Abstract
The Polycomb group (PcG) gene Bmi1 promotes cell proliferation and stem cell self-renewal by repressing the Ink4a/Arf locus. We used a genetic approach to investigate whether Ink4a or Arf is more critical for relaying Bmi1 function in lymphoid cells, neural progenitors, and neural stem cells. We show that Arf is a general target of Bmi1, however particularly in neural stem cells, derepression of Ink4a contributes to Bmi1-/- phenotypes. Additionally, we demonstrate haploinsufficient effects for the Ink4a/Arf locus downstream of Bmi1 in vivo. This suggests differential, cell type-specific roles for Ink4a versus Arf in PcG-mediated (stem) cell cycle control.
Keywords: Bmi1, Ink4a, Arf, Polycomb, stem cell development
The Bmi1 gene, originally identified as a collaborating oncogene in c-Myc induced lymphomagenesis, is a member of the Polycomb group (PcG) gene family of chromatin modifiers and transcriptional repressors (Haupt et al. 1991; Van Lohuizen et al. 1991). Loss of a PcG gene alters Homeobox (Hox) gene expression resulting in skeletal malformations (Van der Lugt et al. 1994; Akasaka et al. 1996; Core et al. 1997; Del Mar Lorente et al. 2000). However, Hox genes are not the only targets of PcG as the tumor suppressor locus Cdkn2a (hereafter Ink4a/Arf locus) is also negatively regulated by Bmi1 and other PcG members (Jacobs et al. 1999; Voncken et al. 2003; Core et al. 2004; Gil et al. 2004).
The Ink4a/Arf locus codes for two proteins, p16ink4a and p19arf (Ink4a and Arf), by use of alternative reading frames (Serrano et al. 1993; Quelle et al. 1995). Ink4a and Arf are important players in the retinoblastoma (pRB) and p53 pathways, respectively, and their activation results in growth arrest, senescence, or apoptosis (for review, see Sharpless and DePinho 1999; Lowe and Sherr 2003). Bmi1-deficient mice suffer from abnormalities of the hematopoietic and nervous system in addition to growth retardation and skeletal malformations (Van der Lugt et al. 1994). Generation of Bmi1;Ink4a/Arf compound mutant mice provided genetic evidence that at least part of these defects are due to derepression of the Ink4a/Arf locus (Jacobs et al. 1999). Interestingly, we and others have recently shown that Bmi1 is essential for the self-renewal of hematopoietic and neural stem cells, and proliferation of cerebellar granule neuron progenitors (Lessard and Sauvageau 2003; Molofsky et al. 2003; Park et al. 2003; Iwama et al. 2004; Leung et al. 2004; Zencak et al. 2005). Loss of Ink4a could partially alleviate the self-renewal defect of Bmi1-deficient neural stem cells, implicating that inappropriate activation of the Ink4a/Arf locus negatively influences stem cell renewal (Molofsky et al. 2003).
However, since Bmi1 is a potent repressor of both Ink4a and Arf it is important to discriminate which one of these genes is crucial for Bmi1 function. Here we took advantage of the various specific knockout mouse models made for the locus. We addressed the relative contribution of Ink4a and Arf deregulation to a variety of Bmi1-/- phenotypes with emphasis on the hematopoietic and central nervous system. We reveal specific dosage effects of Ink4a and Arf in vivo, and demonstrate that Arf is a general target of Bmi1 in lymphoid cells, neural progenitors, and stem cells. Importantly, particularly in neural stem cells deregulated expression of Ink4a contributes to the Bmi1-deficient phenotype, altogether highlighting differential, cell type-specific requirements for Ink4a versus Arf in Bmi1-mediated control of cell proliferation.
Results and Discussion
Differential effects of Ink4a and Arf derepression in Bmi1-deficient lymphoid organs
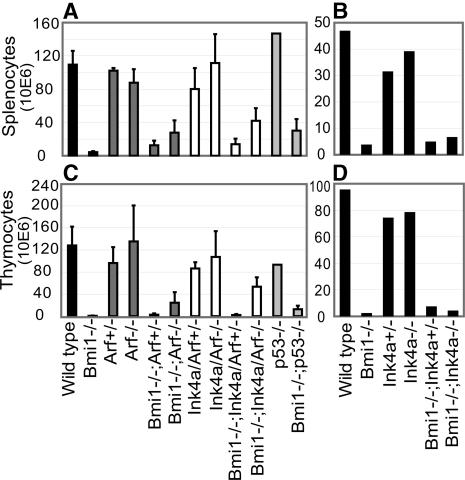
Bmi1-/- mice suffer from progressive hypoplasia of the spleen and thymus as illustrated by a large reduction in lymphocyte counts (Van der Lugt et al. 1994; Fig. 1). We have previously shown that Ink4a/Arf is an in vivo target of Bmi1 in the lymphoid system (Jacobs et al. 1999). Here we investigated the impact of Ink4a versus Arf derepression on lymphocyte counts in Bmi1-/- mice. In the thymus but not in spleen, Arf loss gives a significantly smaller rescue than Ink4a/Arf loss, revealing a role for Ink4a restricting thymocyte growth downstream of Bmi1 (p < 0.01) (Fig. 1A,C). However, loss of Ink4a alone does not alleviate the phenotype showing that reduction of Arf levels is a prerequisite (Fig. 1B,D). Lack of rescue from Bmi1 deficiency upon Ink4a deletion is not unique for splenocytes. Also Bmi1-/- mouse embryonic fibro-blasts (MEFs), which up-regulate p16ink4a and p19arf in a dose-dependent manner (Supplementary Fig. 1A), are only rescued from premature senescence upon loss of Arf (Supplementary Fig. 1B,C). Intriguingly, in the Bmi1-deficient spleen but not in thymus, heterozygosity for either Arf or Ink4a/Arf alleviates the reduced cell counts to a minor yet significant extent (p < 0.001), indicating that specific threshold levels of Ink4a and Arf are required (Fig. 1A,C).
Figure 1.
Differential effects of Ink4a and Arf dosage and derepression in Bmi1-/- lymphoid organs. (A) Splenocyte counts are dramatically reduced in Bmi1-/- mice. Heterozygosity for Arf or Ink4a/Arf partially rescues this phenotype (p < 0.01) but complete loss of Arf or Ink4a/Arf results in a better partial rescue. p53 loss also induces a partial rescue. (B,D) Neither Bmi1-/- splenocyte nor thymocyte counts are significantly restored by loss of Ink4a. (C) Thymocyte counts are dramatically reduced in Bmi1-/- mice. Loss of the complete Ink4a/Arf locus gives a substantially better rescue than loss of Arf alone (p < 0.05). Note that heterozygosity does not lead to a rescue. p53 loss induces a minor rescue in lymphocyte counts.
Similar effects occur in the composition of the splenic B-cell population. Bmi1-/- mice harbor increased thymic populations of CD4-CD8- immature T-cells and a decreased number of splenic mature B220+sIg+ B-cells (Van der Lugt et al. 1994; Jacobs et al. 1999). Applying flow cytometry using standard T- and B-cell differentiation markers, we found that in spleen, heterozygosity for either Arf or Ink4a/Arf completely restores the relative population frequency of mature B-cells to wild-type levels (Supplementary Fig. 2A,B). In contrast in thymus, only complete loss of Arf or Ink4a/Arf results in a relative restoration of wild-type levels of immature CD8-CD4- T-cells (Supplementary Fig. 2A,C).
Lastly, we questioned whether Bmi1 signals to the Arf target p53. p19arf controls the activity of p53 by sequestering Mdm2, an E3 ubiquitin ligase for p53 (for review, see Sherr and Weber 2000). As depicted in Figure 1A,C and Supplementary Figure 1D, both lymphocyte counts and fibroblast proliferation are (partially) rescued upon p53 deletion. Moreover, Mdm2 overexpression rescues the impaired proliferation of Bmi1-/- MEFs as well (Supplementary Fig. 1E). This strongly suggests that at least part of the Bmi1-/- phenotypes are caused by p53 activation through Arf.
Proliferation defects in Bmi1-deficient cerebellum reflect Ink4a and Arf deregulation
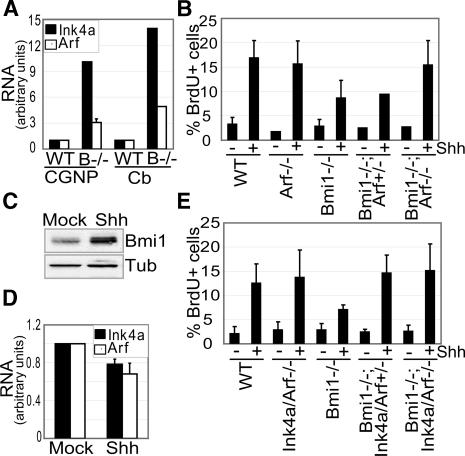
When investigating the neurological defects of Bmi1-/- mice, we recently pinpointed a reduced number of poorly proliferating cells of the cerebellar external granular layer (EGL) to be partially responsible (Leung et al. 2004). During normal early post-natal development, a wave of proliferation of cerebellar granule neuron progenitors (CGNPs) in the EGL is induced by Purkinje neuron secreted Sonic Hedgehog (Shh) (Dahmane and Ruiz-I-Altaba 1999; Wechsler-Reya and Scott 1999). In time, CGNPs differentiate and migrate inwards past the molecular layer (ML) until they reach the internal granular layer (IGL) where they reside as mature granule neurons (for review, see Wang and Zoghbi 2001). We were able to link this process to PcG by showing that Bmi1 is a target of Shh, thus explaining the impaired proliferative response of Bmi1-/- CGNPs upon Shh stimulation (Leung et al. 2004). As Bmi1-/- CGNPs are still partially responsive to Shh, we proposed a model where Shh controls proliferation via at least two routes, i.e., through induction of N-Myc/Cyclin D2 (Kenney et al. 2003) and through Bmi1-mediated repression of Ink4a/Arf.
Analysis by quantitative real-time PCR (qRT-PCR) shows that Ink4a and to a lesser extent Arf expression is increased in Bmi1-/- CGNPs and cerebella (Fig. 2A). Moreover, we demonstrate that stimulation of wild-type CGNPs with Shh leads to an induction of Bmi1 protein expression and a concomitant significant down-regulation of both Ink4a and Arf transcript levels, consistent with the idea of Shh regulating Ink4a/Arf through Bmi1 (Fig. 2C,D). To assess to what extent Ink4a and Arf deregulation underlies the proliferative defects of Bmi1-/- CGNPs, we investigated the Shh response of Bmi1;Arf and Bmi1;Ink4a/Arf doubly deficient CGNPs and their respective controls. Notably, neither Arf nor Ink4a/Arf loss causes enhanced proliferation in absence of Shh in line with the multiple levels of regulation downstream of Shh (Fig. 2B,E). However in the context of Bmi1 deficiency, both Arf loss alone and complete loss of the Ink4a/Arf locus rescues Shh-induced proliferation of Bmi1-/- CGNPs to control levels (p < 0.05) (Fig. 2B,E). Notably, we observe a clear effect of heterozygosity for Ink4a/Arf in Bmi1-/- CGNPs similarly to thymus (Fig. 2E).
Figure 2.
Ink4a/Arf derepression upon Bmi1 loss prevents efficient Shh-induced CGNP proliferation. (A) qRT-PCR shows up-regulation of Ink4a and Arf mRNA expression in Bmi1-/- CGNPs and cerebella. (B) Bmi1; (Cb) cerebellum. (B) Reduced BrdU incorporation in Bmi1-/- CGNPs upon Shh treatment (p < 0.001) is rescued upon subsequent Arf loss (p < 0.01) but not by Arf heterozygosity. (C,D)In wild-type CGNPs, Shh treatment induces an increase in Bmi1 protein levels and a decrease in Ink4a (p < 0.005) and Arf (p < 0.02) mRNA expression. (Tub) Tubulin. (E) Reduced BrdU incorporation in Bmi1-/- CGNPs upon Shh treatment is rescued by Ink4a/Arf loss (p < 0.05) and Ink4a/Arf heterozygosity (p < 0.03).
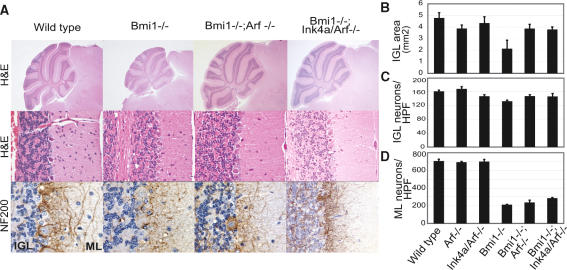
We demonstrated earlier that loss of Ink4a/Arf gives a qualitative rescue of the Bmi1-/- cerebellar defects in vivo (Jacobs et al. 1999). Since we found that Arf loss fully rescued Bmi1-/- CGNPs in vitro, we questioned to what extent this affects in vivo proliferation of cerebellar progenitor cells. Histological analysis revealed that the reduced thickness of the Bmi1-/- granular layer is rescued to a similar extent in either an Arf-/- or Ink4a/Arf-/- background (Fig. 3A,B). Surprisingly, the cell density of the Bmi1-/- IGL is fully rescued in an Ink4a/Arf-/- background, but only partially in an Arf-/- background (p < 0.01) pointing at a subtle role for Ink4a-repression in the IGL in vivo (Fig. 3A,C). Disturbed EGL proliferation can explain multiple abnormalities of the cerebellum. Particularly later during development, granule neurons signal to Purkinje and Basket cells to create the appropriate amount of arborization (Baptista et al. 1994). However, despite proliferation substantially being restored in Bmi1;Arf- and Bmi1;Ink4a/Arf-deficient CGNPs, these mice still display neurological abnormalities. This implies that at least part of the Bmi1-/- cerebellar phenotype is due to defects in cell types other than CGNPs. The cerebellum originates from two germinal layers, the rhombic lip from which the CGNPs are derived, and the ventricular zone which gives birth to the molecular neurons (for review, see Wang and Zoghbi 2001). It is conceivable that Bmi1 also plays a role in cells derived from the ventricular zone, especially since Bmi1 is required for the self-renewal of adult neural stem cells from the subventricular zone, a closely related germinal layer of the cerebrum (see below). Indeed, we observed aberrations such as reduced cellularity of the molecular layer (Fig. 3A,D) and abnormal arborization of basket neurons in Bmi1-/- and Bmi1-/-;(Ink4a/)Arf-/- cerebella (Fig. 3A). Interestingly, the reduction in ML neurons is partially rescued by Ink4a/Arf loss and not significantly by Arf loss (p < 0.05), suggesting a role for Ink4a-repression in molecular layer neurons in vivo (Fig. 3D).
Figure 3.
Ink4a and Arf differentially contribute to histological abnormalities of the Bmi1-/- cerebellum. (A) Morphological analysis of wild-type (left panels), Bmi1-/- (middle left panels), Bmi1-/-;Arf-/- (middle right panels), and Bmi1-/-;Ink4a/Arf-/- (right panels) adult cerebellum. Haematoxylin and eosin (H&E, top and middle panels) staining shows rescue of overall cerebellar size and granular layer thickness in Bmi1-/-;(Ink4a)Arf-/- mice. (Bottom panels) Aberrant arborization of basket neurons (NF200 staining) is observed in all cerebella lacking Bmi1. Final magnification, 5× and 60×. (IGL) Internal granular layer; (ML) molecular layer. (B) IGL area measurements reveal similar significant rescues in Bmi1-/-;Arf-/- and Bmi1-/-;Ink4a/Arf-/- mice. (C) Ink4a/Arf loss induces a complete rescue of the number of Bmi1-/- granule neurons, whereas Arf loss leads to a partial rescue (p < 0.01). (D) The partial rescue in number of Bmi1-/- molecular layer neurons is significantly better in an Ink4a/Arf deficient background (p < 0.05) (HPF, high-power field; n = 4 mice).
Neural stem cell self-renewal critically depends on repression of Ink4a and Arf by Bmi1
A stem cell is defined as a multipotent cell capable of extensive self-renewal. For neural stem cells, multipotency means the cell can give rise to neurons and glial cells. One major neurogenic region in the adult cerebrum harboring stem cells is the subventricular zone (SVZ) of the lateral ventricle wall (for review, see Doetsch 2003). Neural stem cells isolated from the SVZ can be grown as either adherent colonies or as “neurospheres”, floating clusters of stem cells and progeny (Reynolds and Weiss 1992; Morshead et al. 1994).
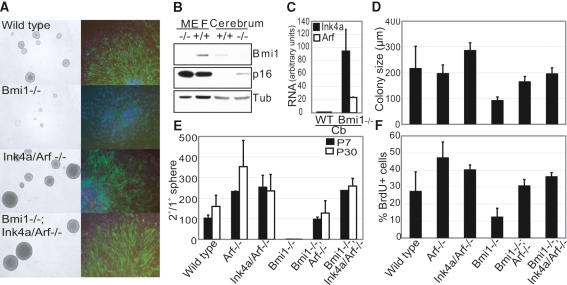
Recently, Molofsky et al. demonstrated that Bmi1 is essential for the self-renewal of SVZ-derived neurospheres and that Ink4a repression is partially mediating this effect (Molofsky et al. 2003). Here we set out to determine whether Arf is required as well. Western blot analysis on wild-type and Bmi1 knockout cerebral tissue (isolated from post-natal day 30 [P30] mice) revealed up-regulated p16ink4a expression in the Bmi1-/- brain (Fig. 4B). In addition, qRT-PCR demonstrated that alike in MEFs and cerebellum, Ink4a and Arf transcripts are up-regulated, establishing Ink4a/Arf as a bona fide in vivo Bmi1 target (Fig. 4C). Next, we prepared adult SVZ neural stem cell cultures at clonal density and assured that the vast majority of primary neurospheres was derived from multipotent stem cells by staining for neuronal and glial markers (Fig. 4A, right panel). Strikingly, Bmi1-/- primary neurospheres not only form less frequently, but also differentiate less efficiently (Fig. 4A right panel) and are much smaller in size than control neurospheres (Fig. 4A [left panel], D). The latter observation suggests that proliferation within a Bmi1-/- neurosphere, reflecting the sum of cell divisions from self-renewing stem cells and their progeny, is impaired. Indeed, Bmi1-/- colonies incorporate far less BrdU than control colonies (Fig. 4F). Notably, this defect in proliferation can be rescued by both loss of Arf and by complete deletion of the Ink4a/Arf locus implicating Arf as an important downstream effector of Bmi1 for stem cell and progenitor proliferation (Fig. 4A,F).
Figure 4.
Accurate repression of Ink4a and Arf is required for neurosphere self-renewal. (A,D) Phase-contrast pictures (A, left panel) and diameter measurements show that Bmi1-/- neurospheres are much smaller than wild types. Loss of either Arf or Ink4a/Arf completely rescues this Bmi1-/- phenotype. (A, right panel) All primary neurospheres are multipotent. Note that Bmi1-/- spheres differentiate less efficiently (GFAP staining in green and β-tubulin-III in red). (B) Western blot analysis reveals increased p16ink4a expression in Bmi1-/- cerebrum. (Tub) Tubulin. (C) qRT-PCR shows increased Ink4a and Arf mRNA expression in Bmi1-/- cerebrum. (Cb) Cerebrum. (E) P7- and P30-derived Bmi1-/- neurospheres are severely impaired in their self-renewal capacity. Arf loss alone gives a partial rescue of this phenotype (p < 0.05). Removal of the complete Ink4a/Arf locus fully restores the self-renewing ability of Bmi1-/- neurospheres. Note that loss of Arf or Ink4a/Arf in a Bmi1+/+ background enhances self-renewal (p < 0.01). (1°) Primary; (2°) secondary. (F) Bmi1-/- SVZ adherent colonies incorporate less BrdU than control colonies, which is completely rescued upon loss of either Arf or Ink4a/Arf.
Next, we performed a neurosphere assay to specifically study stem cell self-renewal. In this assay, the capacity of a primary neurosphere to form new multipotent neurospheres after dissociation is measured. We report a dramatic decrease in the self-renewing capacity of both adult (P30) and P7-derived Bmi1-/- neurospheres in agreement with previous findings (Fig. 4E). Deletion of the Ink4a/Arf locus in Bmi1-/- neurospheres completely rescues this defect. Importantly, loss of Arf alone gives a partial rescue (p < 0.05), reinforcing the earlier observation that proper repression of Ink4a is also required for neurosphere self-renewal. This does not simply reflect a tissue culture phenomenon as is highlighted by Molofsky et al. (2005) who found that Ink4a loss partially restores Bmi1-/- stem cell frequency and neurogenesis in vivo. Upon induction of differentiation, all secondary neurospheres stained positive for neuron and astrocyte specific markers (data not shown). Surprisingly, there is a clear increase in both proliferation and self-renewing capacity of Arf and Ink4a/Arf deficient neurospheres compared with wild types (Fig. 4E,F). This strongly suggests that the Ink4a/Arf locus actively restricts self-renewing cell divisions thus playing a role in controlling the stem cell compartment. Lastly, we tested these neurospheres for long-term self-renewal as, for instance, exhaustion of hematopoietic stem cells sometimes occurs after a prolonged period of time (Park et al. 2003). However, we were able to keep Bmi1-/-;Arf-/- and Bmi1-/-;Ink4a/Arf-/- neurospheres in culture for at least five weekly passages, suggesting that the Bmi1 knockout phenotype is fully reversed.
A specific subset of PcG proteins appears to regulate stem cell self-renewal and progenitor proliferation
How could PcG distinctly regulate its target genes in different cell types? Emerging studies indicate that different “flavors” of PcG complexes may exist (De Napoles et al. 2004; Kuzmichev et al. 2004). An increase in the relative amount of a PcG member such as Bmi1, may alter the affinity of a PcG complex towards the chromatin in such a way that for instance self-renewing divisions are favored (for review, see Valk-Lingbeek et al. 2004). Underscoring a special role for Bmi1, we found that loss of Ring1a or M33, PcG proteins belonging to the same complex as Bmi1, does not affect neurosphere self-renewal or CGNP proliferation (Supplementary Fig. 3A,B). Only loss of Mel18, the closest homolog of Bmi1, has a modest though not significant negative effect on these processes in accordance to recent observations in hematopoietic stem cells (Iwama et al. 2004).
It remains striking that lymphoid cell counts, cerebellar development, skeletal transformations, and overall body growth in Bmi1;Ink4a/Arf doubly deficient mice are at best partially rescued (Figs. 1, 3; Supplementary Table 1; Supplementary Fig. 4). Therefore, other Bmi1 regulated genes must exist. In line with Drosophila not possessing genes resembling the Ink4a/Arf locus, the acquisition of PcG-mediated proliferation control likely evolved later, perhaps reflecting a demand for protection of long lasting stem cells. Candidates for additional Bmi1 targets are the Hox genes. Interestingly, a subset of Hox genes has been implicated in mammalian brain development and control of hematopoiesis. Moreover, several Hox genes are differentially expressed in Bmi1-/- SVZ neurospheres (Molofsky et al. 2003). However, it cannot be excluded that a substantial number of additional targets exists, as PcG proteins as chromatin modifiers are known to act on large gene sets (Kirmizis et al. 2004). Altogether, we have demonstrated profound tissue and cell type specific differences in the effects of Ink4a versus Arf derepression in Bmi1-deficient mice. We propose a general role for Bmi1-mediated Arf/p53 repression in curtailing proliferation. Ink4a repression on the other hand is required only in a subset of cells. Furthermore, we show haploinsufficient effects of the Ink4a/Arf locus in vivo selectively for certain tissues, and suggest that appropriate threshold levels of these two proteins are required to ensure proper proliferation. These observations implicate PcG proteins not only in embryonic developmental fate decisions, but also in discriminative processes between cell cycle control of stem- and more differentiated cells.
Materials and methods
Mice breeding, lymphocyte counts and flow cytometry
Bmi1+/- FVB or C57BL/6 mice (Van der Lugt et al. 1994) were crossed with Ink4a/Arf+/- FVB mice (Serrano et al. 1996), with Arf+/- FVB mice (Kamijo et al. 1997), with p53+/- FVB mice (Donehower et al. 1992), or Ink4a+/- C57BL/6 mice (Krimpenfort et al. 2001). In addition, Ring1a FVB mice (Del Mar Lorente et al. 2000), M33 FVB mice (Core et al. 1997) and Mel18 (mixed background) knockout mice (Akasaka et al. 1996) were used. All mice were genotyped routinely by PCR (list of primers available upon request). Multiple independent animals of the respective genotypes were assayed and all results were subjected to Student's t-tests and Bonferroni correction when appropriate. Preparation of cell suspensions from lymphoid organs, cell counts, and flow cytometric analyses were done as described previously (Jacobs et al. 1999).
Western blot analysis and quantitative real-time PCR
Equal amounts of protein were separated on 13% SDS-PAGE or precast gels (Invitrogen) and blotted onto Immobilon-P membranes (Amersham Biosciences). Bands were visualized using enhanced chemiluminescence (Amersham). Primary antibodies were F6 for Bmi1 (Upstate), M156 for p16ink4a (Santa Cruz), or R562 for p19arf (Abcam). Secondary antibodies were goat-anti-mouse (ZyMed) or goat-anti-rabbit (BioSource), both HRP conjugated. Total RNA was extracted using TRIZOL reagent (Invitrogen) and cDNA was prepared using Superscript II RT and oligod(T)n primers (Invitrogen). qRT-PCR was performed with 50 ng cDNA on an ABI PRISM 7000 using SYBR Green PCR mastermix (Applied Biosystems). For primer sequences and formulas, see Supplemental Material.
CGNP isolation and histology of cerebellar tissue
CGNP cultures were isolated from 7 d-old-mice and cultured according to established protocols (Wechsler-Reya and Scott 1999; Leung et al. 2004). Quantification of Shh-induced proliferation was performed as described previously (Leung et al. 2004). Immunocytochemistry was performed using mouse monoclonal anti-BrdU (DAKO) and Alexa Fluor m488 goat-anti-mouse (Molecular Probes). DAPI (Molecular Probes) was used to visualize nuclei. Immunohistochemistry and histological analysis were performed as described before (Leung et al. 2004).
Neural stem cell isolation, neurosphere proliferation, and self-renewal assays
For neural stem cell isolation and culture conditions, see Supplemental Material. To assess proliferation, primary adherent stem cell colonies were pulsed for 1 h with 1 μM BrdU (7 d after isolation). Immunocytochemistry was performed as for CGNPs. The self-renewal capacity was determined by dissociating 10-d-old primary neurospheres and replating them in six-well ultra-low binding plates at clonal density. After 10 d, the newly generated (secondary) neurospheres were counted. To reveal multipotency, primary and secondary neurospheres were plated onto poly-ornithine and fibronectin- or laminin-coated (Sigma) plates and differentiated for 4 d in neurosphere medium supplemented with 2% FBS (ICN Biochemicals). Cells were labeled with antibodies against GFAP (DAKO) and β-tubulin-III (Sigma). Secondary antibodies were FITC-conjugated goat-anti-rabbit and Cy3-conjugated goat-anti-mouse (Jackson ImmunoResearch). Nuclei were stained with DAPI.
Acknowledgments
We thank the Netherlands Cancer Institute FACS and Microscopy Core Facilities for assistance, and G. Hart for advice on statistics. We thank M. Oren for Mdm2 cDNA, R. DePinho for Ink4a/Arf+/- mice, C. Sherr for Arf+/- mice, A. Berns for Ink4a+/- mice, and L. Donehower for p53+/- mice. We thank J. Deschamps and H. Koseki for Mel18+/- mice, M. Djabali for M33+/- mice, M. Vidal for Ring1a+/- mice, and S. Morrison for sharing unpublished results. S.B., P.S., and J.J. are supported by grants from the Dutch Cancer Society to M.v.L.. M.V.-L. and P.S. are supported by a Pioneer grant from the Netherlands Organization for Scientific Research to M.v.L. S.M. is supported by grants from the Oncosuisse and “Nachwuchsförderung Universität Zürich”. Y.A. is supported by the Swiss National Science Foundation, ProVisu, and Velux Foundations, and AFM.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1299305.
References
- Akasaka T., Kanno, M., Balling, R., Mieza, M.A., Taniguchi, M., and Koseki, H. 1996. A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development 122: 1513-1522. [DOI] [PubMed] [Google Scholar]
- Baptista C.A., Hatten, M.E., Blazeski, R., and Mason, C.A. 1994. Cell-cell interactions influence survival and differentiation of purified Purkinje cells in vitro. Neuron 12: 243-260. [DOI] [PubMed] [Google Scholar]
- Core N., Bel, S., Gaunt, S.J., Aurrand-Lions, M., Pearce, J., Fisher, A., and Djabali, M. 1997. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development 124: 721-729. [DOI] [PubMed] [Google Scholar]
- Core N., Joly, F., Boned, A., and Djabali, M. 2004. Disruption of E2F signaling suppresses the INK4a-induced proliferative defect in M33-deficient mice. Oncogene 23: 7660-7668. [DOI] [PubMed] [Google Scholar]
- Dahmane N. and Ruiz-i-Altaba, A. 1999. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126: 3089-3100. [DOI] [PubMed] [Google Scholar]
- De Napoles M., Mermoud, J.E., Wakao, R., Tang, Y.A., Endoh, M., Appanah, R., Nesterova, T.B., Silva, J., Otte, A.P., Vidal, M., et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7: 663-676. [DOI] [PubMed] [Google Scholar]
- Del Mar Lorente M., Marcos-Gutierrez, C., Perez, C., Schoorlemmer, J., Ramirez, A., Magin, T., and Vidal, M. 2000. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127: 5093-5100. [DOI] [PubMed] [Google Scholar]
- Doetsch F. 2003. A niche for adult neural stem cells. Curr. Opin. Genet. Dev. 13: 543-550. [DOI] [PubMed] [Google Scholar]
- Donehower L.A., Harvey, M., Slagle, B.L., McArthur, M.J., Montgomery Jr., C.A., Butel, J.S., and Bradley, A. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215-221. [DOI] [PubMed] [Google Scholar]
- Gil J., Bernard, D., Martinez, D., and Beach, D. 2004. Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell. Biol. 6: 67-72. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Alexander, W.S., Barri, G., Klinken, S.P., and Adams, J.M. 1991. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E μ-myc transgenic mice. Cell 65: 753-763. [DOI] [PubMed] [Google Scholar]
- Iwama A., Oguro, H., Negishi, M., Kato, Y., Morita, Y., Tsukui, H., Ema, H., Kamijo, T., Katoh-Fukui, Y., Koseki, H., et al. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21: 843-851. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom, K., Marino, S., DePinho, R.A., and van Lohuizen, M. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164-168. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Zindy, F., Roussel, M.F., Quelle, D.E., Downing, J.R., Ashmun, R.A., Grosveld, G., and Sherr, C.J. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91: 649-659. [DOI] [PubMed] [Google Scholar]
- Kenney A.M., Cole, M.D., and Rowitch, D.H. 2003. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130: 15-28. [DOI] [PubMed] [Google Scholar]
- Kirmizis A., Bartley, S.M., Kuzmichev, A., Margueron, R., Reinberg, D., Green, R., and Farnham, P.J. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes & Dev. 18: 1592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P., Quon, K.C., Mooi, W.J., Loonstra, A., and Berns, A. 2001. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413: 83-86. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A., Jenuwein, T., Tempst, P., and Reinberg, D. 2004. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 14: 183-193. [DOI] [PubMed] [Google Scholar]
- Lessard J. and Sauvageau, G. 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423: 255-260. [DOI] [PubMed] [Google Scholar]
- Leung C., Lingbeek, M., Shakhova, O., Liu, J., Tanger, E., Saremaslani, P., Van Lohuizen, M., and Marino, S. 2004. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428: 337-341. [DOI] [PubMed] [Google Scholar]
- Lowe S.W. and Sherr, C.J. 2003. Tumor suppression by Ink4a-Arf: Progress and puzzles. Curr. Opin. Genet. Dev. 13: 77-83. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Pardal, R., Iwashita, T., Park, I.K., Clarke, M.F., and Morrison, S.J. 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.V., He, S., Bydon, M., Morrison, S.J., and Pardal, R. 2005. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Morshead C.M., Reynolds, B.A., Craig, C.G., McBurney, M.W., Staines, W.A., Morassutti, D., Weiss, S., and van der Kooy, D. 1994. Neural stem cells in the adult mammalian forebrain: A relatively quiescent subpopulation of subependymal cells. Neuron 13: 1071-1082. [DOI] [PubMed] [Google Scholar]
- Park I.K., Qian, D., Kiel, M., Becker, M.W., Pihalja, M., Weissman, I.L., Morrison, S.J., and Clarke, M.F. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302-305. [DOI] [PubMed] [Google Scholar]
- Quelle D.E., Zindy, F., Ashmun, R.A., and Sherr, C.J. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83: 993-1000. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A. and Weiss, S. 1992. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255: 1707-1710. [DOI] [PubMed] [Google Scholar]
- Serrano M., Hannon, G.J., and Beach, D. 1993. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366: 704-707. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lee, H., Chin, L., Cordon-Cardo, C., Beach, D., and DePinho, R.A. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85: 27-37. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E. and DePinho, R.A. 1999. The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev. 9: 22-30. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. and Weber, J.D. 2000. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 10: 94-99. [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek M.E. Bruggeman, S.W., and van Lohuizen, M. 2004. Stem cells and cancer: The polycomb connection. Cell 118: 409-418. [DOI] [PubMed] [Google Scholar]
- Van der Lugt N.M., Domen, J., Linders, K., van Roon, M., Robanus-Maandag, E., te Riele, H., van der Valk, M., Deschamps, J., Sofroniew, M., and van Lohuizen, M. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes & Dev. 8: 757-769. [DOI] [PubMed] [Google Scholar]
- Van Lohuizen M., Verbeek, S., Scheijen, B., Wientjens, E., van der Gulden, H., and Berns, A. 1991. Identification of cooperating oncogenes in E μ-myc transgenic mice by provirus tagging. Cell 65: 737-752. [DOI] [PubMed] [Google Scholar]
- Voncken J.W., Roelen, B.A., Roefs, M., de Vries, S., Verhoeven, E., Marino, S., Deschamps, J., and van Lohuizen, M. 2003. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. 100: 2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang V.Y. and Zoghbi, H.Y. 2001. Genetic regulation of cerebellar development. Nat. Rev. Neurosci. 2: 484-491. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R.J. and Scott, M.P. 1999. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22: 103-114. [DOI] [PubMed] [Google Scholar]
- Zencak D., Lingbeek, M., Kostic, C., Tekaya, M., Tanger, E., Hornfeld, D., Jaquet, M., Munier, F.L., Schorderet, D.F., van Lohuizen, M., et al. 2005. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J. Neurosci. (in press). [DOI] [PMC free article] [PubMed]