Abstract
Escherichia coli MutY is an adenine and a weak guanine DNA glycosylase active on DNA substrates containing A/G, A/8-oxoG, A/C or G/8-oxoG mismatches. A truncated form of MutY (M25, residues 1–226) retains catalytic activity; however, the C-terminal domain of MutY is required for specific binding to the 8-oxoG and is critical for mutation avoidance of oxidative damage. Using alkylation interference experiments, the determinants of the truncated and intact MutY were compared on A/8-oxoG-containing DNA. Several purines within the proximity of mismatched A/8-oxoG show differential contact by the truncated and intact MutY. Most importantly, methylation at the N7 position of the mismatched 8-oxoG and the N3 position of mismatched A interfere with intact MutY but not with M25 binding. The electrostatic contacts of MutY and M25 with the A/8-oxoG-containing DNA substrates are drastically different as shown by ethylation interference experiments. Five consecutive phosphate groups surrounding the 8-oxoG (one on the 3′ side and four on the 5′ side) interact with MutY but not with M25. The activities of the truncated and intact MutY are modulated differently by two minor groove-binding drugs, distamycin A and Hoechst 33258. Both distamycin A and Hoechst 33258 can inhibit, to a similar extent, the binding and glycosylase activities of MutY and M25 on A/G mismatch. However, binding and glycosylase activities on A/8-oxoG mismatch of intact MutY are inhibited to a lesser degree than those of M25. Overall, these results suggest that the C-terminal domain of MutY specifies additional contact sites on A/GO-containing DNA that are not found in MutY–A/G and M25–A/8-oxoG interactions.
INTRODUCTION
The Escherichia coli MutY protein, along with MutM and MutT, is involved in defending against the mutagenic effects of 7,8-dihydro-8-oxo-guanine (8-oxoG or GO) lesions, the most stable products known due to oxidative damage to DNA (1,2). The short patch MutY base excision repair pathway specifically repairs A/GO and A/G to C/GO and C·G, respectively, and corrects A/C to G⋅C and G/GO to C/GO at a much lower rate (3–11). The major function of MutY is to reduce mutation frequency caused by GO lesions (4). Adenines are frequently incorporated opposite GO bases during DNA replication (12,13) that subsequently leads to G·C to T·A transversions (13–16). Thus, MutY provides the defense by removing adenines misincorporated opposite GO or G following DNA replication (1,6,17).
MutY, an adenine and a weak guanine glycosylase, is a 39 kDa protein with an iron–sulfur cluster [4Fe–4S] (3–11). The structural and functional analyses of MutY have revealed several interesting aspects. Ethylation interference studies showed that MutY interacts with at least five phosphate groups and covers ~12 bp around the A/G mismatch (17). Methylation interference experiments have demonstrated that MutY specifically interacts with both mispaired A and G as well as the two bases flanking the A/G mismatch (17). The MutY domain structure has been studied through proteolysis (18–20) and site-directed mutagenesis (21–23). The N-terminal domain of MutY, called cdMutY, residues 1–226 (M25) (4,18,24) or residues 1–225 (p26) (19,20), have been shown to retain catalytic activity. Recently, the X-ray crystal structure of a mutant of the cdMutY(D138N) with bound adenine showed that the adenine is buried in the active site of the catalytic domain and suggests that the mismatched adenine must flip out of the DNA helix for the glycosylase action (22). In the active pocket, several amino acids (Glu37, Gln182 and Asp186) are involved in adenine binding. The N-terminal domain of MutY protein shares structural similarity with endonuclease III (endo III), AlkA and human OGG1 (10,22,25–28). This includes the helix–hairpin–helix (HhH) and Gly/Pro…Asp loop motifs. Endonuclease III repairs thymine glycol and oxidized pyrimidines in DNA (29,30), AlkA repairs methylated purines (31,32), and OGG1 repairs GO and formamidopyrimidine (Fapy) when they pair with cytosine (33). However, MutY has an additional C-terminal domain that has no counterpart in other HhH family proteins. The MutY–DNA complex has been modeled by superimposing the protein (22) onto the structure of AlkA complexed to DNA (34,35). The model suggests that cdMutY can bind to and distort DNA in a manner similar to AlkA. The AlkA flips a 1-azaribose abasic nucleotide out of DNA and induces a 66° bend in the DNA with a marked widening of the minor groove (34).
The crystal structure of the N-terminal domain of MutY also suggests some candidate residues (Gln41, Tyr82 and Arg194) for the recognition of the base opposite the adenine. The C-terminal domain of MutY has been shown to play an important role in the recognition of GO lesions (4,18,24). The truncated MutY has >18-fold lower binding affinities with 8-oxoG-containing mismatches than the intact MutY. MutY and M25 have similar binding affinities for an A/G mismatch; however, MutY has a 67-fold greater affinity to A/GO-containing DNA than does M25 (4). Deletion of the C-terminal domain of MutY reduces its catalytic preference for A/GO-containing DNA over A/G-containing DNA (4,24) and confers a mutator phenotype in vivo (4). Thus, repair of 8-oxoG is the major function of MutY. To further investigate the substrate recognition of MutY on A/GO-containing DNA and specific determinants of the C-terminal domain of MutY, methylation and ethylation interference experiments were performed to identify purine and phosphate groups on an A/GO-containing DNA specified by the N-terminal domain of MutY and intact MutY. Alkylation interference experiments show that the C-terminal domain of MutY contributes to the contacts with mismatched A and GO, two bases flanking the A/GO mismatch, and several phosphate groups 5′ to the GO. Thus, the determinants on A/GO-containing DNA of the C-terminal domain of MutY contribute to the 8-oxoG specificity. Our results also hint at the orientation of the C-terminal domain of MutY protein on the bound DNA substrates.
Distamycin A and Hoechst 33258 are drugs that bind to the minor groove of double-stranded DNA at the AT-rich sequences (36–42). Here, we show that the activities of the truncated and intact MutY are modulated differently by these two drugs. Both distamycin A and Hoechst 33258 can inhibit, to a similar extent, the binding and glycosylase activities of MutY and M25 on A/G mismatches. However, binding and glycosylase activities on A/8-oxoG mismatches of intact MutY are less sensitive to inhibition than those of M25. The results suggest that the minor groove 5′ to the mismatched A is the target of these drugs and is involved in the DNA binding of the N-terminal domain of MutY. In conclusion, the C-terminal domain present in the intact MutY provides additional affinity to the A/GO mismatch and MutY activity on this mismatch is less sensitive to drug inhibition than the truncated M25.
MATERIALS AND METHODS
Plasmids containing MutY domains
The expression plasmid pET11a containing the entire mutY gene and the truncated mutY gene corresponding to Met1 to Gln226 (M25) have been described (4,43). The catalytic inactive mutant MutY(D138N) has been constructed by site-directed mutagenesis (43). The same D138N mutation was transferred from the intact MutY to M25 by a cassette replacement of the NdeI–SacII fragment to yield mutant M25(D138N). The expression of these proteins is under the control of the T7 promoter. The expression host of the mutY mutants, PR70 (Su– lacZ X74 galU galK Smr micA68::Tn10Kan) harboring the λDE3 lysogen, was constructed according to the procedures described by Invitrogen.
Protein expression and purification
Escherichia coli strains PR70/DE3 harboring the expression plasmid containing MutY, M25, MutY(D138N) and M25(D138N) were grown in LB broth containing 50 µg/ml of ampicillin at 37°C. The expression of proteins was induced at an OD600 of 0.6 by the addition of isopropyl β-d-thiogalactoside to a final concentration of 0.4 mM to the culture at 20°C. The cells were harvested 16 h later. The purification procedures were carried out as described previously (42). As judged by 12% SDS–polyacrylamide gel electrophoresis, all four proteins were purified to >99% homogeneity (data not shown).
Oligonucleotide substrates
The nucleotide sequences of the mismatch-containing heteroduplexes used in this study were as follows:
19mer 5′-CCGAGGAATTAGCCTTCTG -3′
3′- GCTCCTTAAXCGGAAGACG-5′
40mer 5′-AATTGGGCTCCTCGAGGAATTAGCCTTCTGCAGGCATGCC -3′
3′- CCCGAGGAGCTCCTTAAXCGGAAGACGTCCGTACGGGGCC-5′
(X = G or GO)
The top strand is referred to as the mismatched adenine-containing strand (A-strand) and the bottom strand as the mismatched GO-containing strand (GO-strand). Both heteroduplexes share an 18 bp common sequence centered on the mismatch. Oligonucleotides containing base mismatches were labeled at 3′ or 5′ ends as described by Lu et al. (17). The 40mer heteroduplex containing an A/GO mismatch was labeled at the 5′ end of the A- or GO-strand and used in the alkylation experiments. The 19mers were labeled at the 3′ end on the A-strand and were converted into 20mers after the sticky ends were filled in with the Klenow fragment of DNA polymerase I.
Methylation interference experiments
The labeled 40mer A/GO-containing DNA substrates (2 pmol, 5′ end-labeled) were partially methylated by treatment with 0.05% dimethyl sulfate for 20 min at 25°C as described by Siebenlist and Gilbert (44). The reactions (200 µl) were terminated by the addition of 50 µl of 1.5 M sodium acetate, 1 M β-mercaptoethanol and 250 µg/ml tRNA. DNA was precipitated twice with ethanol and dissolved in water. Purified MutY(D138N) (4 pmol) and M25(D138N) (5 pmol) were incubated with methylated DNA (0.5 pmol) in a 20 µl reaction containing 20 mM Tris–HCl (pH 7.6), 5 mM dithiothreitol, 0.1 mM EDTA, 0.01% NP-40 and 1 µg/ml of poly(dI-dC). Catalytically inactive mutants MutY(D138N) and M25(D138N) were used instead of wild-type enzymes to eliminate the cleavage at the mismatched adenine by the glycosylase/AP lyase activity of the enzymes. A control experiment was performed in the same way except that MutY diluent was used in the binding reaction. After incubating at 37°C for 30 min, the reaction mixture was fractionated on a 4% polyacrylamide gel. Protein-bound and protein-free DNA were electroeluted, ethanol precipitated and subjected to the Maxam–Gilbert A>G cleavage reaction (45) with some modifications. After treatment with 0.1 N HCl at 0°C for 2 h, samples were precipitated with ethanol, resuspended in 1 M piperidine and heated at 90°C for 30 min. Samples were lyophilized to dry twice and analyzed on 14% sequencing gels. The gels were dried and quantitated using a PhosphorImager system (Molecular Dynamic Storm 840). The intensity of bands from the enzyme-free samples was divided over the intensity of corresponding bands from the enzyme-bound samples. These ratios are presented relative to that of G8 on the GO-containing strand and to that of G34 on the A-containing strand. Data were from PhosphorImager quantitative analyses of gel images over more than three experiments.
Ethylation interference experiments
5′ end-labeled 40mer heteroduplex DNA containing an A/GO mismatch was ethylated according to Siebenlist and Gilbert (44) with some modifications. Approximately 2 pmol of DNA in 100 µl of 50 mM sodium cacodylate (pH 7.0) was mixed with an equal volume of freshly prepared ethylnitrosourea (Pfaltz and Bauer) saturated in ethanol and incubated at 45°C for 1 h. The reaction was stopped by adding 20 µl of 3 M sodium acetate (pH 7.0), 5 µg of tRNA and 150 µl of cold ethanol. The ethylated DNA was precipitated with ethanol several times and resuspended in water. Binding reactions were performed as for those in the methylated interference experiments. MutY(D138N) and M25D(138N) were used instead of wild-type enzymes for the same purpose as for the methylation interference. A control experiment was carried out in the same way except that MutY diluent was used in the binding reaction. Strand cleavage at phosphodiester bonds was performed by heating at 90°C for 30 min in 1 M piperidine. Samples were lyophilized to dry twice and analyzed on 14% sequencing gels. The gels were dried and quantitated using the PhosphorImager system. The intensity of the bands from the enzyme-free samples was divided over the intensity of corresponding bands from the enzyme-bound samples. These ratios are presented relative to that of the phosphate at position 9 on the GO-containing strand and to that of the phosphate at position –10 on the A-containing strand. Data were from PhosphorImager quantitative analyses of gel images over more than three experiments.
The effect of distamycin A and Hoechst 33258 on MutY binding and cleavage activities
The binding and cleavage activities of MutY and M25 with A/G- and A/GO-containing 20mer DNA were performed as described by Li et al. (4). Distamycin A and Hoechst 33258 were added to the reaction mixtures to final concentrations of 0.225, 0.45, 0.9, 1.8, 3.6, 7.2, 14.4 and 28.8 µM and pre-incubated at room temperature for 10 min before adding enzymes. MutY and M25 were then added, and the reactions were further incubated at 37°C for 20 min. Protein–DNA complexes were analyzed on 8% polyacrylamide gels in buffer containing 50 mM Tris–borate (pH 8.3) and 1 mM EDTA. Samples after cleavage reactions were lyophilized, resuspended in 3 µl of formamide dye, heated at 90°C for 2 min and loaded onto 14% 7 M urea sequencing gels. The gels were dried and quantitated using a PhosphorImager system. Relative MutY binding or cleavage activities to that of no drug added samples were plotted as a function of drug concentrations. Data were from PhosphorImager quantitative analyses of gel images over three experiments.
RESULTS
Methylation interference
Methylation interference (44) provides a tool to elucidate DNA base determinants of DNA-binding proteins. Dimethyl sulfate can methylate the N7 position of guanines and the N3 position of adenines. Methylation at the particular purines of DNA at the recognition sites will interfere with specific protein binding. Because M25 has much weaker binding affinity and slower glycosylase activity on A/GO-containing DNA than intact MutY (4,24), we expect that M25 and MutY may differentially contact A/GO-containing DNA. A 5′ end-labeled 40mer duplex oligonucleotide containing an A/GO mismatch at position 22 was used in this study to investigate the DNA contact sites of MutY and M25. There are two major problems inherent to these experiments. First, the GO base in DNA is labile to NaOH employed in the Maxam–Gilbert A>G chemical sequencing reaction (45). To minimize this strand cleavage, the A>G Maxam–Gilbert chemical sequencing reaction was slightly modified by replacing NaOH with piperidine. Under this condition, the cleavage at GO was less extensive than the regular procedure but a high background was observed and thus only the strong interference effect could be detected. Second, the AP lyase activity of MutY renders the cleavage at the first phosphodiester bond 3′ to the mismatched adenine. To avoid this cleavage, D138N mutants of MutY and M25 were used in the experiments. The MutY(D138N) mutant has been shown to be catalytically inactive and has a similar binding affinity to A/GO-containing DNA as the wild-type MutY (43). The cdMutY(D138N) mutant is also catalytically inactive but maintains the overall structure as the wild-type domain (22).
Figure 1 (lanes 1–5) shows a gel image by PhosphorImager from methylation interference experiments using 40mer A/GO-containing DNA labeled at the 5′ end of the GO-strand with MutY(D138N) and M25(D138N). Those DNA molecules that have been methylated and have lost specific binding to the protein will be enriched in the free fraction and reduced in the bound fraction. Because of low signal to noise, the effect of methylation interference is not as pronounced. However, some interference effects were observed repeatedly in several experiments as quantitated by PhosphorImager analysis (Fig. 2). The effect of methylation on MutY(D138N) and M25(D138N) binding is represented by white and black bars, respectively. Interference on MutY(D138N) binding was observed upon methylation of mismatched A22 and G23 on the A-strand and A20, A21, mismatched GO22 and G24 on the GO-strand (Fig. 2). Thus, MutY interacts strongly with both mismatched bases and the two bases flanking the mispair. These residues presumably are the key determinants of MutY binding to A/GO-containing DNA. Since N7 groups of G23 and G24 are located in the major groove and N3 groups of A20 and A21 are located in the minor groove, MutY binds DNA in both major and minor grooves. In the stable base pair of A(anti)-GO(syn) configuration (46), the N7 group of mismatched GO22 is located in the minor groove. It appears MutY contacts A/GO-containing DNA in the minor groove 5′ to the mismatched A and in the major groove 3′ to the mismatched A.
Figure 1.

A representative gel image by PhosphorImager of the alkylation interference of MutY and M25. The 40mer heteroduplex DNA containing an A/GO mismatch was 5′ end-labeled on the GO strand, partially methylated with dimethylsulfate (lanes 1–5) or ethylated with N-nitrosoethylurea (lanes 6–10), purified and bound to MutY(D138N) and M25(D138N). After binding, bound (B) and free (F) DNA were separated by 4% native polyacrylamide gel electrophoresis, eluted and chemically cleaved (32). Control samples were DNA treated by the same procedures without adding protein. Cleavage products were then analyzed on a 14% DNA sequencing gel and exposed to a PhosphorImager screen. Arrows mark the position of GO. Alkylation at the underlined nucleotides and backbone phosphates showed significant interference effect with MutY binding.
Figure 2.
Quantitation of methylation interference effects. The 40mer heteroduplex DNA containing an A/GO mismatch (boxed) was 5′ end-labeled on the A-strand (top) or GO-strand (bottom). Numbers between the base pairs mark the positions of the nucleotides from the 5′ end of the A-strand. Interference effects on MutY and M25 are indicated by white and black bars, respectively. Results shown are from PhosphorImager quantitative analyses of gel images similar to Figure 1 from more than three experiments. The extent of interference is expressed as the ratio of the intensity of each band from the enzyme-free DNA to that of the corresponding band in the enzyme-bound samples. The ratios of free:bound at G34 on the A-strand and G8 on the GO-strand are normalized to 1.0. Error bars are ±1 standard deviation. No significant interference was observed for those regions of the 40 base pair DNA, not shown.
In the case of M25(D138N), substantial interference could only be observed upon methylation of the G23 base on the A-strand. Surprisingly, methylation at the mismatched A and GO as well as A20, A21 and G24 had no interference effect on binding of M25(D138N). It is suspected that some weak interactions between some of these groups and M25(D138N) may exist but their observation is hindered by a high background (Fig. 1).
Ethylation interference
Random ethylation at the phosphate groups by ethylnitrosourea provides a tool to study the contact between proteins and DNA phosphate backbone. Because GO is more labile to NaOH than piperidine treatments, strand cleavage at the ethylated sites was carried out with piperidine. Under this condition, a high background was observed and thus only the strong interference effect could be detected. Figure 1 (lanes 6–10) shows a gel image by PhosphorImager from ethylation interference experiments with 40mer A/GO-containing DNA labeled at the 5′ end of the GO-strand with MutY(D138N) and M25(D138N). Those ethylated DNA molecules that cannot be bound specifically by the proteins will be enriched in the free fraction and reduced in the bound fraction. Figure 3 summarizes quantitatively the results of several such experiments on both DNA strands. Ethylation interference patterns on 40mer DNA with A/GO mismatch suggest that MutY interacts strongly with five consecutive phosphate groups surrounding the GO, one on the 3′ side (position –1) and four on the 5′ side (positions 0–3) (Fig. 3, white bars). There were also weak interactions between some phosphate groups on both DNA strands. This may include the phosphates at positions 4–6 and –2 to –4 on the GO-strand and at positions 2, 3 and –2 to –4 on the A-strand. These contribute a large amount of electrostatic interactions between MutY and phosphate groups on the GO-strand. The phosphate groups upstream of the seventh phosphodiester bond 5′ to the mismatched A may not be in contact with MutY due to different degrees of cleavage of enzyme-free and enzyme-bound DNA fractions.
Figure 3.
Quantitation of ethylation interference effects. The phosphate groups at the 5′ and 3′ sides of mismatched bases are labeled with positive and negative numbers, respectively, and the phosphates immediately 5′ to the mismatched bases are labeled as position 0. Interference effects on MutY and M25 are indicated by white and black bars, respectively. Interferences were quantitated and shown as described in Figure 2. The experiments were performed in triplicate. The ratio of free:bound of the phosphate at position 9 on the GO-containing strand and that of the phosphate at position –10 on the A-containing strand are normalized to 1.0. Error bars are ±1 standard deviation. The phosphate groups upstream of the seventh phosphodiester bond 5′ to the mismatched A may not significantly interfere with MutY binding due to different degrees of cleavage of enzyme-free and enzyme-bound DNA fractions.
When ethylation interference experiments were performed with M25(D138N), a completely different pattern was observed (Fig. 3, black bars). Contact at five phosphate groups at positions –1 to 3 on the GO strand was diminished in M25. Surprisingly, only weak contact was observed with M25 to both strands; these probably included the phosphates at positions 2, –2 and –5 on the A-strand and phosphates at positions –4, –3 and 3–6 on the GO strand. Due to the high background (Fig. 1), these potential weak interactions are barely detected by our experiments.
Inhibition of MutY and M25 activity by DNA minor groove binding
Distamycin A and Hoechst 33258 (Fig. 4) are members of compounds that bind to the minor groove of AT-rich regions of double-stranded DNA (36–42). They fit snugly within the DNA minor groove with 3–5 AT base pairs. These drugs have been widely studied as model compounds for sequence-specific recognition of DNA. The 20mer and 40mer DNA substrates used in this study contain a sequence of AATT 5′ to the mismatched A. If MutY or M25 is in contact with the AATT region, the drugs may compete with the enzyme for binding and thus inhibit the enzyme activity.
Figure 4.
Structures of the DNA minor groove binding drugs distamycin A and Hoechst 33258 used in this study.
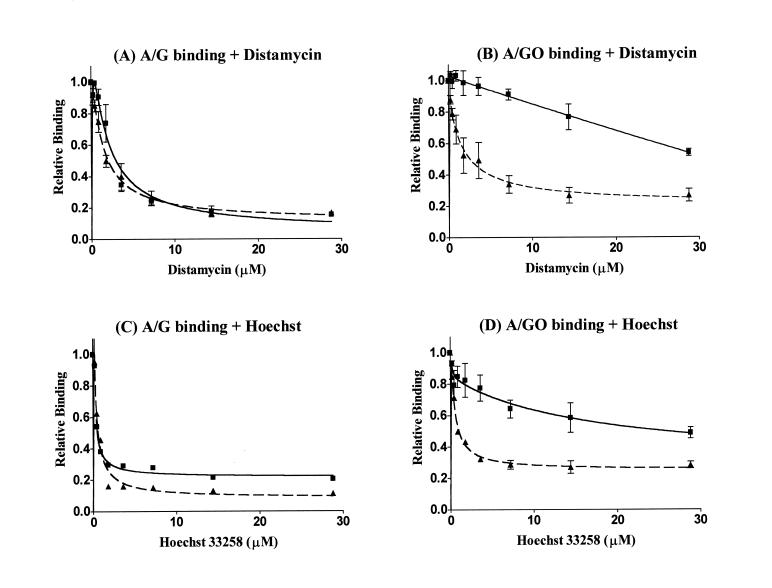
The inhibition of DNA binding activities of MutY and M25 to A/G- and A/GO-containing DNA by distamycin A and Hoechst 33258 was examined by gel-retardation assays. As shown in Figure 5, both distamycin A and Hoechst 33258 inhibited the binding of MutY and M25 on A/G- and A/GO-containing DNA. The extent of inhibition by distamycin A and Hoechst 33258 is quite similar for the MutY and M25 binding with A/G-containing DNA (Fig. 5A and C). When the concentrations of the drugs were increased from 0 to 15 µM, the A/G binding activities of MutY (7.2 nM) and M25 (14.4 nM) decreased sharply to the baseline level. The concentrations of distamycin A required to inhibit 50% of A/G-binding (IC50) for MutY and M25 are 3.0 and 1.9 µM, respectively (Table 1). The IC50 of Hoechst 33258 inhibition on A/G-binding of both enzymes is 0.5 µM. However, the inhibition patterns were quite different for MutY and M25 when distamycin A and Hoechst 33258 were added to the binding reactions with A/GO-containing DNA. As with binding of MutY and M25 on A/G-containing DNA (Fig. 5A and C), the A/GO binding activities of M25 were greatly inhibited by the drugs (Fig. 5B and D, dotted lines). The IC50 values for M25 binding to A/GO mismatch are 2.6 and 0.9 µM for distamycin A and Hoechst 33258, respectively (Table 1). In contrast, the binding activity of MutY with an A/GO mismatch was quite insensitive to both drugs (Fig. 5B and D, solid lines). The IC50 values for MutY binding to A/GO mismatch are >30 and 26 µM for distamycin A and Hoechst 33258, respectively (Table 1). It requires >10-fold of both drugs to achieve the same extent of inhibition on A/GO binding of MutY than for M25. Hoechst 33258 was more effective in inhibiting all binding activities than distamycin A.
Figure 5.
Distamycin A and Hoechst 33258 inhibit the binding activities of MutY and M25 with A/G- and A/GO-containing DNA. Binding activities of MutY are marked by squares and solid lines, and those of M25 are marked by triangles and dotted lines. Relative MutY and M25 binding activities with A/G- (A and C) and A/GO-containing (B and D) DNA to that of no drug added samples were plotted as a function of drug concentrations. Data were from PhosphorImager quantitative analyses of gel images over three experiments.
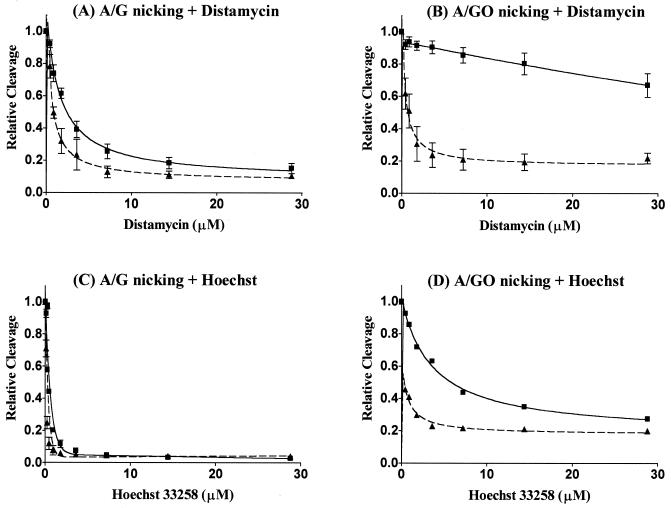
Table 1. Concentrations (µM) of drugs required to inhibit 50% of MutY and M25 activities.
| A/G binding | A/GO binding | A/G cleavage | A/GO cleavage | |||||
|---|---|---|---|---|---|---|---|---|
| Distamycin | Hoescht | Distamycin | Hoescht | Distamycin | Hoescht | Distamycin | Hoescht | |
| MutY | 3.0 | 0.5 | >30 | 26 | 2.5 | 0.4 | >30 | 5.5 |
| M25 | 1.9 | 0.5 | 2.6 | 0.9 | 0.8 | <0.2 | 0.8 | 0.4 |
Similar results were obtained with the inhibition of DNA glycosylase activities of MutY and M25 on A/G- and A/GO-containing DNA by distamycin A and Hoechst 33258 (Fig. 6). The IC50 values of drug inhibition on cleavage activities are summarized in Table 1. The glycosylase activity of MutY on A/GO mismatch was less sensitive to both drugs as compared with MutY and M25 on A/G mismatch and M25 on A/GO mismatch. It requires >15-fold of both drugs to achieve the same extent of inhibition on A/GO cleavage by MutY than for M25 and Hoechst 33258 was more effective in inhibiting all glycosylase activities than distamycin A.
Figure 6.
Distamycin A and Hoechst 33258 inhibit the glycosylase activities of MutY and M25 with A/G- and A/GO-containing DNA. Squares and solid lines mark glycosylase activities of MutY, and that of M25 are marked by triangles and dotted lines. Relative MutY and M25 glycosylase activities with A/G- (A and C) and A/GO-containing (B and D) to that of no drug added samples were plotted as a function of drug concentrations. Data were from PhosphorImager quantitative analyses of gel images over three experiments.
DISCUSSION
The junction between the N-terminal (25 kDa) and the C-terminal (14 kDa) domains is flexible and is easily susceptible to protease digestion (18,19). The N-terminal domain of MutY has catalytic activity (4,18–20) while the C-terminal domain plays an important role in GO recognition and mutation avoidance (4,18). The crystal structure of the catalytic domain of MutY reveals that it belongs to the HhH superfamily of DNA repair enzymes (10,22,25–28,34). The C-terminal domain of MutY has no homology to other members of the HhH superfamily but shares some sequence and structure homology with MutT, which hydrolyzes 8-oxo-dGTP to 8-oxo-dGMP and pyrophosphate (24,35). Among the members of this superfamily, AlkA–DNA and hOGG1–DNA co-crystal structures have been solved (28,34). In the enzyme-bound DNA, the substrate base flips out of the DNA helix, the helix axis is sharply bent and the minor groove is widened. Although the structures of intact MutY and MutY–DNA complex are not available, the cdMutY–DNA model suggests that cdMutY can bind to and distort DNA in a manner similar to AlkA (34). In this study, alkylation interference and DNA minor groove binding drugs were used to define the specific contact sites of different domains of MutY on A/GO-containing DNA.
The potential purine and phosphate contact sites of MutY and M25 on A/GO-containing DNA, as determined by alkylation interference experiments, are summarized in Figure 7B and C, respectively. Here, we assume that the mismatched A flips out from the DNA helix and the DNA is bent according to the DNA structure in AlkA–DNA complex (34). Because there is no information on the MutY–DNA structure, it is not known whether MutY and M25 can kink A/G- or A/GO-containing DNA. Nor is it known whether DNA bending can affect the alkylation interference effects. For example, base methylation may interfere protein binding because of its steric hindrance to DNA bending. MutY covers ~10 bp of the A/GO-containing DNA and has more contacts on the GO-strand (Fig. 7B). MutY specifically interacts with both the mispaired A and GO as well as the two bases flanking the A/GO mismatch. Because the N3 groups of A20 and A21 on the GO-strand are located in the minor groove and the N7 groups of G23 on the A-strand and G24 on the GO-strand are located in the major groove, these findings suggest that MutY recognizes the minor groove 5′ to the mismatched A and the major groove 3′ to the mismatched A. It has been suggested that binding of MutY to the bases flanking the mismatches may reflect the effect of neighboring sequences on repair and cleavage efficiencies (5,47,48). There is a considerable amount of electrostatic interaction between MutY and A/GO-containing DNA. At least 10 phosphate groups (the majority of which are on the GO-strand) are involved in the interactions. In light of the fact that GO may be incorporated into DNA during replication or converted from guanine by oxidation in the random DNA sequences, these backbone contacts are more important than the interactions between MutY and the purines flanking the A/GO mismatch.
Figure 7.
Summary of the alkylation interference on specific complexes between MutY and M25 with DNA containing an A/G or A/GO mismatch. (A–C) MutY with A/G-containing DNA, MutY with A/GO-containing DNA and M25 with A/GO-containing DNA, respectively. The data in (A) are derived from Lu et al. (17). The methylation interference sites on purines are colored in blue for G23, G24 and G25 in the major groove and colored in green for mismatched A and GO as well as A18, A19, A20 and A21 that are believed to be in the minor groove. Methylated purines with strong and weak interference effects on protein binding are in dark and light colors, respectively. Ethylated phosphate groups show strong and weak interference effects on protein binding are shaded in red and purple, respectively. The phosphate groups at the 5′ and 3′ sides of mismatched bases are labeled with positive and negative numbers, respectively, and the phosphates immediately 5′ to the mismatched bases are labeled as position 0. The sequence positions are labeled between base pairs with respect to the A-strand from the 5′ end of the 40mer DNA. GO is marked as O. The DNA shown is bent with the mismatched A flipped-out from the DNA helix according to the DNA structure in AlkA–DNA complex (34). Arrows mark the phosphates that interact with the HhH motif of MutY.
Spatial distributions of the contacts of MutY and M25 with the distorted DNA based on the coordinates of DNA from an AlkA–DNA co-crystal structure (34) are presented in Figure 8. Inspection of this model reveals several interesting features. MutY appears to interact with its DNA substrate on both sides of the double helix as well as in both the major and minor grooves. Most base determinants of MutY are accessible from one side of the helix (Fig. 8C). The N-terminal domain of MutY contacts the widened minor groove mainly on one side of the DNA. Contacts on the other side of the helix at five consecutive phosphates (Fig. 8A) are contributed by the C-terminal domain.
Figure 8.
Spatial distributions of the contacts of MutY and M25 on the distorted DNA based on the coordinates of DNA from an AlkA–DNA co-crystal structure (34). The DNA shown here contains a 1-azaribose abasic base and has a different sequence than the one used in these experiments. (C and D) are viewed at 180° from (A and B) in which the minor groove is on the left and major groove on the right. The color codes are similar to those in Figure 7. The N3 group of mismatched A and the N7 group of mismatched GO (green blocks) are shown in (A) but are at the back of (C). The N7 groups of G23 and G24 (blue cylinders) as well as the N3 groups of A20 and A21 (green blocks) in the minor groove can be viewed in (C). Ethylated phosphate groups show strong, weak and no interference effects on protein binding are represented as red, purple and yellow spheres in different sizes, respectively. The 5′ and 3′ ends of the A-strand, as well as the phosphates that interact with the HhH motif of MutY, are marked.
Using DNA with the same flanking sequences, we observed different patterns of contact of MutY with A/GO mismatch (present results) as compared to MutY with an A/G mismatch as reported by Lu et al. (17) (compare Fig. 7A to B). For the base interactions, methylation at the N3 of adenine (A21) immediately 3′ to the mismatched GO strongly interferes with the MutY–A/GO interactions but only slightly interferes with the MutY–A/G mismatch interactions. MutY interacts weakly with G25 on the G-strand and A18 and A19 on the A-strand of A/G-containing DNA but does not interact with these bases of the A/GO-containing DNA. It appears that MutY covers more base pairs of A/G-containing DNA than of A/GO-containing DNA. There is a substantial difference in the backbone contacts between MutY and the two substrates (Fig. 7A and B). The ethylation patterns suggest that MutY binds to A/G- and A/GO-containing DNA in different conformations and its DNA binding sites on A/GO-containing DNA are shifted toward the 5′ side of GO as compared to A/G-containing DNA. Thus, a large component of the tight binding of MutY to the A/GO mismatch involves electrostatic binding energy. These base and phosphate contacts may account for the 80-fold higher MutY binding with A/GO-containing DNA than with A/G-containing DNA reported by Lu et al. (17). However, a direct comparison of the present results to the data obtained by Lu et al. (17) may be difficult to make because some parameters are different in these two experiments. The present results are derived from the D138N mutant of MutY using a modified Maxam–Gilbert A>G chemical sequencing reaction by replacing NaOH with piperidine. The catalytically inactive D138N mutant of MutY was used in the present experiments to eliminate the cleavage at the phosphodiester bond at the 3′ of mismatched A while wild-type MutY in the presence of methoxyamine was used by Lu et al. (17). Because it has been shown that the D138N mutation does not alter the overall structure of cdMutY (22) and does not change the DNA binding affinity of MutY (43), the D138N mutant of MutY may contact DNA in a manner similar to wild-type MutY. Replacing NaOH with piperidine can reduce the cleavage at GO but may increase the background levels and thus only the strong interference effects could be detected here.
We also demonstrated that by removing the C-terminal domain of MutY, it loses most of the electrostatic interactions with the DNA backbone and the tight contacts with several purine groups flanking the A/GO mismatch (compare Fig. 7B to C). This can partially explain why A/GO binding activity of M25 is ~70-fold lower than MutY (4) and the A/GO glycosylase activity is much faster for MutY than for M25 (4,24). As determined by the methylation interference experiments, the C-terminal domain of MutY influences the contact with mismatched A and GO as well as A20, A21 and G24 on the A-strand. We do not expect that methylation of mismatched A interferes with MutY binding but does not interfere with M25 binding. This result does not seem to be consistent with the data derived from the X-ray crystal structure of cdMutY with bound adenine in that the flipped-out adenine is buried in the active site of the catalytic domain (22). We suggest that the adenine-binding pocket may be large enough to bind the methylated adenine or this pocket may have some conformational change in the intact MutY as compared with M25. It is interesting to note that MutY has high binding affinities to A/GO, G/GO, T/GO and AP/GO (4). Thus, the high affinity to GO specified by the C-terminal domain of MutY may relax the base opposite to GO. Removing the C-terminal domain also alters the ethylation interference pattern. Interestingly, contacts at five consecutive phosphate groups (positions –1 to 3) on the GO-strand by MutY were diminished in the M25–A/GO interaction. These results suggest that the C-terminal domain of MutY may contact DNA directly or that the presence of the C-terminal domain may cause structural changes in the rest of the protein that enhance protein–DNA contacts in the N-terminal region.
The competition experiments of the DNA minor groove binding drugs show that distamycin A and Hoechst 33258 can bind the AATT sequence immediately 5′ to the mismatched A and affect both binding and cleavage activities of MutY. The inhibition is particularly pronounced for MutY–A/G, M25–A/G and M25––A/GO interactions. In contrast, the binding and cleavage activities of MutY with A/GO mismatch were quite insensitive to both drugs. The data suggest that the N-terminal domain of MutY makes some contacts in the minor groove of DNA at the 5′ of the mismatched A. This result is consistent with the X-ray crystal structure of cdMutY that shows the α2–α3 motif is in the DNA minor groove 5′ of the mismatched A (22). However, the methylation interference data indicate the N3 groups of two adenines 3′ to the mismatched GO are not involved in M25–A/GO interaction. As for MutY, the extra contacts on A/GO-containing DNA derived from the C-terminal domain of MutY minimize the inhibition of distamycin A and Hoechst 33258 on the binding and nicking activities of intact MutY. Based on the alkylation interference results, the C-terminal domain may affect the binding to N3 groups of A23 and A24 in the minor groove; however, the majority of the determinants are on the backbone phosphates (compare Fig. 8C and D). Although the two drugs compete with MutY for the minor groove binding sites, not all of the contact sites of MutY are totally blocked.
The co-crystal structures of E.coli AlkA (34) and human OGG1 (28) complexed with DNA show that these HhH proteins interact with the minor groove of DNA. Modeling studies also suggest that other HhH glycosylases including MutY can bind to DNA in a similar manner (22,34,35,49). These models suggest that cdMutY recognizes its DNA substrate through the DNA minor groove and the HhH motif interacts closely with phosphates at positions –2 and –3 on the A-strand (Fig. 7). The Gln42 of MutY is in the proper position to bend the DNA and assist in base flipping. Molecular modeling also suggests that the orientation of the A-strand from 5′ to 3′ is arranged from the [4Fe–4S] domain to the HhH domain, and places the α2–α3 motif in the DNA minor groove 5′ of the mismatched A (22). The crystal structure of the cdMutY also suggests some candidate residues (Gln41, Tyr82 and Arg194) for the recognition of the base opposite the adenine (22). AlkA and OGG1 contact their DNA substrates mainly through interaction on the flipped-out strand. However, MutY has more base and phosphate contacts on the GO-strand. These major determinants of the mismatched GO and electrostatic interactions are dependent on the C-terminal domain of MutY. We have proposed that the DNA is embedded between the catalytic and C-terminal domains of MutY (4). In this model, the C-terminal domain functions like a clamp to hold the GO-containing strand and the clamp is tighter with an A/GO-containing DNA than with an A/G-containing DNA. Our data presented here support this clamp model and specifically point out the orientation of the C-terminal domain on an A/GO substrate.
Volk et al. (35) have attempted to model a MutY–DNA structure based on the X-ray structure of cdMutY (22), the coordinates of DNA from AlkA co-crystal structure (34) and a model for the C-terminal domain based upon the preliminary NMR data (35,50) and the solution structure of MutT (51). Their model is based on the assumption that the structure of the cdMutY remains the same as in the intact MutY and is similar to our clamp model. However, our data do not fully support their model regarding the orientation of the C-terminal domain. In order to contact the five phosphates at positions –1 to 3 on the GO-strand, the C-terminal domain has to be positioned on the opposite side of the HhH motif (Fig. 8). Based on the perspective depicted in Figure 8A, the mismatched adenine (labeled A) flips out into the adenine-binding pocket between the HhH and Fe–S domains with the HhH domain lying above and the Fe–S domain lying below the plane of DNA. The HhH motif binds to the two phosphate groups on the left (marked by arrows). The C-terminal domain begins just after the Fe–S domain near the lower right of the figure and extends from the back of the DNA near the center of the figure to the front of DNA to cover the five phosphate groups (in red) on the GO-strand. In this orientation, the C-terminal domain tilts to some degree toward the HhH motif. In Figure 8C, the DNA is rotated 180° from that in Figure 8A and the cdMutY is located at the top of the figure above the DNA. In this view, the C-terminal domain is on the front of the DNA and wraps around the DNA through the major groove near the center of the figure to reach the five phosphate groups (in red) on the GO-strand. Our results will facilitate the modeling of the MutY–DNA structure when the MutY structure or both domains of MutY become available, but the most convincing evidence requires the solution of the MutY–DNA co-crystal structure.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Patrick Wright for critical reading of the manuscript and helpful comments. This work is supported by Grant GM 35132 from the National Institutes of General Medical Science, National Institutes of Health.
REFERENCES
- 1.Michaels M.L. and Miller,J.H. (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxo-guanine). J. Bacteriol., 174, 6321–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tchou J. and Grollman,A.P. (1993) Repair of DNA containing the oxidatively-damaged base 8-hydroxyguanine. Mutat. Res., 299, 277–287. [DOI] [PubMed] [Google Scholar]
- 3.Au K.G., Clark,S., Miller,J.H. and Modrich,P. (1989) Escherichia coli mutY gene encodes an adenine glycosylase active on G/A mispairs. Proc. Natl Acad. Sci. USA, 86, 8877–8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X., Wright,P.M. and Lu,A.-L. (2000) The C-terminal domain of MutY glycosylase determines the 7,8-dihydro-8-oxo-guanine specificity and is crucial for mutation avoidance. J. Biol. Chem., 275, 8448–8455. [DOI] [PubMed] [Google Scholar]
- 5.Lu A.-L. and Chang,D.-Y. (1988) Repair of single base pair transversion mismatches of Escherichia coli in vitro: correction of certain A/G mismatch is independent of dam methylation and host mutHLS gene functions. Genetics, 118, 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaels M.L., Cruz,C., Grollman,A.P. and Miller,J.H. (1992) Evidence that MutM and MutY combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl Acad. Sci. USA, 89, 7022–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaels M.L., Tchou,J., Grollman,A.P. and Miller,J.H. (1992) A repair system for 8-oxo-7,8-dihydrodeoxyguanine (8-hydroxyguanine). Biochemistry, 31, 10964–10968. [DOI] [PubMed] [Google Scholar]
- 8.Radicella J.P., Clark,E.A. and Fox,M.S. (1988) Some mismatch repair activities in Escherichia coli. Proc. Natl Acad. Sci. USA, 85, 9674–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su S.-S., Lahue,R.S., Au,K.G. and Modrich,P. (1988) Mispair specificity of methyl-directed DNA mismatch correction in vitro. J. Biol. Chem., 263, 6829–6835. [PubMed] [Google Scholar]
- 10.Tsai-Wu J.-J., Liu,H.-F. and Lu,A.-L. (1992) Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A.C and A.G mispairs. Proc. Natl Acad. Sci. USA, 89, 8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q.M., Ishikawa,N., Nakahara,T. and Yonei,S. (1998) Esherichia coli MutY protein has a guanine-DNA glycosylase that acts on 7,8-dihydro-8-oxoguanine:guanine mispair to prevent spontaneous G:C to C:G transversions. Nucleic Acids Res., 26, 4669–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 13.Wood M.L., Dizdaroglu,M., Gajewski,E. and Essigmann,J.M. (1990) Mechanistic studies of ionizing radiation and oxidative mutagenesis: Genetic effects of single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry, 29, 7024–7032. [DOI] [PubMed] [Google Scholar]
- 14.Moriya M. (1993) Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-Oxoguanine in DNA induces targeted G.C to T.A transversions in simian kidney cells. Proc. Natl Acad. Sci. USA, 90, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriya M., Ou,C., Bodepudi,V., Johnson,F., Takeshita,M. and Grollman,A.P. (1991) Site-specific mutagenesis using a gapped duplex vector: A study of translesion synthesis past 8-oxodeoxyguanosine in Escherichia coli. Mutat. Res., 254, 281–288. [DOI] [PubMed] [Google Scholar]
- 16.Cheng K.C., Cahill,D.S., Kasai,H., Nishimura,S. and Loeb,L.A. (1991) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J. Biol. Chem., 267, 166–172. [PubMed] [Google Scholar]
- 17.Lu A.-L., Tsai-Wu,J.-J. and Cillo,J. (1995) DNA determinants and substrate specificities of Escherichia coli MutY. J. Biol. Chem., 270, 23582–23588. [DOI] [PubMed] [Google Scholar]
- 18.Gogos A., Cillo,J., Clarke,N.D. and Lu,A.-L. (1996) Specific recognition of A/G and A/8-oxoG mismatches by Escherichia coli MutY: removal of the C-terminal domain preferentially affects A/8-oxoG recognition. Biochemistry, 35, 16665–16671. [DOI] [PubMed] [Google Scholar]
- 19.Manuel R.C., Czerwinski,E.W. and Lloyd,R.S. (1996) Identification of the structural and functional domains of MutY, an Escherichia coli DNA mismatch repair enzyme. J. Biol. Chem., 271, 16218–16226. [DOI] [PubMed] [Google Scholar]
- 20.Manuel R.C. and Lloyd,R.S. (1997) Cloning, overexpression, and biochemical characterization of the catalytic domain of MutY. Biochemistry, 36, 11140–11152. [DOI] [PubMed] [Google Scholar]
- 21.Golinelli M.-P., Chmiel,N.H. and David,S.S. (1999) Site-directed mutagenesis of the cysteine ligands to the [4Fe-4S] of Escherichia coli MutY. Biochemistry, 38, 6997–7007. [DOI] [PubMed] [Google Scholar]
- 22.Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,S. and Tainer,J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- 23.Lu A.-L., Yuen,D.S. and Cillo,J. (1996) Catalytic mechanism and DNA substrate recognition of Escherichia coli MutY protein. J. Biol. Chem., 271, 24138–24143. [DOI] [PubMed] [Google Scholar]
- 24.Noll D.M., Gogos,A., Granek,J.A. and Clarke,N.D. (1999) The C-terminal domain of the adenine-DNA glycosylase MutY confers specificity for 8-oxoguanine.adenine mispairs and may have evolved from MutT, an 8-oxo-dGTPase. Biochemistry, 38, 6374–6579. [DOI] [PubMed] [Google Scholar]
- 25.Michaels M.L., Pham,L., Nghiem,Y., Cruz,C. and Miller,J.H. (1990) MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res., 18, 3841–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai-Wu J.-J., Radicella,J.P. and Lu,A.-L. (1991) Nucleotide sequence of the Escherichia coli micA gene required for A/G-specific mismatch repair: Identity of MicA and MutY. J. Bacteriol., 173, 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nash H.M., Bruner,S.D., Scharer,O.D., Kawate,T., Addona,T.A., Spooner,E., Lane,W.S. and Verdine,G.L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol., 6, 968–980. [DOI] [PubMed] [Google Scholar]
- 28.Bruner S.D., Norman,D.P. and Verdine,G.L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- 29.Bailly V. and Verly,W.G. (1987) Escherichia coli endonuclease III is not an endonuclease but a yy|-elimination catalyst. Biochem. J., 242, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bor R., Elford,J., Campbell,L., Salt,H., Miller,R., Murray,D. and Johnson,M. (1991) Changing patterns in the workload of a district HIV/AIDS counseling unit 1987–90. Genitourin. Med., 67, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamagata Y., Kato,M., Odawara,K., Tokuno,Y., Nakashima,Y., Matsushima,N., Yasumura,K., Tomita,K., Ihara,K., Fujii,Y. et al. (1996) Three-dimensional structure of a DNA repair enzyme, 3-methyadenine DNA glycosylase II, from Escherichia coli. Cell, 86, 311–319. [DOI] [PubMed] [Google Scholar]
- 32.Labahn J., Scharer,A., Long,A., Ezaz-Nikpay,K., Verdine,G.L. and Ellenberger,T.E. (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- 33.Boiteux S. and Radicella,J.P. (2000) The Human OGG1 Gene: Structure, Functions, and Its Implication in the Process of Carcinogenesis. Arch. Biochem. Biophys., 377, 1–8. [DOI] [PubMed] [Google Scholar]
- 34.Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volk D.E., House,P.G., Thiviyanathan,V., Luxon,B.A., Zhang,S., Lloyd,R.S. and Gorenstein,D.G. (2000) Structural Similarities between MutT and the C-Terminal Domain of MutY. Biochemistry, 39, 7331–7336. [DOI] [PubMed] [Google Scholar]
- 36.Carrondo M.A., Coll,M., Aymami,J., Wang,A.H., van der Marel,G.A., van Boom,J.H. and Rich,A. (1989) Binding of a Hoechst dye to d(CGCGATATCGCG) and its influence on the conformation of the DNA fragment. Biochemistry, 28, 7849–7859. [DOI] [PubMed] [Google Scholar]
- 37.Chen X., Ramakrishnan,B. and Sundaralingam,M. (1997) Crystal structures of the side-by-side binding of distamycin to AT- containing DNA octamers d(ICITACIC) and d(ICATATIC). J. Mol. Biol., 267, 1157–1170. [DOI] [PubMed] [Google Scholar]
- 38.Kopka M.L., Yoon,C., Goodsell,D., Pjura,P. and Dickerson,R.E. (1985) The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl Acad. Sci. USA, 82, 1376–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quintana J.R., Lipanov,A.A. and Dickerson,R.E. (1991) Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G. Biochemistry, 30, 10294–10306. [DOI] [PubMed] [Google Scholar]
- 40.Tabernero L., Bella,J. and Aleman,C. (1996) Hydrogen bond geometry in DNA-minor groove binding drug complexes. Nucleic Acids Res., 24, 3458–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng M.K., Usman,N., Frederick,C.A. and Wang,A.H. (1988) The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res., 16, 2671–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vega M.C., Garcia,S.I, Aymami,J., Eritja,R., van der Marel,G.A., van Boom,J.H., Rich,A. and Coll,M. (1994) Three-dimensional crystal structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor-groove-binding drug Hoechst 33258. Eur. J. Biochem., 222, 721–726. [DOI] [PubMed] [Google Scholar]
- 43.Wright P.M., Yu,J., Cillo,J. and Lu,A.-L. (1999) The active site of the Escherichia coli MutY DNA adenine glycosylase. J. Biol. Chem., 274, 29011–29018. [DOI] [PubMed] [Google Scholar]
- 44.Siebenlist U. and Gilbert,W. (1980) Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc. Natl Acad. Sci. USA, 77, 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maxam A.M. and Gilbert,W. (1980) Sequencing end-labeled DNA with base-specific chemical cleavage. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 46.Kouchakdjian M., Bodepude,V., Shibutani,S., Eisenberg,M., Johnson,F., Grollman,A.P. and Patel,D.J. (1991) NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-oxo-7H-dG(syn):dA(anti) alignment at lesion site. Biochemistry, 30, 1403–1412. [DOI] [PubMed] [Google Scholar]
- 47.Lu A.-L. and Chang,D.-Y. (1988) A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell, 54, 805–812. [DOI] [PubMed] [Google Scholar]
- 48.Radicella J.P., Clark,E.A., Chen,S. and Fox,M.S. (1993) Patch length of localized repair event: role of DNA polymerase I in mutY-dependent mismatch repair. J. Bacteriol., 175, 7732–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mol C.D., Parikh,S.S., Putnam,C.D., Lo,T.P. and Tainer,J.A. (1999) DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys. Biomol. Struct., 28, 101–128. [DOI] [PubMed] [Google Scholar]
- 50.Volk D.E., Thiviyanathan,V., House,P.G., Lloyd,R.S. and Gorenstein,D.G. (1999) 1H, 13C and 15 N resonance assignments of the C-terminal domain of MutY: an adenine glycosylase active on G:A mismatches. J. Biomol. NMR, 14, 385–386. [DOI] [PubMed] [Google Scholar]
- 51.Abeygunawardana C., Weber,D.J., Gittis,A.G., Frick,D.N., Lin,J., Miller,A.F., Bessman,M.J. and Mildvan,A.S. (1995) Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase. Biochemistry, 34, 14997–15005. [DOI] [PubMed] [Google Scholar]