Abstract
A plasmid gap repair assay was used to assess the role of three known nucleases, Exo1, Mre11 and Rad1, in the processing of DNA ends and resolution of recombination intermediates during double-strand gap repair. In this assay, alterations in end processing or branch migration are reflected by the frequency of co-conversion of a chromosomal marker 200 bp from the gap. Gap repair associated with crossing over results in integration at the homologous chromosomal locus, whereas the plasmid remains episomal for non-crossover repair events. In mre11 strains, the frequency of gap repair was reduced 3- to 10-fold and conversion tracts were shorter than in the wild-type strain, consistent with a role for this nuclease in processing double-strand breaks. However, conversion tracts were longer in a strain containing the nuclease deficient allele, mre11-H125N, suggesting increased end processing by redundant nucleases. The frequency of gap repair was reduced 2-fold in rad1 mutants and crossing over was reduced, consistent with a role for Rad1 in cleaving recombination intermediates. The frequency of gap repair was increased in exo1 mutants with a significant increase in crossing over. In exo1 mre11 double mutants gap repair was reduced to below the mre11 single mutant level.
INTRODUCTION
Deoxyribonucleases are involved in many aspects of DNA metabolism, including replication, repair and recombination. Models for the repair of DNA double-strand breaks (DSBs) by recombination invoke nuclease processing at multiple steps (1). Nucleases are predicted to resect the ends at DSBs to generate intermediates with 3′ single-stranded tails. This could occur by the activity of a double-stranded exonuclease, or a single-stranded endo- or exonuclease working in concert with a helicase. Intermediates with 3′ single-stranded tails have been identified during meiotic recombination and at the MAT locus during mating type switching (2,3). Following homologous pairing and strand invasion to produce heteroduplex DNA, the displaced strand (D-loop) may pair with the other side of the break to form an intermediate with Holliday junctions. The Holliday junctions can be resolved by branch migration or endonucleolytic cleavage (4). Alternatively, the D-loop formed by strand invasion could collapse after limited DNA synthesis to displace the invading 3′-end (5), resulting in gap repair without an associated crossover. Mismatches present in heteroduplex DNA are repaired by the mismatch repair system, which involves exonucleolytic processing of one strand of the heteroduplex to the site of the mismatch (6,7).
The nucleases involved in each of these steps in Escherichia coli have been identified by genetic and biochemical methods, however, much less is known of the nucleases with equivalent functions in eukaryotes. Of the genes required for ionizing radiation resistance in yeast, most function at a step subsequent to the processing of DNA ends. The exceptions are the MRE11, RAD50 and XRS2 genes, the products of which function as a complex to regulate the processing of DSBs in vivo. In these mutants there is a delay in the resection of DSBs generated by the HO endonuclease, but repair still occurs (8). The Mre11 protein has endonuclease and 3′→5′ exonuclease activities in vitro (9–12). However, point mutations that abolish the nuclease activity of Mre11 in vitro cause no apparent defect in the processing step in vivo in mitotic cells, although they are defective in processing DSBs produced during meiosis (9,11–13). Furthermore, as the polarity of the Mre11 exonuclease is opposite to that predicted for resection of DSBs, this has raised the issue of whether Mre11 functions directly in the processing of ends. An alternate hypothesis is that the Mre11/Rad50/Xrs2 (MRX) complex unwinds DNA ends for the Mre11 endonuclease to clip off the 5′ strands, and this nuclease activity is redundant with other nucleases in mitotic cells (12).
Exonuclease I was identified from fractionated extracts of Schizosaccharomyces pombe and Saccharomyces cerevisiae as a 5′→3′ double-stranded deoxyribonuclease (14,15). As the polarity of degradation was the same as shown for resection of DSBs in vivo, and the activity was induced 5-fold in meiotic extracts of S.pombe, it appeared to be a good candidate for the nuclease active in processing DSBs (14). However, exo1 mutants are not sensitive to ionizing radiation, are proficient at mating type switching and have only weak defects in meiosis (16,17). Although spore viability is not dramatically reduced in exo1 mutants, there is a significant decrease in intergenic recombination and an increase in meiosis I non-disjunction (18–20). Exo1 interacts with Msh2 and exo1 mutants have a mutator phenotype suggesting a role in mismatch repair (16,21). The role of Exo1 in mitotic recombination is complex: the mutants show a decrease in spontaneous mitotic recombination between direct repeats and an increase in homeologous recombination (17,22). The latter is thought to be due to anti-recombination between diverged sequences observed for many mismatch repair mutants (23).
Rad1 is an essential component of the nucleotide excision repair (NER) pathway and mutants show high sensitivity to UV light (24,25). Rad1 forms a heterodimer with Rad10 that exhibits structure-specific endonuclease activity in vitro (26,27). This nuclease cleaves at the junction between duplex and single-stranded DNA at the 5′ side of a UV photoproduct during NER. In addition to the essential role in NER, Rad1 is required for certain mitotic recombination events to trim recombination intermediates with 3′ heterologous tails and in the repair of loop heteroduplexes formed during meiotic recombination (28,29). rad1 mutants also exhibit defects in mitotic conversion tract length, gene replacement and in plasmid integration (30,31). The Rad1 nuclease has been shown to cleave artificial Holliday junctions generated from oligonucleotides, but the cleavages are not symmetrical like those observed for other Holliday junction resolvases, such as RuvC (32,33). Although rad1 mutants of S.cerevisiae are sporulation proficient, mutation of the Drosophila melanogaster homolog of RAD1, mei-9, causes a defect in meiotic crossing over (34,35). It is possible that the Rad1 nuclease is redundant with another Holliday junction resolvase in yeast, or that the endonuclease activity cleaves other types of branched intermediates formed during recombination.
We have used a plasmid gap repair assay to evaluate the role of these nucleases in mitotic recombination (36,37). A plasmid containing a double-strand break or gap within a region of the plasmid homologous to chromosomal sequences is efficiently repaired when introduced into yeast cells by transformation. Homology-dependent gap repair predominates in recombination proficient cells, but end-joining events can also occur (36,37). If the gapped plasmid contains no origin of replication, only repair events associated with crossing over to integrate the plasmid at the homologous chromosomal locus are recovered. However, if the gapped plasmid contains an origin of replication repair events unassociated with crossing over also occur. Heteroduplex DNA formed adjacent to the gap during plasmid gap repair can be monitored by co-conversion of markers flanking the plasmid-borne gap. Thus, plasmid gap repair provides a convenient system to measure conversion tract lengths and associated crossing over during double-strand gap repair. Here we show that several nucleases implicated in mitotic recombination have both quantitative and qualitative effects on plasmid gap repair.
MATERIALS AND METHODS
Yeast strains and plasmids
All strains are derivatives of W303-1A or W303-1B with the corrected RAD5 allele and are listed in Table 1 (38,39). Strains LSY697 and LSY698 containing the met17-sna and met17::ADE2 alleles, respectively, have been described previously (37). The met17-sna allele results from a single base pair insertion within the SnaBI site that creates a stop codon and destroys the SnaBI site. These strains were crossed to other W303 derivatives containing the exo1::HIS3, mre11::LEU2, mre11-H125N or rad1::LEU2 mutations and the resulting diploids sporulated to generate haploid progeny of the desired genotype. The plasmid substrates for gap repair, pSB101 and pSB110, were described previously (37). Briefly, pSB101 contains the URA3 and MET17 genes and no origin of replication; pSB110 is identical except for insertion of an ARSH4 element.
Table 1. Yeast strains.
| Strain | Relevant genotypea | Source |
|---|---|---|
| LSY697 | MATa met17-sna ADE2 | (37) |
| LSY698 | MATa met17::ADE2 | (37) |
| LSY870-2C | MATa met17-sna ADE2 mre11::LEU2 | This study |
| LSY871-5A | MATa met17::ADE2 mre11::LEU2 | This study |
| LSY889-2D | MATa met17-sna ADE2 mre11::LEU2 exo1::HIS3 | This study |
| LSY894-4C | MATa met17-sna ADE2 exo1::HIS3 | This study |
| LSY895-10A | MATα met17-sna ADE2 rad1::LEU2 | This study |
| LSY918-1 | MATα met17-sna ADE2 mre11-H125N | This study |
| LSY920-20A | MATa met17-sna ADE2 mre11::LEU2 rad1::LEU2 | This study |
| LSY927-1C | MATa met17::ADE2 exo1::HIS3 | This study |
| LSY928-9C | MATa met17::ADE2 rad1::LEU2 | This study |
aAll strains are derivatives of W303-1A or W303-1B (leu2-3,112, trp1-1, can1-100, ura3-1, ade2-1, his3-11,15 RAD5), only differences from this genotype are listed above.
Media and growth conditions
Rich medium (YEPD), synthetic medium (SC) and lead-containing plates were prepared as described previously (40,41). Cultures and plates were incubated at 30°C unless otherwise stated. Methods for yeast transformation and tetrad dissection were as described previously (40).
Plasmid gap repair assays
Plasmids to be used as substrates for gap repair were digested with BspEI and EcoNI, and the large DNA fragment gel purified. Transformation of yeast cells was by the lithium acetate method using 100 ng of gapped plasmid or 100 ng of uncut pSB110 (transformation efficiency control) in the presence of 50 µg of denatured salmon sperm carrier DNA. For the mre11 rad1 and mre11 exo1 double mutants, 300 ng of gapped pSB110 and 600 ng of gapped pSB101 were used. The amounts of DNA used for transformation were determined to be in the linear range. The transformation efficiency using gapped plasmids was improved by inclusion of a 20 min, 30°C incubation step in SOS (1 M sorbitol, 0.3% yeast extract, 0.6% bacto-peptone, 6.5 mM CaCl2) medium following heat shock. The transformed cells were diluted and plated onto SC-Ura and SC-Ura-Met media and colonies counted after 3 days incubation at 30°C. The gap repair frequency was calculated as the number of Met+ Ura+ transformants per microgram of transformed gapped plasmid divided by the number of Met+ Ura+ transformants per microgram of uncut pSB110. The gap repair assay was performed at least three times for each strain and the mean values are presented.
Analysis of gap repair products
Ura+ transformants were picked from transformation plates and patched on SC-Ura plates. After 2 days of growth, the cells were replica plated to SC-Met and YEPD plates to score for the Met phenotype and to allow non-selective growth. After 2 days of growth on YEPD, the transformants were replica plated onto 5-FOA and Pb2+ plates to assess the mitotic stability phenotype. Confluent growth on 5-FOA indicated that the Ura+ phenotype was unstable. Integration of the plasmid results in a direct repeat and a stable Ura+ phenotype due to the low rate of excision of the plasmid from the chromosomal duplication. Met– cells are pigmented dark brown due to the accumulation of PbS when grown on plates containing Pb2+. Transformants that grew on SC-Met, but were dark brown on Pb plates following non-selective growth, were scored as unstable Met+. The data in Table 3 were pooled from three independent transformations of each strain with the exception of the mre11 rad1 strain, for which two independent analyses were performed. Within each strain, there was little variation in the ratios of Met+ to Met– and integrated to non-integrated between transformations. Significance was determined by chi-squared tests.
Table 3. Distribution of Ura+ products from pSB110 gap repair.
| Relevant genotype | Total Ura+a | Met+b | Short tract non-crossover (Ura+u Met+u) | Long tract non-crossover (Ura+u Met–) | Short tract corssover (Ura+s Met+s) | Long tract corssover (Ura+s Met–) | Othersc | % Crossovers |
|---|---|---|---|---|---|---|---|---|
| RAD1 EXO1 MRE11 | 926 | 50.9 | 40.3 | 36.4 | 9.5 | 12.6 | 1.2 | 22.1 |
| mre11Δ | 675 | 62.9d | 55.4 | 29.3 | 7.6 | 7.7 | 0 | 15.3d |
| mre11-H125N | 516 | 34.9d | 28.1 | 52.3 | 6.8 | 12.6 | 0.2 | 19.4 |
| rad1Δ | 510 | 52.5 | 48.0 | 40.6 | 4.3 | 6.9 | 0.2 | 11.2d |
| mre11Δ rad1Δ | 406 | 66.5d | 61.1 | 26.1 | 5.4 | 7.4 | 0 | 12.8d |
| exo1Δ | 415 | 54.9 | 37.6 | 28.9 | 17.1 | 16.1 | 0.2 | 33.2d |
| mre11Δ exo1Δ | 477 | 65.0d | 40.9 | 23.5 | 24.1 | 11.5 | 0 | 35.6d |
aNumber of Ura+ transformants analyzed; the other numbers are given as percentages of the total Ura+.
bPercentages of Ura+ transformants that were Met+.
cPercentages of transformants that were classified as Ura+s Met+u and Ura+u Met+s.
dSignificantly different from wild-type (P < 0.001).
The efficiency of gene replacement was determined by transforming strains containing the met17::ADE2 allele with a 2.5 kb SpeI fragment containing MET17 derived from pSB101 and selecting Met+ transformants. Correct replacement of the met17::ADE2 allele results in a Met+ Ade– phenotype; the rare Met+ Ade+ transformants were deducted from the total number of Met+ to calculate recombination efficiencies. The number of Met+ transformants was normalized to the transformation efficiency using uncut pSB110.
RESULTS
Plasmid gap repair assay
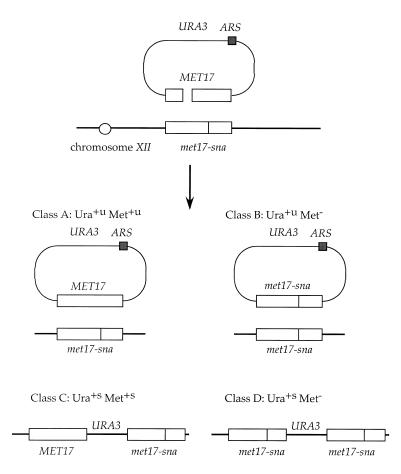
We previously described a gap repair assay based on a series of plasmids containing the selectable/counter-selectable marker, URA3, and the selectable, colony-color marker, MET17 (37). A 238 bp double-strand gap was made within the plasmid MET17 gene by digestion with BspEI and EcoNI endonucleases. The gapped plasmid DNA was introduced into host cells containing a nonsense mutation within the chromosomal MET17 gene, met17-sna, by transformation. The efficiency of repair was determined from the number of Met+ transformants relative to the transformation efficiency with an uncut replicating plasmid. We assume linear and circular molecules enter cells with equal efficiency and this ratio is not altered in any of the mutants tested. To determine the spectrum of repair events, Ura+ transformants were selected and analyzed for the Met phenotype and also for the mitotic stability of the plasmid markers. Gap repair unassociated with crossing over results in a Ura+ phenotype that is mitotically unstable (Ura+u), whereas gap repair associated with crossing over (integration) generates a stable Ura+ phenotype (Ura+s). Although direct repeat recombination can result in excision of the plasmid, these events occur at much lower frequency than plasmid mis-segregation. Four major classes of events were recovered from wild-type cells using the gapped ARS-containing plasmid (Fig. 1): Ura+u Met+u, Ura+u Met–, Ura+s Met+s and Ura+s Met–. Forty-nine percent of the Ura+ transformants obtained from the wild-type strain were Met– and for both the integrated and episomal events the Met– phenotype was due to conversion of the plasmid MET17 gene to met17-sna (37). Conversion from the chromosomal marker to the plasmid could occur by extension of the gap, or heteroduplex DNA (hDNA) formation adjacent to the gap, followed by mismatch repair (Fig. 2). For both HO-induced mitotic recombination and DSB-induced meiotic recombination the 3′-end at break sites remains intact and conversion events are thought to occur by mismatch correction of asymmetric hDNA (3,42). The frequency of conversion of a marker adjacent to the break or gap is distance-dependent and reflects the length of the 3′ single-stranded tail formed by resection of the 5′ strand (3). Although we assume the ratio of Met+ to Met– transformants is directed by the length of the 3′ single-stranded tail, post-synaptic processing and mismatch repair could also influence co-conversion.
Figure 1.
The gap repair assay. Repair of the gap within the MET17 gene of pSB110 occurs by interaction with the homologous chromosomal locus. The chromosomal met17-sna mutation (elimination of the SnaBI site) is located 216 bp from the EcoNI site bordering the gap. Repair of the gap unassociated with crossing over generates an unstable Ura+ (Ura+u) phenotype, whereas repair associated with crossing over results in a stable Ura+ (Ura+s) phenotype. Co-conversion of the met17-sna mutation during gap repair results in a Met– phenotype, whereas repair events that do not extend to the met17-sna site, or heteroduplexes repaired in favor of the wild-type sequences, are Met+. Repair of pSB101 can only occur by crossing over because the plasmid has no ARS element (yeast replication origin) for maintenance as an episome.
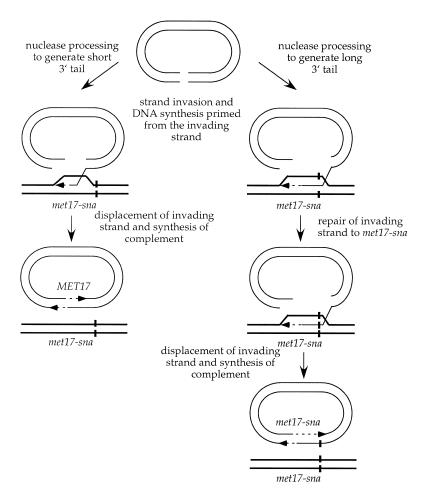
Figure 2.
Origin of the Met+ and Met– recombinants. Extensive processing of the ends flanking the plasmid gap generates long 3′ single-stranded tails including the MET17 SnaBI site. Heteroduplexes resulting from strand invasion will contain the mutant and wild-type SnaBI sites. Mismatch repair of the invading strand will result in met17-sna information on the invading strand. Following DNA synthesis from the invading 3′-end, the invading strand is displaced and pairs with the 3′ tail at the other side of the break. DNA synthesis initiated from the other 3′-end, or by coupled lagging strand synthesis during the initial invasion, will duplicate the met17-sna allele. Less extensive processing of the break will result in the MET17 SnaBI site remaining double-stranded and it will not be incorporated into hDNA during strand invasion.
Decreased efficiency of gap repair and reduction in conversion tract length in the mre11 mutant
The Mre11 nuclease is implicated in the end-processing step based on physical studies of DNA intermediates formed during DSB repair. We expected to see an increase in the ratio of Met+ to Met– transformants in mre11 mutants if the Mre11 nuclease in involved in the processing reaction. The efficiency of gap repair with gapped pSB110 was reduced 3-fold in the mre11Δ strain with a significant increase in the number of Met+ transformants and a decrease in the number of integration events (P < 0.001) (Tables 2 and 3). The class of transfomants showing the greatest alteration in number was the Ura+u Met+u class, which represented 55% of the total Ura+ transformants in the mre11Δ strain, compared with 40% in the wild-type strain (Table 3). To determine whether the nuclease activity of Mre11, or some other function of Mre11, is responsible for the alteration in the spectrum of recombination products, the gap repair assay was performed in an mre11-H125N strain, which contains a point mutation abolishing the Mre11 nuclease activity (12). The efficiency of gap repair was only slightly reduced compared with the wild-type strain and was significantly higher than the mre11Δ strain (Table 2). Surprisingly, the percentage of Ura+ transformants that were Met+ was much lower than the wild-type and mre11Δ strains (P < 0.001) (Table 3). The mre11-H125N strain was the only one examined in which the Ura+u Met– class exceeded 50% of the total and the Ura+u Met+u class was <29% of the total. The mre11Δ strain showed a significant reduction in the number of integration events, suggesting that the defect in gap repair was likely to be enhanced using the non-replicating vector, pSB101. Ura+ transformants derived from pSB101 are all stable and result from integration of the plasmid. There was a 10-fold decrease in gap repair efficiency of pSB101 in the mre11Δ strain, compared with wild-type, an even greater decrease than predicted.
Table 2. Gap repair efficiency in exo1, mre11 and rad1 strains.
| Relevant genotype | Transformants /µg ×104 uncut pSB110 | pSB110a gap repair (×10–2)b | Fold increase | pSB110a gap repair (×10–2)b | Fold increase | Gene replacement (×10–3)c | Fold increase |
|---|---|---|---|---|---|---|---|
| EXO1 MRE11 RAD1 | 17.6 ± 2.8 | 24.3 ± 7.4 | 4.4 ± 1.3 | 8.2 ± 1.6 | |||
| exo1Δ | 16.2 ± 0.8 | 46.3 ± 9.6 | 1.9 | 9.8 ± 3.0 | 2.2 | 23.1 ± 2.9 | 2.8 |
| mre11Δ | 4.4 ± 1.8 | 7.4 ± 2.8 | 0.3 | 0.41 ± 0.1 | 0.09 | 21.9 ± 7.7 | 2.7 |
| mre11-H125N | 19.1 ± 6.5 | 18.4 ± 5.4 | 0.8 | NDd | ND | ||
| rad1Δ | 13.2 ± 3.9 | 16.0 ± 5.0 | 0.7 | 1.9 ± 0.8 | 0.4 | 2.6 ± 1.1 | 0.3 |
| exo1Δ mre11Δ | 0.9 ± 0.6 | 2.8 ± 1.6 | 0.1 | 0.08 | 0.02 | ND | |
| rad1Δ mre11Δ | 3.3 ± 1.6 | 4.8 ± 1.6 | 0.2 | 0.26 ± 0.03 | 0.06 | ND |
apSB110 is ARS+; pSB101 contains no ARS element.
bGap repair frequencies were determined by the number of Ura+ Met+ transformants obtained from the gapped plasmid divided by the number of Ura+ Met+ transformants obtained from an equivalent amount of uncut pSB110 DNA.
cGene replacement frequencies were determined by the number of Met+ transformants obtained from the MET17 fragment divided by the number of Met+ transformants obtained from an equivalent amount of uncut pSB110 DNA.
dND, not determined.
Plasmid integration is reduced in rad1 mutants
Mutation of the RAD1 gene has previously been shown to cause severe defects in plasmid integration and one-step gene replacement (30). The frequency of gap repair of pSB110 was reduced less than 2-fold in the rad1 strain (Table 2). However, there was a significant reduction in the number of integration events recovered (P < 0.001) (Table 3). As expected, based on the low frequency of integration of pSB110, there was a greater decrease in gap repair efficiency using the non-replicating gapped plasmid. Because there was a reduction in crossing over (integration) in both the mre11 and rad1 strains, a rad1 mre11 mutant was also tested to see if these nucleases function in the same or different pathways. The effect of the two mutations on the frequency of gap repair was additive suggesting both nucleases contribute to efficient gap repair. Ura+ transformants from the mre11 rad1 strain showed the same ratio of Met+:Met– as observed for the mre11Δ strain, and the same ratio of integrated to episomal events as the rad1 strain.
Gap repair efficiency and integration are increased by mutation of EXO1
The efficiency of gap repair was increased by almost 2-fold in the exo1 strain (Table 2) and analysis of the Ura+ products revealed a significant increase in the number of integration events (P < 0.001) (Table 3). The ratio of Met+:Met– events was not significantly different from the wild-type strain, suggesting that Exo1 does not normally affect the processing of DSBs. Strains deleted for both MRE11 and EXO1 show enhanced sensitivity to methyl methane sulfonate (MMS) and slower processing of an HO-induced DSB than mre11 single mutants (20). To determine whether the Mre11 and Exo1 nucleases act independently in processing plasmid DSBs, the gap repair assay was repeated in an exo1 mre11 double mutant. The efficiency of gap repair of pSB110 was reduced 19-fold compared with the exo1 strain, 10-fold compared with wild-type and 3-fold compared with the mre11 strain, consistent with the synergism observed in other DNA repair assays. Analysis of Ura+ products revealed the same distribution of Met+:Met– transformants as observed for the mre11 strain and the high level of crossing over characteristic of the exo1 strain. In this strain, 24% of the Ura+ transformants were stable Met+. Although the gap repair efficiency of pSB110 was reduced 10-fold, one-third of the products contained integrated plasmids suggesting that gap repair of pSB101 might be reduced only 3-fold compared with pSB110. However, the gap repair efficiency of pSB101 was reduced 100-fold in the double mutant, compared with the exo1 strain, 50-fold compared with wild-type and 5-fold compared with the mre11 mutant. The recombination frequency of gapped pSB101 in the exo1 mre11 strain may be even lower than presented in Table 2 because only one Met+ Ura+ colony was obtained from four independent transformations.
The frequency of gene replacement is elevated in exo1 and mre11 mutants
Plasmid gap repair is thought to be similar to the repair of chromosomal DSBs in the requirement for templated repair of the gap from a homologous donor duplex. In contrast, gene replacement could occur by assimilation of a single strand from the linear duplex, or by two independent end invasions involving both ends of the targeting fragment (43). Previous studies revealed no defect in gene replacement in rad50 mutants, and a 50-fold decrease in rad1 mutants (30,44). We measured the efficiency of replacement of the chromosomal met17::ADE2 insertion allele by a 2.5 kb fragment containing the MET17 gene. Each end of the targeting fragment has 600 bp of homology shared with the chromosomal locus and there are no heterologies present at the ends that could present barriers to efficient recombination. Correct gene replacement results in a Met+ Ade– phenotype. The efficiency of targeting was increased almost 3-fold in the exo1 and mre11 strains and reduced 3-fold by the rad1 mutation (Table 2).
DISCUSSION
We have used a plasmid gap repair assay to determine the efficiency of double-strand gap repair, as well as alterations in processing of recombination intermediates, in several nuclease-deficient strains of yeast. Most of the mutants examined were proficient at plasmid gap repair, but showed alterations in either conversion tract length or plasmid integration. These results are discussed in detail below.
Role of the Mre11 nuclease in end-processing events
Null mutations of MRE11 cause severe defects in meiotic recombination and in survival to a variety of DNA damaging agents, including ionizing radiation, MMS and inter-strand crosslinking agents (45,46). Although the DNA repair defect is generally attributed to defective homologous recombination, mre11Δ mutants show elevated rates of spontaneous heteroallelic recombination and only a small reduction in sister chromatid recombination (47). Kinetic studies of mating type switching revealed a reduction in the rate of resection of the 5′-end at HO-induced DSBs, and a delay in repair (48). Although Mre11 has endonuclease and exonuclease activity in vitro, mutations that abolish the nuclease activity do not alter the rate of resection of HO-induced DSBs (12). This suggests alternate roles for Mre11 (or the MRX complex) in regulation of the processing activity, or functional redundancy for the nuclease-processing step in mitotic cells. We found a 3-fold reduction in the efficiency of plasmid gap repair in mre11Δ mutants, but the mre11-H125N strain, defective for the nuclease function, showed almost wild-type levels of gap repair. In contrast, rad52 and rad51 mutants show 500- and 100-fold decreases, respectively, in the efficiency of gap repair (37,49). Therefore, the mre11 deficiency in gap repair is much less severe than for other mutants of the RAD52 epistasis group, but is consistent with studies of mating type switching and spontaneous mitotic recombination.
The altered ratio of Met+:Met– events is consistent with decreased processing of the 5′-ends of break sites (Fig. 2). The SnaBI site is less likely to be incorporated into hDNA if the 3′ tails are shorter and, consequently, less likely to be co-converted with the gapped region. This alteration in the length of hDNA could possibly contribute to the hyper-recombination phenotype observed for heteroallelic recombination in mre11 diploids. During heteroallelic recombination hDNA is likely to span both alleles and co-repair of both mismatches would not generate a prototroph. However, if hDNA tracts are shorter so that only one of the two alleles is incorporated then prototrophs will be generated at higher frequencies. Surprisingly, repaired products were biased to Met– from the strain containing the nuclease defective mre11-H125N allele (12). Our previous studies indicated no defect in mating type switching in this strain, but the physical analysis may not have detected an increase in the length of the 3′ single-stranded tails. We previously suggested redundancy for the nuclease-processing step in mitotic cells based on the weak DNA repair defects of the mre11-H125N strain. Based on the results presented here, we propose the MRX complex unwinds ends to provide access to the Mre11 endonuclease, or other nucleases that degrade single-stranded DNA (Fig. 3). In the absence of Mre11, the complex is absent and ends are processed inefficiently by a double-stranded DNA exonuclease, such as Exo1. In mre11-H125N strains, the ends are still unwound by the MRX complex, but are degraded by a more processive single-stranded DNA exonuclease or endonuclease, resulting in longer 3′ tails and increased co-conversion of the SnaBI site.
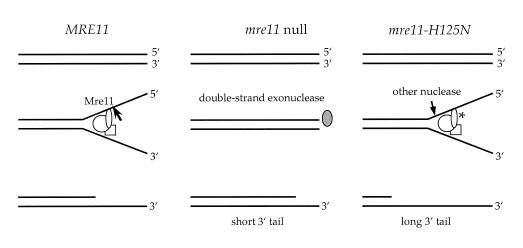
Figure 3.
Models for end processing in mre11 strains. In wild-type strains, the MRX complex binds to double-strand breaks and unwinds the ends producing single-stranded DNA, the substrate for the Mre11 endonuclease. In mre11Δ strains, the MRX complex is absent and no unwinding occurs, but the ends can be inefficiently processed by other exonucleases, such as Exo1. We imagine the MRX complex still forms in mre11-H125N strains and is able to unwind duplex ends, but the single-stranded tails are cleaved by other endo- or exonucleases. To account for apparent increase in gene conversion tract length, we propose that the redundant activities are more processive, or that unwinding by the MRX complex is normally coupled with the Mre11 endonuclease activity.
Previous studies of spontaneous mitotic recombination have found a correlation between conversion tract length and crossing over (50). We found a bias towards Met+ (short heteroduplex) among the non-crossover recombinants and a bias towards Met– (long heteroduplex) among the crossover recombinants in the wild-type strains (Table 3). Therefore, the decrease in crossing over observed in mre11Δ strains could be a consequence of shorter heteroduplex tracts.
Rad1 influences crossing over during gap repair
The Rad1 endonuclease is reported to cleave artificial Holliday junctions generated from oligonucleotides (33). Although this result has been contested (51), the rad1 mutation clearly affects plasmid integration, a process thought to occur by resolution of a Holliday junction intermediate. There are several possible explanations for this observation. First, as mentioned above, shorter heteroduplex tracts are associated with lower crossing over. This seems unlikely to be the cause of the integration defect in the rad1 strain because co-conversion of the met17-sna marker is the same as observed in the wild-type strain. Second, Rad1 could resolve some Holliday junction intermediates, especially if they contain single-stranded DNA near the junction. Third, Rad1 might cleave the D-loop formed by strand invasion (Fig. 4). This cleavage could possibly stabilize the intermediate by allowing pairing of the cleaved strand with the other side of the break to form a Holliday structure. The resulting double Holliday junction intermediate could then be resolved to form either crossover or non-crossover products. In the absence of Rad1, more intermediates would be resolved by D-loop collapse leading to fewer crossover events. Chiu et al. identified a structure-specific nuclease activity from E.coli extracts that stabilizes paranemic joints formed by recA (52); Rad1/10 could potentially act in a similar way to stabilize strand invasion intermediates made by Rad51. Finally, the D-loop could be cleaved by another endonuclease, and the tail subsequently removed by Rad1 to prevent competition for pairing with the invading strand.
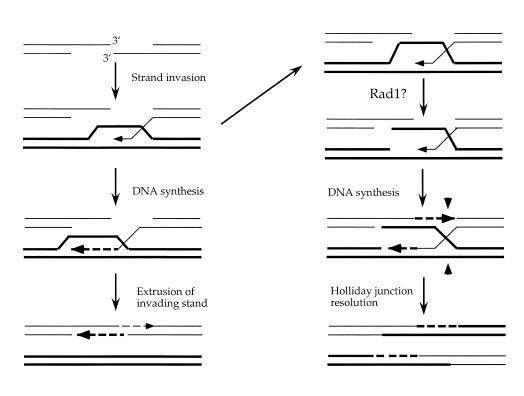
Figure 4.
A model for Rad1 cleavage of recombination intermediates. Strand invasion occurs to form a D-loop structure and the invading 3′-end serves as a primer for DNA synthesis. If the elongated invading strand is extruded it can then pair with the single-strand extension at the other side of the break to repair the break with no associated crossover. Alternatively, the displaced strand of the unbroken duplex could pair with the single-strand tail on the other side of the break to form a double Holliday junction. We suggest Rad1 could cleave the D-loop structure to allow more efficient pairing or stabilization of the D-loop with the single-strand tail on the other side of the break. In the absence of Rad1 intermediates would be more likely to be channeled through the pathway on the left and thus less likely to be resolved as crossovers.
Most of the previous studies demonstrating a defect in mitotic recombination in rad1 mutants utilized substrates with heterologies at the site of a DSB that would require removal by the Rad1/10 endonuclease to complete the recombination event (28). In our studies of plasmid integration and gene replacement there are no heterologies present at the ends of the plasmid/fragment that might prevent efficient recombination. The defect in gene replacement observed in rad1 mutants could be due to a requirement for Rad1 in processing the loop structure predicted to arise by single-strand assimilation, or by defective resolution of a strand invasion intermediate, as suggested above for plasmid gap repair.
exo1 mutants exhibit a hyper-recombination phenotype for gap repair and gene replacement
Strains deleted for EXO1 have a complex phenotype indicative of defects in recombination and mismatch repair pathways (16,17,20–22). Although we detected no effect on end-processing events, the exo1 strains showed a significant increase in both the frequency of gap repair and associated crossing over. Consistent with this observation, no alteration in heteroduplex tract length was observed at the HIS4 locus during meiotic recombination (19). The increase in gap repair efficiency could be due to elimination of a degradative enzyme. However, this seems unlikely because we have not detected an increase in plasmid end joining in exo1 strains (J.Ferguson and L.S.Symington, unpublished data) and a large excess of carrier DNA is used during transformation.
The substrates for gap repair are homologous except for a single base pair insertion to create the met17-sna allele, 200 bp from the gapped region. This may be sufficient to trigger anti-recombination by the mismatch repair system. A recent study showed elevated rates of spontaneous recombination between homologous sequences in exo1 strains. Therefore, it seems likely that the increased levels of gap repair and gene replacement observed in the exo1 strains are due to a defect in the mismatch repair system. The increase in plasmid integration may also be attributed to a defect in mismatch repair as several studies have documented increased crossing over in mismatch repair defective strains (53,54). During meiosis, intergenic recombination is reduced almost 2-fold in exo1 homozygotes in contrast to the increase in crossing over observed for plasmid gap repair (18–20). Exo1 is induced during meiosis and may interact with other meiosis-specific proteins to form complexes with altered specificity for recombination intermediates (20).
The exo1 mre11 double mutant has slow growth and the efficiency of transformation was reduced ~20-fold compared with the wild-type strain. The frequency of plasmid gap repair was reduced in the double mutant, consistent with studies showing an enhancement of the mre11 mutant MMS sensitivity by an exo1 mutation (20). Although physical studies at the MAT locus revealed a greater stability of the cut fragment in exo1 mre11 strains, the ratio of Met+:Met– transformants during gap repair was not significantly different from the mre11 single mutant. Like the exo1 strain, the exo1 mre11 double mutant showed a significant increase in crossing over of the ARS-containing plasmid, even though conversion tracts were shorter. Because integration events were common with the ARS-containing plasmid, we expected to observe only a 3-fold reduction in the gap repair frequency using the integrating plasmid, compared with the ARS-containing plasmid. However, the frequency was much lower than expected. This was also found for the mre11 single mutant. Based on this, we suggest the ARS element plays an important role during gap repair in mre11 strains. It is possible that gap repair can occur by an alternate mechanism, such as break-induced replication from the two ends of the plasmid (55,56), and these events are dependent on close proximity of an ARS element.
Acknowledgments
ACKNOWLEDGEMENTS
We thank William Holloman and Lance Langston for critical reading of the manuscript. This work was supported by Public Health Service grant NIH GM54099 from the National Institutes of Health.
REFERENCES
- 1.Szostak J.W., Orr-Weaver,T.L., Rothstein,R.J. and Stahl,F.W. (1983) Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- 2.White C.I. and Haber,J.E. (1990) EMBO J., 9, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun H., Treco,D. and Szostak,J.W. (1991) Cell, 64, 1155–1161. [DOI] [PubMed] [Google Scholar]
- 4.West S.C. (1997) Annu. Rev. Genet., 31, 213–244. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson D.O. and Holloman,W.K. (1996) Proc. Natl Acad. Sci. USA, 93, 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper D.L., Lahue,R.S. and Modrich,P. (1993) J. Biol. Chem., 268, 11823–11829. [PubMed] [Google Scholar]
- 7.Kolodner R.D. and Marsischky,G.T. (1999) Curr. Opin. Genet. Dev., 9, 89–96. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov E.L., Sugawara,N., White,C.I., Fabre,F. and Haber,J.E. (1994) Mol. Cell. Biol., 14, 3414–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuse M., Nagase,Y., Tsubouchi,H., Murakami-Murofushi,K., Shibata,T. and Ohta,K. (1998) EMBO J., 17, 6412–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paull T.T. and Gellert,M. (1998) Mol. Cell., 1, 969–979. [DOI] [PubMed] [Google Scholar]
- 11.Usui T., Ohta,T., Oshiumi,H., Tomizawa,J., Ogawa,H. and Ogawa,T. (1998) Cell, 95, 705–716. [DOI] [PubMed] [Google Scholar]
- 12.Moreau S., Ferguson,J.R. and Symington,L.S. (1999) Mol. Cell. Biol., 19, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nairz K. and Klein,F. (1997) Genes Dev., 11, 2272–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szankasi P. and Smith,G.R. (1992) J. Biol. Chem., 267, 3014–3023. [PubMed] [Google Scholar]
- 15.Huang K.N. and Symington,L.S. (1993) Mol. Cell. Biol., 13, 3125–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szankasi P. and Smith,G.R. (1995) Science, 267, 1166–1169. [DOI] [PubMed] [Google Scholar]
- 17.Fiorentini P., Huang,K.N., Tishkoff,D.X., Kolodner,R.D. and Symington,L.S. (1997) Mol. Cell. Biol., 17, 2764–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khazanehdari K.A. and Borts,R.H. (2000) Chromosoma, 109, 94–102. [DOI] [PubMed] [Google Scholar]
- 19.Kirkpatrick D., Ferguson,J., Petes,T.D. and Symington,L.S. (2000) Genetics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsubouchi H. and Ogawa,H. (2000) Mol. Biol. Cell, 11, 2221–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tishkoff D.X., Boerger,A.L., Bertrand,P., Filosi,N., Gaida,G.M., Kane,M.F. and Kolodner,R.D. (1997) Proc. Natl Acad. Sci. USA, 94, 7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson A., Hendrix,M., Jinks-Robertson,S. and Crouse,G.F. (2000) Genetics, 154, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta A., Hendrix,M., Lipsitch,M. and Jinks-Robertson,S. (1997) Proc. Natl Acad. Sci. USA, 94, 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox B.S. and Parry,J.M. (1968) Mutat. Res., 6, 37–55. [DOI] [PubMed] [Google Scholar]
- 25.Game J.C. and Cox,B.S. (1971) Mutat. Res., 12, 328–331. [Google Scholar]
- 26.Bailly V., Sommers,C.H., Sung,P., Prakash,L. and Prakash,S. (1992) Proc. Natl Acad. Sci. USA, 89, 8273–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardwell A.J., Bardwell,L., Tomkinson,A.E. and Friedberg,E.C. (1994) Science, 265, 2082–2085. [DOI] [PubMed] [Google Scholar]
- 28.Fishman-Lobell J. and Haber,J.E. (1992) Science, 258, 480–484. [DOI] [PubMed] [Google Scholar]
- 29.Kirkpatrick D.T. and Petes,T.D. (1997) Nature, 387, 929–931. [DOI] [PubMed] [Google Scholar]
- 30.Schiestl R.H. and Prakash,S. (1988) Mol. Cell. Biol., 8, 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aguilera A. and Klein,H.L. (1989) Genetics, 123, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett R.J., Dunderdale,H.J. and West,S.C. (1993) Cell, 74, 1021–1031. [DOI] [PubMed] [Google Scholar]
- 33.Habraken Y., Sung,P., Prakash,L. and Prakash,S. (1994) Nature, 371, 531–534. [DOI] [PubMed] [Google Scholar]
- 34.Dowling E.L., Maloney,D.H. and Fogel,S. (1985) Genetics, 109, 283–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekelsky J.J., McKim,K.S., Chin,G.M. and Hawley,R.S. (1995) Genetics, 141, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orr-Weaver T.L. and Szostak,J.W. (1983) Proc. Natl Acad. Sci. USA, 80, 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartsch S., Kang,L.E. and Symington,L.S. (2000) Mol. Cell. Biol., 20, 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas B.J. and Rothstein,R. (1989) Genetics, 123, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan H.Y., Cheng,K.K. and Klein,H.L. (1996) Genetics, 142, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman F., Fink,G. and Hicks,J. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Cost G.J. and Boeke,J.D. (1996) Yeast, 12, 939–941. [DOI] [PubMed] [Google Scholar]
- 42.Haber J.E., Ray,B.L., Kolb,J.M. and White,C.I. (1993) Proc. Natl Acad. Sci. USA, 90, 3363–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung W., Malkova,A. and Haber,J.E. (1997) Proc. Natl Acad. Sci. USA, 94, 6851–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiestl R.H., Zhu,J. and Petes,T.D. (1994) Mol. Cell. Biol., 14, 4493–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ajimura M., Leem,S.H. and Ogawa,H. (1993) Genetics, 133, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McHugh P.J., Sones,W.R. and Hartley,J.A. (2000) Mol. Cell. Biol., 20, 3425–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bressan D.A., Baxter,B.K. and Petrini,J.H. (1999) Mol. Cell. Biol., 19, 7681–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsubouchi H. and Ogawa,H. (1998) Mol. Cell. Biol., 18, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orr-Weaver T.L., Szostak,J.W. and Rothstein,R.J. (1981) Proc. Natl Acad. Sci. USA, 78, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jinks-Robertson S., Michelitch,M. and Ramcharan,S. (1993) Mol. Cell. Biol., 13, 3937–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies A.A., Friedberg,E.C., Tomkinson,A.E., Wood,R.D. and West,S.C. (1995) J. Biol. Chem., 270, 24638–24641. [DOI] [PubMed] [Google Scholar]
- 52.Chiu S.K., Low,K.B., Yuan,A. and Radding,C.M. (1997) Proc. Natl Acad. Sci. USA, 94, 6079–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selva E.M., New,L., Crouse,G.F. and Lahue,R.S. (1995) Genetics, 139, 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inbar O., Liefshitz,B., Bitan,G. and Kupiec,M. (2000) J. Biol. Chem., 275, 30833–30838. [DOI] [PubMed] [Google Scholar]
- 55.Malkova A., Ivanov,E.L. and Haber,J.E. (1996) Proc. Natl Acad. Sci. USA, 93, 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrow D.M., Connelly,C. and Hieter,P. (1997) Genetics, 147, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]