriptomic, proteomic, epigenomic, and integrated multi-‘omic signatures in human cord blood through benchmark dose modeling (Rager et al., 2017). Thus, data integration from ‘omics in population-based studies may provide direct human-relevant reference values to quantify biological effects from chemical exposures. The systematic evidence map presented here identifies the existing evidence that may be used for including ‘omics information from epidemiological studies for quantitative and qualitative risk assessments and also informs where more data may be needed for specific chemical(s). This systematic evidence map will be updated on an annual basis as an ongoing resource for researchers investigating chemical exposures and ‘omics data in epidemiological studies.
Supplementary Material
Acknowledgements
The authors would like to thank the following individuals from U.S. Environmental Protection Agency: Jeffry Dean, Erin Yost, Kris Thayer, and Barbara Glenn for internal review.
Funding and Disclaimer
Authors declare no financial conflict of interest and no competing interests. Funding was provided through author’s employment as listed above with no additional external funding. The views expressed are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency.
Footnotes
CRediT authorship contribution statement
Stephanie Kim: Conceptualization, Methodology, Software, Writing. Hillary Hollinger: Methodology, Writing – original draft. Elizabeth G. Radke: Visualization, Software, Writing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2022.107243.
References
- Ameer SS, Engström K, Hossain MB, Concha G, Vahter M, Broberg K, 2017. Arsenic exposure from drinking water is associated with decreased gene expression and increased DNA methylation in peripheral blood. Toxicol. Appl. Pharmacol. 321, 57–66. 10.1016/j.taap.2017.02.019. [DOI] [PubMed] [Google Scholar]
- Andrew AS, Jewell DA, Mason RA, Whitfield ML, Moore JH, Karagas MR, 2008. Drinking-water arsenic exposure modulates gene expression in human lymphocytes from a U.S. population. Environ. Health Perspect. 116 (4), 524–531. 10.1289/ehp.10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M, Chen L, Jasmine F, Tong L, Pierce BL, Roy S, Paul-Brutus R, Gamble MV, Harper KN, Parvez F, Rahman M, Rakibuz-Zaman M, Slavkovich V, Baron JA, Graziano JH, Kibriya MG, Ahsan H, 2015. Gene-specific differential DNA methylation and chronic arsenic exposure in an epigenome-wide association study of adults in Bangladesh. Environ. Health Perspect. 123 (1), 64–71. 10.1289/ehp.1307884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, García-Vargas G, Del Razo L-M, Drobná Z, Stýblo M, Fry RC, 2013. Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. J. Biochem. Mol. Toxicol. 27, 106–115. 10.1002/jbt.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Zacharewski TR, 2006. Toxicogenomics in risk assessment: applications and needs. Toxicol. Sci. Off. J. Soc. Toxicol. 89, 352–360. 10.1093/toxsci/kfj018. [DOI] [PubMed] [Google Scholar]
- Bragge P, Clavisi O, Turner T, Tavender E, Collie A, Gruen RL, 2011. The Global Evidence Mapping Initiative: Scoping research in broad topic areas. BMC Med. Res. Methodol. 11, 92. 10.1186/1471-2288-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg K, Ahmed S, Engström K, Hossain MB, Jurkovic Mlakar S, Bottai M, Grandér M, Raqib R, Vahter M, 2014. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J. Dev. Orig. Health Dis. 5 (4), 288–298. 10.1017/S2040174414000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmeier EK, Hodges G, Hutchinson TH, Butler E, Hecker M, Tollefsen KE, Garcia-Reyero N, Kille P, Becker D, Chipman K, Colbourne J, Collette TW, Cossins A, Cronin M, Graystock P, Gutsell S, Knapen D, Katsiadaki I, Lange A, Marshall S, Owen SF, Perkins EJ, Plaistow S, Schroeder A, Taylor D, Viant M, Ankley G, Falciani F, 2017. The role of omics in the application of adverse outcome pathways for chemical risk assessment. Toxicol. Sci. Off. J. Soc. Toxicol. 158, 252–262. 10.1093/toxsci/kfx097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesen R, Chorley BN, da Silva Lima B, Daston G, Deferme L, Ebbels T, Gant TW, Goetz A, Greally J, Gribaldo L, Hackermüller J, Hubesch B, Jennen D, Johnson K, Kanno J, Kauffmann H-M, Laffont M, McMullen P, Meehan R, Pemberton M, Perdichizzi S, Piersma AH, Sauer UG, Schmidt K, Seitz H, Sumida K, Tollefsen KE, Tong W, Tralau T, van Ravenzwaay B, Weber RJM, Worth A, Yauk C, Poole A, 2017. Applying ‘omics technologies in chemicals risk assessment: report of an ECETOC workshop. Regul. Toxicol. Pharmacol. 91, S3–S13. 10.1016/j.yrtph.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CJ, LaKind JS, Mattison DR, Alcala CS, Branch F, Castillo J, Clark A, Clougherty JE, Darney SP, Erickson H, Goodman M, Greiner M, Jurek AM, Miller A, Rooney AA, Zidek A, 2019. A matrix for bridging the epidemiology and risk assessment gap. Glob. Epidemiol. 1, 100005. 10.1016/j.gloepi.2019.100005. [DOI] [Google Scholar]
- Cardenas A, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, Wright RO, Christiani DC, Kile ML, 2015. In utero arsenic exposure and epigenome-wide associations in placenta, umbilical artery, and human umbilical vein endothelial cells. Epigenetics 10 (11), 1054–1063. 10.1080/15592294.2015.1105424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Rifas-Shiman SL, Agha G, Hivert M-F, Litonjua AA, DeMeo DL, Lin X, Amarasiriwardena CJ, Oken E, Gillman MW, Baccarelli AA, 2017. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci. Rep. 7, 288. 10.1038/s41598-017-00384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona JJ, Sofer T, Hutchinson J, Cantone L, Coull B, Maity A, Vokonas P, Lin X, Schwartz J, Baccarelli AA, 2014. Short-term airborne particulate matter exposure alters the epigenetic landscape of human genes associated with the mitogen-activated protein kinase network: a cross-sectional study. Environ. Health Glob. Access Sci. Source 13, 94. 10.1186/1476-069X-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S.u., Huang M, Kelly R, Benedetti E, Siddiqui J, Zeleznik O, Pereira A, Herrington D, Wheelock C, Krumsiek J, McGeachie M, Moore S, Kraft P, Mathé E, Lasky-Su J, 2019. Integration of metabolomic and other omics data in population-based study designs: an epidemiological perspective. Metabolites 9 (6), 117. 10.3390/metabo9060117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeson A-S, Gouveia-Figueira S, Häggström J, Fowler CJ, Nording ML, 2017. Levels of oxylipins, endocannabinoids and related lipids in plasma before and after low-level exposure to acrolein in healthy individuals and individuals with chemical intolerance. Prostaglandins Leukot. Essent. Fatty Acids 121, 60–67. 10.1016/j.plefa.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Clifford RL, Jones MJ, MacIsaac JL, McEwen LM, Goodman SJ, Mostafavi S, Kobor MS, Carlsten C, 2017. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J. Allergy Clin. Immunol. 139 (1), 112–121. 10.1016/j.jaci.2016.03.046. [DOI] [PubMed] [Google Scholar]
- Dai L, Mehta A, Mordukhovich I, Just AC, Shen J, Hou L, Koutrakis P, Sparrow D, Vokonas PS, Baccarelli AA, Schwartz JD, 2017. Differential DNA methylation and PM2.5 species in a 450K epigenome-wide association study. Epigenetics 12 (2), 139–148. 10.1080/15592294.2016.1271853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster S, van Leeuwen DM, Jennen DGJ, Koppen G, Den Hond E, Nelen V, Schoeters G, Baeyens W, van Delft JHM, Kleinjans JCS, van Larebeke N, 2013. Gender-specific transcriptomic response to environmental exposure in Flemish adults. Environ. Mol. Mutagen. 54 (7), 574–588. 10.1002/em.21774. [DOI] [PubMed] [Google Scholar]
- Drizik E, Corbett S, Zheng Y, Vermeulen R, Dai Y, Hu W, Ren D, Duan H, Niu Y, Xu J, Fu W, Meliefste K, Zhou B, Zhang X, Yang J, Bassig B, Liu H, Ye M, Liu G, Jia X, Meng T, Bin P, Zhang J, Silverman D, Spira A, Rothman N, Lenburg ME, Lan Q, 2020. Transcriptomic changes in the nasal epithelium associated with diesel engine exhaust exposure. Environ. Int. 137, 105506. 10.1016/j.envint.2020.105506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta SK, Mitra PS, Ghosh S, Zang S, Sonneborn D, Hertz-Picciotto I, Trnovec T, Palkovicova L, Sovcikova E, Ghimbovschi S, Hoffman EP, 2012. Differential gene expression and a functional analysis of PCB-exposed children: understanding disease and disorder development. Environ. Int. 40, 143–154. 10.1016/j.envint.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A, Sakurai K, Watanabe M, Mori C, 2017. Exploration of potential biomarkers and related biological pathways for PCB exposure in maternal and cord serum: A pilot birth cohort study in Chiba. Japan. Environ. Int. 102, 157–164. 10.1016/j.envint.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Engström K, Wojdacz TK, Marabita F, Ewels P, Käller M, Vezzi F, Prezza N, Gruselius J, Vahter M, Broberg K, 2017. Transcriptomics and methylomics of CD4-positive T cells in arsenic-exposed women. Arch. Toxicol. 91 (5), 2067–2078. 10.1007/s00204-016-1879-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín-Pérez A, Krauskopf J, de Kok TM, Kleinjans JC, 2014. ‘OMICS-based’ Biomarkers for Environmental Health Studies. Curr. Environ. Health Rep. 1 (4), 353–362. 10.1007/s40572-014-0028-6. [DOI] [Google Scholar]
- Estill M, Hauser R, Nassan FL, Moss A, Krawetz SA, 2019. The effects of di-butyl phthalate exposure from medications on human sperm RNA among men. Sci. Rep. 9, 12397. 10.1038/s41598-019-48441-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everson TM, Marsit CJ, 2018. Integrating -omics approaches into human population-based studies of prenatal and early-life exposures. Curr. Environ. Health Rep. 5 (3), 328–337. 10.1007/s40572-018-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everson TM, Punshon T, Jackson BP, Hao K.e., Lambertini L, Chen J, Karagas MR, Marsit CJ, 2018. Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of two U.S. birth cohorts. Environ. Health Perspect. 126 (1), 017010. 10.1289/EHP2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Kim JH, Jones TR, McEachin RC, Nahar MS, Dolinoy DC, Sartor MA, 2015. Bisphenol A-associated alterations in genome-wide DNA methylation and gene expression patterns reveal sequence-dependent and non-monotonic effects in human fetal liver. Environ. Epigenetics 1. 10.1093/eep/dvv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest MS, Lan Q, Hubbard AE, Zhang L, Vermeulen R, Zhao X, Li G, Wu Y-Y, Shen M, Yin S, Chanock SJ, Rothman N, Smith MT, 2005. Discovery of novel biomarkers by microarray analysis of peripheral blood mononuclear cell gene expression in benzene-exposed workers. Environ. Health Perspect. 113 (6), 801–807. 10.1289/ehp.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PW, Pomares-Millan H, 2020. Next-generation epidemiology: the role of high-resolution molecular phenotyping in diabetes research. Diabetologia 63 (12), 2521–2532. 10.1007/s00125-020-05246-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, Bhattacharya S, Kandjanapa K, Soontararuks S, Nookabkaew S, Mahidol C, Ruchirawat M, Samson LD, 2007. Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers. PLoS Genet. 3, e207. 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Rager JE, Bauer R, Sebastian E, Peden DB, Jaspers I, Alexis NE, 2014. Air toxics and epigenetic effects: ozone altered microRNAs in the sputum of human subjects. Am. J. Physiol. Lung Cell. Mol. Physiol. 306 (12), L1129–L1137. 10.1152/ajplung.00348.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Colicino E, Shen J, Kioumourtzoglou M-A, Just AC, Nwanaji-Enwerem JC, Coull B, Lin X, Vokonas P, Zheng Y, Hou L, Schwartz J, Baccarelli AA, 2019. Impacts of air pollution, temperature, and relative humidity on leukocyte distribution: an epigenetic perspective. Environ. Int. 126, 395–405. 10.1016/j.envint.2019.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi S, Khateri S, Soroush MR, Shamsara M, Naeli P, Najafi A, Korsching E, Mowla SJ, Amendola R, 2018. MicroRNA expression in serum samples of sulfur mustard veterans as a diagnostic gateway to improve care. PloS One 13 (3), e0194530. 10.1371/journal.pone.0194530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Bernstein JA, Khurana Hershey GK, Rothenberg ME, Mersha TB, 2018. Leveraging multilayered “Omics” data for atopic dermatitis: a road map to precision medicine. Front. Immunol. 9, 2727. 10.3389/fimmu.2018.02727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Loffredo CA, Mitra PS, Trnovec T, Palkovicova Murinova L, Sovcikova E, Hoffman EP, Makambi KH, Dutta SK, 2018. PCB exposure and potential future cancer incidence in Slovak children: an assessment from molecular finger printing by ingenuity pathway analysis (IPA®) derived from experimental and epidemiological investigations. Environ. Sci. Pollut. Res. Int. 25 (17), 16493–16507. 10.1007/s11356-017-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JM, Reddy P, Naidoo RN, Asharam K, Batterman S, Dolinoy DC, 2016. Prenatal exposures and DNA methylation in newborns: a pilot study in Durban. South Africa. Environ. Sci. Process. Impacts 18 (7), 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia-Figueira S, Karimpour M, Bosson JA, Blomberg A, Unosson J, Pourazar J, Sandström T, Behndig AF, Nording ML, 2017. Mass spectrometry profiling of oxylipins, endocannabinoids, and N-acylethanolamines in human lung lavage fluids reveals responsiveness of prostaglandin E2 and associated lipid metabolites to biodiesel exhaust exposure. Anal. Bioanal. Chem. 409 (11), 2967–2980. 10.1007/s00216-017-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia-Figueira S, Karimpour M, Bosson JA, Blomberg A, Unosson J, Sehlstedt M, Pourazar J, Sandström T, Behndig AF, Nording ML, 2018. Mass spectrometry profiling reveals altered plasma levels of monohydroxy fatty acids and related lipids in healthy humans after controlled exposure to biodiesel exhaust. Anal. Chim. Acta 1018, 62–69. 10.1016/j.aca.2018.02.032. [DOI] [PubMed] [Google Scholar]
- Green BB, Karagas MR, Punshon T, Jackson BP, Robbins DJ, Houseman EA, Marsit CJ, 2016. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the new hampshire birth cohort study (USA). Environ. Health Perspect. 124 (8), 1253–1260. 10.1289/ehp.1510437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzieva O, Xu C-J, Breton CV, Annesi-Maesano I, Antó JM, Auffray C, Ballereau S, Bellander T, Bousquet J, Bustamante M, Charles M-A, de Kluizenaar Y, den Dekker HT, Duijts L, Felix JF, Gehring U, Guxens M, Jaddoe VVW, Jankipersadsing SA, Merid SK, Kere J, Kumar A, Lemonnier N, Lepeule J, Nystad W, Page CM, Panasevich S, Postma D, Slama R, Sunyer J, Söderhäll C, Yao J, London SJ, Pershagen G, Koppelman GH, Melén E, 2017. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ. Health Perspect. 125 (1), 104–110. 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzieva O, Xu C-J, Yousefi P, Relton C, Merid SK, Breton CV, Gao L.u., Volk HE, Feinberg JI, Ladd-Acosta C, Bakulski K, Auffray C, Lemonnier N, Plusquin M, Ghantous A, Herceg Z, Nawrot TS, Pizzi C, Richiardi L, Rusconi F, Vineis P, Kogevinas M, Felix JF, Duijts L, den Dekker HT, Jaddoe VWV, Ruiz JL, Bustamante M, Antó JM, Sunyer J, Vrijheid M, Gutzkow KB, Grazuleviciene R, Hernandez-Ferrer C, Annesi-Maesano I, Lepeule J, Bousquet J, Bergström A, Kull I, Söderhäll C, Kere J, Gehring U, Brunekreef B, Just AC, Wright RJ, Peng C, Gold DR, Kloog I, DeMeo DL, Pershagen G, Koppelman GH, London SJ, Baccarelli AA, Melén E, 2019. Prenatal particulate air pollution and DNA methylation in newborns: an epigenome-wide meta-analysis. Environ. Health Perspect. 127 (5), 057012. 10.1289/EHP4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Chen X, Wang J, Liu Z, Gaile D, Wu H, Yu G, Mao G, Yang Z, Di Z, Guo X, Cao L, Chang P, Kang B, Chen J, Gao W, Ren X, 2018. Multigenerational impacts of arsenic exposure on genome-wide DNA methylation and the implications for arsenic-induced skin lesions. Environ. Int. 119, 250–263. 10.1016/j.envint.2018.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Vargas H, Castelino J, Silver MJ, Dominguez-Salas P, Cros M-P, Durand G, Calvez-Kelm FL, Prentice AM, Wild CP, Moore SE, Hennig BJ, Herceg Z, Gong YY, Routledge MN, 2015. Exposure to aflatoxin B1 in utero is associated with DNA methylation in white blood cells of infants in The Gambia. Int. J. Epidemiol. 44 (4), 1238–1248. 10.1093/ije/dyv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach K, van Leeuwen DM, Gmuender H, Gottschalk RW, Stølevik SB, Nygaard UC, Løvik M, Granum B, Namork E, Meltzer HM, Kleinjans JC, van Delft JHM, van Loveren H, 2012. Toxicogenomic profiles in relation to maternal immunotoxic exposure and immune functionality in newborns. Toxicol. Sci. Off. J. Soc. Toxicol. 129, 315–324. 10.1093/toxsci/kfs214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard BE, Phillips J, Tandon A, Maharana A, Elmore R, Mav D, Sedykh A, Thayer K, Merrick BA, Walker V, Rooney A, Shah RR, 2020. SWIFT-active screener: accelerated document screening through active learning and integrated recall estimation. Environ. Int. 138, 105623. 10.1016/j.envint.2020.105623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo W-A, Sul D, Lee D-Y, Lee E, Kim C-W, 2004. Proteomic analysis of plasma proteins of workers exposed to benzene. Mutat. Res. 558 (1–2), 35–44. 10.1016/j.mrgentox.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Snyder MP, 2018. Integrative omics for health and disease. Nat. Rev. Genet. 19 (5), 299–310. 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey KT, Rytel M, Dere E, Butler R, Eliot M, Huse SM, Houseman EA, Koestler DC, Boekelheide K, 2019. Serum dioxin and DNA methylation in the sperm of operation ranch hand veterans exposed to Agent Orange. Environ. Health Glob. Access Sci. Source 18, 91. 10.1186/s12940-019-0533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibriya MG, Jasmine F, Argos M, Verret WJ, Rakibuz-Zaman M, Ahmed A, Parvez F, Ahsan H, 2007. Changes in gene expression profiles in response to selenium supplementation among individuals with arsenic-induced pre-malignant skin lesions. Toxicol. Lett. 169 (2), 162–176. 10.1016/j.toxlet.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Rozek LS, Soliman AS, Sartor MA, Hablas A, Seifeldin IA, Colacino JA, Weinhouse C, Nahar MS, Dolinoy DC, 2013. Bisphenol A-associated epigenomic changes in prepubescent girls: a cross-sectional study in Gharbiah. Egypt. Environ. Health Glob. Access Sci. Source 12, 33. 10.1186/1476-069X-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippler M, Engström K, Mlakar SJ, Bottai M, Ahmed S, Hossain MB, Raqib R, Vahter M, Broberg K, 2013. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics 8 (5), 494–503. 10.4161/epi.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ, 2013. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ. Health Perspect. 121 (8), 971–977. 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassowski M, Das V, Sahu SK, Misra BB, 2020. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front. Genet. 11, 610798. 10.3389/fgene.2020.610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf J, de Kok TM, Hebels DG, Bergdahl IA, Johansson A, Spaeth F, Kiviranta H, Rantakokko P, Kyrtopoulos SA, Kleinjans JC, 2017. MicroRNA profile for health risk assessment: environmental exposure to persistent organic pollutants strongly affects the human blood microRNA machinery. Sci. Rep. 7, 9262. 10.1038/s41598-017-10167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrtopoulos SA, 2013. Making sense of OMICS data in population-based environmental health studies. Environ. Mol. Mutagen. 54 (7), 468–479. 10.1002/em.21778. [DOI] [PubMed] [Google Scholar]
- Lam J, Howard BE, Thayer K, Shah RR, 2019. Low-calorie sweeteners and health outcomes: a demonstration of rapid evidence mapping (rEM). Environ. Int. 123, 451–458. 10.1016/j.envint.2018.11.070. [DOI] [PubMed] [Google Scholar]
- Leroy P, Tham A, Wong H, Tenney R, Chen C, Stiner R, Balmes JR, Paquet AC, Arjomandi M, Fu J, 2015. Inflammatory and repair pathways induced in human bronchoalveolar lavage cells with ozone inhalation. PloS One 10 (6), e0127283. 10.1371/journal.pone.0127283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Y-K, Ouyang B, Niu L, Xie C, Ying J, Medvedovic M, Chen A, Weihe P, Valvi D, Grandjean P, Ho S-M, 2018. Identification of sex-specific DNA methylation changes driven by specific chemicals in cord blood in a Faroese birth cohort. Epigenetics 13 (3), 290–300. 10.1080/15592294.2018.1445901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang M, Liang Q, Jin S, Sun X, Jiang Y, Pan X, Zhou Y, Peng Y, Zhang B, Zhou A, Zhang Y, Chen Z, Cao J, Zhang H, Xia W, Zheng T, Cai Z, Li Y, Xu S, 2017. Urinary metabolomics revealed arsenic exposure related to metabolic alterations in general Chinese pregnant women. J. Chromatogr. A 1479, 145–152. 10.1016/j.chroma.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Ljunggren SA, Helmfrid I, Norinder U, Fredriksson M, Wingren G, Karlsson H, Lindahl M, 2017. Alterations in high-density lipoprotein proteome and function associated with persistent organic pollutants. Environ. Int. 98, 204–211. 10.1016/j.envint.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Llorach R, Urpi-Sarda M, Jauregui O, Monagas M, Andres-Lacueva C, 2009. An LC-MS-based metabolomics approach for exploring urinary metabolome modifications after cocoa consumption. J. Proteome Res. 8 (11), 5060–5068. 10.1021/pr900470a. [DOI] [PubMed] [Google Scholar]
- Maccani JZJ, Koestler DC, Lester B, Houseman EA, Armstrong DA, Kelsey KT, Marsit CJ, 2015. Placental DNA methylation related to both infant toenail mercury and adverse neurobehavioral outcomes. Environ. Health Perspect. 123 (7), 723–729. 10.1289/ehp.1408561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale CM, Zhang L, Lan Q, Li G, Hubbard AE, Forrest MS, Vermeulen R, Chen J, Shen M, Rappaport SM, Yin S, Smith MT, Rothman N, 2009. Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross-comparison on two microarray platforms. Genomics 93 (4), 343–349. 10.1016/j.ygeno.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale CM, Zhang L, Lan Q, Vermeulen R, Li G, Hubbard AE, Porter KE, Thomas R, Portier CJ, Shen M, Rappaport SM, Yin S, Smith MT, Rothman N, 2011. Global gene expression profiling of a population exposed to a range of benzene levels. Environ. Health Perspect. 119 (5), 628–640. 10.1289/ehp.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miake-Lye IM, Hempel S, Shanman R, Shekelle PG, 2016. What is an evidence map? a systematic review of published evidence maps and their definitions, methods, and products. Syst. Rev. 5, 28. 10.1186/s13643-016-0204-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz A, Chervona Y, Hall M, Kluz T, Gamble MV, Costa M, 2015. Sex-specific ˜ patterns and deregulation of endocrine pathways in the gene expression profiles of Bangladeshi adults exposed to arsenic contaminated drinking water. Toxicol. Appl. Pharmacol. 284 (3), 330–338. 10.1016/j.taap.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobakht BF, Arefi Oskouie A, Rezaei-Tavirani M, Aliannejad R, Taheri S, Fathi F, Taghi Naseri M, 2017. NMR spectroscopy-based metabolomic study of serum in sulfur mustard exposed patients with lung disease. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 22 (5), 413–419. 10.1080/1354750X.2016.1203995. [DOI] [PubMed] [Google Scholar]
- Nwanaji-Enwerem JC, Dai L, Colicino E, Oulhote Y, Di Q, Kloog I, Just AC, Hou L, Vokonas P, Baccarelli AA, Weisskopf MG, Schwartz JD, 2017. Associations between long-term exposure to PM2.5 component species and blood DNA methylation age in the elderly: The VA normative aging study. Environ. Int. 102, 57–65. 10.1016/j.envint.2016.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennie W, Pettit SD, Lord PG, 2004. Toxicogenomics in risk assessment: an overview of an HESI collaborative research program. Environ. Health Perspect. 112 (4), 417–419. 10.1289/ehp.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera F, Herbstman J, 2011. Prenatal environmental exposures, epigenetics, and disease. Reprod. Toxicol. Elmsford N 31 (3), 363–373. 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz A, Peck EC, Bammler TK, Beyer RP, Sullivan JH, Trenga CA, Srinouanprachnah S, Farin FM, Kaufman JD, 2007. Diesel exhaust inhalation and assessment of peripheral blood mononuclear cell gene transcription effects: an exploratory study of healthy human volunteers. Inhal. Toxicol. 19 (14), 1107–1119. 10.1080/08958370701665384. [DOI] [PubMed] [Google Scholar]
- Rager JE, Auerbach SS, Chappell GA, Martin E, Thompson CM, Fry RC, 2017. Benchmark dose modeling estimates of the concentrations of inorganic arsenic that induce changes to the neonatal transcriptome, proteome, and epigenome in a pregnancy cohort. Chem. Res. Toxicol. 30 (10), 1911–1920. 10.1021/acs.chemrestox.7b00221. [DOI] [PubMed] [Google Scholar]
- Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, Drobná Z, Currier J, Douillet C, Olshan AF, Rubio-Andrade M, Stýblo M, García-Vargas G, Fry RC, 2014. Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen. 55 (3), 196–208. 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager JE, Tilley SK, Tulenko SE, Smeester L, Ray PD, Yosim A, Currier JM, Ishida MC, González-Horta MDC, Sánchez-Ramírez B, Ballinas-Casarrubias L, Gutiérrez-Torres DS, Drobná Z, Del Razo LM, García-Vargas GG, Kim WY, Zhou Y-H, Wright FA, Stýblo M, Fry RC, 2015. Identification of novel gene targets and putative regulators of arsenic-associated DNA methylation in human urothelial cells and bladder cancer. Chem. Res. Toxicol. 28 (6), 1144–1155. 10.1021/tx500393y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana HK, Akhtar MR, Ahmed MB, Lio P, Quinn JMW, Huq F, Moni MA, ` 2019. Genetic effects of welding fumes on the progression of neurodegenerative diseases. Neurotoxicology 71, 93–101. 10.1016/j.neuro.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Rana HK, Akhtar MR, Islam MB, Ahmed MB, Lió P, Huq F, Quinn JMW, Moni MA, 2020. Machine learning and bioinformatics models to identify pathways that mediate influences of welding fumes on cancer progression. Sci. Rep. 10, 2795. 10.1038/s41598-020-57916-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockett JC, Burczynski ME, Fornace AJ, Herrmann PC, Krawetz SA, Dix DJ, 2004. Surrogate tissue analysis: monitoring toxicant exposure and health status of inaccessible tissues through the analysis of accessible tissues and cells. Toxicol. Appl. Pharmacol. 194 (2), 189–199. 10.1016/j.taap.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Rojas D, Rager JE, Smeester L, Bailey KA, Drobná Z, Rubio-Andrade M, Stýblo M, García-Vargas G, Fry RC, 2015. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol. Sci. Off. J. Soc. Toxicol. 143, 97–106. 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner P, Tulupova E, Rossnerova A, Libalova H, Honkova K, Gmuender H, Pastorkova A, Svecova V, Topinka J, Sram RJ, 2015. Reduced gene expression levels after chronic exposure to high concentrations of air pollutants. Mutat. Res. 780, 60–70. 10.1016/j.mrfmmm.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Rynning I, Arlt VM, Vrbova K, Neča J, Rossner P Jr, Klema J, Ulvestad B, Petersen E, Skare Ø, Haugen A, Phillips DH, Machala M, Topinka J, Mollerup S, 2019. Bulky DNA adducts, microRNA profiles, and lipid biomarkers in Norwegian tunnel finishing workers occupationally exposed to diesel exhaust. Occup. Environ. Med. 76 (1), 10–16. 10.1136/oemed-2018-10544510.1136/oemed-2018-105445.supp1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AP, Burris HH, Just AC, Motta V, Amarasiriwardena C, Svensson K, Oken E, Solano-Gonzalez M, Mercado-Garcia A, Pantic I, Schwartz J, Tellez-Rojo MM, Baccarelli AA, Wright RO, 2015. Altered miRNA expression in the cervix during pregnancy associated with lead and mercury exposure. Epigenomics 7 (6), 885–896. 10.2217/epi.15.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A, Smeester L, Rojas D, DeBussycher T, Wu M, Wright F, Zhou Y-H, Laine J, Rager J, Swamy G, Ashley-Koch A, Lynn Miranda M, Fry R, 2014. Cadmium exposure and the epigenome: Exposure-associated patterns of DNA methylation in leukocytes from mother-baby pairs. Epigenetics 9 (2), 212–221. 10.4161/epi.26798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis R, Brown PO, 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270 (5235), 467–470. 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schiffman C, McHale CM, Hubbard AE, Zhang L, Thomas R, Vermeulen R, Li G, Shen M, Rappaport SM, Yin S, Lan Q, Smith MT, Rothman N, Peddada SD, 2018. Identification of gene expression predictors of occupational benzene exposure. PloS One 13 (10), e0205427. 10.1371/journal.pone.0205427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Cingolani P, Senut M-C, Land S, Mercado-Garcia A, Tellez-Rojo MM, Baccarelli AA, Wright RO, Ruden DM, 2015a. Lead exposure induces changes in 5-hydroxymethylcytosine clusters in CpG islands in human embryonic stem cells and umbilical cord blood. Epigenetics 10 (7), 607–621. 10.1080/15592294.2015.1050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Heredia N, Senut M-C, Land S, Hollocher K, Lu X, Dereski MO, Ruden DM, 2015b. Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren. Sci. Rep. 5, 14466. 10.1038/srep14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeester L, Rager JE, Bailey KA, Guan X, Smith N, García-Vargas G, Del Razo L-M, Drobná Z, Kelkar H, Stýblo M, Fry RC, 2011. Epigenetic changes in individuals with arsenicosis. Chem. Res. Toxicol. 24 (2), 165–167. 10.1021/tx1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Xue J, Li J, Luo F, Chen X, Liu Y, Wang Q, Qi C, Zou Z, Zhang A, Liu Q, 2017. Circulating miRNAs and their target genes associated with arsenism caused by coal-burning. Toxicol. Res. 6 (2), 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowiec I, Karimpour M, Gouveia-Figueira S, Wu J, Unosson J, Bosson JA, Blomberg A, Pourazar J, Sandström T, Behndig AF, Trygg J, Nording ML, 2016. Multi-platform metabolomics assays for human lung lavage fluids in an air pollution exposure study. Anal. Bioanal. Chem. 408 (17), 4751–4764. 10.1007/s00216-016-9566-0. [DOI] [PubMed] [Google Scholar]
- Tham A, Lullo D, Dalton S, Zeng S, van Koeverden I, Arjomandi M, 2017. Modeling vascular inflammation and atherogenicity after inhalation of ambient levels of ozone: exploratory lessons from transcriptomics. Inhal. Toxicol. 29 (3), 96–105. 10.1080/08958378.2017.1310333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breda SGJ, Wilms LC, Gaj S, Jennen DGJ, Briedé JJ, Kleinjans JCS, de Kok TMCM, 2015. The exposome concept in a human nutrigenomics study: evaluating the impact of exposure to a complex mixture of phytochemicals using transcriptomics signatures. Mutagenesis 30, 723–731. 10.1093/mutage/gev008. [DOI] [PubMed] [Google Scholar]
- van den Dungen MW, Murk AJ, Kampman E, Steegenga WT, Kok DE, 2017. Association between DNA methylation profiles in leukocytes and serum levels of persistent organic pollutants in Dutch men. Environ. Epigenetics 3, dvx001. 10.1093/eep/dvx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen R, Lan Q, Zhang L, Gunn L, McCarthy D, Woodbury RL, McGuire M, Podust VN, Li G, Chatterjee N, Mu R, Yin S, Rothman N, Smith MT, 2005. Decreased levels of CXC-chemokines in serum of benzene-exposed workers identified by array-based proteomics. Proc. Natl. Acad. Sci. U. S. A. 102 (47), 17041–17046. 10.1073/pnas.0508573102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DI, Uppal K, Zhang L, Vermeulen R, Smith M, Hu W, Purdue MP, Tang X, Reiss B, Kim S, Li L, Huang H, Pennell KD, Jones DP, Rothman N, Lan Q, 2016. High-resolution metabolomics of occupational exposure to trichloroethylene. Int. J. Epidemiol. 45 (5), 1517–1527. 10.1093/ije/dyw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DD, Shams-White M, Bright OJM, Parrott JS, Chung M, 2016. Creating a literature database of low-calorie sweeteners and health studies: evidence mapping. BMC Med. Res. Methodol. 16, 1. 10.1186/s12874-015-0105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yan R, Wang B, Meng P, Tan W, Guo X, 2019. The functional analysis of selenium-related genes and magnesium-related genes in the gene expression profile microarray in the peripheral blood mononuclear cells of keshan disease. Biol. Trace Elem. Res. 192 (1), 3–9. 10.1007/s12011-019-01750-2. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang T, Xu M, Yu H, Ding C, Wang Z, Pan X, Li Y, Niu Y, Yan R, Song J, Yan H, Dai Y, Sun Z, Su W, Duan H, 2020. Independent effect of main components in particulate matter on DNA methylation and DNA methyltransferase: a molecular epidemiology study. Environ. Int. 134, 105296. 10.1016/j.envint.2019.105296. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zheng Y, Zhao B, Zhang Y, Liu Z, Xu J, Chen Y, Yang Z, Wang F, Wang H, He J, Zhang R, Abliz Z, 2015. Human metabolic responses to chronic environmental polycyclic aromatic hydrocarbon exposure by a metabolomic approach. J. Proteome Res. 14 (6), 2583–2593. 10.1021/acs.jproteome.5b00134. [DOI] [PubMed] [Google Scholar]
- Wolffe TAM, Whaley P, Halsall C, Rooney AA, Walker VR, 2019. Systematic evidence maps as a novel tool to support evidence-based decision-making in chemicals policy and risk management. Environ. Int. 130, 104871. 10.1016/j.envint.2019.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Estill MS, Shershebnev A, Suvorov A, Krawetz SA, Whitcomb BW, Dinnie H, Rahil T, Sites CK, Pilsner JR, 2017. Preconception urinary phthalate concentrations and sperm DNA methylation profiles among men undergoing IVF treatment: a cross-sectional study. Hum. Reprod. Oxf. Engl. 32, 2159–2169. 10.1093/humrep/dex283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M-M, Chiou H-Y, Ho I-C, Chen C-J, Lee T-C, 2003. Gene expression of inflammatory molecules in circulating lymphocytes from arsenic-exposed human subjects. Environ. Health Perspect. 111 (11), 1429–1438. 10.1289/ehp.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jurkovic-Mlakar S, Li Y, Wahlberg K, Scott K, Pineda D, Lindh CH, Jakobsson K, Engström K, 2020a. Association between serum concentrations of perfluoroalkyl substances (PFAS) and expression of serum microRNAs in a cohort highly exposed to PFAS from drinking water. Environ. Int. 136, 105446. 10.1016/j.envint.2019.105446. [DOI] [PubMed] [Google Scholar]
- Xu Y, Jurkovic-Mlakar S, Lindh CH, Scott K, Fletcher T, Jakobsson K, Engström K, 2020b. Associations between serum concentrations of perfluoroalkyl substances and DNA methylation in women exposed through drinking water: a pilot study in Ronneby. Sweden. Environ. Int. 145, 106148. 10.1016/j.envint.2020.106148. [DOI] [PubMed] [Google Scholar]
- Yang T-Y, Hsu L-I, Chiu AW, Pu Y-S, Wang S-H, Liao Y-T, Wu M-M, Wang Y-H, Chang C-H, Lee T-C, Chen C-J, 2014. Comparison of genome-wide DNA methylation in urothelial carcinomas of patients with and without arsenic exposure. Environ. Res. 128, 57–63. 10.1016/j.envres.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Yu SY, Paul S, Hwang SY, 2016. Application of the emerging technologies in toxicogenomics: an overview. BioChip J. 10 (4), 288–296. 10.1007/s13206-016-0405-3. [DOI] [Google Scholar]
- Zhai R, Su S, Lu X, Liao R, Ge X, He M, Huang Y, Mai S, Lu X.i., Christiani D, 2005. Proteomic profiling in the sera of workers occupationally exposed to arsenic and lead: identification of potential biomarkers. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 18 (6), 603–613. 10.1007/s10534-005-3001-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.