Abstract
The DNA-binding DNA polymerase (gp43) of phage T4 is also an RNA-binding protein that represses translation of its own mRNA. Previous studies implicated two segments of the untranslated 5′-leader of the mRNA in repressor binding, an RNA hairpin structure and the adjacent RNA to the 3′ side, which contains the Shine–Dalgarno sequence. Here, we show by in vitro gp43–RNA binding assays that both translated and untranslated segments of the mRNA contribute to the high affinity of gp43 to its mRNA target (translational operator), but that a Shine–Dalgarno sequence is not required for specificity. Nucleotide sequence specificity appears to reside solely in the operator’s hairpin structure, which lies outside the putative ribosome-binding site of the mRNA. In the operator region external to the hairpin, RNA length rather than sequence is the important determinant of the high binding affinity to the protein. Two aspects of the RNA hairpin determine specificity, restricted arrangement of purine relative to pyrimidine residues and an invariant 5′-AC-3′ in the unpaired (loop) segment of the RNA structure. We propose a generalized structure for the hairpin that encompasses these features and discuss possible relationships between RNA binding determinants of gp43 and DNA binding by this replication enzyme.
INTRODUCTION
The replicative DNA polymerase of bacteriophage T4, product of phage gene 43 (gp43), is a DNA-binding protein that also binds its own mRNA and regulates its own biosynthesis at the translational level (1–4). Relationships between the DNA and RNA binding functions of this replication enzyme are not clearly defined. The protein binds specifically and with high affinity (Kd = 1–2 nM) to a ribonucleotide sequence (termed ‘translational operator’) that overlaps the ribosome-binding site (RBS) of the translation-initiation region (TIR) of gene 43 mRNA (1,4,5). In contrast, binding of the protein to DNA is weak (Kd = 70–100 nM) and exhibits no dependence on primary or higher-order structure of the nucleic acid (6). Ribonuclease-sensitivity assays and other types of studies implicate an RNA stem–loop (hairpin) structure and its adjoining 3′-terminal 13-nt sequence (3′-tail) in recognition of the mRNA by gp43. Both of these components are located in the untranslated 5′-leader segment of the mRNA (Fig. 1; 2,4). Some variants of this sequence are known that bind T4 gp43 with similar affinity to the natural mRNA target (2,7). It appears that some of the gp43 determinants for recognition of the operator are also involved in non-specific binding to nucleic acids because operator RNA is a potent inhibitor of gp43–DNA interactions and DNA is a weak inhibitor of gp43–operator interactions (4,6). Possibly, selectivity of gp43 to its mRNA target in vivo utilizes both nucleotide-sequence-dependent and -independent interactions. In the work described here we assessed the contributions of these two types of interactions to the affinity of gp43 to its mRNA.
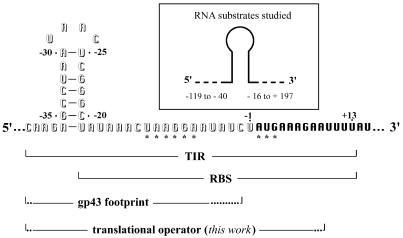
Figure 1.
Ribonucleotide sequence of the TIR for phage T4 gene 43 displaying the untranslated 5′ leader segment (outlined letters) and other landmarks (1,4) of the mRNA. The open reading frame is shown in bold type and the Shine–Dalgarno sequence (UAAGGA) and initiator AUG are marked with asterisks. The boundaries of the putative RBS, the gp43-footprint (2,4) and proposed translational operator region (this work) are also marked. The operator hairpin loop has been previously represented as the 8-nt sequence from position –31 to –24 (1,4,7). The tetralooped structure depicted here is consistent with structures derived from NMR analysis (14,15). The tetra- and octa-loop configurations may co-exist. The ‘TIR’ is defined according to McCarthy and Gualerzi (5) as “that region determining both the site and efficiency of initiation”; this definition would include the translational operator.
We used mutant constructs of the T4 gene 43 translational operator as gp43 targets in binding studies which identified four components of the RNA that determine its high affinity to the protein: (i) stability of the operator hairpin structure (which is determined by base-pairing in the stem); (ii) a 5′-AC-3′ dinucleotide sequence in the hairpin loop (which could not be substituted without loss of specificity); (iii) the positioning of purine and pyrimidine residues in the polyribonucleotide (which probably determines the three-dimensional placement of the 5′-AC-3′ sequence relative to the protein); and (iv) length, although not the nucleotide sequence, of the single-stranded region to the 3′ side of the operator hairpin structure, i.e., the 3′-tail domain of the operator. Our results suggest that gp43 carries separate clusters of determinants for RNA sequence-dependent (hairpin structure) and sequence-independent (3′-tail segment) interactions with its translational operator and that some of these determinants overlap the sequence-independent binding determinants for DNA. The results also suggest a model to explain the ability of T4 gp43 to bind and translationally repress a range of related RNA sequences.
MATERIALS AND METHODS
Materials for gp43 binding studies
Purified T4 DNA polymerase, synthetic oligodeoxynucleotides and in vitro transcribed RNA preparations were made as described earlier (6,8). RNA samples were resolved by electrophoresis in urea-containing gels and subsequently eluted from the gels, concentrated by evaporation, and desalted by centrifugation through a spun column. Prior to use in gel retardation assays, purified RNA samples exceeding 200 nt in length (at ∼0.5 µM) were subjected to several cycles of freezing (in a dry-ice bath) and thawing (at room temperature) to disaggregate the nucleic acids and minimize their heterogeneous banding patterns during subsequent electrophoretic analyses. The duplex RNA·DNA sample used in the experiment in Table 2 was obtained by annealing a 1:1 mixture (at 3 µM each) of the wild-type T4 operator RNA (WT-RNA, 68 nt) and ssDNA of complementary sequence (NS-DNA, 54 nt) constructs described previously (6). Synthetic oligoribonucleotides were produced on an Applied Biosystems Instruments (ABI) Model 394 oligonucleotide synthesizer, purified by anion exchange HPLC (Waters, Millipore) using a Poros 10Q resin (PerSeptive Biosystems), desalted on OPC syringe microcolumns (Glenn Research), extracted with chloroform, and lyophilized before reconstitution in sterile water (at 3 µM) for use in binding experiments. Polyuridylic acid (polyU) was purchased from Pharmacia Biotech and used without further purification. Conditions for 5′ end-labeling of synthetic RNAs (including polyU) with 32P were as described elsewhere for oligodeoxyribonucleotides (6), except that the RNase inhibitor RNasin (Promega Life Science) was added (at 2 U/µl) in the kinase reaction mix. All 5′ end-labeled oligoribonucleotides used, except polyU, were purified by electrophoresis before use in gel retardation assays (6).
Table 2. In vitro and in vivo effects of operator RNA mutations.
| Operator Mutant type | Name | Relative Kd (KdMut/KdWT ± S.E.) | Repression in vivo | Summary |
|---|---|---|---|---|
| Loop: single substitutions; Figure 3A | GA23 | 0.6 ± 0.3 | – | C26 and A27 appear to be invariant. Nucleotide positions: –24,–25, –28, –29, –30, and –32 are permissive to transition but not transversion mutations. |
| GU23 | – | Yesa | ||
| CU24 | 1.1 ± 0.4 | – | ||
| UA25 | 10.1 ± 4.5 | No (2)b | ||
| UC25 | 2.6 ± 1.1 | – | ||
| CU26 | 11.1 ± 3.8 | – | ||
| CA26 | 8.2 ± 5.4 | No (2)b | ||
| AG27 | 11.2 ± 2.4 | – | ||
| AG28 | 1.3 ± 1.2 | – | ||
| UA29 | 9.5 ± 1.9 | No (2)b | ||
| AG31 | 1.7 ± 0.7 | Yesa | ||
| UC32 | 1.4 ± 0.2 | – | ||
| Loop: multiple substitutions; Figures 3B and 4Bc | MJV | 0.7 ± 0.1 | Yesa | Two or more base substitutions at certain positions decrease affinity to that of non-specific RNA. |
| RL | 10.9 ± 5.1 | Noa | ||
| SAU | 13.4 ± 5.9 | Noa | ||
| NoU | 23.4 ± 13 | Noa | ||
| LA | 25.3 ± 6.5 | Noa | ||
| LU | 40.4 ± 15 | Noa | ||
| ASd | 32.0 ± 8.3 | – | ||
| Stem: single and multiple substitutions; Figure 4 | CG20 | 1.4 ± 1.2 | – | Stem integrity is essential but sequence is less critical for binding. |
| GA21 | 0.4 ± 0.2 | – | ||
| ST20 | 1.3 ± 0.7 | – | ||
| ST21 | 2.6 ± 1.1 | – | ||
| ST22 | 0.8 ± 0.6 | – | ||
| WTRB69 (2) | 14.7 ± 0.5 | No (7)b | ||
| SR | 14.9 ± 0.7 | No (2)b | ||
| SD | 29.0 ± 7.4 | No (2)b | ||
| 3′ tail: Figure 5 | UA17 | 1.8 ± 0.9 | – | Nucleotide sequence of the 3′-tail domain is not a determinant of affinity to T4 gp43. |
| AU18 | 1.2 ± 0.1 | Yes (2)b | ||
| AAUU67 | 1.4 ± 0.4 | – | ||
| GGAA89 | 1.1 ± 0.4 | – | ||
| PA 3P | 1.1 ± 0.5 | – | ||
| INS1520 | 1.4 ± 0.4 | – | ||
| Non-specific RNAs: Materials and Methods | PolyU | 31.0 ± 15.1 | – | |
| WT-RNA–DNA duplex | 27.1 ± 11.2 | – |
aUnpublished observations.
bNumbers in parenthesis are reference numbers.
cFor MJV.
dAS, antisense RNA, complementary to WT-RNA (Fig. 3).
32P-labeled RNA products were prepared by in vitro transcription as described in the Materials and Methods. Kd values for binding reactions between purified T4 gp43 and RNA targets were determined by gel retardation assays, as previously described (6). The KdMut/KdWT ratio represents the relative Kd for mutant (Mut) RNA constructs as compared to WT-RNA having the same 3′-tail length.
Gel retardation assays
These assays were carried out as described by Pavlov and Karam (6) except that scanning of the gels for radioactive bands and quantitating densities of the bands were carried out on a FUJIX BAS 1000 (Fuji) phosphorimager rather than by standard X-ray film autoradiography. Binding reactions were carried out at 25°C for 1 h in 10 µl of solution containing 42 mM Tris–acetate (pH 7.8), 200 mM potassium acetate, 5 mM dithiothreitol, 3 mM EDTA, and a range of concentrations of 32P-labeled operator RNA construct (6). Reactions were started by introducing T4 gp43 at 10 nM. In addition, in the assay of operator hairpin with 2-nt 3′-tail (Table 1), the RNA was titrated with protein. In the assays for 32P-labeled polyU (which yielded a smear of radioactivity rather than discrete bands on the gels), we integrated the profiles of radioactivity along gel lanes by using the computer software provided with the phosphorimager. Profiles recorded for controls containing polyU alone were digitally subtracted from the profiles for binding mixtures containing gp43 plus polyU to determine the amount of radioactive RNA bound by the protein. The average length of the polyU was determined by electrophoretic analyses in 12% polyacrylamide gels containing 7 M urea. In these analyses, 5′-end labeled synthetic oligoribonucleotides and uniformly labeled in vitro transcribed RNA preparations (size range of 34–316 nt; Fig. 1) served as length markers. The average length of polyU was calculated as the mean length distribution of 32P-labeled sample components separated along the gel. The relationship used was:
Table 1. Dependence of the affinity of WT operator RNA to T4 gp43 on length of the 3′ tail segment.
| RNA length (nt) | Length of 3′ tail (nt) | Kd ± S.E. (nM) | |
|---|---|---|---|
| WT | UA29 | ||
| 34 | 2 | 537 ± 61a | 4800 ± 780a |
| 29 | 7 | 249 ± 96b | – |
| 36 | 14 | 30.9 ± 11b | – |
| 33 NFc | 14 | 92.1 ± 45b | – |
| 56 | 23 | 3.6 ± 0.4a | – |
| 62 | 23 | 4.9 ± 1.3d | – |
| 63 | 30 | 2.5 ± 1.9a | – |
| 68 | 32 | 2.4 ± 0.3a | 26 ± 7a |
| 74 | 32 | 2.1 ± 0.3a | – |
| 69 | 36 | 0.7 ± 0.8a | – |
| 75 | 36 | 1.2 ± 0.9a | – |
| 207 | 98 | – | 11 ± 4d |
| 316 | 215 | 0.9 ± 0.8d | – |
aPrepared by in vitro transcription under SP6 promoter.
bPrepared by chemical synthesis.
cNF, no 5′-leader to the hairpin.
dPrepared by in vitro transcription under T7 promoter.
–, not determined.
The length of the 5′-leader sequence for the RNA varied depending on the type and position of the promoter used in transcription reactions; it ranged between 5 and 53 nt proximal to the 5′-GCC-3′ of the hairpin stem (Fig. 1). The 33 NF construct lacked unpaired nucleotides to the 5′-side of the hairpin. RNA substrates were prepared as described in the Materials and Methods.
lav = ∫l(x)p(x)dx/∫p(x)dx
where x is the migration distance from the sample loading well, the function l(x) relates the oligonucleotide length to its position on the gel (as determined by migration of length standards), and the function p(x) describes the radioactivity profile (the density of radioactivity at the specific migration distance x).
Computations and statistical analysis
RNA secondary structures of wild-type and mutant operator constructs were predicted by using the computer program PCFold 4.0 (9), which applies Zuker’s algorithm (10,11). Computations of lowest free-energy RNA folding were carried out by using the energy sets of Freier et al. (12). Equilibrium parameters (binding capacity and Kd values) were determined from gel retardation experiments as described previously (6).
RESULTS
RNA length and the affinity of T4 gp43 to its translational operator
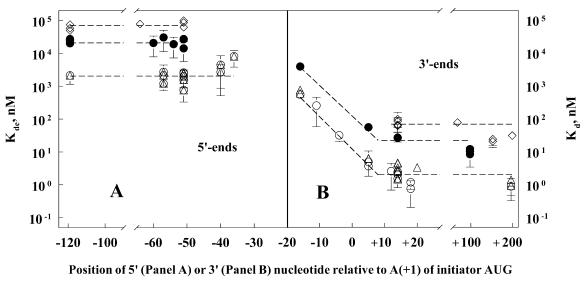
The segment of wild-type gene 43-specific mRNA that is protected by T4 gp43 in RNase and chemical sensitivity (RNA footprinting) assays is 35–40 nt long, including the 18-nt long hairpin sequence shown in Figure 1 (1,4). We have found that operator binding to the protein varies with length of the unprotected 3′-terminal segment of the mRNA. This is demonstrated by the results summarized in Table 1. In the experiments for Table 1, Kd values for gp43–operator complexes were measured by using in vitro transcribed and chemically synthesized RNA substrates bearing different 3′-terminal, but similar 5′-terminal lengths of nucleotide sequence. The lowest Kd values (∼1 nM) were obtained with wild-type operator sequences that contained no less than an ∼26 nt 3′-tail to the operator hairpin structure depicted in Figure 1. The Kd did not change with additional increases in length of the 3′-terminal tail segment, although we were unable to obtain reliable measurements of affinity for substrates longer than 350 nt because the RNA electrophoretic banding patterns became very complex in this size range (see also 2,6). The binding affinity was much lower when RNA targets with 3′-tails shorter than ∼26 nt were used; however, specificity of the RNA to the protein could be demonstrated irrespective of 3′-tail length (e.g., compare the Kd values for the WT and UA29 RNA constructs carrying 2-nt 3′-tails, Table 1). In contrast to the observed dependence of the Kd on length of the RNA 3′-tail segment, RNA length at the 5′-side of the hairpin appeared to be much less critical, although constructs bearing fewer than 2–3 nt immediately 5′-proximal to the hairpin structure exhibited Kd values that were ~3-fold higher than constructs with longer 5′ leaders (construct 33NF, Table 1). Results comparing the dependence on RNA length to the 5′- and 3′-sides of the hairpin are summarized in Figure 2. These results, like those of the footprinting experiments reported elsewhere (1,4), localize the major determinants of operator specificity and affinity to the RNA hairpin and the adjacent 3′-tail (Fig. 1). Most of the binding studies to be described in the remainder of this report utilized RNA targets ranging in length between 56 and 316 nt, including a 19-nt 5′-leader segment to the hairpin and sufficient 3′-tail length to include at least the first five codons of the gene 43 open reading frame.
Figure 2.
Effects of operator RNA length on the binding affinity to T4 gp43. Kd values are plotted for T4 wild-type RNA (open circles; WT in Table 2 and Fig. 3B) and for mutant derivatives that either lowered (closed circles and open diamonds), or did not affect (open triangles) binding affinity of the RNA to T4 gp43. The ‘closed-circle set’ included UA25, CU26, CA26, AG27 and UA29 (Table 2 and Fig. 3A); the ‘closed-diamond set’ included AS, SD, LU, LA, NoU, polyU and a WTRNA–DNA duplex (Table 2 and Figs 3B and 4A); the ‘open-triangle set’ included GA23, CU24, UC25, AG28, AG31, UC32, MJV, CG20, GA21, ST20, ST21, ST22, UA17, AU18, AAUU67, GGAA89, PA 3P and INS1520 (Table 2 and Figs 3–5). The 0 point on the abscissa corresponds to nucleotide position +1 on the sequence shown in Figure 1. (A) Plots of the expected Kd (Kde) values for RNA targets carrying different 5′-leader lengths, but the same 3′-tail length of ‘zero’. These values were calculated by extrapolation from the linear relationship between the logKd and 3′-tail length shorter than ∼26 nt, i.e., as depicted in (B).
Mutational fine-structure analysis of the gene 43 translational operator: an overview
In addition to being dependent on 3′-terminal RNA length (Table 1), affinity of the operator to T4 gp43 depends on the nucleotide sequence and higher-order structure of the RNA (2,4,6); however, several studies have established that the requirement for a specific nucleotide sequence is not very stringent since some variants of the operator bind T4 gp43 with similar affinity to wild-type RNA (2,7,13). We used a mutational analysis to explore the stringency to nucleotide sequence further and to distinguish between operator determinants that are required for high affinity from those that are also required for specificity of binding to the gp43 repressor.
Our mutational analysis took into consideration several observations that were previously derived from phylogenetic studies (7,8,13), other mutational analyses (2,4) and NMR structural determinations (14,15) of the gp43–operator interaction. Collectively, these studies suggested certain unifying features among the limited number of operator variants that have been observed to bind T4 gp43 with wild-type (or near wild-type) affinity. In particular, these variants share similar sequences for the central 4 nt of the hairpin loop, especially the 5′-AC-3′ dinucleotide unit (2,7,13; Fig. 1). Where they differ in nucleotide sequence, they do so through base transitions rather than transversions (2,7). Also, computer-assisted predictions (results not shown) suggested that the NMR structures that were determined for two variant operators by Mirmira and Tinoco (14,15) cannot be achieved if transversions are introduced at any nucleotide position of the hairpin. In fact, several transversion mutants had been examined previously and shown to inactivate the operator (2). Based on the considerations summarized above, we limited our mutational analysis here mostly to operator constructs bearing transition mutations, although some transversions were also examined in order to assess the importance of primary structure in the base-paired stem segment of the hairpin.
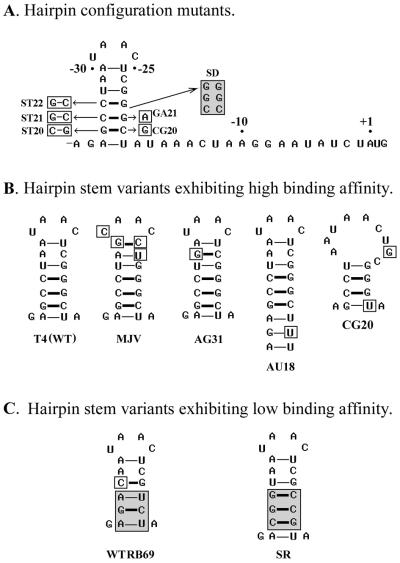
We targeted three operator subdomains to mutational fine-structure analysis: (a) the unpaired hairpin loop, (b) the base-paired stem, and (c) the 3′-tail. Figures 3–5 list the operator mutant groups we constructed, and Table 2 compares their binding affinities to T4 gp43. With some constructs it was possible to introduce the operator mutations into the T4 genome and measure their effects on autogenous control of gp43 biosynthesis in vivo, e.g., as described previously (2). Results from the physiological experiments are also summarized in Table 2. The paragraphs to follow highlight the main observations from our analyses.
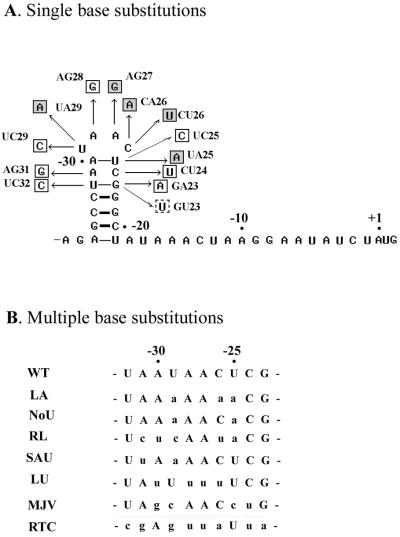
Figure 3.
Single (A) and multiple (B) base substitutions that were introduced in the loop segment of the T4 gene 43 translational operator in this study. Synthetic oligodeoxynucleotide templates for the RNAs (made as complementary DNA duplexes) were cloned in T7/SP6 expression vectors and transcribed in vitro in the presence of [α-32P]UTP to produce the desired substrates for binding studies (Materials and Methods). Single-base substitutions that resulted in reduced binding affinity of the RNA to T4 gp43 are shaded in (A). Multiple substitutions are shown in lower case in (B). See Table 2 for results of binding experiments with these operator mutants; the Kd for the GU23 mutant (dashed box) was not measured.
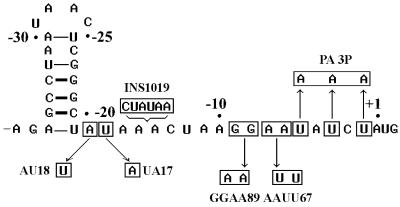
Figure 5.
Mutant constructs of the 3′-tail segment of the T4 gene 43 translational operator. The chart shows the positions of single-base substitutions (UA17 and AU18), 2-base substitutions (GGAA89 and AAUU67), a 3-base substitution (PA3P) and a 6-base insertion (INS1019).
Specificity determinants in the operator hairpin loop
As summarized in Table 2, mutational analysis of operator loop nucleotides (see also Fig. 3) identified two positions, –26 (C26) and –27 (A27), as most probably invariant. Other loop residues were tolerant to transitions, but not transversions. The 4-base transition mutant MJV (Fig. 3B), which bears the same hairpin sequence to the SELEX-generated ‘major variant’ described by Tuerk and Gold (7), bound T4 gp43 with wild-type affinity in vitro and, when crossed into the T4 genome, exhibited a wild-type phenotype (it was repressed by gp43) in phage infected cells as well (Table 2). On the basis of these observations we suspect that positioning of purine and pyrimidine residues relative to one another is important for establishing an operator geometry that is compatible with specific recognition by T4 gp43 (Discussion).
Results from the analysis of additional mutant operator constructs, including multiple-base and single-base substitutions, also implicated purine vis a vis pyrimidine positioning in the nucleotide sequence of the loop as an important criterion for specificity. For example, the introduction of a CU26 substitution next to a UA25 substitution (construct RL; Fig. 3) did not cause any additional change in binding affinity than was observed with the UA25 single mutant. Similarly, a UA31 base change in addition to a UA29 substitution did not decrease binding any further than was observed with either single mutant (compare constructs SAU and UA29 in Table 2). In contrast, results with the NoU and LA constructs showed that a double substitution at positions –29 and –26 of the operator results in loss of specific binding. Also, the LU construct (which combines the UA27, UA28 and CU26 substitutions) lacks specific binding to gp43. In summary, it appears that substitutions at –26 or –29 render these and adjacent positions unavailable for contacts with gp43. Transversion mutations at both of these sites (i.e. results with constructs NoU, LA, LU and AS; Table 2) increased the Kd values to levels similar to those observed for non-specific nucleic acids without affecting the predicted hairpin structure.
Specificity determinants in the hairpin stem
To assess the importance of primary structure in the operator stem segment for determination of specificity of the gp43–RNA interaction, we measured affinities of T4 gp43 to a series of operator mutant constructs in which nucleotide substitutions were predicted to either disrupt or conserve hairpin structure. The constructs we examined are illustrated in Figure 4 and results of binding assays are summarized in Table 2. Here also, we observed a dependence of the Kd on purine versus pyrimidine positioning; however, this dependence was less stringent than in the case of base substitutions in the loop segment. Note for example the effects of reversing the positions of purines and pyrimidines in individual G-C base pairs of the stem on binding (i.e., constructs ST20, ST21 and ST22; Fig. 4 and Table 2). Interestingly, some single base substitutions that we predicted would destabilize the stem did not lead to reductions in binding affinity to gp43 (e.g., G to A substitution at position –21 and C to G substitution at position –20); however, reversal of the positions of purine relative to pyrimidine in base pairs at these two sites (i.e., at the –21:–34 and –22:–33 base pairs) did decrease the binding affinity (compare the results with SD, SR and WTRB69 constructs, Figure 4). We suspect that these decreases may be due to changes in loop orientation that misalign the operator with its binding site on the protein (Discussion).
Figure 4.
Mutant constructs of the base-paired (stem) segment of the T4 gp43 translational operator. Base substitutions that reduced the binding affinity (Table 2) to the protein are shaded. (A) Positions of single (GA21, CG20), double (ST20, ST21, ST22) and multiple (SD and SR) base substitutions that were predicted to affect configuration of the RNA operator stem. (B) The predicted secondary structures of operator variants that exhibited wild-type affinities to T4 gp43. (C) The predicted secondary structures of operator variants (WTRB69 and SR) that exhibited reduced affinities to the T4 protein. See Table 2 for results of binding measurements.
The 3′ tail: an operator domain for sequence-independent binding to gp43
The untranslated leader segment of the gene 43 TIR includes the Shine–Dalgarno sequence (nucleotide positions –6 to –12 in Fig. 1), which constitutes part of the gp43 footprint on the mRNA (1,4). To test if primary structure in this segment is critical for operator specificity, we introduced nucleotide changes therein that obliterated the complementarity to 16S RNA and/or altered the spacing between the operator hairpin and initiator AUG. Figure 5 illustrates the mutants we analyzed and Table 2 summarizes their relative binding affinities to T4 gp43. Remarkably, changes in primary structure of the 3′-tail (ranging from single-base substitutions to multiple replacements and insertions) all yielded binding affinities to gp43 similar to those observed with wild-type operator constructs, i.e., Kd values of 1–4 nM (Table 2). That is, it appears that binding of gp43 to its operator does not depend on the presence of nucleotide sequence determinants that are characteristic of the typical prokaryotic RBS. Unfortunately, because such determinants are essential for translation, we were unable to correlate the in vitro binding affinities of 3′-tail mutants with potential effects on translational repression.
DISCUSSION
Previous studies defined the T4 gene 43 translational operator as the mRNA sequence that could be protected (footprinted) by the purified gene product in RNase or chemical sensitivity assays (1,4). By such criteria, the operator mapped exclusively in the untranslated 5′ leader segment of the mRNA (Fig. 1). In contrast, the gp43–RNA binding studies described in this report suggest that differential recognition of the mRNA by its repressor also involves the participation of nucleotide determinants outside the gp43-footprinted region, including some of the codons that specify the N-terminal segment of the autogenous repressor. The highest gp43–RNA binding affinities we were able to measure in the purified system used here ranged between Kd values of 1 and 4 nM and required the gp43-footprinted sequence as well as an additional length of 11–13 nt for the 3′-tail segment (Table 1). We detected little contribution to the affinity from the RNA segment to the 5′-terminal side of the operator hairpin structure (Fig. 2). The absence of an effect from the 5′-terminal sequence is consistent with observations from in vivo studies which showed that most gp43 regulated T4 gene 43 transcripts are either initiated or processed at points close to the 5′ end of the operator’s hairpin domain (1,16,17). Thus, functionally, the translational operator for this system of autogenous translational repression probably includes all of the mRNA determinants required for ribosome binding (i.e., nucleotide positions –20 to +13) as well as the adjacent RNA sequence to the 5′ side, which forms the operator hairpin structure (Fig. 1). By the criteria used to define elements of the prokaryotic RBS (18), the operator hairpin for this T4 cistron lies outside the region bound by the initiating ribosome.
Results of our mutational analyses (Table 2) are also consistent with findings from other studies which have shown that T4 gp43 can bind variants of the T4 natural operator with affinities similar to that of this operator (2,4). For example, compare the results we obtained with the WT and MJV constructs (Table 2; see also ref. 13 for a summary of other examples). At least some of these RNA variants can be shown to be repressible by T4 gp43 in vivo when they are placed in the sequence context of an otherwise wild-type gp43-encoding mRNA (e.g., the MJV construct listed in Table 2 and construct AU18 in ref. 2). On the basis of the few examples where we have been able to compare in vitro and in vivo responses to T4 gp43 for the same operator constructs, we estimate that in vivo repressible RNA targets for this protein would yield Kd values that are no higher than twice the value obtained with the natural T4 operator, when tested under the type of abbreviated conditions represented in the in vitro assays used here, i.e., with RNA targets that are 62–207 nt in length. The highest binding affinities we were able to measure for gp43–operator complexes under these conditions were only ~20–50-fold higher than the affinities of gp43 to non-specific nucleic acids of similar polynucleotide length e.g., polyU binding data in Table 2; see also ref. 6). This difference appears to be too low to account for in vivo observations, which indicate that T4 gp43 selectively represses its mRNA in the presence of a vast excess of non-specific nucleic acids. Possibly, additional interactions with the ∼3000 nt mRNA for the protein as well as other factors (‘translational corepressors’) enhance the selectivity of gp43 for its translational target in the phage-infected cell. At this time, there is no evidence that such enhancers of gp43-mediated translational repression in vivo exist.
Another conclusion we can draw from our studies is that although ribosome-binding determinants are involved in the gp43–mRNA interaction, specificity to the ribonucleotide sequence appears to be directed at the RNA hairpin domain only. We observed that 3′-terminally truncated versions of the operator, which exhibited sharply reduced affinities to the protein, still exhibited specificity to the repressor (compare WT and UA29 for the 2 nt 3′-tail length in Table 1). In contrast, alterations in the sequence (rather than length) of the 3′-tail, including substitutions at the Shine–Dalgarno sequence, had only small effects on affinity of the protein to hairpin-bearing RNA targets (Table 2). These observations suggest that the 3′-tail contributes essential sequence-independent interactions that enhance differential binding to the specificity domain of the RNA, i.e., the hairpin. We envisage that in vivo, T4 gp43 may use nucleotide-sequence-independent interactions to diffuse on the nucleic acid until it encounters its specific target (the RNA hairpin), where it can establish tight binding through both sequence-dependent and -independent interactions. Sequence-specific hairpin binding would fix the orientation of the 3′-tail domain in relation to the protein surface and as a consequence, limit the number of choices that the 3′-tail domain can use to make contacts with the protein. In this context, the covalent linkage that exists between the hairpin and 3′-tail can be regarded as a specificity determinant. Consistent with this explanation is the observation that increasing the length of the nucleic acid to the 5′-side of the hairpin does not substitute non-specific interactions for a 3′-tail segment (Fig. 2A). The lack of sequence specificity in a discrete segment of the operator may mean that T4 DNA polymerase bears two discrete sets of RNA-binding determinants, one set for the specific RNA sequence and structure of the hairpin and the other set for non-specific sequence of the 3′-tail segment. Phylogenetic studies have implicated a segment of the Palm domain of gp43 in determination of specificity to the operator (8), and some genetic analyses have suggested that the N-terminal domain of the protein is also involved in RNA binding (19). In addition, studies of crystals of the gp43 variant from phage RB69 detected rGMP in the N-terminal domain of the protein, complexed with a module of protein secondary structure that resembles the βαββαβ architecture of some RNA-binding domains in other proteins (20). The same architecture has recently been observed and discussed for two DNA polymerases from the archae whose structures resemble that of RB69 gp43 (21,22); however, it is not known if the archaeal enzymes bind RNA or if the motif in gp43 is indeed involved in operator binding. Possibly, the Palm and N-terminal domains of gp43 harbor separate clusters of RNA-binding determinants, some of which overlap with the nucleotide-sequence-independent determinants of DNA binding.
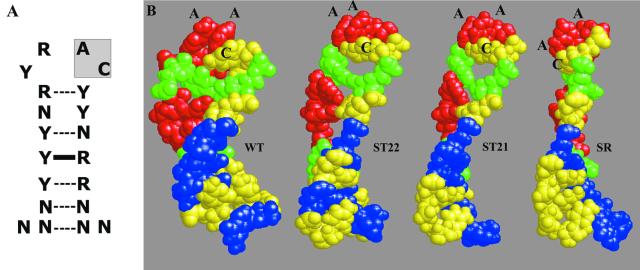
The three-dimensional structure of the gp43–operator complex is not known, although some generalizations about this structure are possible, based on the current study as well as other studies that examined binding of T4 gp43 to operator variants (2,4,6,14,15). A unifying feature among members of the range of RNA primary structures that can be repressed by gp43 may be their propensity to form very similar (perhaps identical) higher-order structures as they establish complexes with the protein. In regard to specificity to the ribonucleotide sequence, our studies define two aspects of the operator hairpin that are essential for recognition of this RNA by T4 gp43: (i) the apparently invariant 5′-AC-3′ dinucleotide sequence in the unpaired (loop) region of the hairpin and (ii) the restricted positioning of purine and pyrimidine residues in both the hairpin stem and loop regions. It has been noted previously (2) that all of the tight-binding RNA ligands that were selected by T4 gp43 in the first application of the SELEX method (7) had either the wild-type T4 operator hairpin sequence or contained purine-to-purine and pyrimidine-to-pyrimidine (i.e., transition type) differences from the wild type. Also, the study reported here (Table 2) and our previously reported mutational analyses (2) have shown that transition mutations anywhere in the hairpin except for the 5′-AC-3′ site of the loop segment exhibit minor effects on operator function, whereas transversion mutations of this domain drastically reduce operator binding to gp43. We present the models shown in Figure 6 to explain how purine vis à vis pyrimidine positioning may affect protein access to the invariant 5′-AC-3′ site of the operator. The figure shows a generalized nucleotide sequence for the hairpin of gp43-repressible RNAs (Fig. 6A) and depicts the predicted loop structures of four operator constructs (Fig. 6B) that we compared for effects of alterations in the stem sequence on in vitro binding to gp43 (Table 2). Of the four constructs, only ‘stem restored’ construct SR exhibited diminished binding to T4 gp43, despite its predicted stable hairpin structure. We suggest that in this construct, the two C-G to G-C base-pair switches at positions –34/–21 and –33/–22 of the base-paired region lead to a displacement of the location of the 5′-AC-3′ sequence in the unpaired region of the hairpin, such that essential contacts are precluded from forming between this dinucleotide element and its complementary amino acid residues in the hairpin-loop recognition region of gp43. The other two constructs (ST21 and ST22) bound T4 gp43 with nearly wild-type affinity (Table 2). The NMR structures for facsimiles of the WT and MJV hairpins suggest that the A and C residues of the 5′-AC-3′ unit are oriented differently with respect to the body of the hairpin (14,15). The A points to the outside and the C is located inside the loop, making an internal hydrogen bond with the pyrimidine at position –29 (as depicted in Fig. 6B); however, it is not known if this structure is actually formed when the operator is bound to gp43. A more direct structural analysis of gp43–operator complexes is required to better understand the geometry of the interaction between T4 DNA polymerase and its translational operator, especially with regard to the roles of hairpin loop residues and the non-specific sequence of the 3′-tail.
Figure 6.
Predicted hairpin structures for RNA substrates used in this study. (A) Generalized RNA hairpin configuration that contains the nucleotide sequence elements required for differential binding of the operator to T4 gp43. R, purine; Y, pyrimidine; N, any base. The 5′-AC-3′ loop sequence (positions –26 and –27) is shaded. (B) Space-filled structural models for the hairpin domains of the WT, ST22, ST21 and SR RNA constructs used in this study (Fig. 4). The hairpin ribonucleotides are colored in red (A), green (U), yellow (C) and blue (G). Prediction of three-dimensional structures of the operator hairpins shown in (B) was done by using Insight II and Discover software (Biosym Technologies) on IRIS INDIGO XS24 workstation (SiliconGraphics). For the initial NMR structure of WT-RNA reported by Mirmira and Tinoco (14), we performed an energy minimization procedure under simulated vacuum conditions and 25°C. No structural restraint was introduced during the energy minimization. To predict structures of mutant RNAs, we replaced nucleotide bases in the final WT-structure, explored molecular dynamics of the new structures at 50°C and 100 ps, and repeated the optimization.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Vasiliy Petrov and James Nolan for many helpful discussions and Dr Ignacio Tinoco for providing unpublished information about NMR structures for RNA hairpins that bind T4 gp43. We also thank Carol Carlton for synthesis of oligonucleotides, Dr Mark Andrake, Dr Elena Tourkina and Mr Jesse Guidry for cloning synthetic DNAs, and Jill Barbay for help with manuscript preparation. This work was supported by grants GM18842 and GM54627 from the NIGMS.
REFERENCES
- 1.Andrake M., Guild,N., Hsu,T., Gold,L., Tuerk,C. and Karam,J. (1988) Proc. Natl Acad. Sci. USA, 85, 7942–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrake M.D. and Karam,J.D. (1991) Genetics, 128, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russel M. (1973) J. Mol. Biol., 79, 83–94. [DOI] [PubMed] [Google Scholar]
- 4.Tuerk C., Eddy,S., Parma,D. and Gold,L. (1990) J. Mol. Biol., 213, 749–761. [DOI] [PubMed] [Google Scholar]
- 5.McCarthy J.E. and Gualerzi,C. (1990) Trends Genet., 6, 78–85. [DOI] [PubMed] [Google Scholar]
- 6.Pavlov A.R. and Karam,J.D. (1994) J. Biol. Chem., 269, 12968–12972. [PubMed] [Google Scholar]
- 7.Tuerk C. and Gold,L. (1990) Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 8.Wang C.C., Pavlov,A. and Karam,J.D. (1997) J. Biol. Chem., 272, 17703–17710. [DOI] [PubMed] [Google Scholar]
- 9.Zuker M. (1989) Methods Enzymol., 180, 262–288. [DOI] [PubMed] [Google Scholar]
- 10.Jacobson A.B., Good,L., Simonetti,J. and Zuker,M. (1984) Nucleic Acids Res., 12, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuker M. and Stiegler,P. (1981) Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freier S.M., Kierzek,R., Jaeger,J.A., Sugimoto,N., Caruthers,M.H., Neilson,T. and Turner,D.H. (1986) Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller E.S., Karam,J.D. and Spicer,E. (1994) In Karam,J.D. (ed.), Molecular Biology of Bacteriophage T4. ASM Press, Washington, DC, pp. 193–208.
- 14.Mirmira S.R. and Tinoco,I.,Jr (1996) Biochemistry, 35, 7675–7683. [DOI] [PubMed] [Google Scholar]
- 15.Mirmira S.R. and Tinoco,I.,Jr (1996) Biochemistry, 35, 7664–7674. [DOI] [PubMed] [Google Scholar]
- 16.Guild N., Gayle,M., Sweeney,R., Hollingsworth,T., Modeer,T. and Gold,L. (1988) J. Mol. Biol., 199, 241–258. [DOI] [PubMed] [Google Scholar]
- 17.Hsu T. and Karam,J.D. (1990) J. Biol. Chem., 265, 5303–5316. [PubMed] [Google Scholar]
- 18.Gold L. (1988) Annu. Rev. Biochem., 57, 199–233. [DOI] [PubMed] [Google Scholar]
- 19.Hughes M.B., Yee,A.M., Dawson,M. and Karam,J. (1987) Genetics, 115, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Sattar,A.K., Wang,C.C., Karam,J.D., Konigsberg,W.H. and Steitz,T.A. (1997) Cell, 89, 1087–1099. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez A.C., Park,H.W., Mao,C. and Beese,L.S. (2000) J. Mol. Biol., 299, 447–462. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y., Jeruzalmi,D., Moarefi,I., Leighton,L., Lasken,R. and Kuriyan,J. (1999) Structure Fold Des., 7, 1189–1199. [DOI] [PubMed] [Google Scholar]