Abstract
Replication protein A (RPA) is a three-subunit protein complex with multiple functions in DNA replication. Previous study indicated that human RPA (h-RPA) could not be replaced by Schizosaccharomyces pombe RPA (sp-RPA) in simian virus 40 (SV40) replication, suggesting that h-RPA may have a specific function in SV40 DNA replication. To understand the specificity of h-RPA in replication, we prepared heterologous RPAs containing the mixture of human and S.pombe subunits and compared these preparations for various enzymatic activities. Heterologous RPAs containing two human subunits supported SV40 DNA replication, whereas those containing only one human subunit poorly supported DNA replication, suggesting that RPA complex requires at least two human subunits to support its function in SV40 DNA replication. All heterologous RPAs effectively supported single-stranded (ss)DNA binding activity and an elongation of a primed DNA template catalyzed by DNA polymerase (pol) α and δ. A strong correlation between SV40 DNA replication activity and large tumor antigen (T-ag)-dependent RNA primer synthesis by pol α–primase complex was observed among the heterologous RPAs. Furthermore, T-ag showed a strong interaction with 70- and 34-kDa subunits from human, but poorly interacted with their S.pombe counterparts, indicating that the specificity of h-RPA is due to its role in RNA primer synthesis. In the SV40 replication reaction, the addition of increasing amounts of sp-RPA in the presence of fixed amount of h-RPA significantly reduced overall DNA synthesis, but increased the size of lagging strand, supporting a specific role for h-RPA in RNA primer synthesis. Together, these results suggest that the specificity of h-RPA in SV40 replication lies in T-ag-dependent RNA primer synthesis.
INTRODUCTION
Eukaryotic replication protein A [RPA; also known as human single-stranded (ss)DNA binding protein, hSSB] was originally identified as an essential factor in simian virus 40 (SV40) DNA replication in vitro (1–3). RPA contains three subunits of 70, 34 and 11 kDa (p70, p34 and p11, respectively), which are tightly associated with each other in various eukaryotic RPA preparations (2,4), suggesting its functional structure is highly conserved. Genetic studies in yeast (4) and in vitro replication and repair studies (5–8) in mammalian systems strongly suggest that all three subunits are required for cell survival and its function in DNA metabolism. RPA complexes assemble in an ordered process (9,10) and the cellular localization of the individual subunits varies during the cell cycle (11).
In replication, RPA functions during both the initiation and elongation stages. In the initiation stage, it is involved in the SV40 large tumor antigen (T-ag)-dependent unwinding of SV40 origin-containing DNA. During initiation, RPA interacts with SV40 T-ag and polymerase (pol) α primase (12), resulting in the formation of an initiation complex at the replication origin. Physical interactions between RPA, T-ag and pol α primase are necessary for the initiation of SV40 DNA replication (13) and for its function in the elongation stage (14,15). Recent studies indicate that the 34-kDa subunit of RPA contacts pre-Okazaki fragments most likely RNA–DNA hybrid, suggesting a specific role for RPA in lagging strand synthesis (16). RPA from other sources, such as Escherichia coli, adenovirus and yeast, do not replace human RPA (h-RPA) in SV40 replication reactions, even though they support the T-ag-dependent unwinding reaction (17–19). Moreover, initiation of DNA synthesis, determined by the synthesis of small RNA primers by DNA pol α–primase complex, requires h-RPA which cannot be replaced by yeast RPA (20), suggesting a specific role for h-RPA in replication. In replication, RPA mediates unwinding of SV40 origin-containing DNA in the presence of SV40 T-ag and topoisomerase. It interacts with SV40 T-ag and pol α–primase (12,13), which is necessary for the initiation of SV40 DNA replication (13–15). RPA is also likely to function in elongation because it stimulates the elongation of primed DNA templates catalyzed by pol α, pol δ and pol ɛ (7,21). RPA also stimulates the 3′–5′ exonuclease activity of pol δ, which suggests a possible role in proofreading activity (22).
In this report, we examined the specificity of h-RPA and its subunits in SV40 DNA replication in vitro using heterologous RPAs containing subunits from human and Schizosaccharomyces pombe. Heterologous RPAs containing two human subunits supported SV40 DNA replication, whereas those containing only one human subunit poorly supported DNA replication, suggesting that at least two human subunits are necessary for RPA to function in replication. A strong correlation between SV40 DNA replication activity and T-ag-dependent RNA primer synthesis was observed among the heterologous RPAs, which may be due to the fact that an efficient interaction of T-ag with p70 and p34 subunits from human, but not with those from S.pombe. Collectively our study suggests that the specificity of h-RPA is due to its interaction with T-ag in RNA primer synthesis.
MATERIALS AND METHODS
Cell extracts, proteins, antibodies and DNA
SV40 origin-containing circular duplex DNA (pUC-ori+), SV40 T-ag, human pol α–primase complex, topoisomerase I, human DNA pol δ, replication factor C (RF-C) and proliferating cell nuclear antigen (PCNA) were purified as described previously (21). Ammonium sulfate fractions (35–65%) of HeLa cell cytosolic extracts were prepared as described (1). Various heterologous RPA containing either human or S.pombe subunits were purified from Sf-9 lysates using the established procedure (23). Polyclonal antibodies to h-RPA subunits, p70, p34 and p11, were from rabbits (9) and polyclonal antibodies to S.pombe RPA (sp-RPA) p70, and p34 were described previously (19).
SV40 DNA replication in vitro
The reactions were carried out as described previously (24). Briefly, the reaction mixtures (40 µl) contained 40 mM creatine phosphate-di-Tris salt (pH 7.7), 1 µg of creatine kinase, 7 mM MgCl2, 0.5 mM DTT, 4 mM ATP, 200 µM UTP, GTP and CTP, 100 µM dATP, dGTP and dCTP, 25 µM [methyl-3H]dTTP (300 c.p.m./pmol), 0.6 µg of SV40 T-ag, 0.3 µg of SV40 pUC-ori+, and 100 µg of the ammonium sulfate fraction from HeLa cell extracts (1). In dipolymerase reactions, where replication of SV40 ori+ DNA was carried out in the presence of both DNA pol α and pol δ, the indicated amounts of PCNA and RF-C were also added to the reactions. The reactions were incubated for 90 min at 37°C after which time acid-insoluble radioactivity was measured. Replication products were analyzed using [α-32P]dATP (30 000 c.p.m./pmol) in lieu of [3H]dTTP in the reactions described above. After incubation, reactions were stopped by the addition of 40 µl of a solution containing 20 mM EDTA, 1% SDS, and E.coli tRNA (0.5 mg/ml). One-tenth of the reaction mixture was used to measure the acid-insoluble radioactivity and the rest used to isolate DNA after phenol extraction and ethanol precipitation. The labeled DNA formed was analyzed by electrophoresis through a 1.0 or 1.2% neutral agarose gel in 1× TBE buffer for 6 h at 5 V/cm or by alkaline-agarose gel (40 mM NaOH and 1 mM EDTA) for 12–14 h at 2 V/cm. Gels were subsequently dried and exposed to X-ray film.
SDS–PAGE and western blot analysis
Protein samples were separated by 12% SDS–PAGE as described previously (9). Proteins were then transferred to nitrocellulose (BA83; Bio-Rad), immunoblotted with an anti-h-p70, h-p34 and h-p11 (or anti-sp-p70 and sp-p34) polyclonal antibodies from rabbits, and detected by [125I]protein A (ICN) addition.
RNA primer synthesis
The reaction mixtures (40 µl) contained 40 mM creatine phosphate di-Tris salt (pH 7.7), 1 µg of creatine kinase, 7 mM MgCl2, 0.5 mM DTT, 4 mM ATP, 10 U of human placental RNase inhibitor, 200 µM GTP and CTP, 10 µM [α-32P]UTP (10 c.p.m./fmol), 0.8 µg of SV40 T-ag, 0.3 µg of SV40 pUC-ori+, 1200 U of topoisomerase I, pol α (0.6 U)–primase (3 U), and the indicated amount of RPA (20). After incubation at 37°C for 30 min, products were extracted with a phenol/chloroform mixture, precipitated with ethanol and the precipitates were dissolved in 10 µl of loading buffer [10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 90% (v/v) formamide]. The mixtures were heated for 5 min at 90°C and loaded onto a 20% polyacrylamide gel containing 7 M urea, 1 mM EDTA and 89 mM Tris borate (pH 8.0). Electrophoresis was carried out at 800 V for 7 h with size markers. Gels were then subjected to autoradiography.
Coupled DNA pol α–primase assay
Reactions were performed using unprimed M13 ssDNA under conditions similar to those used in the SV40 DNA replication assay described above. Briefly, reaction mixtures (40 µl) contained 40 mM creatine phosphate-di-Tris salt (pH 7.7), 1 µg of creatine kinase, 7 mM MgCl2, 0.5 mM DTT, 4 mM ATP, 200 µM UTP, GTP and CTP, 100 µM dATP, dGTP and dCTP, 25 µM [α-32P]dTTP (30 000 c.p.m./pmol), 40 ng unprimed M13 ssDNA, 0.1 U of human pol α–primase complex, 0.4 µg of RPA, and the indicated amounts of SV40 T-ag. After incubation at 37°C for 30 min, one-tenth of the sample was used for measuring acid-insoluble radioactivity and the rest of the reaction products were extracted with a phenol/chloroform mixture following the procedure described earlier. Reaction products were analyzed by 20% PAGE.
Metabolic labeling of Sf-9 cells with [35S]methionine and immunoprecipitation
[35S]methionine-labeled cell lysates were prepared as described previously (9). Sf-9 cells (2.0 × 106) were plated on a 60-mm dish and infected with individual recombinant baculoviruses encoding RPA subunits at a multiplicity of infection of 15 for ~40 h at 27°C. The insect cells were then labeled with trans-35S-labeled methionine at 200 µCi/ml (1200–1600 Ci/mmol) for 4 h in methionine-free medium containing 5% dialyzed fetal calf serum. Cells were briefly washed with PBS, and lysed for 1 h on ice in 0.5 ml of lysis buffer [50 mM Tris–HCl, pH 8.0, 250 mM NaCl, 0.5% NP-40, 1 mM DTT, 1 mM EDTA, 0.1 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 0.1 mM phenylmethyl sulfonyl fluoride (PMSF), 0.1 mg/ml leupeptin and 0.2 mg/ml antipain]. For immunoprecipitation, cleared cell lysates (50 µl) were incubated with the indicated polyclonal antibody (5 µl) in the presence of bovine serum albumin (200 µg/ml) at 4°C with rocking. Protein A–Sepharose was then added, and the mixture incubated for 1 h at 4°C. Immunoprecipitates were collected by centrifugation, washed five times with cell lysis buffer, and analyzed by 12% SDS–PAGE.
RESULTS
Formation of heterologous RPA complex containing human and S.pombe subunits
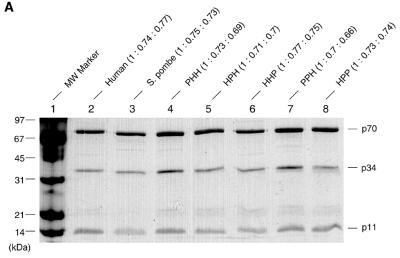
h-RPA is essential for SV40 DNA replication and cannot be replaced by RPA from S.pombe and Saccharomyces cerevisiae (18,19), suggesting that h-RPA is likely to govern specificity of SV40 DNA replication. h-RPA and sp-RPA share ~30% homology in their primary amino acid sequences (19) and a strong homology is found in the C-domains of h-p70 and sp-p70. In the case of h-RPA, the C-terminus region of the p70 subunit was shown to interact with p34 and p11 (23,25), suggesting that the RPA subunits derived from these two species likely interact to form a heterologous RPA complex. In an effort to find a role for h-RPA as a specificity factor in replication, we constructed recombinant baculoviruses expressing individual subunits of sp-RPA and examined whether h-RPA subunit(s) forms a complex with those from S.pombe. Insect cells (Sf-9) were coinfected with recombinant baculoviruses (9) encoding RPA subunits from human and S.pombe and examined for the formation of heterologous RPA complexes between the subunits. Immunoprecipitation with an anti-p34 polyclonal antibody (against either h-p34 or sp-p34) revealed that h-RPA subunits formed stable complexes with those from S.pombe (data not shown). SDS–PAGE of various heterologous RPAs showed that the subunit stoichiometry among all the heterologous RPA complex was very similar (Fig. 1A, top), suggesting that RPA subunits from human and S.pombe formed a stable complex. Western blot analysis using anti-h-RPA antibodies (Fig. 1B) and anti-sp-RPA antibodies (Fig. 1C) showed that the subunits from heterologous RPAs formed a stable complex with each other. Surprisingly, of the eight different permutations possible, we did not detect the formation of RPA containing the structure PHP. Since all other heterologous RPAs form stable complex, it is highly likely that the formation of sub-complex between p70 (S.pombe)–p34 (human) or p34 (human)–p11 (S.pombe) occurs. The reasons for this are not clear.
Figure 1.
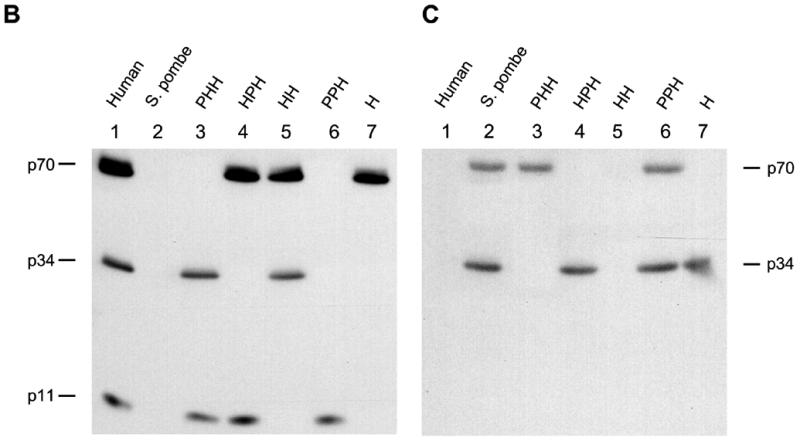
SDS–PAGE and western blot of heterologous RPA from h- and sp-RPA was purified using a previously described protocol (23) and analyzed by 12% SDS–PAGE (A) or immunoblotting procedure using mixtures of anti-p70, -p34, -p11 antibodies raised against h-RPA subunits (B) or the mixtures of anti-p70- and -p34 antibodies against sp-RPA subunits (C). The subunit stoichiometry of various RPA complex (p70: p34: p11) was measured by densitometric scanning and is indicated at the top of (A). The three-letter symbols at the top of the figure represent the source of RPA subunits [either from human (H) or from S.pombe (P)] in the order of p70, p34 and p11. For example, PHH represents RPA complex containing sp-p70, h-p34 and h-p11.
Effect of heterologous RPA on SV40 DNA replication in vitro
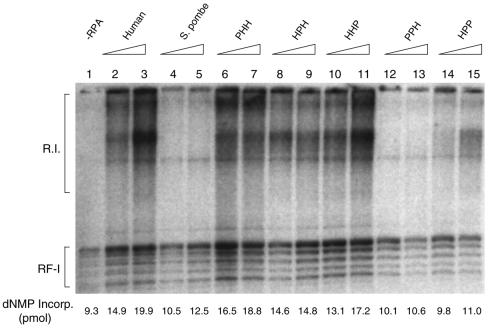
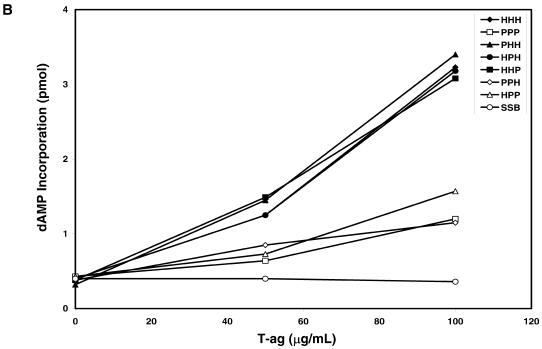
We first examined the biological activity of the different RPAs in the in vitro SV40 DNA replication using crude extracts depleted of RPA by ammonium sulfate fractionation (1). As reported previously (19), sp-RPA poorly supported SV40 DNA replication under the conditions where h-RPA worked efficiently (Fig. 2, lanes 2 and 3 versus lanes 4 and 5). Heterologous RPA, PHH (sp-p70, h-p34 and h-p11), HPH (h-p70, sp-p34 and h-p11) and HHP (h-p70, h-p34 and sp-p11) efficiently supported DNA replication (Fig. 2, lanes 6–11), whereas PPH (sp-p70, sp-p34 and h-p11) and HPP (h-p70, sp-p34 and sp-p11) did not (Fig. 2, lanes 12–15). This result suggests that (i) all three h-RPA subunits are involved in species specificity of h-RPA in supporting SV40 DNA replication in vitro, and (ii) at least two human subunits are necessary to retain its function in replication.
Figure 2.
The effect of heterologous RPA on SV40 DNA replication reconstituted with purified proteins in vitro. Replication reactions contained SV40 pUC-ori+, 0.6 µg of SV40 T-ag and 100 µg of ammonium sulfate fraction (35–65%) from HeLa cell extracts. Indicated amounts of RPA (150 ng in lanes 2, 4, 6, 8, 10, 12 and 14, and 300 ng in lanes 3, 5, 7, 9, 11, 13 and 15) were added to the reaction. The composition of the RPA is indicated by the three-letter symbol at the top of the figure. Reaction mixtures were incubated for 90 min at 37°C, and the reaction products were analyzed by agarose gel electrophoresis (1.2%) and acid-insoluble radioactivity.
Analysis of heterologous RPAs for their biochemical activities
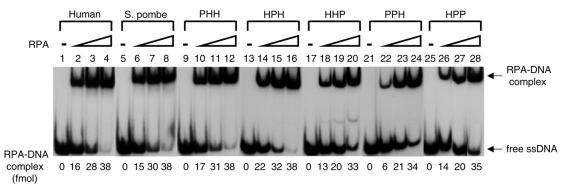
We examined heterologous RPAs for their fundamental biochemical function, ssDNA binding activity. Although the affinity of ssDNA binding appears somewhat diminished in several heterologous RPAs (HHP, PPH and HPP) in the presence of low RPA, all RPAs formed stable complexes with oligo(dT)50 which appeared as bands in the polyacrylamide gel (Fig. 3). This result suggests that the species specificity of h-RPA in SV40 DNA replication was not due to its ssDNA binding activity. RPA stimulates the elongation of primed DNA templates catalyzed by pol α and pol δ (17), indicating its role in the elongation stage. Therefore we examined heterologous RPAs for their ability to stimulate pol α activity using primed DNA templates. The addition of increasing amounts of various heterologous RPAs stimulated pol α activity 2–3-fold, comparable to that observed with h-RPA (data not shown). This finding is not surprising since the stimulation of pol α activity by RPA is not essential for SV40 DNA replication (23).
Figure 3.
The ssDNA binding activity of various heterologous RPAs. Three concentrations (5, 10 and 20 ng) of RPA containing either the human or S.pombe subunit, as indicated at the top of the figure, were incubated with 100 fmol of 5′-32P-labeled (dT)50 for 15 min at room temperature. The RPA–DNA complexes were separated from free DNA by 5% PAGE (acrylamide:bisacrylamide = 79:1). For quantitation, the protein–DNA complex bands were excised and analyzed by liquid scintillation counting.
Effect of heterologous RPAs on RNA primer synthesis
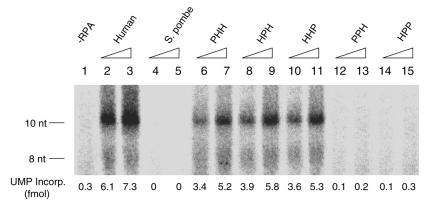
The specificity of h-RPA in the SV40 replication reaction may be due to the pol α–primase-mediated synthesis of oligoribonucleotides since the RPA requirement for this reaction was not replaced by other RPAs (20), even though these RPAs supported the SV40 T-ag-catalyzed unwinding of the SV40 origin-containing plasmid DNA (17–19). We therefore examined whether the different heterologous RPAs supported oligoribonucleotide synthesis with SV40 replication origin-containing circular duplex DNA. As reported previously, small RNA synthesis was dependent on the presence of RPA (Fig. 4). Heterologous RPA (PHH, HPH and HHP) containing two human subunits supported oligonucleotide synthesis, whereas poor or no RNA synthesis was observed in the presence of sp-RPA (PPP) or heterologous RPA containing two S.pombe subunits (PPH and HPP) (Fig. 4, lanes 4, 5 and 12–15). h-RPA and heterologous RPA containing two human subunits supported a very similar level of DNA replication activity (Fig. 2), whereas none of the heterologous RPA appears to reach to the level of h-RPA in RNA primer synthesis (Fig. 4). This may be explained in part by the fact that DNA replication reaction requires both RNA primer synthesis and its elongation. h-RPA may be more efficient in supporting RNA synthesis, whereas the elongation reaction is equally supported by both h-RPA and the heterologous RPA.
Figure 4.
Effects of heterologous RPAs on RNA-primer synthesis during the initiation of SV40 DNA replication. Reaction mixtures containined 0.3 µg of SV40 pUC-ori+, 0.8 µg of SV40 T-ag, 1200 U of topoisomerase I, pol α (0.6 U)–primase (3 U) complex, and rNTPs but no dNTPs (see Materials and Methods for details). Where indicated, increasing amounts of RPA (150 ng in lanes 2, 4, 6, 8, 10, 12 and 14; 300 ng in lanes 3, 5, 7, 9, 11, 13 and 15) were added to the reaction. The composition of the RPA is indicated by the three-letter symbol at the top of the figure. After incubation at 37°C for 30 min, reaction products were processed as described in Materials and Methods and analyzed by 20% PAGE containing 7 M urea, 1 mM EDTA and 89 mM Tris–borate (pH 8.0).
We also examined the influence of heterologous RPAs on DNA primase activity using unprimed M13 ssDNA. As shown in Figure 6, the coupled synthesis of RNA primers followed by dNTP incorporation catalyzed by the pol α–primase was significantly inhibited by h-RPA (Fig. 5, lane 2), sp-RPA (Fig. 5, lane 5) or heterologous RPA (Fig. 5, lanes 8, 11, 14, 17, 20 and 23). This inhibition can be reversed by the addition of SV40 T-ag, presumably reflecting the formation of a primosome complex between RPA–T-ag and pol α–primase complex (14). The inhibitory effects of h-RPA and the heterologous RPAs containing two human subunits (PHH, HPH and HHP) were reversed 75–85% by the addition of SV40 T-ag (Fig. 5A and B), whereas the coupled synthesis inhibited by sp-RPA or heterologous RPAs containing two S.pombe subunits (PPH and HPP) was poorly reversed by T-ag (Fig. 5A and B). This result is not only in keeping with those observed in the SV40 DNA replication system (Figs 2 and 4), but also supports a notion that the formation of a primosome complex between RPA, T-ag and pol α–primase is essential for RNA primer synthesis.
Figure 6.
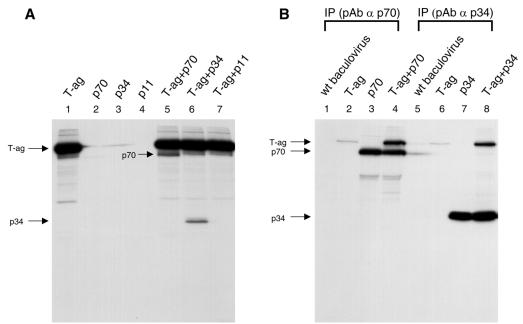
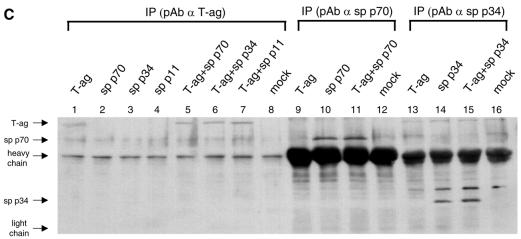
Interaction of SV40 T-ag and RPA subunits in vivo. (A and B) Singly or coinfected with recombinant baculoviruses encoding T-ag and h-RPA subunit are indicated at top of panels. Lysates prepared from [35S]methionine-labeled insect cells were incubated with anti-T-ag antibody (A), anti-human p70 polyclonal antibody (B, lanes 1–4), or anti-human p34 polyclonal antibody (B, lanes 5–8). The immune complexes were precipitated with protein A–Sepharose and analyzed by 12% SDS–PAGE. (C) Singly or coinfected with recombinant baculoviruses encoding T-ag and sp-RPA subunit are indicated at top of panel. Lysates prepared from insect cells (without [35S]methionine) were incubated with anti-T-ag antibody (lanes 1–8), anti-sp-p70 polyclonal antibody (lanes 9–12), or anti-sp-p34 polyclonal antibody (lanes 13–16). The immune complexes were precipitated with protein A–Sepharose and analyzed by 12% SDS–PAGE followed by western blot using the mixture of anti-sp-p70 and sp-p34 polyclonal antibodies. Immunoglobulin heavy chain and light chain are indicated by arrows.
Figure 5.
Effect of heterologous RPA on the coupled DNA pol α–primase activity on ssM13 DNA. (A) The coupled DNA pol α–primase reactions containing 40 ng of unprimed M13 ssDNA template and 0.1 U of human pol α–primase complex were incubated at 37°C for 30 min in the presence of 0.4 µg RPA (except lane 1) and the increasing amount of T-ag (0 µg, lanes 1, 2, 5, 8, 11, 14, 17, 20 and 23; 0.4 µg, lanes 3, 6, 9, 12, 15, 18, 21 and 24; 0.8 µg, lanes 4, 7, 10, 13, 16, 19, 22 and 25). The heterologous RPAs used are indicated by the three-letter symbol. After incubation, reaction products were isolated as described in Materials and Methods and analyzed by 20% PAGE containing 7 M urea, 1 mM EDTA and 89 mM Tris–borate (pH 8.0). (B) For quantitation, regions containing reaction products were excised and analyzed by liquid scintillation counting.
To further analyze the species-specificity of hRPA in SV40 DNA replication, we examined the physical interaction of T-ag with individual subunits of RPA from either human or S.pombe. As reported previously, both p70 and p34 subunits from human interacted with T-ag (Fig. 6A). Strong interaction between T-ag and h-RPA subunits (p34 or p70) was observed (Fig. 6B, lanes 4 and 8), whereas a poor or undetectable interaction was observed between SV40 T-ag and RPA subunits (p34 or p70) from S.pombe (Fig. 6C, lanes 5 and 6). Together, these results strongly suggest that (i) the specificity of RPA in DNA replication lies in RNA primer synthesis, and (ii) h-RPA subunits are likely to be involved in primosome assembly and DNA priming events through protein–protein interaction (14,15). At least two h-RPA subunits may be required in the RPA complex to support the latter event.
Effect of heterologous RPA on the leading and lagging strand synthesis
The poor SV40 replication activity and RNA primer synthesis observed with sp-RPA would most likely have severe affects on the synthesis of lagging strand because this reaction requires the continued synthesis of RNA primers (26,27). For this reason, we examined the effects of sp-RPA on the synthesis of leading and lagging strands using the SV40 replication system with purified replication proteins (Fig. 7). In this experiment, increasing amounts of sp-RPA were added to replication reactions that contained a constant amount (240 ng) of h-RPA (Fig. 7). The addition of an increasing amount of sp-RPA to replication reactions decreased the synthesis of both leading and lagging strands, whereas it significantly increased the size of lagging strand and not the leading strand (Fig. 7). This result strongly supports a specific role for hRPA in T-ag-dependent RNA primer synthesis that cannot be replaced by sp-RPA.
Figure 7.
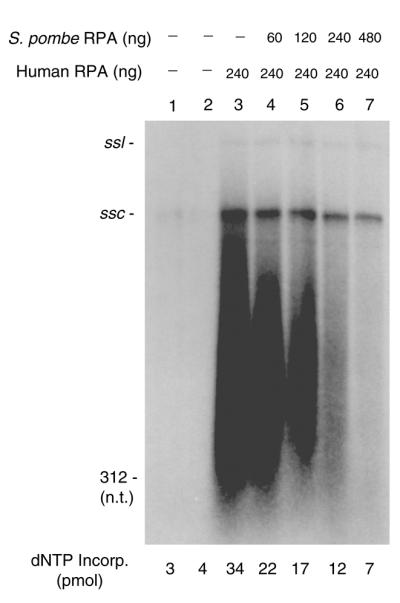
Effect of sp-RPA on the size of replication products formed in the in vitro SV40 replication system. Reaction mixtures (40 µl) contained SV40 pUC-ori+, 0.6 µg of SV40 T-ag, 1200 U of topoisomerase I, pol α (0.1 U)–primase (0.5 U) complex, RF-C (0.1 µg), PCNA (0.2 µg) and pol δ (0.05 U), and the indicated amounts of sp- and h-RPA. After incubation at 37°C for 90 min, replication products were isolated and analyzed by 1.2% alkaline agarose gel electrophoresis. ssl and ssc indicate the position of single-stranded linear and single-stranded circular DNA, respectively.
DISCUSSION
Eukaryotic RPA, composed of three tightly associated subunits, plays an important role in DNA replication, repair and recombination (1–3,6,7). Nonetheless, h-RPA cannot be replaced by sp-RPA in SV40 DNA replication, suggesting a specific role for h-RPA in DNA replication (19). In this study, we have examined the specificity of h-RPA in the SV40 DNA replication reaction using heterologous RPAs that contain various combinations of subunits from both human and S.pombe.
h-RPA and sp-RPA, and those containing both human and S.pombe subunits, all supported ssDNA binding activity (Fig. 3), the stimulation of pol α activity (data not shown), and T-ag-mediated unwinding of SV40 DNA (19, data not shown). However, the initiation of DNA replication, measured by RNA primer synthesis, was not supported by sp-RPA (PPP) or heterologous RPAs containing two S.pombe subunits (PPH and HPP), whereas those containing two human subunits (PHH, HPH and HHP) supported oligoribonucleotide synthesis (Fig. 4). Similar results were observed in the coupled primosome assay using unprimed M13 ssDNA (Fig. 6), suggesting that the specificity of h-RPA in DNA replication lies in the T-ag-dependent synthesis of RNA primers. It is conceivable that the primosome assembly requires a specific protein–protein interaction among T-ag, h-RPA and pol α–primase since sp-RPA failed to interact with T-ag (Fig. 6C). These results are consistent with previous studies by others which demonstrated that RPA strongly inhibited the primase activity of pol α–primase complex (14,15), and T-ag reversed the inhibition of DNA pol α–primase activity on h-RPA-coated ssDNA. T-ag was unable to reverse the inhibitory effect of S.cerevisiae RPA or E.coli SSB, suggesting a specific role for h-RPA in primosome assembly (14,15).
The addition of increasing amounts of sp-RPA to the reconstituted replication reactions significantly affected the size of Okazaki fragments (Fig. 7), suggesting that the specificity of h-RPA in replication is on the lagging strand synthesis that heavily relies on the synthesis of RNA primers. Based on the results presented here, we suggest that all three RPA subunits are involved in the T-ag-dependent synthesis of RNA primer catalyzed by DNA primase and that minimally two h-RPA subunits are required for this event. As shown above, the third subunit can be derived from sp-RPA. Interestingly, the sp-RPA subunit can replace either the 70-, 34- or 11-kDa of h-RPA.
Recent findings also suggest a complex role for RPA in lagging strand synthesis. Mass et al. (16) found that the 34-kDa subunit of RPA could be directly cross-linked to nascent RNA–DNA primers during lagging strand synthesis. However, at later stages, only the 70-kDa subunit was cross-linked to lagging strands. RPA has been shown to stabilize the association of the pol α–primase complex to primed DNA templates. RF-C, due to its interaction with the 70-kDa subunit, can displace the pol α–primase complex from such sites (28). Thus, RPA plays an important role in contributing to the polymerase switch in the maturation of Okazaki fragments (28). Interestingly, the displacement of pol α–primase complex by RF-C was not observed at the stage where the complex synthesized only RNA but was observed after the complex synthesized RNA–DNA chains.
In light of the results reported here, that at least two subunits of RPA derived from h-RPA support SV40 DNA replication, it is possible that the species-specific effects of h-RPA involve minimally two of the three subunits of h-RPA. Identity and homology between the h- and sp-RPA subunits are highest for the large subunits (37 and 64%), less for the middle subunits (24 and 49%) and least for the smallest subunits (19 and 50%). Possibly, by virtue of forming stable chimeric three subunit complexes, the conserved regions present in S.pombe and human subunits lead to a structure capable of interacting with pol α–primase and SV40 T-ag. Our findings suggest that there may be multiple sites within the three subunits of RPA that contribute to the observed species specificity.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr D.-K. Kim for the help in the beginning of the project, Ms E. Stigger for preparing recombinant baculoviruses encoding S.pombe RPA subunits, and Ms E.-J. Oh for preparation of figures. This work was supported by the Indiana University Cancer Center and grants from National Institutes of Health, GM52358 (S.-H.L.) and GM38559 (J.H.). M.W. was supported by a postdoctoral fellowship from the NIH (F32 GM20167).
REFERENCES
- 1.Wobbe C.R., Weissbach,L., Borowiec,J.A., Dean,F.B., Murakami,Y., Bullock,P. and Hurwitz,J. (1987) Proc. Natl Acad. Sci. USA., 82, 5710–5714. [Google Scholar]
- 2.Fairman M.P. and Stillman,B. (1988) EMBO J., 7, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wold M.S. and Kelly,T.J. (1988) Proc. Natl Acad. Sci. USA, 85, 2523–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brill S.J. and Stillman,B. (1991) Genes Dev., 5, 1589–1600. [DOI] [PubMed] [Google Scholar]
- 5.Erdile L.F., Heyer,W.-D., Kolodner,R. and Kelly,T.J. (1991) J. Biol. Chem., 266, 12090–12098. [PubMed] [Google Scholar]
- 6.Coverly D., Kenny,M.K., Lane,D.P. and Wood,R.D. (1992) Nucleic Acids Res., 20, 3873–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenny M.K., Schlegel,U., Furneaux,H. and Hurwitz,J. (1990) J. Biol. Chem., 265, 7693–7700. [PubMed] [Google Scholar]
- 8.Erdile L.F., Wold,M.S., Kelly,T.J. (1990) J. Biol. Chem., 265, 3177–3182. [PubMed] [Google Scholar]
- 9.Stigger E., Dean,F.B., Hurwitz,J. and Lee,S.-H. (1994) Proc. Natl Acad. Sci. USA, 91, 579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henricksen L.A., Umbricht,C.B. and Wold,M.S. (1994) J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- 11.Murti K.G., He,D.C., Brinkley,B.R., Scott,R. and Lee,S-H. (1996) Exp. Cell Res., 223, 279–289. [DOI] [PubMed] [Google Scholar]
- 12.Dornreiter I., Erdile,L.F., Gilbert,I.U., von Winkler,D., Kelly,T.J. and Fanning,E. (1992) EMBO J., 11, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S.-H. and Kim,D.K. (1995) J. Biol. Chem., 270, 12801–12807. [DOI] [PubMed] [Google Scholar]
- 14.Melendy T. and Stillmam,B. (1993) J. Biol. Chem., 268, 3389–3395. [PubMed] [Google Scholar]
- 15.Collins K.L. and Kelly,T.J. (1991) Mol. Cell. Biol., 11, 2108–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mass G., Nethanel,T. and Kaufmann,G. (1998) Mol. Cell. Biol., 18, 6399–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenny M.K., Lee,S.-H. and Hurwitz,J. (1989) Proc. Natl Acad. Sci. USA, 86, 9757–9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brill S.J. and Stillman,B. (1989) Nature, 342, 92–95. [DOI] [PubMed] [Google Scholar]
- 19.Ishiai M., Sanchez,J.P., Amin,A.A., Murakami,Y. and Hurwitz,J. (1996) J. Biol. Chem., 271, 20868–20878. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto T., Eki,T. and Hurwitz,J. (1990) Proc. Natl Acad. Sci. USA, 87, 9712–9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S.-H., Kwong,A.D., Pan,Z.Q. and Hurwitz,J. (1991) J. Biol. Chem., 266, 594–602. [PubMed] [Google Scholar]
- 22.Lee S.-H. (1993) Nucleic Acids Res., 21, 1935–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D.-K., Stigger,S. and Lee,S.-H. (1996) J. Biol. Chem., 271, 15124–15129. [DOI] [PubMed] [Google Scholar]
- 24.Wobbe C.R., Dean,F., Weissbach,L. and Hurwitz,J. (1985) Proc. Natl Acad. Sci. USA, 82, 5710–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes X.V. and Wold,M.S. (1995) J. Biol. Chem., 270, 4534–4543. [DOI] [PubMed] [Google Scholar]
- 26.Tsurimoto T., Melendy,T. and Stillman,B. (1990) Nature, 346, 534–539. [DOI] [PubMed] [Google Scholar]
- 27.Hurwitz J., Dean,F.B., Kwong,A.D. and Lee,S.-H. (1990) J. Biol. Chem., 265, 18043–18046. [PubMed] [Google Scholar]
- 28.Yuzhakov A., Kelman,Z., Hurwitz,J. and O’Donnell,M. (1999) EMBO J., 18, 6189–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]