Abstract
The tail structures of bacteriophages infecting gram-positive bacteria are largely unexplored, although the phage tail mediates the initial interaction with the host cell. The temperate Lactococcus lactis phage TP901-1 of the Siphoviridae family has a long noncontractile tail with a distal baseplate. In the present study, we investigated the distal tail structures and tail assembly of phage TP901-1 by introducing nonsense mutations into the late transcribed genes dit (orf46), talTP901-1 (orf47), bppU (orf48), bppL (orf49), and orf50. Transmission electron microscopy examination of mutant and wild-type TP901-1 phages showed that the baseplate consisted of two different disks and that a central tail fiber is protruding below the baseplate. Evaluation of the mutant tail morphologies with protein profiles and Western blots revealed that the upper and lower baseplate disks consist of the proteins BppU and BppL, respectively. Likewise, Dit and TalTP901-1 were shown to be structural tail proteins essential for tail formation, and TalTP901-1 was furthermore identified as the tail fiber protein by immunogold labeling experiments. Determination of infection efficiencies of the mutant phages showed that the baseplate is fundamental for host infection and the lower disk protein, BppL, is suggested to interact with the host receptor. In contrast, ORF50 was found to be nonessential for tail assembly and host infection. A model for TP901-1 tail assembly, in which the function of eight specific proteins is considered, is presented.
The initial interaction between a tailed phage and its bacterial host is mediated by the distal part of the phage tail, and studies of the distal tail structures thus contribute to a fundamental understanding of phage-host interactions. Moreover, determinations of the underlying mechanisms of phage-host interactions at the level of single proteins depend on a prior knowledge of the interacting part of the phage structure. Investigations of the tail structures of phages infecting lactic acid bacteria (LAB) and of gram-positive phages in general are scarce compared to those of the Escherichia coli phages, which have been studied extensively. Therefore, increased knowledge of the distal tail structures and the tail assembly processes of LAB phages would greatly enhance our understanding of the infection processes of gram-positive phages.
Tailed phages are classified into the families of Myoviridae, Siphoviridae, and Podoviridae, which are characterized by a long contractile tail, a long noncontractile tail, and a short noncontractile tail, respectively (1). Minor tail structures, such as fibers and baseplates, are also found among members of these families. A long phage tail is usually assembled from the distal end proceeding towards the head-proximal end until a mature tail structure is formed, and the tail is ultimately joined to a mature and independently assembled head, thereby forming an infectious phage virion (12, 13). Tail formation of the E. coli Siphoviridae phage λ is initiated from an initiator complex, which is composed of the tail fiber protein, the tape measure protein, and four other proteins (36, 37). The major tail protein polymerizes onto the λ initiator complex to form the tail tube, which is the channel for translocation of the phage genomic DNA during infection (34). The tail fiber protein constitutes the very distal structure of the λ tail and is responsible for the recognition of the host receptor (57, 58; for a comprehensive review on λ tail structure and assembly, see reference 35) The E. coli T5 phage has a distal tail structure similar to phage λ, but the straight tail fiber of phage T5 is presumed to act as an injection needle, which enables transfer of the phage genome through the host envelope. This process is preceded by host interactions of both the long tail fibers and the receptor binding protein, located in a conical structure just above the straight tail fiber (5, 24, 26). In contrast to the Siphoviridae phages λ and T5, a much more complex tail structure on the E. coli T4 phage of the Myoviridae family is observed. The T4 tail formation is initiated from the assembly of a distal baseplate, which is composed of at least 16 different proteins constituting six wedges joined around a central hub, and the remaining tail components are assembled onto the baseplate (for reviews on T4 tail structure and assembly, see references 16 and 41).
The temperate Lactococcus lactis phage TP901-1 belongs to the P335 species of the Siphoviridae family, the predominant family of LAB phages (10, 30). The TP901-1 virion contains a small isometric head, a collar, whiskers, and a long noncontractile tail with a distal baseplate (33). Many aspects of the TP901-1 life cycle have been examined in detail, such as prophage integration and excision, replication, transcription, and regulation of the lytic/lysogenic switch (7, 8, 15, 31, 46, 47, 51); moreover, the complete 37.7-kbp genome sequence has been determined (GenBank accession no. NC_002747) (9). The functions of three tail gene products have previously been investigated experimentally. In these studies, open reading frame 42 (orf42) was found to encode the major tail protein (MTP) that forms the tail tube; orf45 was shown to encode the tape measure protein (TMP), which determines the length of the phage tail; and orf49 was shown to encode a structural protein (BPP) proposed to constitute the baseplate (32, 33, 52).
In this work, we investigate the distal tail structures of phage TP901-1 through mutagenesis of five putative tail genes, orf46, orf47, orf48, orf49, and orf50. Analysis of these mutant phages with respect to morphology, protein content, and infection efficiency facilitated the elucidation of novel structures associated with the TP901-1 tail. These findings, in conjunction with previously published data, have enabled us to propose a model for TP901-1 tail assembly which considers the structural roles fulfilled by individual proteins, rendering this the best-characterized tail structure among LAB phages.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. E. coli strains were cultured with agitation at 37°C in Luria-Bertani broth (54) supplemented with 50 to 150 μg/ml ampicillin, 12.5 μg/ml tetracycline, 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and 40 μg/ml isopropyl-β-d-thiogalactoside where appropriate. Chemical-competent or electrocompetent cells were prepared and transformed essentially as described by Sambrook and Russell (54). Lactococcus lactis strains were cultured without agitation at 28°C or 30°C in M17 broth (Oxoid Ltd., Basingstoke, Hampshire, England) supplemented with 0.5% (wt/vol) glucose (GM17) (56) and 2 μg/ml tetracycline or 5 μg/ml chloramphenicol where appropriate. Electroporation of L. lactis was performed essentially as described by Holo and Nes (28), except that glycine and sucrose concentrations in the propagation medium were reduced to 1% (wt/vol) and 0.2 M, respectively.
TABLE 1.
Bacterial strains, plasmids, and phages
| Bacterial strain, plasmid, or phage | Relevant featurea | Source or reference |
|---|---|---|
| L. lactis subsp. cremoris strain | ||
| 901-1 | Lysogenic for phage TP901-1 | 6 |
| 3107 | Indicator strain for phage TP901-1 | 15 |
| 3107 + pAK89 | Indicator strain for phage TP901-1 + supD | 52 |
| MG1363 | Transformational strain | 22 |
| CSV57-1 | 901-1 lysogenic for TP901-1orf46(Am) | This study |
| CSV59-2 | 901-1 lysogenic for TP901-1orf47(Am) | This study |
| CSV61-1 | 901-1 lysogenic for TP901-1orf48(Am) | This study |
| CSV63-1 | 901-1 lysogenic for TP901-1orf49(Am) | This study |
| CSV65-1 | 901-1 lysogenic for TP901-1orf50(Am) | This study |
| E. coli strain | ||
| DH5α | Transformational strain | Invitrogen |
| XL1-Blue MRF′ | Transformational strain, Tcr | Stratagene |
| Plasmid | ||
| pBluescipt II SK | Cloning and positive selection vector, lacZ, Apr | Stratagene |
| pGhost8 | Ts replicon, Tcr | 48 |
| pCI372 | Parental plasmid for pAK89, Cmr | 23 |
| pAK89 | supD amber suppressor, Cmr | 19 |
| pCSV01-22 | pBluescript::TP901-1(25.124-26.927)orf46(Am) (codon no. 22)b | This study |
| pCSV02-10 | pBluescript::TP901-1(26.180-28.168)orf47(Am) (codon no. 163)b | This study |
| pCSV03-3 | pBluescript::TP901-1(28.822-30.632)orf48(Am) (codon no. 84)b | This study |
| pCSV04-1 | pBluescript::TP901-1(29.561-31.417)orf49(Am) (codon no. 31)b | This study |
| pCSV06-4 | pBluescript::TP901-1(30.141-31.883)orf50(Am) (codon no. 39)b | This study |
| pCSV28-1 | pGhost8::TP901-1(25.124-26.927)orf46(Am) (codon no. 22)b | This study |
| pCSV29-2 | pGhost8::TP901-1(26.180-28.168)orf47(Am) (codon no. 163)b | This study |
| pCSV30-1 | pGhost8::TP901-1(28.822-30.632)orf48(Am) (codon no. 84)b | This study |
| pCSV31-1 | pGhost8::TP901-1(29.561-31.417)orf49(Am) (codon no. 31)b | This study |
| pCSV32-1 | pGhost8::TP901-1(30.141-31.883)orf50(Am) (codon no. 39)b | This study |
| Phage | ||
| TP901-1 | Temperate phage. Isolated following induction of L. lactis 901-1 | 6 |
| 46− | TP901-1orf46(Am) | This study |
| 47− | TP901-1orf47(Am) | This study |
| 48− | TP901-1orf48(Am) | This study |
| 49− | TP901-1orf49(Am) | This study |
| 50− | TP901-1orf50(Am) | This study |
Am, amber mutation; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Ts, temperature sensitive.
Amber-mutated codon.
Induction and purification of phages.
Phages used in this study are listed in Table 1. Wild-type (wt) and mutant TP901-1 phages were induced from the respective lysogenic L. lactis 901-1 strains by 3 μg/ml mitomycin C (Sigma-Aldrich, St. Louis, Missouri) or by UV light essentially as described previously (15). For phage purification following bacterial lysis, 1 M sodium chloride was added and the lysates were incubated at 4°C overnight. Cellular debris was removed by 15 min of centrifugation at 10,000 × g, and the phage particles were precipitated with 10% (wt/vol) PEG 6000 and resuspended in SM buffer (100 mM sodium chloride, 10 mM magnesium sulfate, 50 mM Tris [pH 7.5], and 0.01% [wt/vol] gelatin). Following treatment with 10 μg/ml of DNase I and RNase A (Sigma-Aldrich), phage particles were purified by isopycnic centrifugation for 22 h through cesium chloride equilibrium gradients, essentially as described for phage λ by Sambrook and Russell (54). Phage infection efficiency was determined by plaque assay on the indicator strain L. lactis 3107 and the amber-suppressing derivative (Table 1) in GM17 medium supplemented with 5 mM calcium chloride and agarose (Cambrex Bio Science, Rockland, Maine) as described by Lillehaug (44).
DNA technology and sequencing.
The plasmids used in this study are listed in Table 1. Plasmid DNA was isolated from E. coli and L. lactis with a QIAprep Spin Miniprep kit or a Plasmid Midi kit (QIAGEN GmbH, Hilden, Germany) using the manufacturer's instructions, except that the lysis step of L. lactis cultures was carried out for 20 min at 37°C in the presence of 20 mg/ml lysozyme (Sigma-Aldrich). Chromosomal DNA was extracted from L. lactis strains with a GenElute Bacterial Genomic DNA kit (Sigma-Aldrich). TP901-1 phage DNA was isolated from induced and purified wt phages by phenol-chloroform extraction as described by Sambrook and Russell for phage λ (54). L. lactis colonies were resuspended in water and treated with 10 units of mutanolysin (Sigma-Aldrich) prior to PCR amplifications, while plaque suspensions, if necessary, were pretreated with 2 units of TURBO DNase (Ambion Ltd., Cambridgeshire, United Kingdom) for 30 to 60 min at 37°C. PCR amplifications of phage DNA used for cloning were performed with Pwo DNA polymerase (Roche, Mannheim, Germany), while other amplifications were performed with a Taq DNA polymerase. Restriction endonuclease enzymes supplied by New England BioLabs (Beverly, Mass.), shrimp alkaline phosphatase and T4 DNA Ligase supplied by USB (Cleveland, Ohio), and other enzymes were used as recommended by the suppliers. Phage DNA sequences were determined by using a Thermo Sequenase fluorescent dye-labeled primer cycle sequencing kit with an ALF Express DNA sequencer (Pharmacia Biotech), by using a CEQ 2000 dye terminator cycle sequencing kit with a Beckman Coulter CEQ 2000 DNA analysis system (Beckman Coulter Inc., Fullerton, California), or by MWG (Ebersberg, Germany). Sequences were aligned and assembled with the software from the Genetics Computer Group at the University of Wisconsin (18). Multiple sequence alignments of phage sequences amplified from plaques were aligned with the program ClustalW at the website of the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/index.html).
Construction and verification of phage mutants.
The splicing by overlap extension (SOEing) PCR technique (29) was used together with the double sets of mismatch oligonucleotides, which are listed in Table 2, to amplify 1.7- to 2.0-kbp TP901-1 fragments with in-frame amber mutations and restriction sites introduced into orf46, orf47, orf48, orf49, and orf50, using isolated TP901-1 wt DNA as a template. The mutated fragments, each containing an amber mutation doubly flanked by approximately 0.9-kbp wt sequences, were digested with BamHI and SalI, and each was cloned into an equally digested pBluescript II SK vector. E. coli DH5α or XL1-Blue MRF′ cells were transformed with the constructs, and the TP901-1 inserts were sequenced to ensure that only the desired mutations had been incorporated. The inserts were subsequently subcloned into the pGhost8 vector and, following L. lactis MG1363 transformation, propagation, and plasmid purification, the plasmids were finally introduced into the TP901-1 lysogenic strain L. lactis 901-1 by transformation. Mutations were transferred from the pGhost8 vectors to the TP901-1 genome by homologous recombination, as described previously (3, 52). TP901-1 lysogenic strains with mutations in orf46, orf47, orf48, orf49, and orf50, respectively, were verified by DNA sequencing or by Southern blotting with an ECL direct nucleic acid labeling and detection system (Amersham Bioscience, Uppsala, Sweden). Briefly, chromosomal DNA from potential lysogenic mutants was isolated, digested with the restriction enzymes EcoRV and AvrII or EcoRV and XbaI, transferred to a Hybond-N+ nylon membrane (Amersham Bioscience), probed with appropriate peroxidase-labeled TP901-1 fragments of approximately 2 kbp, and detected with chemiluminescence. Lysogenic mutants with an included restriction site correctly introduced into the TP901-1 genome were selected for further studies and subsequently transformed individually with the plasmid pCI372 or the amber suppressor derivative pAK89.
TABLE 2.
Oligonucleotides used for construction of specific amber mutations in orf46, orf47, orf48, orf49, and orf50 of phage TP901-1a
| Oligonucleotide | Sequence |
|---|---|
| 46am1F | 5′ CGCGGATCCGTTGTAAATACGATTCAAAGTCTTTGGGGAG |
| 46am1R | 5′ CATCATAACTCATGGCctaGGTTGGAATAAAAG |
| 46am2F | 5′ CCTTTTATTCCAACCtagGCCATGAGTTATG |
| 46am2R | 5′ ACGCGTCGACAACTCTTGTTCTTTCGTATCATTGACTGC |
| 47am1F | 5′ CGCGGATCCTGACGATAAAATATAAGCTTGAGGATAG |
| 47am1R | 5′ GTAATTTCAATctAgAAGGTAAACTCTGCACC |
| 47am2F | 5′ GCAGAGTTTACCTTcTagATTGAAATTACAGG |
| 47am2R | 5′ ACGCGTCGACAACGAATCTTGACCCCTTGAAGCGTCC |
| 48am1F | 5′ CGCGGATCCGACACAATCACAGCCCAATTTAAGCTC |
| 48am1R | 5′ CTGCAAAGCATTATCctaGGCAACATATTTC |
| 48am2F | 5′ GAAATATGTTGCCtagGATAATGCTTTGCAGTTTG |
| 48am2R | 5′ ACGCGTCGACAATGATCCCACCTCCGAGTTCTCCAGACC |
| 49am1F | 5′ CGCGGATCCATGCGGATGTCAATAGTCAAGCCATTGTTG |
| 49am1R | 5′ TGAACTACATTTCCACCctaGGTTAATTCAG |
| 49am2F | 5′ CTGAATTAACCtagGGTGGAAATGTAGTTC |
| 49am2R | 5′ ACGCGTCGACCTGTCATCTCAGTAGGAATTATAAGCGTG |
| 50am1F | 5′ CGCGGATCCATGCAATTGGCACTGAAACTGATGGATTT |
| 50am1R | 5′ AGTTTTGCATTctATTGGGCTATTAAAAGC |
| 50am2F | 5′ TTTTAATAGCCCAATagAATGCAAAACTTA |
| 50am2R | 5′ ACGCGTCGACGCTGTAAACTTTCTAAATCTGTTTGATTAG |
TP901-1 DNA fragments with overlapping amber mutations were amplified with oligonucleotide sets 1 and 2, and the fragments were spliced by the SOEing PCR technique. AvrII, BamHI, SalI, and XbaI restrictions sites are underlined, and mismatch nucleotides are indicated with lowercase letters.
Electron microscopy and immunological detection.
Purified phage preparations were dialyzed for 10 to 15 min against SM buffer for negative staining or TGB buffer (200 mM Tris, 500 mM glycine, 2% [vol/vol] butanol [pH 7.5]) for immunogold labeling. For negative staining, a carbon film was floated from a mica sheet into a suspension of dialyzed phages and incubated for 10 min. The film was subsequently rinsed in demineralized water and was finally stained with 2% (wt/vol) uranyl acetate (Agar Scientific, Stansted, United Kingdom) for 30 s. The carbon film was picked up with 400-mesh copper grids (Agar Scientific) and examined in a transmission electron microscope (Tecnai 10; FEI, Eindhoven, The Netherlands) at an acceleration voltage of 80 kV. Micrographs were taken with a MegaView II CCD camera (SIS, Münster, Germany). For colloidal gold immunolabeling, dialyzed phages were incubated overnight at room temperature with primary antibody solution (polyclonal anti-Tal2009 [C-terminal] raised in rabbits [38]), which was diluted 1:300 in TGB buffer. A carbon film was floated from a mica sheet into the suspension and incubated for 30 min. Alternatively, phages were first adsorbed to a carbon film for 30 min and then incubated overnight in primary antibody solution. The carbon films were subsequently washed in TGB buffer and incubated for 1 h at room temperature in goat anti-rabbit immunoglobulin G 5-nm gold conjugate solution (BBI, Cardiff, United Kingdom), diluted 1:40 (vol/vol) with TGB buffer. After fixation for 20 min at room temperature in 0.25% (vol/vol) glutaraldehyde in phosphate-buffered saline buffer (54), negative staining was done as described before except that 400-mesh nickel grids (Agar Scientific) were used instead of copper grids.
Polyclonal antibody preparation.
The phage Tuc2009 tail structure-encoding genes orf49Tuc2009, Tal2009 (N terminal), and orf51Tuc2009 were cloned, overexpressed, and purified using a QIAexpressionist system (QIAGEN) as recommended by the manufacturer (Mc Grath et al., unpublished data). Polyclonal antibodies against these proteins were raised in rabbits by Harlan Sera-Lab (Leicestershire, England). Initial immunizations of individual proteins were complemented with Freund's complete adjuvant with five subsequent booster injections of protein. The final serum samples were acquired 11 weeks after the initial immunizations.
SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Approximately 1010 PFU of purified phages, dialyzed against SM buffer, were mixed with sample buffer and reducing agent (Invitrogen, Carlsbad, California), boiled for 15 to 20 min, and loaded on 10% or 4 to 12% NuPAGE Novex Bis-Tris polyacrylamide gels (thickness, 1 mm; Invitrogen). Electrophoresis was performed at 200 V in either MES (morpholineethanesulfonic acid) or MOPS (morpholinepropanesulfonic acid) sodium dodecyl sulfate (SDS) running buffer (Invitrogen), and protein bands were stained with a silver staining kit protein (Amersham Bioscience). Western blots were performed with phage proteins separated by electrophoresis and subsequently electroblotted onto polyvinylidene difluoride membranes using an XCell II Blot module and NuPAGE transfer buffer (Invitrogen). Membranes were incubated for 1 h in Tris-buffered saline (TBS) blocking buffer (20 mM Tris, 140 mM sodium chloride [pH 8.0], and 1% [wt/vol] skim milk powder) and washed twice in TBST (TBS-0.05% [wt/vol] Tween 20). TP901-1 proteins were subsequently identified using polyclonal antibodies raised in rabbits against homologous proteins of the related L. lactis phage Tuc2009. These antibodies were diluted 1:1000 in TBST-1% (wt/vol) bovine serum albumin (Sigma-Aldrich), and the incubation step was performed overnight. Following two washes with TBST, the membranes were incubated for 1 h with the secondary antibody, anti-rabbit IgG antibody conjugated with alkaline phosphate (Sigma-Aldrich) diluted 1:30,000 in TBST-1% (wt/vol) bovine serum albumin. The membranes were finally washed twice in TBST and once in TBS before antibody reactions were visualized with 5-bromo-4-chloro-3-indolyl phosphate/Nitro Blue Tetrazolium tablets (Sigma-Aldrich) dissolved in pure water.
RESULTS
Construction of TP901-1 tail mutants.
In order to investigate the distal tail structures and assembly of phage TP901-1, five late transcribed genes (orf46, orf47, orf48, orf49, and orf50) were selected for mutagenic analysis. These genes, located downstream from the tape measure protein gene, are genomically positioned equivalent to the distal tail protein genes of the E. coli phage λ. Moreover, the gene products are homologous to several proteins encoded by related L. lactis phages (Table 3). Defined mutants were obtained by introducing site-specific in-frame amber mutations into each of the selected genes by homologous recombination between the TP901-1 prophage genome present in L. lactis 901-1 and specific mutated TP901-1 sequences cloned in the temperature-sensitive pGhost8 vector. The mutagenesis was carried out with TP901-1 prophages to facilitate the generation of potentially noninfectious phage mutants without the use of an amber suppressor, due to previous problems obtaining suppression using a supD amber suppressor plasmid (52). In fact, we were unsuccessful in obtaining suppression of the generated prophage amber mutants (results not shown), which is consistent with prior observations (52) but not understood at this point.
TABLE 3.
Homologous proteins of ORF46, ORF47, ORF48, ORF49, and ORF50 from phage TP901-1
| ORF | Function and alternative name | Size
|
Homologous protein of L. lactis phageb | % Identity (no. of aa)c | Reference | |
|---|---|---|---|---|---|---|
| aa | kDaa | |||||
| 46 | Distal tail protein; Dit | 253 | 29.1 (30) | ORF49 of phage Tuc2009 | 95 (253) | 55 |
| ORF253 structural protein of phage ul36 | 94 (253) | 39 | ||||
| 47 | Tail fiber; Tal | 918 | 102.1 (95 + 63*) | Tal2009 structural protein of phage Tuc2009 | 93 (918) | 38 |
| ORF908 of phage ul36 | 87 (920) | 39 | ||||
| ORF54 of phage bIL285 | 79 (545) | 14 | ||||
| Conserved domain in peptidase family | 2e-21d | |||||
| M23/M37 | ||||||
| 48 | Upper baseplate disc; BppU | 299 | 33.8 (35) | ORF51 of phage Tuc2009 | 80 (296) | 55 |
| ORF322 structural protein of phage ul36 | 79 (296) | 39 | ||||
| 49 | Lower baseplate disc; BppL | 163 | 17.2 (17) | ORF343 of L. lactis φLC3 | 70 (151) | 4 |
| ORF53 structural protein of phage Tuc2009 | 56 (62) | 55 | ||||
| ORF165B structural protein of phage ul36 | 33 (65) | 39 | ||||
| Receptor binding proteins of phages jw30, jw32, fd13, and jw31 | 28-26 (112) | 20, 21 | ||||
| 50 | Unknown | 74 | 8.7 (-)e | ORF74 of phage φLC3 | 98 (74) | 4 |
| Putative holin of phage BK5-T | 98 (73) | 49 | ||||
| ORF58 of phage bIL286 | 98 (74) | 14 | ||||
| ORF60 of phage bIL285 | 95 (74) | 14 | ||||
| ORF54 of phage Tuc2009 | 95 (74) | 55 | ||||
Predicted molecular weight from aa sequence and the experimentally measured molecular weight by SDS-PAGE in parentheses. *, processed protein.
Identified from BLASTp search (2).
The percentage identity followed by the number of amino acids (aa) over which the identity is determined in parentheses.
E value from BLAST conserved domain alignment.
Not observed by SDS-PAGE analysis.
In each of the five genes, orf46-50, a serine codon was replaced by an amber stop codon (TAG) and, where possible, an AvrII (CC TAG G) or an XbaI (TC TAG A) restriction site was introduced with the amber mutation without changing the encoded amino acids of the flanking codons. In orf50, a restriction site could not be introduced without changing the amino acid sequence, and the amber mutation was therefore inserted alone into the first serine codon of the gene. The prophages mutated in orf46, orf47, orf48, and orf49 were verified in Southern blots by recognition of the inserted restriction sites in chromosomal DNA from the lysogenic strains, while the prophage mutated in orf50 was verified by sequencing (results not shown). The Southern blots also revealed that the L. lactis strain 901-1, lysogenic for TP901-1, contains other prophage sequences with homology to the mutated tail genes of TP901-1, since additional and unexpected bands in blots with five different probes covering TP901-1 orf46-50 were observed.
TP901-1 tail morphology.
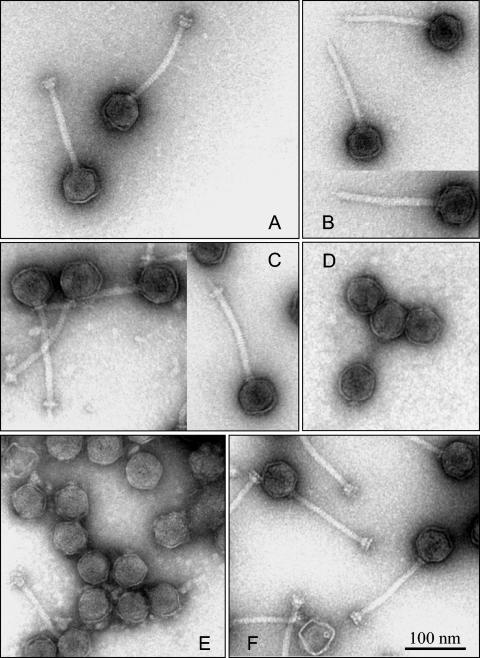
With the purpose of determining the morphological phenotypes of the mutant phages, i.e., analyzing the effect of removing individual proteins from the tail assembly process and resolving detailed features of the distal tail structure, we investigated the purified TP901-1 wt and mutant phages by transmission electron microscopy (TEM).
Analysis of the wt phage revealed that the TP901-1 virion has a central tail fiber protruding below the baseplate and that the two baseplate disks have different dimensions (upper disk diameter, 23 ± 3 nm [n = 19]; lower disk diameter, 28 ± 3 nm [n = 19]) (Fig. 1A). This indicates that the two disks consist of different proteins and not only ORF49 as suggested previously (52). When the mutant phages were examined, we found that the 48− mutant completely lacked the baseplate structure, thus clearly exposing the central tail fiber (length, 16 ± 3 nm [n = 26]) and a conical structure (length, 7 ± 2 [n = 33]) between the fiber and the tail tube (Fig. 1B). While the 48− mutant completely lacked the baseplate, the 49− mutant had a partial baseplate structure, which contained only the upper disk assembled to the proximal part of the conical structure (Fig. 1C), thus supporting our hypothesis that the baseplate consists of at least one other protein in addition to ORF49. The single baseplate disk of the 49− mutant was, moreover, found to dissociate from the phage tail upon storage of the phage preparation for several months (data not shown). The microscopy analysis of the 46− and 47− mutants showed that these phages exhibited only head structures (Fig. 1D and E), thus lacking the entire tail and hence demonstrating that the mutated gene products are essential for the TP901-1 tail formation. Finally, the microscopy examination of the 50− mutant showed that this mutant was morphologically indistinguishable from the wt phage (Fig. 1F), which indicates that ORF50 is a protein of minor or no significance for tail formation.
FIG. 1.
Transmission electron micrographs of TP901-1 phages negatively stained with uranyl acetate. A, TP901-1 wt; B, 48− mutant; C, 49− mutant; D, 46− mutant; E, 47− mutant; F, 50− mutant.
Identification of ORF47 as the tail fiber protein.
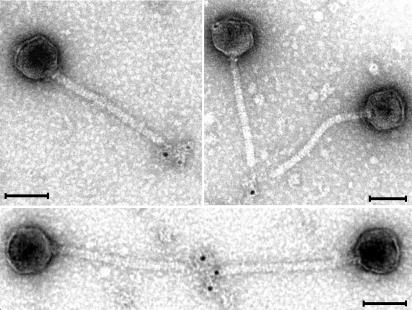
The observations of a central tail fiber in the TP901-1 virion lead us to speculate about which protein could constitute this structure. The tail fiber of the phage λ is formed from gpJ, which is the largest protein (124 kDa) encoded by the phage, and the phenotype of a λ J− mutant is tailless (35, 37). In phage TP901-1, these findings points towards ORF47 as the TP901-1 tail fiber protein, since orf47 is the largest gene in the phage genome and the 47− mutant is tailless. ORF47 has high identity with the structural protein Tal2009 from the related L. lactis phage Tuc2009 (93% identity over the entire amino acid sequence; Table 3). Tal2009 has been shown to be a self-processing protein with a lytic activity located within the C-terminal portion (38). Moreover, it has been demonstrated by immunoelectron microscopy with polyclonal antibodies raised against a purified C-terminal fragment of Tal2009 that the protein is located at the tip of the Tuc2009 tail (38). To determine if ORF47 constitutes the tail fiber of TP901-1, we used the same anti-Tal2009 (C-terminal) antibodies together with a secondary gold-conjugated antibody to label ORF47 in the 48− mutant. The 48− mutant was used for these experiments, because this mutant's lack of baseplate structure facilitated the examination and access of the tail fiber. As depicted in Fig. 2, it was possible to obtain gold labeling of the 48− mutant tail fiber with the anti-Tal2009 (C-terminal) antibodies. Furthermore, it was also illustrated that the antibodies formed aggregates with phages joined by the fibers, thus proving that ORF47 does, in fact, constitute the tail fiber of TP901-1.
FIG. 2.
Transmission electron micrographs of immunogold-labeled TP901-1 tail fiber protein ORF47. 48− mutant phages were incubated with anti-Tal2009 (C-terminal) antibodies and labeled with secondary antibodies conjugated to 5-nm gold particles. (ORF47TP901-1 and Tal2009 share 93% identity.) Bars, 50 nm.
Identification of structural TP901-1 tail proteins.
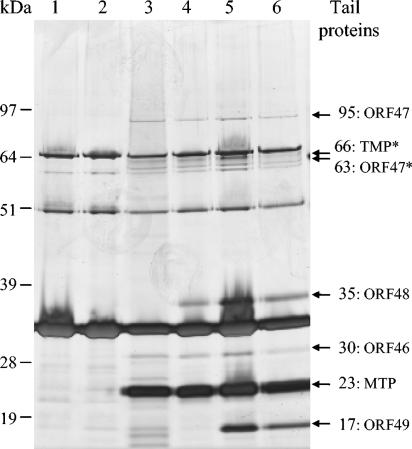
In order to determine the protein contents and profiles of the TP901-1 mutants, the proteins of purified and denatured phage particles were analyzed by SDS-PAGE (Fig. 3). The TEM analysis showed that the 46− and 47− mutants were tailless, and the protein bands from these mutants (Fig. 3, lanes 1 and 2) must therefore originate from head proteins. Tail protein bands can consequently be identified as those bands present in the wt phage but absent in the tailless 46− and 47− mutants, and hence it could be estimated that the wt protein bands of approximately 95, 66, 63, 35, 30, 23, and 17 kDa originated from tail proteins (Fig. 3). In previous studies of TP901-1, the protein bands of approximately 66, 23, and 17 kDa were identified as processed TMP, the MTP, and the baseplate protein (ORF49), respectively, although the precise values of molecular weight varied a little from the values observed in this study (33, 52). No protein band with the predicted 8.7-kDa size of ORF50 was observed in the wt profile, even when the phage proteins were separated in gel and buffer systems facilitating the separation of proteins of low molecular weight (results not shown). Moreover, the protein profile of the 50− mutant was indistinguishable from the wt profile (Fig. 3, lanes 5 and 6), which indicates that ORF50 is either a nonstructural protein or a structural protein present in very low amounts.
FIG. 3.
Silver-stained 10% SDS-PAGE gel with protein profiles of TP901-1 phages. Lane 1, 46− mutant; lane 2, 47− mutant; lane 3, 48− mutant; lane 4, 49− mutant; lane 5, 50− mutant; lane 6, TP901-1 wt. Molecular masses are indicated to the left of the gel. Tail protein bands and estimated molecular masses are indicated with arrows to the right of the gel. *, processed protein.
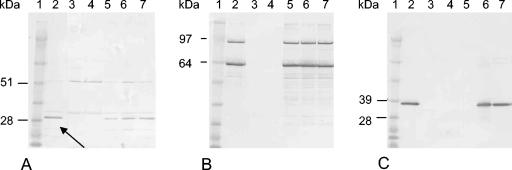
To determine whether the remaining yet unidentified 95, 63, 35, and 30 kDa tail protein bands could be encoded by orf46, orf47, and orf48, the separated phage proteins were transferred to polyvinylidene difluoride membranes and analyzed by Western blotting. ORF46TP901-1, ORF47TP901-1, and ORF48TP901-1 are highly identical to the phage Tuc2009 proteins ORF49Tuc2009, Tal2009, and ORF51Tuc2009, respectively (Table 3), and polyclonal antibodies raised against these Tuc2009 proteins could therefore be used for the Western blot analysis. The anti-ORF49Tuc2009 antibodies reacted with the TP901-1 tail protein band of approximately 30 kDa (Fig. 4A). Because ORF49Tuc2009 and ORF46TP901-1 show 95% aa sequence identity and the predicted size of ORF46TP901-1, furthermore, is 29 kDa, we conclude that the 30-kDa protein band is ORF46TP901-1. Minor unspecific reactions to head protein bands of approximately 51 and 33 kDa, however, were also observed. The anti-Tal2009 (N-terminal) antibodies reacted strongly with the two tail protein bands of approximately 95 and 63 kDa (Fig. 4B), thus demonstrating that the corresponding proteins both originate from ORF47TP901-1. This finding was not unexpected, since it has been determined that both the unprocessed and the processed N-terminal fragments of Tal2009 are incorporated into the Tuc2009 virion (38). A motif similar to the Tal2009 self-cleavage site (GGSSG↓GG) (38) is found with only one mismatch (GGNSGGG) in the ORF47TP901-1 sequence; cleavage of ORF47TP901-1 at this position will result in an N-terminal fragment of 67 kDa and a C-terminal fragment of 35 kDa. The Western blot in Fig. 4B illustrates that ORF47TP901-1 is, in fact, processed and that both the unprocessed protein and the processed N-terminal fragment are assembled into the phage virion, equivalent to Tal2009 in the Tuc2009 phage. Finally, the 35-kDa tail protein band was detected with the anti-ORF51Tuc2009 antibodies, and since ORF51Tuc2009 and ORF48TP901-1 display 80% aa sequence identity (Table 3), this demonstrates that the 35-kDa tail protein band is ORF48TP901-1 (Fig. 4C).
FIG. 4.
Western blot analysis for detection of ORF46TP901-1, ORF47TP901-1, and ORF48TP901-1 from purified wt and mutant TP901-1 phages. (A) Phage proteins detected with anti-ORF49Tuc2009 (ORF49Tuc2009 and ORF46TP901-1 share 95% identity). (B) Phage proteins detected with anti-Tal2009 (N-terminal) (Tal2009 and ORF47TP901-1 share 93% identity). (C) Phage proteins detected with anti-ORF51Tuc2009 (ORF51Tuc2009 and ORF48TP901-1 share 80% identity). Lane 1, molecular marker; lane 2, TP901-1 wt; lane 3, 46− mutant; lane 4, 47− mutant; lane 5, 48− mutant; lane 6, 49− mutant; lane 7, 50− mutant. Relevant molecular masses are indicated to the left of the blots. Arrow indicates band of ORF46TP901-1.
In summary, the TP901-1 tail protein bands of approximately 95, 66, 63, 35, 30, 23, and 17 kDa are, from this and previous studies, identified as ORF47, TMP*, N-terminal ORF47*, ORF48, ORF46, MTP, and ORF49, respectively, and these proteins are concluded to be the predominant structural proteins of the TP901-1 tail (Fig. 3).
The identified structural tail proteins of TP901-1 can be associated to specific tail structures by comparing and evaluating the SDS-PAGE mutant protein profiles with the TEM results. It thus appears that the essential tail proteins TMP (66 kDa), ORF46 (30 kDa), and ORF47 (95 and 63 kDa) are present in all of the TP901-1 derivatives with tail shafts, independent of baseplate structure (Fig. 1 and 3). The tail tube formation is addressed to the 23-kDa MTP (33) and, therefore, TMP, ORF46, and ORF47 must be components of the remaining tail shaft structures, such as the fiber and the conical structure, which is in agreement with the above identification of ORF47 as the tail fiber protein. Likewise, it is observed that the 48− mutant (revealing a baseplate-free phenotype) is lacking both the 35-kDa ORF48 and the 17-kDa ORF49 protein bands, while the 49− mutant, which is missing only the lower baseplate disk, is lacking solely the 17-kDa ORF49 protein band compared to the wt phage (Fig. 3, lanes 3, 4, and 6). We therefore conclude that ORF48 constitutes the upper baseplate disk, while ORF49 constitutes the lower baseplate disk. Other proteins may, of course, be involved in the baseplate formation, but these proteins must be present in significantly lower amounts, since they were not detected by SDS-PAGE. The structural functions of the individual proteins are listed in Table 3.
Infection efficiency.
Lysates of induced TP901-1 phages were tested by plaque assays on the indicator strain L. lactis 3107 in order to determine the titer and, hence, the infection efficiency of the mutants (Table 4). For this experiment, the concentrations of phages in the individual lysates were considered to be comparable, since the wt and mutant phages were estimated to be induced to approximately the same concentration from the phage purifications and the subsequent electron microscopy examination. The titer of the 50− lysate was found to be at the level of the wt phage, whereas the titers of the 48− and 49− lysates were reduced 104- and 105-fold, respectively, which in the case of the 49− mutant is consistent with prior observations (52). In contrast, the titers of the 46− and 47− lysates were reduced only 103-fold compared to the wt phage, which was unexpected, as these mutants are tailless.
TABLE 4.
Titration and sequencing of plaque-forming phages
| Induced TP901-1 phage | Titer on L. lactis 3107 (PFU/ml) | No. of plaque-derived DNA sequences | Length of sequence alignment (nucleotides)a | Sequenceb |
|---|---|---|---|---|
| Wild type | 2 × 109 | NDd | ||
| 46− mutant | 1 × 106 | 10 | 794 | wt |
| 47− mutant | 2 × 106 | 0c | ||
| 48− mutant | 4 × 105 | 9 | 624 | wt |
| 49− mutant | 4 × 104 | 2 | 360 | wt |
| 50− mutant | 5 × 109 | 2 | 574 | Amber mutation |
Sequences were aligned with ClustalW at the website of the European Bioinformatics Institute; lengths of alignments are stated.
Sequence at site of introduced amber mutation.
Amplified phage fragments from plaques contaminated with DNA from L. lactis.
dND, not determined.
In order to examine the mutant phages that were able to form plaques on the indicator strain, TP901-1 fragments were amplified from individual plaques of mutant and wt phages and subjected to sequencing. From the obtained and aligned sequences (Table 4), it was observed that, while the 50− mutant phages did contain the amber mutation, the plaque-forming phages from the 46−, 48−, and 49− lysates all had reverted to the wt sequence and consequently did not contain the introduced amber mutations (sequences not shown). We were not able to obtain DNA sequences of phages from the 47− lysate, but based on the similarity to the 46− mutant, we presume that the plaque-forming phages from the 47− lysate have also suffered a reversion of the amber mutation.
Within the alignment of phage DNA sequences from the 48− lysate, there were several indications of homologous recombination, i.e., that the mutant phages had obtained a sequence which was different but highly similar to the wt sequence instead of the inserted mutation. This is in accordance with the Southern blot analysis, which revealed that the TP901-1 lysogenic strain L. lactis 901-1 has additional prophage sequences homologous to TP901-1 orf46-50. The sequenced regions of the phages originating from the remaining mutant lysates were identical to wt TP901-1 sequences; however, we find it most likely that these mutational reversions also are a consequence of homologous recombination between the mutant phage and prophage sequences identical to TP901-1.
From the sequencing of plaque-forming phages, it is concluded that ORF50 is unessential for infection of L. lactis 3107, since the 50− mutant could infect the indicator strain with the same efficiency as the wt phage. The 46−, 48−, and 49− tail mutants were, however, unable to infect L. lactis 3107 unless the phages had obtained the wt sequence in place of the amber mutation, hence verifying that the tail and baseplate structure are fundamental for host infection.
DISCUSSION
The infection processes of LAB phages and phages infecting gram-positive bacteria are, in general, poorly understood compared to those of phages infecting gram-negative bacteria (25, 27). The elucidation of the tail structure of a typical LAB-infecting phage is a prerequisite for unraveling the mechanisms of phage-host receptor recognition and phage DNA injection of these phages. This report describes the investigation of the distal tail structures of the L. lactis phage TP901-1 through the construction and analysis of phage mutants, which were mutated in five late transcribed genes.
Our analysis demonstrated that ORF46 and ORF47 are structural tail proteins and that ORF47 constitutes a central tail fiber. Because ORF46 and ORF47 are essential for tail formation, as is the TMP (52), we propose that these three proteins form a tail initiator complex constituting the tail fiber and the conical structure, which we have discovered in the distal part of the TP901-1 tail. It has previously been argued that the genomic organization of TP901-1 is comparable to the λ genome (9, 11). A comparison of the genetic organization of the structural modules of TP901-1 and λ supports this assumption, since the structural proteins constituting the λ tail initiator complex are encoded by genes positioned from the tape measure protein gene to the tail fiber protein gene, corresponding to tmp, orf46 and orf47 in the TP901-1 genome.
The TP901-1 baseplate was shown to consist of at least two proteins, ORF48 and the previously identified ORF49 (52), which form the upper and lower disk, respectively. Since the 48− mutant lacks both disks, while the 49− mutant has the upper disk unstably assembled to the broader part of the conical structure, we conclude that the assembly of the upper disk is required for the lower disk assembly and stabilization and that the baseplate disks are assembled onto the conical fiber structure. The baseplate is not required for tail shaft formation, as the 48− mutant displays a complete tail shaft without a baseplate structure. The baseplate was, however, proven essential for TP901-1 infection, as all tested plaque-forming phages from 48− and 49− lysates were shown to contain the wt sequence in place of the amber mutations.
In this study, we have also analyzed the function of ORF50, which is encoded between a baseplate gene and the putative neck passage structure gene. The protein is highly conserved among several L. lactis phages (Table 3 and data not shown) and, moreover, contains a putative transmembrane domain. It was anticipated that ORF50 could be involved in the infection process, but the 50− mutant was determined to be indistinguishable from the wt phage with regard to morphology, protein profile, and infection efficiency. We therefore conclude that ORF50 is unessential for TP901-1 host infection and tail assembly. It is unclear whether ORF50 is independently expressed and constitutes part of the phage structure, but work is currently under way to resolve this matter.
It is possible that proteins other than those examined in this study and earlier studies are involved in the TP901-1 tail assembly pathway. Two λ proteins, gpG and gpGT, assist the λ tail assembly process and are not part of the λ phage virion. The proteins are encoded immediately upstream of the λ TMP, and a slippery sequence in gene G results in a sporadic translational frameshift and, hence, the production of gpGT (43). At the corresponding genomic position, an identical slippery sequence has been found in the 3′ end of TP901-1 orf43 (52), and it has recently been demonstrated that the slippery sequence connecting the λ genes G and T is strongly conserved among tail assembly genes of double-stranded DNA phages (59). It is therefore very likely that TP901-1 ORF43 and ORF43-44 are assisting proteins of the TP901-1 tail assembly processes.
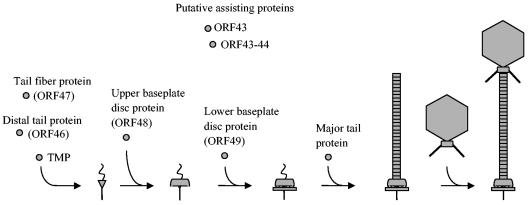
The data presented here facilitate the formulation of a novel hypothesis for TP901-1 tail assembly, which is based on the identification of several new tail proteins and the finding that the baseplate disks are assembled individually onto the conical fiber structure (Fig. 5). In this hypothesis, the N-terminal part of the TMP is expected to protrude from the initiator complex in the opposite direction of the fiber, since this part of the TMP determines the length of the tail tube (52). Furthermore, in accordance with the general consideration that phage tails are assembled from the distal to the proximal end (12), we propose that the baseplate is assembled onto the initiator complex before the tail tube is formed from polymerization of the major tail protein (Fig. 5). The baseplate assembly is, however, not fundamental for tail shaft formation, as the 48− mutant contained the tail shaft without a baseplate.
FIG. 5.
TP901-1 tail assembly hypothesis. An initiator complex is assumed to be formed from TMP, ORF46, and the tail fiber protein ORF47. The tail length determining the N-terminal part of TMP is expected to protrude from the complex. The baseplate is assembled onto the conical fiber structure by independent assembly of the upper and lower disks constituted from ORF48 and ORF49, respectively, and the tail tube is formed from polymerization of the major tail protein. It is currently unknown whether the baseplate disks are assembled before, during, or after tail tube formation. Finally, the tail is joined to an independently assembled head structure. ORF43 and ORF43-44 (expected to be formed from a translational frameshift at a slippery sequence in orf43) are putative assisting proteins equivalent to gpG and gpGT in the λ tail assembly process.
The present detailed analysis of the distal tail structures of phage TP901-1 makes it possible to consider the function of the individual structural elements. We have shown that the baseplate, and specifically the lower baseplate disk, is crucial for TP901-1 infection as the phage is unable to infect the indicator strain when the baseplate genes are mutated, but what is the function of the baseplate? The lower disk protein, ORF49, is the most unique protein of the TP901-1 tail module, since it displays full-length similarity to only a single protein encoded by phage φLC3 (Table 3). Because TP901-1 and φLC3 have overlapping host ranges (they both infect the L. lactis strains 3107 and Wg2 [4, 15]), this suggests that the baseplate is a host interaction element and that the lower disk protein, ORF49, is responsible for host receptor recognition. In an effort to prove this thesis experimentally, further studies of ORF49 and TP901-1 host interaction are in progress. Another notable structure of the TP901-1 tail is the central tail fiber, which is the most distal part of the tail. The corresponding fiber structure of the λ phage is responsible for binding of the host receptor during infection (57); however, since the TP901-1 baseplate is expected to possess this function, the TP901-1 tail fiber protein ORF47 must have a function other than that of λ gpJ. ORF47 was found to share many characteristics regarding sequence similarity, processing, and virion incorporation with the lytic active Tal2009 protein from the Tuc2009 phage (38); therefore, we expect that ORF47 has a lytic activity equal to Tal2009 and that the two proteins share the same function. Kenny et al. (38) suggested that Tal2009 participates in the phage infection process by degradation of the peptidoglycan (PG) in the cell wall. This suggestion is supported by the present discovery of ORF47 constituting the tail fiber, which must be inserted into the PG layer in order to bring the baseplate in contact with the host cell receptor. Proteins with muralytic activity, considered to be involved in phage infection, are widespread among phages infecting both gram-negative and gram-positive bacteria (40, 50). Moreover, Moak and Molineux (50) claimed that the enzymatic activity of these proteins must be constrained, e.g., by combination with a structural component, in order to prevent enzymatic diffusion and, hence, extensive degradation of the host PG layer, which would lead to premature lysis. The construction of the distal TP901-1 tail is comparable to the distal tail structures of the E. coli T5 phage, since the T5 receptor binding protein is localized above a straight tail fiber. Upon T5 binding to the host receptor, the straight tail fiber is considered to traverse the cell envelope, thus allowing transfer of the phage genome into the host cell (5, 42). It is, however, unlikely that the TP901-1 tail fiber, which is only approximately one-third the length of the T5 fiber, can traverse the thick PG layer of a gram-positive host cell, and it is therefore more probable that the TP901-1 tail fiber participates in the host infection process by promoting access to the host receptor or by assisting other, as-yet-unidentified DNA transporting mechanisms.
The findings of this study have resulted in a thorough characterization of the distal tail structure of TP901-1 and, furthermore, have led to reflections on host interacting mechanisms. Based on this analysis and the proposed functions of the tail proteins, we will use the following names in future studies: Dit (distal tail protein) for ORF46, TalTP901-1 for ORF47 (because of high identity with Tal2009 from phage Tuc2009), BppU (baseplate protein upper disk) for ORF48, and BppL (baseplate protein lower disk) for ORF49. The analyzed TP901-1 proteins show high identity to other phage proteins (Table 3), and the structural functions are thus most likely also identified for the homologous proteins of the L. lactis phages ul36, Tuc2009, and bIL285. Furthermore, proteins and genomic organization among LAB phages are highly conserved (17, 45, 53), and the results of this study may therefore also be applied to other phages.
Acknowledgments
This work was supported by the Royal Veterinary and Agricultural University of Denmark. Stephen Mc Grath is the recipient of an Embark postdoctoral fellowship from the Irish Research Council for Science, Engineering and Technology. Douwe van Sinderen is the recipient of an investigator grant from Science Foundation Ireland (02/IN1/B198).
We thank John Kenny for supplying the anti-Tal2009 (C terminal) antibodies. The help of Bernd Fahrenholz (FRCNF, Kiel, Germany) in the preparation of phages for electron microscopy is also acknowledged.
REFERENCES
- 1.Ackermann, H. W. 1998. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blatny, J. M., L. Godager, M. Lunde, and I. F. Nes. 2004. Complete genome sequence of the Lactococcus lactis temperate phage ϕLC3: comparative analysis of ϕLC3 and its relatives in lactococci and streptococci. Virology 318:231-244. [DOI] [PubMed] [Google Scholar]
- 5.Böhm, J., O. Lambert, A. S. Frangakis, L. Letellier, W. Baumeister, and J. L. Rigaud. 2001. FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr. Biol. 11:1168-1175. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., Jr., S. Hertwig, H. Neve, A. Geis, and M. Teuber. 1989. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. J. Gen. Microbiol. 135:2551-2560. [Google Scholar]
- 7.Breüner, A., L. Brøndsted, and K. Hammer. 1999. Novel organization of genes involved in prophage excision identified in the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 181:7291-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brøndsted, L., M. Pedersen, and K. Hammer. 2001. An activator of transcription regulates phage TP901-1 late gene expression. Appl. Environ. Microbiol. 67:5626-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brøndsted, L., S. Østergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 10.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 11.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casjens, S., and R. Hendrix. 1988. Control mechanisms in dsDNA bacteriophage assembly, p. 15-91. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y.
- 13.Casjens, S., and J. King. 1975. Virus assembly. Annu. Rev. Biochem. 44:555-611. [DOI] [PubMed] [Google Scholar]
- 14.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christiansen, B., M. G. Johnsen, E. Stenby, F. K. Vogensen, and K. Hammer. 1994. Characterization of the lactococcal temperate phage TP901-1 and its site-specific integration. J. Bacteriol. 176:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coombs, D. H., and F. Arisaka. 1994. T4 tail structure and function, p. 259-281. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 17.Desiere, F., S. Lucchini, C. Canchaya, M. Ventura, and H. Brüssow. 2002. Comparative genomics of phages and prophages in lactic acid bacteria. Antonie Leeuwenhoek 82:73-91. [PubMed] [Google Scholar]
- 18.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickely, F., D. Nilsson, E. B. Hansen, and E. Johansen. 1995. Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol. Microbiol. 15:839-847. [DOI] [PubMed] [Google Scholar]
- 20.Dupont, K., F. K. Vogensen, and J. Josephsen. 2005. Detection of lactococcal 936-species bacteriophages in whey by magnetic capture hybridization PCR targeting a variable region of receptor-binding protein genes. J. Appl. Microbiol. 98:1001-1009. [DOI] [PubMed] [Google Scholar]
- 21.Dupont, K., F. K. Vogensen, H. Neve, J. Bresciani, and J. Josephsen. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ. Microbiol. 70:5818-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes, F., C. Daly, and G. F. Fitzgerald. 1990. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl. Environ. Microbiol. 56:202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heller, K. J. 1984. Identification of the phage gene for host receptor specificity by analyzing hybrid phages of T5 and BF23. Virology 139:11-21. [DOI] [PubMed] [Google Scholar]
- 25.Heller, K. J. 1992. Molecular interaction between bacteriophage and the Gram-negative cell envelope. Arch. Microbiol. 158:235-248. [DOI] [PubMed] [Google Scholar]
- 26.Heller, K. J., and H. Schwarz. 1985. Irreversible binding to the receptor of bacteriophages T5 and BF23 does not occur with the tip of the tail. J. Bacteriol. 162:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henning, U., and S. Hashemolhosseini. 1994. Receptor recognition by T-even-type coliphages, p. 291-298. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 28.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 30.Jarvis, A. W. 1989. Bacteriophages of lactic acid bacteria. J. Dairy Sci. 72:3406-3428. [Google Scholar]
- 31.Johansen, A. H., L. Brøndsted, and K. Hammer. 2003. Identification of operator sites of the CI repressor of phage TP901-1: evolutionary link to other phages. Virology 311:144-156. [DOI] [PubMed] [Google Scholar]
- 32.Johnsen, M. G., K. F. Appel, P. L. Madsen, F. K. Vogensen, K. Hammer, and J. Arnau. 1996. A genomic region of lactococcal temperate bacteriophage TP901-1 encoding major virion proteins. Virology 218:306-315. [DOI] [PubMed] [Google Scholar]
- 33.Johnsen, M. G., H. Neve, F. K. Vogensen, and K. Hammer. 1995. Virion positions and relationships of lactococcal temperate bacteriophage TP901-1 proteins. Virology 212:595-606. [DOI] [PubMed] [Google Scholar]
- 34.Katsura, I. 1976. Morphogenesis of bacteriophage lambda tail. Polymorphism in the assembly of the major tail protein. J. Mol. Biol. 107:307-326. [DOI] [PubMed] [Google Scholar]
- 35.Katsura, I. 1983. Tail assembly and injection, p. 331-346. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), LAMBDA II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Katsura, I., and R. W. Hendrix. 1984. Length determination in bacteriophage lambda tails. Cell 39:691-698. [DOI] [PubMed] [Google Scholar]
- 37.Katsura, I., and P. W. Kühl. 1975. Morphogenesis of the tail of bacteriophage lambda. III. Morphogenetic pathway. J. Mol. Biol. 91:257-273. [DOI] [PubMed] [Google Scholar]
- 38.Kenny, J. G., S. Mc Grath, G. F. Fitzgerald, and D. van Sinderen. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J. Bacteriol. 186:3480-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labrie, S., and S. Moineau. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308-320. [DOI] [PubMed] [Google Scholar]
- 40.Lehnherr, H., A.-M. Hansen, and T. Ilyina. 1998. Penetration of the bacterial cell wall: a family of lytic transglycosylases in bacteriophages and conjugative plasmids. Mol. Microbiol. 30:454-457. [DOI] [PubMed] [Google Scholar]
- 41.Leiman, P. G., S. Kanamaru, V. V. Mesyanzhinov, F. Arisaka, and M. G. Rossmann. 2003. Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 60:2356-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letellier, L., L. Plançon, M. Bonhivers, and P. Boulanger. 1999. Phage DNA transport across membranes. Res. Microbiol. 150:499-505. [DOI] [PubMed] [Google Scholar]
- 43.Levin, M. E., R. W. Hendrix, and S. R. Casjens. 1993. A programmed translational frameshift is required for the synthesis of a bacteriophage λ tail assembly protein. J. Mol. Biol. 234:124-139. [DOI] [PubMed] [Google Scholar]
- 44.Lillehaug, D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83:85-90. [DOI] [PubMed] [Google Scholar]
- 45.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madsen, P. L., and K. Hammer. 1998. Temporal transcription of the lactococcal temperate phage TP901-1 and DNA sequence of the early promoter region. Microbiology 144:2203-2215. [DOI] [PubMed] [Google Scholar]
- 47.Madsen, P. L., A. H. Johansen, K. Hammer, and L. Brøndsted. 1999. The genetic switch regulating activity of early promoters of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 181:7430-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahanivong, C., J. D. Boyce, B. E. Davidson, and A. J. Hillier. 2001. Sequence analysis and molecular characterization of the Lactococcus lactis temperate bacteriophage BK5-T. Appl. Environ. Microbiol. 67:3564-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moak, M., and I. J. Molineux. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51:1169-1183. [DOI] [PubMed] [Google Scholar]
- 51.Østergaard, S., L. Brøndsted, and F. K. Vogensen. 2001. Identification of a replication protein and repeats essential for DNA replication of the temperate lactococcal bacteriophage TP901-1. Appl. Environ. Microbiol. 67:774-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pedersen, M., S. Østergaard, J. Bresciani, and F. K. Vogensen. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315-328. [DOI] [PubMed] [Google Scholar]
- 53.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desiere, and H. Brüssow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Seegers, J. F., S. Mc Grath, M. O'Connell-Motherway, E. K. Arendt, M. van de Guchte, M. Creaven, G. F. Fitzgerald, and D. van Sinderen. 2004. Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology 329:40-52. [DOI] [PubMed] [Google Scholar]
- 56.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, J., M. Hofnung, and A. Charbit. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, J., V. Michel, M. Hofnung, and A. Charbit. 1998. Cloning of the J gene of bacteriophage lambda, expression and solubilization of the J protein: first in vitro studies on the interactions between J and LamB, its cell surface receptor. Res. Microbiol. 149:611-624. [DOI] [PubMed] [Google Scholar]
- 59.Xu, J., R. W. Hendrix, and R. L. Duda. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11-21. [DOI] [PubMed] [Google Scholar]